Abstract
POT1 is a single-copy gene in yeast and humans that encodes a single-strand telomere binding protein required for chromosome end protection and telomere length regulation. In contrast, Arabidopsis harbors multiple, divergent POT-like genes that bear signature N-terminal OB-fold motifs, but otherwise share limited sequence similarity. Here, we report that plants null for AtPOT1 show no telomere deprotection phenotype, but rather exhibit progressive loss of telomeric DNA. Genetic analysis indicates that AtPOT1 acts in the same pathway as telomerase. In vitro levels of telomerase activity in pot1 mutants are significantly reduced and are more variable than wild-type. Consistent with this observation, AtPOT1 physically associates with active telomerase particles. Although low levels of AtPOT1 can be detected at telomeres in unsynchronized cells and in cells arrested in G2, AtPOT1 binding is significantly enhanced during S-phase, when telomerase is thought to act at telomeres. Our findings indicate that AtPOT1 is a novel accessory factor for telomerase required for positive telomere length regulation, and they underscore the coordinate and extraordinarily rapid evolution of telomere proteins and the telomerase enzyme.
Keywords: G-overhang, OB-fold, telomere, TRAP
Introduction
Telomeres stabilize eukaryotic genomes by facilitating the complete replication of the chromosome terminus, and sequestering the ends from recognition by DNA damage checkpoint machinery that would otherwise lead to inappropriate engagement of recombination and DNA repair activities. Telomeres typically consist of simple G-rich repeats that terminate in a single-strand 3′ extension, termed the G-overhang. The G-overhang serves as the substrate for the telomerase ribonucleoprotein (RNP) reverse transcriptase, which replenishes telomeric DNA. How telomerase engages the telomere is unknown, but its activity is modulated in cis by cell cycle-regulated interactions with resident telomeric DNA binding proteins (reviewed in Collins, 2006).
G-strand binding proteins play a crucial role in regulating telomerase access to telomeres, and in controlling other telomere-associated activities. The best characterized of these is Cdc13p from budding yeast (Nugent et al, 1996). Typical of this class of proteins, Cdc13p binds telomeric DNA via an oligonucleotide–oligosaccharide binding fold (OB-fold) (Mitton-Fry et al, 2002). Cdc13p is a multifunctional protein (reviewed in Lustig, 2001). It dynamically interacts with other constituents of the chromosome terminus and contributes to positive and negative regulation of telomerase, coupling of leading and lagging strand DNA synthesis, and protection of the C-rich strand of the chromosome terminus against nucleolytic attack.
In Schizosaccharomyces pombe and in higher eukaryotes, the presumed ortholog of Cdc13p is POT1 (Protection Of Telomeres) (Baumann and Cech, 2001). Although most organisms harbor only a single POT1 gene, ciliates, mouse and Arabidopsis possess at least two of these (Wang et al, 1992; Shakirov et al, 2005; Hockemeyer et al, 2006; Jacob et al, 2006; Wu et al, 2006). POT1 binds telomeric DNA in vitro (Baumann and Cech, 2001), but its attachment to the chromosome terminus in vivo is mediated primarily through protein interactions in the Shelterin complex (Loayza and de Lange, 2003; de Lange, 2005). Recent studies indicate that POT1 function is conveyed through its association with TPP1, another OB-fold containing protein (Houghtaling et al, 2004; Liu et al, 2004; Ye et al, 2004; Wang et al, 2007; Xin et al, 2007).
The co-crystal structure of human POT1 bound to its DNA substrate indicates that the 3′ terminal residues of the DNA are sequestered within the protein binding pocket, implying that hPOT1 functions to protect against nucleases and limit accessibility to telomerase (Lei et al, 2004). Consistent with this prediction, hPOT1 negatively regulates telomerase activity in vitro; this inhibition requires the DNA binding activity of hPOT1 (Kelleher et al, 2005; Lei et al, 2005). In vitro studies suggest that hPOT1 could also promote telomerase action at the chromosome terminus. Disruption of G-quartet structures by hPOT1 facilitates elongation by telomerase in vitro (Zaug et al, 2005). Moreover, hPOT1 stimulates unwinding of telomeric DNA by WRN and BLM helicases (Opresko et al, 2005), and depending on the location of POT1 binding site, hPOT1 can improve telomerase activity and processivity in vitro (Lei et al, 2005).
Genetic analysis of POT1 in fission yeast and vertebrates reveals a complex role for this protein in telomere length control. Human cells with reduced levels of POT1 display telomere elongation (Veldman et al, 2004; Ye et al, 2004; Yang et al, 2005) as do mice conditionally null for POT1a (Wu et al, 2006). Similarly, reduction of telomere-bound POT1 in S. pombe results in dramatic telomere elongation (Bunch et al, 2005). In contrast, overexpression studies implicate S. pombe and human POT1 in the positive regulation of telomere length (Colgin et al, 2003; Armbruster et al, 2004; Liu et al, 2004; Bunch et al, 2005). Thus, like Cdc13p, POT1 may contribute to both positive and negative regulation of telomere length.
POT1 is also necessary for chromosome end protection. S. pombe pot1− mutants suffer immediate and catastrophic loss of telomeric repeats, erosion of subtelomeric DNA, and chromosome mis-segregation (Baumann and Cech, 2001). Depletion of vertebrate POT1 leads to a DNA damage response at telomeres (Hockemeyer et al, 2005; Churikov et al, 2006), and in chicken cells results in a rapid G2 cell cycle arrest (Churikov et al, 2006). Other studies on POT1-depleted mammalian cells reveal genome instability, senescence and apoptosis (Veldman et al, 2004; Yang et al, 2005). The mouse POT1a and POT1b genes appear to be partially redundant for chromosome end protection. Although POT1b mutants are viable (Hockemeyer et al, 2006), conditional knockout of POT1a results in embryonic lethality (Hockemeyer et al, 2006; Wu et al, 2006). Single POT1a or double POT1a POT1b mutants exhibit a strong telomere DNA damage response, low levels of telomere fusions and endoreduplication, along with proliferative arrest and senescence (Hockemeyer et al, 2006). A second study implicated POT1a and POT1b in repression of non-homologous end joining and homologous recombination at telomeres (He et al, 2006; Wu et al, 2006).
Arabidopsis encodes two POT-like proteins, AtPOT1 and AtPOT2 (Shakirov et al, 2005), and possibly a third, AtPOT3 (Surovtseva et al, in preparation). In contrast to the mouse POT1a and POT1b proteins, which share 72% similarity (Hockemeyer et al, 2006), the plant POT proteins are more divergent and display only 49% overall sequence similarity. AtPOT2 is implicated in chromosome end protection, as overexpression of the N-terminal portion of the protein leads to severe growth and developmental defects, telomere shortening, and a high incidence of anaphase bridges and chromosome mis-segregation. AtPOT1, by contrast, contributes to telomere length regulation. Overexpression of a C-terminal fragment of AtPOT1 lacking the OB-fold motifs results in modest telomere shortening, but plants are wild type in appearance and show no signs of genome instability (Shakirov et al, 2005).
In this study, we examined the fate of Arabidopsis mutants null for AtPOT1. We found no evidence that AtPOT1 contributes to chromosome end protection or genome stability. Instead, AtPOT1 is required for positive regulation of telomere length: pot1 mutants display progressive telomere shortening at the same rate as telomerase-null plants. Notably, in vitro telomerase activity levels are reduced in pot1 mutants, but not abolished. Finally, we show that AtPOT1 physically associates with the telomerase RNP, and is enriched at telomeres during S-phase. Thus, AtPOT1 appears to be a novel telomerase accessory factor that promotes its activity in vitro and in vivo.
Results
Plants null for AtPOT1 do not exhibit genome instability
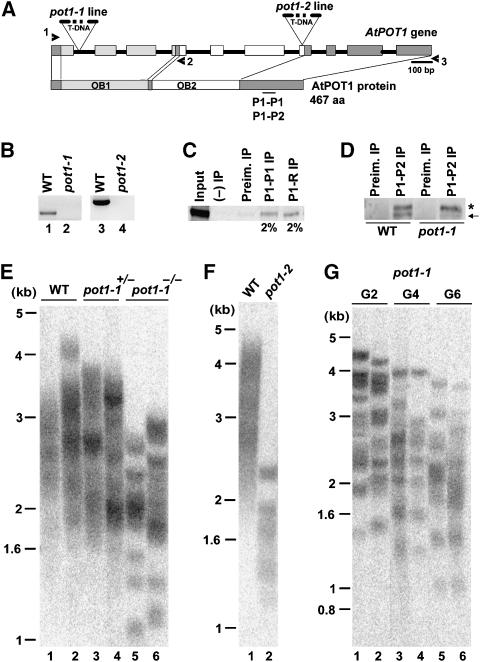
To identify an Arabidopsis line null for AtPOT1, we screened T-DNA collections from the University of Wisconsin Arabidopsis Knock-out Facility. Analysis of the ALPHA population uncovered a mutant with an insertion in the first intron of AtPOT1 (Figure 1A and Supplementary Figure 1). This line was designated pot1-1. In the Weigel collection, we found a second AtPOT1 allele (pot1-2), bearing a T-DNA in the seventh exon (Figure 1A and Supplementary Figure 1). To determine if these insertions disrupt AtPOT1 gene expression, RT–PCR was performed using primers flanking the insertion sites (Figure 1B and Supplementary Figure 1). No PCR products were generated in reactions with cDNA from the mutant plants (Figure 1B, lanes 2 and 4), confirming that expression of the full-length AtPOT1 mRNA was abolished in pot1-1 and pot1-2 mutants.
Figure 1.
Telomere phenotypes in AtPOT1-deficient Arabidopsis. (A) Genomic map and coding region of the AtPOT1 locus. Rectangles are exons; black lines represent introns. The position of T-DNA insertions in the pot1-1 and pot1-2 alleles are shown. OB1 and OB2 indicate two predicted OB-folds in the AtPOT1 protein. The position of the peptide used to raise P1-P1 and P1-P2 antibodies is indicated. Both peptides were raised against a similar AtPOT1 region, but P1-P2 peptide is slightly longer (see Materials and Methods). (B) RT–PCR analysis of the AtPOT1 gene expression in pot1-1 and pot1-2 mutants. Primer pairs 1–2 and 1–3 (shown as arrowheads in panel A) were used to analyze gene expression. (C) IP of recombinant 35S-labeled AtPOT1 protein. IP efficiencies for P1-P1 and P1-R antibodies are indicated. (D) Detection of endogenous AtPOT1 protein in wild-type and pot1-1 callus. AtPOT1 was immunoprecipitated and detected by Western blot analysis using P1-P2 antibody. The arrow indicates the 55 kDa endogenous AtPOT1 protein immunoprecipitated from wild-type callus. The asterisk indicates a nonspecific cross-reacting protein. (E) TRF analysis of DNA from six siblings segregating from a heterozygous pot1-1 parent. (F) TRF analysis of a pot1-2 mutant. (G) Multi-generational TRF analysis of pot1-1. DNA samples from two individual pot1-1 plants from the second (G2), fourth (G4), and sixth (G6) generation of self-pollination were analyzed. Blots shown in panels E, F and G were hybridized with a radiolabeled telomeric DNA probe. Molecular weight markers are indicated. Plants analyzed in panel G are from the WS ecotype, plants in panel E are from a Columbia-WS cross, and plants in panel F are from Columbia. Telomeres in wild-type WS plants are typically longer than those in Columbia (Shakirov and Shippen, 2004).
To monitor AtPOT1 protein, antibodies were raised against two peptides corresponding to a segment in the C-terminus of AtPOT1 protein (P1-P1 and P1-P2) (Figure 1A), and against a full-length recombinant AtPOT1 protein (P1-R). All three antibodies detected recombinant AtPOT1 by Western blotting (data not shown), and each immunoprecipitated the recombinant protein with ∼2% immunoprecipitation (IP) efficiency (Figure 1C; data not shown). Importantly, P1-P1 and P1-P2 detected a 55 kDa protein that corresponds to endogenous AtPOT1 protein in extracts from wild-type seedlings and callus, but not from pot1-1 mutants (Figure 1D; data not shown). We conclude that pot1-1 and likely pot1-2 (see below) are null for AtPOT1.
In striking contrast to yeast and vertebrate cells deficient in POT1, Arabidopsis pot1 mutants appeared morphologically indistinguishable from wild-type and showed no decrease in fertility or perturbation in growth and development for the six generations they were propagated. Furthermore, chromosome ends were refractory to nuclease attack and non-homologous end joining in the absence of AtPOT1. No anaphase bridges were observed in first (G1) or second (G2) generations of pot1-1 mutants (Supplementary Table 1; data not shown). The more sensitive telomere fusion PCR assay (Heacock et al, 2004) also failed to detect an increased frequency in chromosome end joining reactions in pot1-1 (data not shown). Thus, AtPOT1 is dispensable for chromosome end protection in Arabidopsis.
AtPOT1 is required for telomere length maintenance in vivo
To examine telomere length in pot1 mutants, terminal restriction fragment (TRF) analysis was performed on plants segregated from self-pollination of a heterozygous pot1-1 parent. As expected, telomeres in wild-type siblings appeared as a homogeneous smear of products ranging from 1.6 to 4.5 kb (Figure 1E, lanes 1 and 2). As for AtTERT (Fitzgerald et al, 1999), AtPOT1 is not haploinsufficient for telomere maintenance in Arabidopsis; plants heterozygous for the T-DNA insertion exhibited a wild-type telomere profile (Figure 1E, lanes 3 and 4).
Strikingly, telomere tracts in pot1-1 were much shorter than in wild-type and showed a more discrete banding pattern (Figure 1E, lanes 5 and 6). To determine whether disruption of the AtPOT1 gene was responsible for the telomere phenotypes, TRF analysis was performed on pot1-2 mutants. Telomeres were significantly shorter in pot1-2 than in wild-type, or even pot1-1 (Figure 1F, lane 2). Since the pot1-2 mutant was homozygous when we identified it, we suspect that this line had already been propagated at the Arabidopsis stock center for several generations in the absence of AtPOT1 prior to our analysis, leading to more substantial loss of telomeric DNA than in pot1-1. Complementation experiments provided further verification that AtPOT1 depletion caused telomere shortening. Plants heterozygous for pot1-1 were transformed with the wild-type AtPOT1 coding region under control of the constitutive CaMV 35S promoter. In plants expressing the 35S∷AtPOT1 transgene telomeres, particularly the shortest ones in the population, were returned to the wild-type length (Supplementary Figure 2A, lanes 2 and 3). A second complementation experiment performed with pot1 ku70 double mutants confirmed that AtPOT1 is required for telomere maintenance (Supplementary Figure 2B; see below).
Telomeres in pot1 mutants shorten at the same rate as in tert mutants
We followed the fate of telomeres in pot1 mutants for several plant generations and found that telomere length in pot1-1 decreased progressively with each generation (Figure 1G). The ever-shorter-telomere phenotype and sharp TRF banding profile were strikingly similar to the phenotype associated with tert mutants, which lose 200–500 bp of telomeric DNA per plant generation (Fitzgerald et al, 1999; Riha et al, 2001).
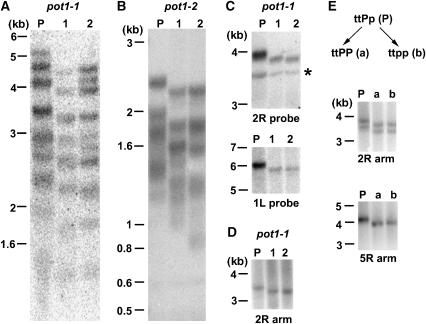
To determine if the rate of telomere shortening in pot1-1 was the same as in tert, we used a parent-progeny analysis to measure the rate of bulk telomere loss. DNA extracted from first generation (G1) parents (P) homozygous for the pot1-1 or pot1-2 allele, and their progeny (G2) was subjected to TRF analysis. For pot1-1, bulk telomeres declined by approximately 200–500 bp, while for pot1-2, a loss of approximately 200 bp was observed (Figure 2A and B). To obtain a more accurate estimate of the telomere shortening rate, individual telomeres were examined using subtelomeric TRF analysis (Shakirov and Shippen, 2004). Using probes specific for the South (right) arm of chromosome two (2R), or the North (left) arm of chromosome one (1L) (Figure 2C), only a single discrete band was detected in the parent and its progeny, possibly reflecting the coordinate regulation of telomere length on homologous chromosomes throughout plant development (Shakirov and Shippen, 2004). For both telomeres, the length decreased by approximately 200–300 bp relative to the parent (Figure 2C).
Figure 2.
Parent–progeny analysis reveals the same rate of telomere shortening in pot1-1, pot1-2, and tert mutants. (A, B) TRF analysis of bulk telomeric DNA from pot1-1 and pot1-2 parents (P) and two progeny (1 and 2) using a telomeric probe. (C) Subtelomeric TRF analysis of DNA from a pot1-1 parent and two progeny. DNA blots were hybridized with a probe corresponding to unique subtelomeric regions on 2R and 1L chromosome arms. The asterisk indicates a cross-hybridizing band. (D) PETRA analysis of the 2R telomere in a pot1-1 parent and two progeny. (E) PETRA analysis of the 2R and 5R telomeres in a parent homozygous for tert and heterozygous for pot1-1 (ttPp), and its tert (ttPP) (a) and pot1-1 tert (ttpp) (b) progeny. The two PETRA bands detected in the 2R reaction may represent different size telomeres on homologous chromosomes or two populations of cells (Shakirov and Shippen, 2004). A telomeric probe was used to detect PETRA products.
To further examine the rate of telomere shortening in pot1 mutants, we performed primer extension telomere repeat amplification (PETRA) (Heacock et al, 2004). In this assay, telomeres are amplified in a PCR reaction using primers directed at the G-overhang and a unique subtelomeric sequence. PETRA showed a decline of approximately 200 bp on the 2R and 5R telomeres in both pot1-1 and tert mutants (Figure 2D and E; data not shown). The same degree of shortening occurred in pot1-2 (data not shown). We conclude that disruption of AtPOT1 leads to a progressive loss of telomeric DNA that proceeds at the same rate as in tert mutants.
Telomeres become critically shortened in G6 tert mutants, giving rise to end-to-end fusions and genome instability (Riha et al, 2001). Because our pot1-1 mutants were derived from the WS ecotype, which naturally has longer telomeres than the Columbia ecotype (Shakirov and Shippen, 2004), from which the tert mutant was obtained, it is not surprising that G6 pot1-1 mutants do not yet show signs of genome instability. Assuming a telomere shortening rate of 200 bp/plant generation, we expect two or three additional generations are required for some pot1-1 telomeres to become critically shortened (Heacock et al, 2004).
AtPOT1 and AtTERT act in the same genetic pathway
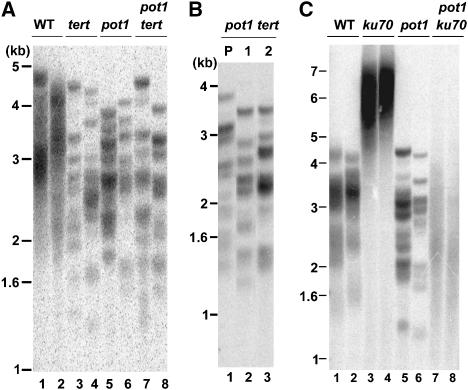
To determine whether AtPOT1 and AtTERT act in the same genetic pathway, plants heterozygous for pot1-1 were crossed to plants heterozygous for tert. Double heterozygous mutants from F1 were allowed to self-pollinate to generate F2 progeny. As shown in Figure 3A, telomeres of the same length and sharp banding profile were found in pot1-1 tert, as in their tert and pot1-1 siblings. PETRA and TRF parent–progeny analysis confirmed that telomeres in pot1-1 tert mutants shortened at the same rate as in either single mutant (Figures 2E and 3B).
Figure 3.
AtPOT1 functions in the telomerase pathway. (A) TRF analysis of pot1-1 tert mutants. Results for eight progeny (two for each genotype) that were segregated from a parent heterozygous for pot1-1 and tert are shown. (B) TRF analysis of telomeres in pot1-1 tert parent (P) and its two progeny (1 and 2) are shown. (C) TRF analysis of pot1-1 ku70 mutants. Results for progeny segregated from a parent heterozygous for pot1-1 and ku70 are shown. Two different progeny were analyzed for each genotype. The blot was hybridized with a telomeric DNA probe.
If AtPOT1 is required for telomerase function in vivo, its contribution should be especially obvious in a genetic background, where telomerase generates ultra-long telomere tracts. KU is a negative regulator of telomere length in Arabidopsis, and telomeres in mutants deficient in KU70 or KU80 undergo telomerase-dependent expansion to more than twice the normal length in a single generation (Riha et al, 2002; Gallego et al, 2003) (Figure 3C, lanes 3 and 4). In contrast, ku70 tert double mutants display accelerated telomere shortening and a precocious onset of genome stability (Riha and Shippen, 2003). To further investigate the role of AtPOT1, we generated pot1 ku70 mutants. In contrast to their ku70 siblings, pot1 ku70 mutants failed to elongate their telomeres (Figure 3C, lanes 7 and 8), and exhibited a heterogeneous profile of TRF products similar to that seen in ku70 tert (Supplementary Figure 3B, lane 3). Notably, telomeres in pot1 ku70 mutants were significantly elongated when an exogenous copy of AtPOT1 was introduced (Supplementary Figure 2B), confirming that AtPOT1 is required for telomere elongation in the absence of KU.
We also found that degree of telomere shortening was the same in pot1 ku70 and ku70 tert double mutants as in triple pot1 ku70 tert mutants. Parent–progeny analysis confirmed that plants from all three genotypes lost approximately the same amount of telomeric DNA from G1 (Supplementary Figure 3B, lanes 3, 4, 6) to G2 (Supplementary Figure 3C). All three mutants reached the terminal phenotype in G3. The incidence of anaphase bridges in pot1 ku70, tert ku70 and pot1 ku70 tert mutants in G3 were the same (Supplementary Table 1). These genetic data reinforce the notion that AtPOT1, like AtTERT, does not contribute to chromosome end protection. We conclude that AtPOT1 acts in the same genetic pathway as telomerase and is specialized for telomere length maintenance in vivo.
AtPOT1 is a component of the telomerase RNP required for maximal activity in vitro
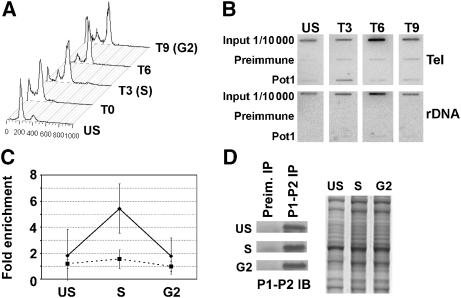
We considered the possibility that AtPOT1 is required for telomerase enzyme activity. TRAP assays were performed in parallel with extracts from wild-type and pot1 seedlings. As shown in Figure 4A (lanes 1 and 3–5), robust telomerase activity was reproducibly detected in extracts from wild type plants and from suspension culture. In contrast, telomerase levels were reduced and more variable in both pot1-1 and pot1-2 mutants (Figure 4A, lanes 6–12). Extract mixing experiments indicated that the reduction in enzyme activity was not due to the presence of a soluble PCR inhibitor (data not shown). Interestingly, although the in vitro levels of telomerase activity varied among pot1 mutants, the progressive telomere shortening phenotype observed in vivo was extremely consistent among the dozens of pot1-1 and pot1-2 mutant plants we examined.
Figure 4.
AtPOT1 interacts with the telomerase RNP. (A) TRAP assay results for wild-type (WT), pot1-1, and pot1-2 flowers. Results for extracts prepared from 10 different plants are shown in lanes 3–12. (B) TRAP assay results for wild-type and pot1-2 mutant flowers. 10 × and 100 × dilutions of protein extracts were used for TRAP as indicated. In panels A and B, extract prepared from Arabidopsis suspension culture (lane 1) served as the positive control (+). (C) TRAP assay following AtPOT1 IP from suspension culture using P1-P1 or P1-R antibodies. (D) TRAP assays with P1-P2 immunoprecipitates from wild-type and 4-day-old pot1-2 seedlings extracts. (E) TRAP assay results for P1-P1 antibody immunoprecipitates. IP from suspension culture extract was performed with no addition of peptide (−), 100 × excess of P1-P1 peptide (P1), or 100 × excess of a nonspecific AtPOT2 peptide (P2). (F) TRAP assays with eluates from P1-R IP in the presence of NaCl. NaCl concentrations are indicated. (G) TRAP assays with P1-R immunoprecipitates from unsynchronized suspension culture (unsyn.) or S-phase and G2-phase synchronized cells. The relative amount of active telomerase precipitated in the reaction is indicated. In all panels, IP with no antibody added ((−) IP), or with preimmune serum (Preim. IP), was used as a negative control.
Most TRAP reactions carried out with pot1 mutants showed reduced, but detectable levels of enzyme activity (Figure 4A, lanes 6–8 and 10–12). Titration of such samples suggested that telomerase activity was decreased by approximately 10-fold (Figure 4B, compare lanes 3 and 6). A more detailed understanding of AtPOT1's role in Arabidopsis telomerase biochemistry will require the development of a conventional (non-PCR based) primer extension assay to monitor the catalytic properties of the enzyme, a goal that has thus far proven elusive. Nevertheless, our current data allow us to conclude that AtPOT1 promotes telomerase action in vitro, but is not absolutely essential for its biochemical activity.
Next we looked for a physical interaction between AtPOT1 and the telomerase RNP. We could not detect a direct association between recombinant AtPOT1 and AtTERT by co-IP or by yeast two-hybrid assay (Y Surovtseva, M Jasti, and D Shippen, unpublished data). However, since AtTERT is the only Arabidopsis telomerase subunit isolated so far, AtPOT1 could contact another component of the RNP. To test this idea, IPs were performed with POT1 antibody on extracts from wild-type seedlings and asynchronous Arabidopsis cell culture (Menges and Murray, 2002). We confirmed that telomeres in this cell line fall within the wild-type range (Supplementary Figure 4A and B), and chromosome ends are protected against end joining reactions (Supplementary Figure 4C). Moreover, a 55 kDa band corresponding to endogenous AtPOT1 was observed by Western blotting following IP of cell culture extracts (Supplementary Figure 4D).
TRAP assays conducted on immunoprecipitates in the absence of antibody or with preimmune serum did not generate PCR products (Figure 4C, lanes 2 and 3; Figure 4D, lanes 2 and 5). However, telomerase activity could be immunoprecipitated from both seedlings and cell culture using all three of the POT1 antibodies (Figure 4C, lanes 4 and 5; Figure 4D, lane 3). The specificity of the AtPOT1 interaction with telomerase was demonstrated in three ways. First, IP of extracts from pot1 mutant seedlings failed to precipitate telomerase activity (Figure 4D, lane 6). Second, addition of a 100-fold excess of the P1-P1 peptide to cell culture extracts during the IP with the P1-P1 antibody dramatically decreased the TRAP signal, while a nonspecific AtPOT2 peptide of a similar length failed to compete (Figure 4E, lanes 2 and 3). Third, the AtPOT1 interaction with telomerase was stable in high salt; the association persisted in up to 400 mM NaCl (Figure 4F, lane 6). Since telomerase activity is strongly inhibited in salt concentrations greater than 450 mM (Figure 4F, lane 7), the AtPOT1 interaction may be even more robust.
To investigate whether AtPOT1 association with telomerase is cell cycle regulated, IP was performed on synchronized cell extracts (Menges and Murray, 2002). The level of telomerase activity is approximately the same in unsynchronized cultured cells as in cells arrested in S or G2 (Supplementary Figure 4E). While POT1 antibodies immunoprecipitated telomerase activity at all of the time points examined (Figure 4G, lane 3 in all panels), in three separate experiments TRAP products precipitated from S-phase cell extracts were significantly increased relative to unsynchronized cells (average=4.5-fold). Hence, AtPOT1 shows a dynamic interaction with the telomerase RNP.
AtPOT1 dynamically associates with telomeres in vivo
Mammalian and fission yeast POT1 bind telomeric DNA in vitro (Baumann and Cech, 2001; Wu et al, 2006). To investigate AtPOT1 interactions with telomeric DNA, gel-shift experiments were performed. AtPOT1 is extremely insoluble when expressed in E. coli, but soluble full-length AtPOT1 or the N-terminal domain containing the OB-folds can be obtained from rabbit reticulocyte lysate (data not shown). Under the same conditions that the OB-fold containing N-terminus of mouse POT1a binds its cognate telomere sequence (Wu et al, 2006), AtPOT1 failed to bind Arabidopsis telomeric DNA (Supplementary Figure 5). Thus, under these in vitro assay conditions, AtPOT1 does not interact with telomeric DNA in the same manner as its mammalian counterpart.
Human POT1 binds telomeres throughout the cell cycle, showing a transient decrease in binding late in G2 (Verdun et al, 2005). To investigate AtPOT1 interaction with telomeres in vivo, chromatin IP (ChIP) was employed. Since we failed to detect AtPOT1 binding to telomeres in chromatin preparations from plant cell extracts, ChIP assays were performed on suspension culture cells. Slight (1.8-fold) enrichment of AtPOT1 at telomeres was observed in unsynchronized cells relative to the preimmune sera control (Figure 5B and C). Therefore, we asked whether AtPOT1 localization at telomeres was regulated during the cell cycle. Our synchronization protocol did not allow us to examine cells blocked in G1, however more than 75% of the unsynchronized cells are in this phase of the cell cycle (Figure 5A). Using aphidicolin, we could enrich for cells in S-phase and in G2. In four separate experiments, AtPOT1 interaction with telomeres significantly increased in S-phase cells; the enhancement ranged from 3.4 to 7.1-fold (average=5.4-fold) over the preimmune sera control (Figure 5B and C). As an additional control, we monitored the ratio of the rDNA signal immunoprecipitated by the POT1 antibody relative to the preimmune control. As expected, no significant enrichment in S-phase was observed (Figure 5B and C). The AtPOT1 association with telomeres decreased dramatically as cells transitioned into G2 (down to 1.8-fold enrichment), indicating that AtPOT1 association with telomeres in S-phase does not reflect an increased number of binding sites after telomere replication. Furthermore, AtPOT1 protein levels were unchanged during the cell cycle (Figure 5D), arguing that its interaction with telomeres is dynamic and peaks in S-phase.
Figure 5.
AtPOT1 is associated with telomeric chromatin in S-phase. (A) FACS analysis of Arabidopsis suspension culture cells synchronized with aphidicolin. Data are shown for unsynchronized (US) and synchronized cells at 0, 3, 6, and 9 h after release from aphidicolin arrest. (B) Example of ChIP analysis on synchronized suspension culture extracts using P1-P2 antibody or preimmune serum. Immunoprecipitated DNA was monitored on slot blot using a radiolabeled telomeric or rDNA probe. (C) Quantitation of AtPOT1 association with telomeric DNA. The average of results from four independent experiments is shown. The solid black line indicates the ratio of the telomeric DNA signal obtained with the POT1 antibody relative to the preimmune sera control. As a negative control, the rDNA signal obtained with the POT1 antibody relative to the preimmune sera (gray dashed line) is shown. (D) Western blot analysis of AtPOT1 protein. Extracts from synchronized cells were precipitated with P1-P2 antibody, followed by P1-P2 Western blot analysis. Commassie-stained inputs (right) are shown as loading controls.
Discussion
Cdc13p and POT1 from yeast and vertebrates are multifunctional gatekeepers at the chromosome terminus, performing the crucial functions of distinguishing the ends from double-strand breaks, protecting against inappropriate recombination and nucleolytic attack, and controlling telomerase activity (de Lange, 2005; Baumann, 2006). Here, we demonstrate that AtPOT1 exhibits distinctly different interactions with telomeres. Unlike human POT1, disruption of the AtPOT1 gene is not lethal and Arabidopsis pot1 mutants display no evidence of chromosome end deprotection. Even in a ku70 tert background, where Arabidopsis telomeres are severely compromised (Riha and Shippen, 2003), the loss of AtPOT1 does not exacerbate telomere erosion or increase the frequency of chromosome end joining. While it is conceivable that AtPOT1 acts redundantly with another component of the telomere complex to protect the terminus, the dynamic interaction of AtPOT1 with telomeres is inconsistent with a primary role in this pathway.
Further distinguishing AtPOT1 from the previously described POT1 proteins is the ever-shorter-telomere phenotype displayed by Arabidopsis null mutants. Depletion of POT1 in mammals leads to telomere elongation (Veldman et al, 2004; Ye et al, 2004; Yang et al, 2005; Wu et al, 2006), implying a role in the negative regulation of telomere length. Although it is possible that the mammalian POT1 contribution to positive telomere length regulation is obscured by the other more severe phenotypes associated with POT1 depletion (Churikov et al, 2006; Hockemeyer et al, 2006; Wu et al, 2006), our data argue that the AtPOT1 protein has evolved a pivotal and highly specialized role in promoting telomerase action at telomeres by working in the context of the telomerase RNP.
Four lines of genetic and biochemical evidence strongly implicate AtPOT1 in the telomerase pathway. First, telomeres shorten at the same rate in tert and pot1 mutants, and depletion of telomere tracts is not accelerated in plants with a deficiency in both genes. Second, AtPOT1 is required for the telomerase-dependent elongation of telomeres in ku70 mutants. Third, in vitro telomerase activity levels are significantly reduced in pot1 mutants. Fourth, enzymatically active telomerase is specifically immunoprecipitated with AtPOT1 antibodies.
How AtPOT1 interacts with the telomerase RNP to facilitate telomere maintenance is unknown. Transient transfection experiments in tobacco with GFP-tagged AtPOT1 and AtTERT show that the two proteins co-localize in the nucleolus (N Kato, E Lam, E Shakirov, and D Shippen, unpublished data), where telomerase RNP biogenesis occurs in both yeast and mammals (Etheridge et al, 2002; Teixeira et al, 2002). Intriguingly, we found that AtPOT1 association with enzymatically active telomerase is regulated in the cell cycle, increasing by an average of ∼4.5-fold in S-phase relative to unsynchronized cells (predominantly G1) and cells arrested in G2. Thus, AtPOT1 may stabilize an enzymatically active form of the RNP. In support of this model, telomerase activity levels are more variable in the absence of AtPOT1. Although Arabidopsis shows no haploinsufficiency with respect to AtTERT (Fitzgerald et al, 1999) or AtPOT1 (this study), the more compromised telomerase enzyme found in pot1-null mutants may be unable to solve the end replication problem.
It is also possible that AtPOT1 functions to promote telomerase action on its telomeric DNA substrate. Notably, the variability of in vitro telomerase activity levels in pot1 mutants is incongruent with the highly consistent ever-shorter-telomere phenotype displayed by these plants. By virtue of its two OB-folds, AtPOT1 is predicted to directly bind telomeric DNA. However, under conditions where S. pombe and mammalian POT1 associate with telomeric DNA in vitro (Baumann and Cech, 2001; Wu et al, 2006), AtPOT1 showed no binding. While AtPOT1 may simply need different biochemical reaction conditions to associate with telomeric DNA, another more interesting possibility is that AtPOT1 requires a binding partner. Recent studies reveal that the mammalian POT1 binding partner, TPP1, which cannot bind telomeric DNA on its own (Wang et al, 2007; Xin et al, 2007), not only greatly enhances the affinity of POT1 for telomeric DNA in vitro, but also stimulates telomerase activity and processivity (Wang et al, 2007). Moreover, like AtPOT1, TPP1 can assume a canonical OB-fold (Wang et al, 2007; Xin et al, 2007) and physically interacts with the telomerase RNP (Xin et al, 2007). Mice deficient in TPP1 show profound developmental defects and animals that survive to adulthood are infertile (Keegan et al, 2005). Thus, TPP1 contrasts with Arabidopsis POT1 in that it appears to possess additional functions besides stimulating telomerase activity (Xin et al, 2007). Altogether our observations underscore the extraordinarily rapid evolution of the telomeric complex, and indicate that OB-fold bearing proteins, such as AtPOT1, are co-evolving with the telomerase RNP.
Materials and methods
Mutant lines
The pot1-1 allele was identified in ALPHA population of T-DNA insertion lines at the University of Wisconsin Arabidopsis Knock-out Facility. The collection was screened using primers 5′-TTTGTACTGGCCTCTCCAAGGTTCACCAT-3′ and 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′, according to the protocol available at http://www.biotech.wisc.edu/Arabidopsis/Index2.asp. The pot1-2 line was identified by screening a pooled genomic DNA collection (ABRC #CD10-A) from the Weigel T-DNA lines using primers 5′-CGGGATCCCACCCAGAAGATACTAAGATG-3′ and 5′-TTGACCATCATACTCATTGCTG-3′. tert and ku70 mutants and plant growth conditions are as described (Riha et al, 2001, 2002). All crosses were made between plants heterozygous for the desired mutations. Double- and triple-heterozygous F1 plants were identified by PCR genotyping and then self-propagated to F2 to obtain single-, double-, and triple-homozygous mutants and their wild-type siblings. F2 plants (G1) were self-propagated for several generations. Independent lines from at least two F2 plants were established and analyzed for each genotype. Complementation was performed as described in Supplementary Figure 2. Wild-type and G3 pot1-1 were used to establish callus. Callus initiation and maintenance were performed as described (Watson et al, 2005), with slight modifications. Seeds were germinated on 0.5 × Murashige and Scoog (MS) medium plates supplemented with 3% sucrose and 2.8 g/l phytagel. Roots were harvested at 3 weeks, finely chopped, and placed on 1 × MS medium plates supplemented with 2 mg/l 2,4-D, 0.05 mg/l kinetin, 3% sucrose, and 2.8 g/l phytagel (CIM). Callus was grown on CIM at 25°C in the dark and transferred to fresh medium every 4 weeks.
RT–PCR analysis, telomere analysis, TRAP assays, and cytogenetics
Total RNA was extracted from plant tissue using Tri Reagent solution (Sigma). Reverse transcription was performed using Superscript III reverse transcriptase (Invitrogen), as described (Shakirov et al, 2005). To evaluate expression of the regions flanking the T-DNA insertion in the pot1-1 allele, primer 1 (5′-GGATCCATGGCGAAGAAGAGAGAGAGTCCC AAGCTCATCA-3′) and primer 2 (5′-GCTCTAGACTTGATCTCTCTCAAGAAGGA-3′) were used. To analyze the pot1-2 allele, we used primer 1 and primer 3 (5′-TACCTCGAGCTAGATTAGGCTATCAGAGA-3′). DNA from individual whole plants was extracted as described (Cocciolone and Cone, 1993). TRF analysis was performed using Tru1I (Fermentas, Hanover, MD) restriction enzyme and a [32P] 5′ end-labeled (T3AG3)4 oligonucleotide probe (Fitzgerald et al, 1999). Subtelomeric TRF analysis was conducted using a 2R probe (Shakirov and Shippen, 2004), or a probe for 1L generated with 5′-ACGCTTGTCATCTCATCTCT-3′ and 5′-CGGGATCTTTGGTGTTTCTC-3′. Telomere fusion PCR and PETRA were performed as described (Heacock et al, 2004). TRAP assays were conducted on plant tissues as described in Fitzgerald et al (1996), using protein extracts prepared from a single wild-type or pot1 inflorescence, unless otherwise indicated. For the anaphase, spreads were prepared from pistils and stained with DAPI (4′,6′-diamidino-2-phenylindole), as discussed in Riha et al (2001).
Antibodies, Western blotting, and IP
The P1-R polyclonal antibody was raised in rabbits against full-length recombinant AtPOT1 protein expressed in E. coli (Covance). The P1-P1 and P1-P2 peptide antibodies were raised in rabbits against the N′-CSDENRRHHQVLLTLEDST and N′-AAYPWQVEDFCSDENRRHHQVLLT peptides, respectively, and affinity purified (Covance). Western blots were conducted with primary antibodies and peroxidase-conjugated light chain-specific mouse anti-rabbit secondary antibodies (Jackson Immunoresearch). For IP of endogenous proteins from suspension culture, protein extracts were prepared in buffer containing 50 mM Tris–HCl, pH 7.4, 10 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM PMSF, 1 mM DTT, and protease inhibitors (Roche). For IP of endogenous proteins from 4-day-old seedlings, protein extracts were prepared according to Fitzgerald et al (1996). Extracts were diluted at 1:5 in buffer W100 (20 mM TrisOAc, pH 7.5, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 200 mM NaCl, 100 mM KGlu, 1% NP-40, 0.5 mM Na deoxycholate, 1 mM DTT), precleared with protein A agarose (Pierce), and subjected to IP with POT1 antibodies. Following IP, the beads were washed three times with buffer W300 (same as buffer W100, but 300 mM KGlu) and two times with TMG buffer (10 mM TrisOAc, pH 8.0, 1 mM MgCl2, 10% glycerol, 1 mM DTT), and used for TRAP assay. In peptide competition experiments, a 100 × molar excess of the P1-P1 peptide or AtPOT2 peptide (N′-DDYKFLRIQDAFKALHLHVNC) was added during the IP step. For salt stability experiments, the NaCl concentration in the input was adjusted by addition of 5 M NaCl to suspension culture extracts. For IP of recombinant proteins, AtPOT1 and AtPOT2 were expressed in rabbit reticulocyte lysate (Promega) in the presence of 35S-labeled methionine. Following IP, the signal was quantified using ImageQuant Software. IP efficiency was calculated as a ratio of the immunoprecipitated signal versus input.
Cell culture synchronization and FACS
MM2d Arabidopsis cell suspension culture was maintained as described in Menges and Murray (2002). For synchronization, the original protocol was employed with the following modifications: 100 ml of 4-day-old culture was divided into five 20 ml aliquots, diluted at 1:5 with fresh medium and then blocked in G1/early S-phase for 21 h with 12 μg/ml of aphidicolin (AG Scientific). Cells were filtered through Miracloth (Calbiochem), washed twice with 500 ml of fresh media, and resuspended in 100 ml of fresh media. Aliquots were taken at various time points for DNA content analysis and immediately frozen. DNA content was analyzed by flow cytometry. The cultured cells were chopped with a razor blade, resuspended in homogenization buffer, filtered through a 20 μm nylon mesh, treated with 10 μg/ml of RNase A, and stained with 50 μg/ml of propidium iodide. Samples were run on a Becton-Dickinson FACSCalibur at 488 nm and analyzed using CellQuest (Becton-Dickinson) and ModFit LT (Verity) programs.
ChIP
ChIP was performed essentially as described by Leibfried et al (2005), with some modifications. The equivalent of 100 ml of 4-day-old unsynchronized or synchronized cell suspension culture (∼3 g of dry material) was fixed for 1 h in 1% formaldehyde, followed by quenching for 10 min in 125 mM glycine. After vacuum filtration through Miracloth, cells were resuspended and washed in 500 ml of PBS, and filtered again before storage at −80°C. Cells were ground in liquid N2 and then ChIP was performed using P1-R or P1-P2 antibody at a 1:100 dilution. The elution products were subjected to slot blot (Hybond N+, Amersham) and hybridized using a (TTTAGGG)4 telomeric probe. Blots were stripped and rehybridized with a combination of radiolabeled 5S (5′-TTGCAGAATCCCGTGAACCATCGAGT-3′) and 18S rDNA (5′-TGGAGCCTGCGGCTTAATTTGACTCA-3′) oligo-probes to monitor the specificity of IP. For quantification, the fold enrichment was determined as a ratio of the hybridization signal obtained with the POT1 antibody versus the preimmune sera control. rDNA sequences were used as a negative control.
Note added in proof
Since three different groups previously published on Arabidopsis POT-like genes and gave them different names (Kuchar and Fajkus, 2004; Shakirov et al, 2005; Tani and Murata, 2005), it is our joint decision to employ a unifying nomenclature that more closely follows the general trend in the telomere field. Therefore, At2g05210 (AtPOTl throughout this article) will hereafter be designated AtPOTla, while At5g06310 (AtPOT2) will be AtPOTl b.
Supplementary Material
Supplementary Informations
Acknowledgments
We thank Sandy Chang for the mouse POT1aN clone and helpful advice for gel-shift experiments. We are also grateful to Kalpana Kannan for performing some of the TRAP assays and Roger Smith for FACS analysis. Jeff Kapler, Tom McKnight, and members of the Shippen laboratory provided insightful comments on the manuscript. This work was supported by NIH grant GM65383 and NSF grant MCB0349993 to DES.
References
- Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter CM (2004) Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol Cell Biol 24: 3552–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P (2006) Are mouse telomeres going to pot? Cell 126: 33–36 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bunch JT, Bae NS, Leonardi J, Baumann P (2005) Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability. Mol Cell Biol 25: 5567–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Wei C, Price CM (2006) Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol Cell Biol 26: 6971–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone SM, Cone KC (1993) Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR (2003) Human POT1 facilitates telomere elongation by telomerase. Curr Biol 13: 942–946 [DOI] [PubMed] [Google Scholar]
- Collins K (2006) The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol 7: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Etheridge KT, Banik SS, Armbruster BN, Zhu Y, Terns RM, Terns MP, Counter CM (2002) The nucleolar localization domain of the catalytic subunit of human telomerase. J Biol Chem 277: 24764–24770 [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE (1996) Characterization and developmental patterns of telomerase expression in plants. Proc Natl Acad Sci USA 93: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE (1999) Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA 96: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego ME, Jalut N, White CI (2003) Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell 15: 782–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Multani AS, Cosme-Blanco W, Tahara H, Ma J, Pathak S, Deng Y, Chang S (2006) POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J 25: 5180–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M, Spangler E, Riha K, Puizina J, Shippen DE (2004) Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J 23: 2304–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T (2006) Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T (2005) POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 24: 2667–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S (2004) A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol 14: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Jacob NK, Lescasse R, Linger BR, Price CM (2006) Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol 27: 1592–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan CE, Hutz JE, Else T, Adamska M, Shah SP, Kent AE, Howes JM, Beamer WG, Hammer GD (2005) Urogenital and caudal dysgenesis in adrenocortical dysplasia (acd) mice is caused by a splicing mutation in a novel telomeric regulator. Hum Mol Genet 14: 113–123 [DOI] [PubMed] [Google Scholar]
- Kelleher C, Kurth I, Lingner J (2005) Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol 25: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchar M, Fajkus J (2004) Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants. FEBS Lett 578: 311–315 [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR (2004) Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol 11: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR (2005) Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem 280: 20449–20456 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z (2004) PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Lustig AJ (2001) Cdc13 subcomplexes regulate multiple telomere functions. Nat Struct Biol 8: 297–299 [DOI] [PubMed] [Google Scholar]
- Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS (2002) Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147 [DOI] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA (2005) POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem 280: 32069–32080 [DOI] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE (2001) Living with genome instability: plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE (2003) Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc Natl Acad Sci USA 100: 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE (2002) Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J 21: 2819–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Shippen DE (2004) Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16: 1959–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Surovtseva YV, Osbun N, Shippen DE (2005) The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol 25: 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani A, Murata M (2005) Alternative splicing of Pot1 (Protection of telomere)-like genes in Arabidopsis thaliana. Genes Genet Syst 80: 41–48 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Forstemann K, Gasser SM, Lingner J (2002) Intracellular trafficking of yeast telomerase components. EMBO Rep 3: 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman T, Etheridge KT, Counter CM (2004) Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol 14: 2264–2270 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J (2005) Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M (2007) The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wang W, Skopp R, Scofield M, Price C (1992) Euplotes crassus has genes encoding telomere-binding proteins and telomere-binding protein homologs. Nucleic Acids Res 20: 6621–6629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Bulankova P, Riha K, Shippen DE, Vyskot B (2005) Telomerase-independent cell survival in Arabidopsis thaliana. Plant J 43: 662–674 [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S (2006) Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z (2007) TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Yang Q, Zheng YL, Harris CC (2005) POT1 and TRF2 cooperate to maintain telomeric integrity. Mol Cell Biol 25: 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T (2004) POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR (2005) Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA 102: 10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Informations