Abstract
A critical step of neuronal terminal differentiation is the permanent withdrawal from the cell cycle that requires the silencing of genes that drive mitosis. Here, we describe that the α isoform of the heterochromatin protein 1 (HP1) protein family exerts such silencing on several E2F-targeted genes. Among the different isoforms, HP1α levels progressively increase throughout differentiation and take over HP1γ binding on E2F sites in mature neurons. When overexpressed, only HP1α is able to ensure a timed repression of E2F genes. Specific inhibition of HP1α expression drives neuronal progenitors either towards death or cell cycle progression, yet preventing the expression of the neuronal marker microtubule-associated protein 2. Furthermore, we provide evidence that this mechanism occurs in cerebellar granule neurons in vivo, during the postnatal development of the cerebellum. Finally, our results suggest that E2F-targeted genes are packaged into higher-order chromatin structures in mature neurons relative to neuroblasts, likely reflecting a transition from a ‘repressed' versus ‘silenced' status of these genes. Together, these data present new epigenetic regulations orchestrated by HP1 isoforms, critical for permanent cell cycle exit during neuronal differentiation.
Keywords: E2F, heterochromatin, HP1α, neuronal terminal differentiation, transcription
Introduction
In the mammalian central nervous system, differentiated neurons are devoid of replicative capability. Neurons withdraw from the cell cycle immediately after their differentiation from proliferative neuroepithelial cells. The continuous repression of cell cycle regulators is a prerequisite to achieve a permanent quiescent state. In fully differentiated neurons, the regulation of these factors is of prime importance, as inappropriate re-activation of cell cycle genes leads to neuronal apoptosis and progression of neurodegenerative diseases (Herrup et al, 2004). Although the fundamental mechanisms, which operate at the interface between growth arrest and neuronal differentiation, should be tightly linked; they are still poorly understood.
E2F transcription factors are critical regulators of cell cycle genes required for G1/S transition (reviewed by Frolov and Dyson, 2004). They regulate transcription via E2F-responsive elements (E2F-RE) found in genes involved in cell division and apoptosis. To maintain cells in a quiescent state, E2F-targeted genes have to be silenced, in part through p130 and E2F4 binding to E2F-REs (Ghosh and Harter, 2003). More particularly, an E2F-dependent repressive role was demonstrated to be essential in neuroprotection (Boutillier et al, 2003; Liu et al, 2005).
To produce active repression, proteins of the Rb and E2F families recruit chromatin remodeling factors to the targeted sequences. For example, Rb proteins play crucial roles in both cell proliferation and differentiation, by recruiting histone deacetylases (HDACs) (see review Frolov and Dyson, 2004), histone H3 methyltransferase (Suv39H) (Nielsen et al, 2001b; Nicolas et al, 2003) and DNA methyltransferase 1 (Robertson et al, 2000). Interestingly, Rb targets Suv39H to promoters causing methylation of histone H3, thus providing a binding site for heterochromatin protein 1 (HP1) (Nielsen et al, 2001b).
HP1 is a class of multifunctional chromatin-associated adapter protein, which is present in constitutive heterochromatin where it plays an essential role in establishing and maintaining heterochromatin-mediated gene silencing (Hiragami and Festenstein, 2005). In mammalian cells, there are three HP1 protein isoforms: α, β and γ, encoded by three different genes (Jones et al, 2001). HP1 proteins contain a conserved amino-terminal region called the chromodomain, followed by a variable hinge region and a conserved carboxy-terminal chromoshadow domain. The multipartite organization of HP1 proteins allows several proteins to bind simultaneously. Whereas the chromodomain targets HP1 molecules to methylated lysine 9 from histone H3 (Bannister et al, 2001; Lachner et al, 2001), the chromoshadow domain forms homo- and heteromers between the different isoforms. HP1 variants display different subnuclear localizations: HP1α and β are concentrated at pericentric heterochromatin, although HP1β is also present at more diffuse nucleoplasmic sites, whereas HP1γ is predominantly localized in euchromatin (Hiragami and Festenstein, 2005).
If different binding properties and localization have been attributed to each HP1 isoforms, so far, no physiological function on each individual isoform has been clearly defined. Herein, we report that HP1α presence is absolutely needed in the terminal differentiation process of neurons and define HP1α as a guide of neuronal fate, ensuring cell cycle gene sequestration into heterochromatin.
Results
Silencing of e2f-targeted genes during NTD
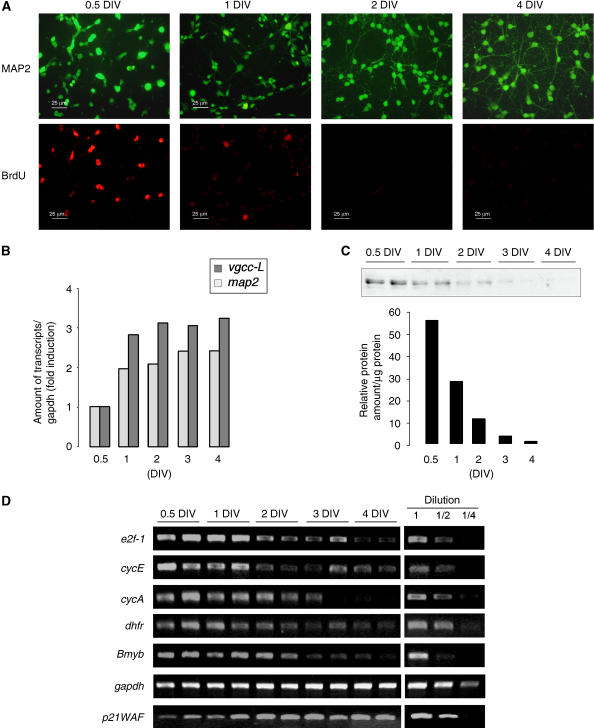
In this report, we investigated the fate of e2f1 gene expression during in vitro terminal differentiation of immature cerebellar granule neurons (CGN) obtained from 7-day postnatal (PN) mice. At this in vivo time, granule cells are undergoing their last mitotic rounds before migration in the different cerebellar layers (Wang and Zoghbi, 2001). Once plated in specific culture conditions (25 mM KCl, serum), CGN presented a proliferation activity during the first day in vitro (DIV), and further developed differentiated features such as neurite extension within 4 DIV (Figure 1A). Under these conditions, gene expression of neuronal maturation markers such as microtubule-associated protein 2 (MAP2) or L-type voltage-gated calcium channels (vgcc-L) increased over time (Figure 1B). E2F1 protein expression and transcriptional level were robustly repressed after 1 DIV (Figure 1C and D). Consistent with this, the expression of direct E2F1-target genes (i.e. that display E2F-RE within their promoter) were strongly and synchronously repressed (Figure 1D). In contrast, a non-E2F-targeted gene, the cyclin-dependent kinase inhibitor p21WAF gene, displayed increased expression in mature neurons (Figure 1D). These data clearly show a selective and dramatic transcriptional repression of several E2F-targeted genes in a simplified model of NTD.
Figure 1.
Synchronous silencing of e2f-targeted genes during NTD. (A) CGN morphology in primary cultures was visualized at different DIV by a MAP2 immunostaining (upper panel). Mitotic activity was evidenced by BrdU incorporation followed by immunodetection (lower panel). (B) Expression of the neuronal markers MAP2 and the L-type voltage-gated calcium channel (vgcc-L) was measured by semiquantitative RT–PCR. Bands were quantified relative to gapdh amounts and results are represented as fold induction. (C) E2F1 protein levels were measured by western blot analyses at different DIV. A typical experiment is shown and quantified (D) Expression of several e2f-targeted genes: e2f1, cyclin E, cyclin A, dhfr, Bmyb was analyzed by semiquantitative RT–PCR. A series of dilutions (1/2 and 1/4) was performed to ensure the linear range of amplification. Experiments have been repeated 3–5 times.
Differential recruitment of the HP1 protein family at E2F sites
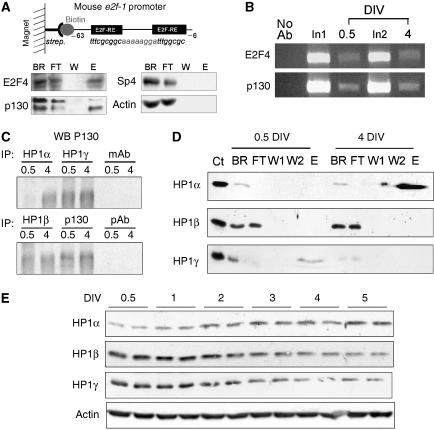
We previously showed that the two E2F-REs within the e2f1 promoter were required to maintain low levels of e2f1 expression in mature CGN (Boutillier et al, 2002) and wished to characterize the protein complexes ensuring these repressions. The mouse e2f1 promoter region from –63 to –6 was used as a probe to isolate specific E2F-RE-binding proteins from nuclear extracts with microcolumns (Figure 2A). After elution, bound complexes were analyzed by western blot. E2F4 and p130 were present at E2F sites, similar to that reported for the Bmyb promoter (Liu et al, 2005), whereas nonrelated proteins (actin or the transcription factor SP4) were not detected (Figure 2A). A mismatched probe did not bind to E2F4 or p130 (data not shown). Direct binding of E2F4 and p130 to the e2f1 promoter (Figure 2B), cyclin A and dhfr (data not shown), was further confirmed by chromatin immunoprecipitation (ChIP) using primers surrounding E2F-RE. We next tested the ability of p130 to recruit the members of the HP1 family of proteins by co-immunoprecipitation. Interestingly, p130 was able to bring down all the three HP1 members, α, β and γ (Figure 2C). The binding affinities of the three different isoforms of HP1 for the mouse E2F probe (used in 2A) were then measured at the beginning and at the end of differentiation. Eluates were enriched with HP1α when microcolumns were loaded with CGN nuclear extracts obtained at 4 DIV, whereas HP1γ binding was only detected at 0.5 DIV. As far as HP1β was concerned, no binding at all was detected (Figure 2D). We further checked the expression of the three HP1 isoforms in CGN cultures. HP1α protein levels clearly increased upon neuronal maturation, an event that likely accounts for the lack of HP1α binding to p130 at 0.5 DIV (Figure 2C). Both HP1β and γ isoforms decreased (Figure 2E). This points to a primary regulation of HP1 isoforms at the level of protein expression in vitro.
Figure 2.
Recruitment of HP1 proteins at E2F sites during NTD. (A) A biotinylated fragment of the mouse e2f1 gene promoter (−63/−6) containing two E2F-RE is bound to Streptavidin MicroBeads (strep.) and retained in the magnetic field. Nuclear extracts from mature neurons (4 DIV) were tested. All fractions obtained from columns were analyzed by western blot (BR, binding reaction, which represents the input material; E, eluate; FT, flow through; W, washes). (B) ChIP assays were performed on CGN at 0.5 and 4 DIV with antibodies against E2F4 and p130. Precipitated DNA were analyzed by semiquantitative PCR with primers to the mouse e2f1 promoter. Control PCR experiments were performed with total DNA (input) and DNA isolated in the absence of primary antibody. (C) Immunoprecipitation experiments were performed on CGN extracts from 0.5 and 4 DIV with antibodies against HP1 proteins and p130 as indicated. Precipitated proteins were analyzed by western blot for p130. mAb and pAb: control IPs performed with, respectively, unrelated monoclonal (actin) and polyclonal (sp4) primary antibodies. (D) In vitro binding assay as in (A), performed with nuclear extracts obtained from mature CGN (4 DIV) or nondifferentiated neurons (0.5 DIV). All fractions were analyzed by western blot for the three different variants of HP1 as noted (W1 and W2: two washes steps). Ct represents total CGN extracts (positive control). (E) Expression of HP1 isoforms were measured at different DIV in cultured CGN by Western blot analyses. Equal loadings were confirmed by actin immunodetection.
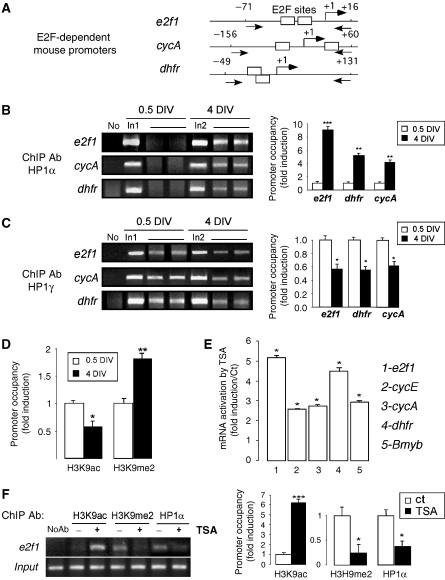
To evaluate the occupancy of E2F sites by the different HP1 proteins in context, we performed ChIP assays in CGN at both 0.5 and 4 DIV on different E2F-targeted genes: e2f1, cyclin A and dhfr. The regions of the mouse promoters tested are represented in Figure 3A. The e2f1 promoter was associated with HP1α at 4 DIV, but this was not observed in neuronal progenitors (Figure 3B). Cyclin A and dhfr also showed an interaction with HP1α protein at 4 DIV only. In contrast, all three gene promoters were found to be strongly associated with HP1γ at 0.5 DIV, the binding of which was reduced in postmitotic neurons (Figure 3C). These data clearly evidence a molecular switch between HP1α and HP1γ isoforms during NTD at E2F sites. We then investigated the acetylation/methylation status of histone H3 lysine 9 (H3K9) at the e2f1 promoter by ChIP experiments during NTD. Figure 3D shows that acetylated H3K9 levels associated with E2F sites decreased in mature neurons relative to cycling cells. By contrast, methylated H3K9 levels were significantly higher once neurons were differentiated. Interestingly, a 5-h treatment of mature neurons with the HDAC inhibitor trichostatin A (TSA) forced acetylation of H3K9 (Figure 3F) and resulted in a marked increase of E2F-targeted gene transcription (Figure 3E). In this case, H3K9 methylation level decreased concomitantly with HP1α binding to E2F sites (Figure 3F). We previously reported that such treatment induced neuronal death at this concentration (Boutillier et al, 2003) and these data underline that HP1α binding is displaceable in mature neurons, an event that may contribute to cell cycle reactivation during neuronal death.
Figure 3.
HP1α binding is associated with H3K9 methylation and is displaceable by H3K9 hyperacetylation. (A) Schematics of promoters analyzed in these studies. Boxes represent E2F-REs, arrows indicate primer positions. (B) Binding of HP1α during neuronal maturation to e2f1, cyclin A and dhfr gene promoters was measured by ChIP experiments from immature (0.5 DIV) or mature (4 DIV) CGN. Immunoprecipitated DNA were analyzed by semiquantitative PCR. Control PCR experiments were performed with total DNA (input) and DNA isolated in the absence of primary antibody. In1, input DNA at 0.5 DIV; In2, input DNA at 4 DIV. Bands were quantified relative to the corresponding input DNA sample. Promoter occupancy at 0.5 DIV was arbitrary set at 1. *, **, *** when P<0.01, P<0.05, P<0.001. (C) Binding of HP1γ during neuronal maturation to e2f1, cyclin A and dhfr gene promoters was measured by ChIP experiments and quantified as in (B). (D) ChIP experiments were performed and quantified on CGN at 0.5 and 4 DIV with antibodies against histone H3 dimethylated on lysine 9 (H3K9me2) or acetylated on lysine 9 (H3K9ac), as in (B). (E) TSA treatment induced e2f-targeted gene transcription. Semiquantitative PCR were run on total RNA from mature CGN treated with 100 nM TSA for 5 h, for different genes as noted. Bands were quantified relative to gapdh amounts. Histograms represent fold induction relative to nontreated cells arbitrarily set at 1. * when P<0.001. (F) After 4 DIV, CGN were treated with 100 nM of TSA for 5 h or not (Ct). ChIP experiments were performed as described above with H3K9me2 or H3K9ac and HP1α antibodies. Promoter occupancy in control was arbitrarily set at 1. *, *** when P<0.01, P<0.001.
Function of HP1 during NTD
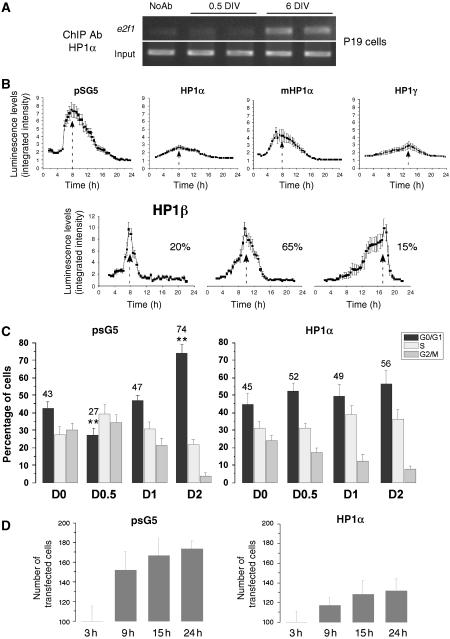
To better understand the function of each HP1 isoform on E2F-dependent transcription, we measured the dynamics and the levels of E2F-dependent transcription in response to overexpression of HP1 isoforms. To this aim, we used the P19 embryonic carcinoma cell line that can be differentiated into neurons upon retinoic acid (RA) stimulation (Jones-Villeneuve et al, 1982). We first confirmed that HP1α also differentially bound E2F sites in these cells by ChIP experiments with e2f1 primers (Figure 4A). P19 cells were then co-transfected with an e2f1 promoter reporter vector together with the different HP1 expression vectors at the embryonic stage. One day after transfection, the differentiation process was initiated and the transcription activity was followed in single living cell by measurement of photon emission using a highly sensitive CCD camera. The cells were highly synchronous in the cell cycle progression, as they all displayed cyclin D1- (Supplementary Figure S1) and E2F (pSG5, Figure 4B)-dependent transcription in a narrow time period. E2F-dependent transcription peaked 8 h after RA induction. In the presence of HP1α, this transcription was highly damped, but the cells remained synchronous (Figure 4B). Transcriptional activity was partly recovered in presence of an HP1α isoform mutated on its chromodomain, a mutation that diminishes its binding ability to methylated-H3K9 (Nielsen et al, 2001a). Interestingly, expression of HP1γ not only strongly reduced E2F-dependent transcription but also delayed it (from 8 to 13 h). Surprisingly, not only the expression of HP1 β had no effect on the levels of E2F-dependent transcription, but also it abolished the synchrony of the transcription (Figure 4B). To average the timing of the transcription of the cell population, we split cells into three categories. The early-responding cells (20%) peaked as other cells normally did and represent 20% of the whole cell population (Figure 4B). The middle (65%)- and the late (15%)-responding cells represent together 80% of cells that are not synchronous compared to the control-transfected cell population. Taken together, this series of experiments shows that each of the HP1 isoform has a specific role either on the level or on the timing of E2F-dependent transcription, HP1α and HP1γ both acting as repressors.
Figure 4.
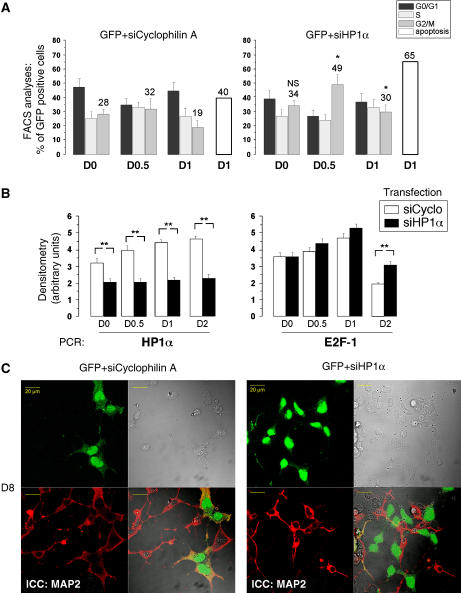
HP1α controls E2F-dependent transcription and the last cell division before P19 embryonic cell differentiation. (A) ChIP experiments were performed on nondifferentiated (0.5 DIV) and differentiated (6 DIV) P19 cells with an HP1α antibody. Control PCR experiments were performed with total DNA (input) and DNA isolated in the absence of primary antibody, no Ab (B) P19 cells were co-transfected with HP1α, HP1α mutated in the chromodomain (mHP1α), HP1β, HP1γ, or the empty pSG5 vector as a control, and the E2F-luc reporter plasmid. Neuronal differentiation was induced 24 h after transfection, using 1 μM RA. Simultaneously, firefly luciferin (Biosynth, Switzerland) was added to the medium and luminescence imaging was carried out. Images were acquired using 30 min integration times. Results are expressed as the integrated intensity of light in the analyzed area. For HP1β, the percentage of cells averaged in the peak is given on the right. (C) Cells were co-transfected with EGFP and HP1α expression vector or the empty pSG5 plasmid as a control and neuronal differentiation was induced 24 h post-transfection. Cell cycle stage was determined by FACS at indicated times after RA treatment. Cell cycle analysis was specifically performed on GFP-expressing cells. One-way ANOVA test followed by Bonferroni means comparison test was performed only for G0/G1 values. For pSG5 transfected cells, ** indicates statistical significance compared with control cells at day 0 (D0). For HP1α-transfected cells, the G0/G1 values for the different time points are not statistically different compared to D0. (D) P19 cells were co-transfected as in C and the number of transfected cells were counted in six different fields for each transfection conditions at indicated time points. The total number of transfected cells counted are represented on the histograms.
The role of HP1α on cell cycle progression during the differentiation process was further investigated by flow cytometry, in presence or absence of a transfected HP1α expression vector introduced before differentiation. In the control situation, 74% of cells had accumulated in the G0/G1 phase within 2 days after differentiation (Figure 4C and Supplementary Figure S2), indicating the end of the cell cycle progression. The progression observed in transfected cells with the pSG5 vector was the same as in untransfected cells (data not shown). In contrast, in cells overexpressing HP1α, the final cell cycle was not induced: at day 0.5 of RA treatment, about 50% of the cells were still in G0/G1 phase, throughout the 2 days (Figure 4C). Indeed, whereas the number of cells in absence of HP1α almost doubled (Figure 4D, left panel), the number of HP1α-transfected cells only varied by 30% (Figure 4D, right panel). These results show that only the few HP1α-overexpressing cells that were engaged in S+G2M phases at day 0.5 could reach the G0/G1 phase at day 2. Nevertheless, HP1α-overexpressing cells differentiated properly thereafter (data not shown).
Taken together, these data show a clear role of the different HP1 isoforms in the timing and the levels of E2F-dependent gene transcription.
HP1α orchestrates NTD
Altogether, our data point to a major role of HP1α in timing the last cell cycle of neuroblasts before engaging NTD. We then investigated the repercussions of an HP1α knockdown on the differentiation process. Validation of specificity has been performed in NIH3T3 and differentiated P19 cells cells (Supplementary Figure S3). Cell cycle phases were measured by FACS analyses on GFP-positive cells after co-transfection of siRNA for HP1α together with GFP. Results were compared with the transfection of an unrelated siRNA-blocking cyclophilin A (siCycA) production. We found that after HP1α knockdown, almost 50% of cells were already cycling in G2/M phase at day 0.5, relative to the 32% in the control experiment. At day 1, the difference was reduced (30% against 19%) but still significant and the percentage of cell death increased (65% versus 40%). So, if HP1α is not expressed, neuroblasts will continue to cycle, but may not be able to prolong this process and die. We then looked at the fate of neuroblasts, in which HP1α was blocked at the beginning of differentiation. HP1α labeling performed on cells transfected with siHP1α revealed that the protein expression level is recovered at least after 5 days of differentiation (Supplementary Figure S3E). Strikingly, at day 8, the neuronal marker MAP2 was no longer expressed by the transfected cells (Figure 5C). Indeed, the left panel shows that all cells express MAP2 after the control siRNA transfection, whereas after siHP1α all GFP-positive cells were MAP2-negative, as well as some GFP-negative cells (Figure 5C, right panel). Altogether, these results demonstrate for the first time a clear role for the α isoform of HP1 in NTD, in controlling the timing of neuroblast cell cycle exit, a phenomenon that is likely to be a prerequisite before the expression of neuronal markers and differentiation.
Figure 5.
HP1α knockdown impairs proper neuronal differentiation and activates e2f-1 expression. P19 cells were co-transfected with pGFP and siRNA directed against cyclophilinA as control or HP1α, 72 h before differentiation induction. (A) Time course analyses of the transfected cell population by FACS. One-way ANOVA test followed by Bonferroni means comparison test was performed. The G2/M values for each time points of siHP1α/GFP-transfected cells were compared to the corresponding times of siCyclophilin A/GFP-transfected cells. * indicates statistical difference compared to control cells with P<0.05 (B). Expressions of e2f1 and hp1α genes in P19 cells transfected with control siRNA against cyclophilin A and HP1α were measured by semiquantitative RT–PCR. Bands were quantified and results are presented as arbitrary units of densitometry. One-way ANOVA test followed by Bonferroni means comparison test was performed. The values for each time points of siHP1α-transfected cells were compared to the corresponding times of siCyclophilin A-transfected cells. ** indicates statistical significance with P<0.01. (C) Immunocytochemistry at day 8 (D8) after differentiation with MAP2 antibody (red) and visualization of GFP-expressing cells (green). The GFP-transfected cells (that previously showed a transient knockdown for HP1α at D1, see Supplementary Figure S3D,E) are not MAP2-positive.
Expression and binding of HP1 isoforms in vivo
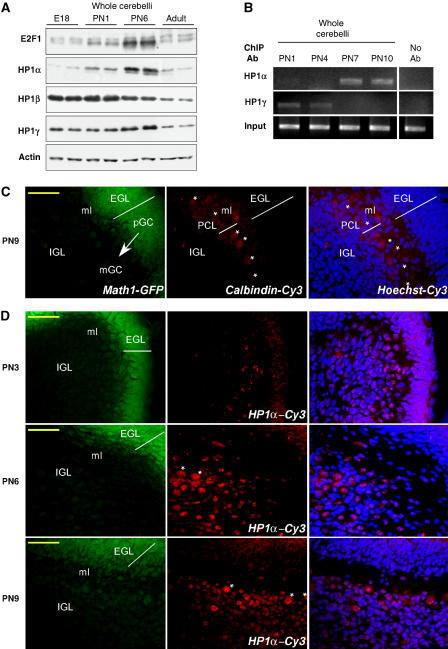
To support our results obtained on CGN cultured in vitro, we next measured E2F1 and HP1 protein levels in total protein extracts from cerebellum obtained at four different stages of development (between embryonic E18 to adult). Unlike most organogenesis, the cerebellum is largely formed after birth. A massive proliferation of granule neuron progenitor cells is observed in the external granule layer (EGL) at the early PN stages, after which CGN begin their differentiation (Wang and Zoghbi, 2001). As shown in Figure 6A, high levels of E2F1 protein expression are indeed detected at postnatal day 6 (PN6). HP1α protein levels were clearly increased at that time, suggesting that HP1α must be present to time cell cycle arrest. HP1β levels decreased over time, whereas HP1γ levels slightly peaked at PN6. Altogether, HP1 levels were decreased in the adult compared to the different PN stages. To define which isoform is indeed bound to E2F sites in vivo, we performed ChIP experiments on cerebelli dissected at critical time points of HP1α and γ expression. Figure 6B shows that the occupancy of the e2f1 gene promoter by the two HP1 isoforms α and γ followed a similar pattern to that observed on isolated CGN (Figure 3B and C). Notably, there was no detectable association of HP1α with the promoter at the early stages (PN1-4), whereas its binding was predominant at PN7-10, when granule neurons are mostly differentiated. In contrast, HP1γ was more strongly associated with the e2f1 promoter at early stages of NTD. Thus, in vivo ChIP experiments demonstrate a dynamic occupancy pattern of the e2f1 promoter, γ isoforms switching to α isoforms during the critical stages of granule neuron development.
Figure 6.
Expression and binding of HP1 proteins in vivo. (A) Cerebelli were dissected at different developmental stages of the mice: embryonic stage 18 (E18), PN1, PN6 and adult (PN50). Western blot analysis was performed with antibodies against E2F1, HP1α, β, γ and actin as indicated. (B) Cerebelli were dissected from mice at PN1, PN4, PN7 and PN10 and ChIP experiments were performed on fresh tissue with antibodies against HP1α or HP1γ with primers specific to the e2f1 gene promoter. Control PCR experiments were performed with total DNA (input) and DNA isolated in the absence of primary antibody, no Ab. A typical experiment is shown. (C) Cerebellar sections from Math1-GFP transgenic mice at PN9 were immunostained for calbindin (middle panel). Most intense GFP was found in the EGL reflecting the normal expression pattern of Math1 in pGC (left panel). An overlay of Hoechst staining and Cy3 immunolabeling is presented on the right panel. The arrow indicates pGC migration through ml to IGL where mGC are localized. Asterisks show single Purkinje cells in PCL. Scale bar 50 μm. (D) Cy3-coupled HP1α immnunostaining was performed on cerebellar sections from Math1-GFP transgenic mice at different PN as noted. Indications are the same as in (C). Note the increased population of HP1α-positive mGC in the IGL at PN6.
Next, we checked for HP1α presence throughout CGN migration in vivo. Once exit from cell cycle, CGN extend axons and migrate more deeply into the internal granule layer (IGL), passing through the developing molecular layer (ml) and the Purkinje cell layer (PCL) (Wang and Zoghbi, 2001). We performed HP1α immunohistochemistry on cerebelli sections at different PN stages, using transgenic animals bearing Math1-driven GFP (Lumpkin et al, 2003). Math1 is normally expressed in progenitor granule cells (pGC) and not in mature granules (mGC), so that progenitors will be readily detected in green (Figure 6C). PCL was located by calbindin immunohistochemistry. PC display very large nuclei (Hoechst dye) at PN9. According to our in vitro studies, we hypothesize that HP1α levels increase must be concomitant with the last cell cycle burst in vivo. Figure 6D shows that a large population of cells are labeled with HP1α, especially at PN6, following the predicted time course of developing CGN. Remarkably, none of the pGC (green cells from the EGL) displays HP1α labeling, a result in line with the fact that they are still cycling. These cells bear small nuclei and are not likely to be PC. Of note, differentiated PC also display high HP1α levels that likely interfered with our western blot studies presented in Figure 6A. At PN9, once in the IGL, HP1α immunoreactivity is decreased, a result in line with western blot analyses performed at that time (data not shown) or at later stages (see levels in the adult, Figure 6A).
Altogether, these results favor a timed increase in HP1α expression in vivo when cerebellar granules stop cycling and begin their differentiation and migration.
HP1α binding links E2F-dependent genes to higher-order structures of chromatin in mature neurons
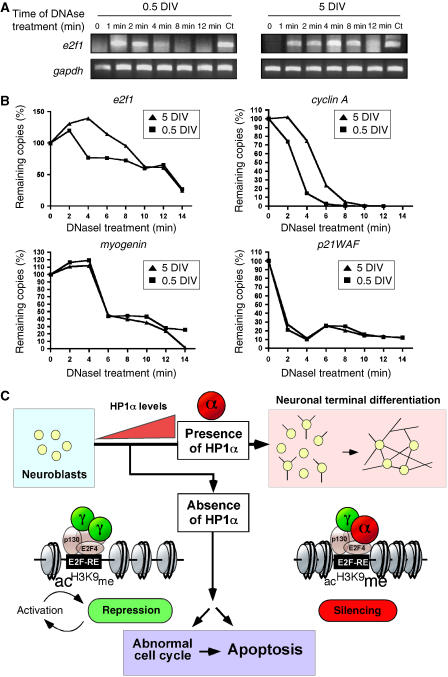
The finely tuned regulation of E2F-dependent genes by HP1α at the onset of NTD led us to postulate that these genes could be more tightly linked to heterochromatin structures in mature neurons than in proliferating neuroblasts. The ability of several gene promoters to be degraded by nucleases was then studied at both stages in primary granule neurons. DNaseI treatment of nuclei isolated at 0.5 and 5 DIV, was followed by amplification of specific promoters by semiquantitative PCR. E2f1 and cyclin A promoters were representative of E2F-dependent genes, whereas myogenin and p21WAF represented non-neuronal and cell cycle regulator gene promoters, respectively. A typical gel obtained for the e2f1 gene is represented Figure 7A. The time course of DNA digestion showed a rapid loss of copy number at 0.5 DIV compared with 5 DIV for both e2f1 and cyclin A promoters, suggesting that these promoters are less accessible to DNaseI treatment in mature neurons (Figure 7B). The myogenin and p21WAF promoters were equally susceptible to DNaseI digestion at both 0.5 and 5 DIV. These data suggest that E2F-targeted genes are indeed packaged into a higher-order chromatin structure in differentiated neurons, a result reminiscent with the fact that HP1α is preferentially bound to their promoter at that time.
Figure 7.
Promoter accessibility tested by DNaseI protection assays in CGN. (A, B) A time course of DNaseI sensitivity was assayed on nuclei isolated from 0.5 and 5 DIV neurons at the time indicated (in minutes). Semiquantitative PCR were run to check the presence of different promoters as noted. Equal loadings were confirmed by control PCR with primers against gapdh. A typical experiment is shown for the e2f1 promoter (A) and quantification relative to gapdh amounts is shown for e2f1, cyclin A, myogenin and p21WAF1 promoters (B). (C) A model showing that, in neuroblasts reaching the quiescent state, transcriptional activation/repression of E2F-targeted genes occurs through p130 and E2F4 under HP1γ control. Lysine9 from histone H3 (H3K9) surrounding E2F-RE within promoters are rather acetylated (ac) than methylated (me). During maturation, HP1α levels increase and take control at E2F-REs. H3K9 on E2F promoters become more methylated, bringing a favorable environment to HP1α binding. Effective silencing of E2F-dependent genes thus ensures neuronal survival throughout the differentiation process. In the absence of HP1α, neuroblasts will either enter an abnormal cell cycle and/or die.
Altogether, these data reveal a new role for HP1 isoforms, more particularly, as a prime regulator allowing NTD onset.
Discussion
Differential binding of HP1 isoforms at E2F-dependent promoters during NTD
Differentiated neurons are completely devoid of replicative activity and the effective silencing of cell cycle genes is an indispensable step of neuronal differentiation. In this paper, we show that several E2F-dependent genes are synchronously transcriptionally downregulated during NTD. In line with previous studies demonstrating that p130/E2F4 complex are needed to maintain cells in a quiescent state (Ghosh and Harter, 2003) and promote E2F-depending genes repression in neurons (Liu et al, 2005), we show that both E2F4 and p130 participate in these events. Earlier studies from Kouzarides and colleagues demonstrated that HP1 is able to bind the Rb protein at the cyclin E promoter (Nielsen et al, 2001b). Herein, we found that the Rb family member p130 can physically interact with the three mammalian HP1 isoforms in neurons, making it a potential candidate to participate in HP1 recruitment at E2F sites. Strikingly, the three HP1s were yet not able to bind E2F sites to the same extent; we found a change in the HP1γ/α isoforms ratio associated with the promoters during NTD. p130 was bound to E2F sites at both stages of differentiation and HP1β, despite displaying binding affinity for p130, did not bind to E2F sites, so fine mechanisms that account for this switch remain to be established. Supporting our data, the recent report from Smallwood et al (2007) describes that HP1γ is present on the survivin gene when it is active, whereas α/β are recruited on silencing.
Our results indicate that modification of HP1 expression levels during NTD is a straightforward mechanism to underlie HP1α presence at E2F sites. HP1α levels were not only markedly increased on the time of differentiation in a homogenous population of CGN in culture, but also HP1α levels decreased, suggesting that the molecular switch between isoforms could at first rely on a simple dose-dependent process. In vivo ChIP experiments also demonstrated differential HP1 binding to e2f1 promoter. Moreover, using Math1-GFP transgenic mice, we provided compelling evidence that the HP1α isoform was localized in postmitotic granular cells migrating towards the IGL of the cerebellum, but not in progenitor cells forming the EGL. These findings confirm that HP1α levels increase in differentiating granule neurons in vivo and strongly support our data showing that HP1α is needed for establishment of the postmitotic status of a mature neuron.
Molecular mechanisms of HP1α binding
We found that H3K9 associated with E2F sites became more methylated during NTD, thus favoring HP1 binding in mature neurons. Interestingly, HP1γ strongly bound these sites in undifferentiated cells, a stage at which H3K9 acetylation was found in larger proportions than H3K9 methylation, likely because neuroblasts are cycling. It is noteworthy that H3K9 methylation is found in the coding regions of actively transcribed genes (Vakoc et al, 2005) and HP1γ could respond to a dynamic release from methylated H3K9 as described during the cell cycle (Dormann et al, 2006), despite the overall abundance in H3K9 acetylation found in neuroblasts. Nevertheless, recent observations indicate that HP1 binding is not necessarily linked to H3K9 methylation (reviewed by Hiragami and Festenstein, 2005) and the presence of alternate factors can ultimately account for HP1 binding dependency (Maison and Almouzni, 2004; Eskeland et al, 2007).
How the binding of each HP1 isoform is targeted at a specific time point of differentiation remains to be fully elucidated. Recent studies demonstrated that HP1 proteins can be extensively modified, similarly to histones, suggesting the existence of an ‘HP1-mediated subcode' (Lomberk et al, 2006). HP1γ was recently shown to associate with transcriptional elongation (Vakoc et al, 2005) through impairment of its silencing activity by phosphorylation (Lomberk et al, 2006). All mammalian HP1 variants display several phosphorylation sites that affect their localization, function (Zhao et al, 2001), self-dimerization and methyl H3K9-binding activity (Badugu et al, 2005), so that a timed phosphorylation modification of one of the HP1 isoforms could influence its binding. Interestingly, transcriptional intermediate factor β, that has been shown to phosphorylate all mammalian HP1 isoforms in vitro (Nielsen et al, 1999), is an HP1 transcriptional co-repressor for progression through differentiation (Cammas et al, 2004).
Alternatively, HP1 has preferred DNA-binding sites (Greil et al, 2003). The occurrence of certain AT-rich motifs could facilitate binding of the different HP1 molecules and representative sequence motifs that could either bind HP1 or HP1c, the Drosophila HP1α and γ homologs, with different affinities have been defined. Interestingly, the mouse e2f1 promoter presents two of these potential ‘HP1-binding boxes' along its E2F-REs. Further work is needed to understand whether the different HP1 isoforms can differentially bind to the e2f promoter sequences, with or without the help of other chromatin modifiers.
Biological meaning of the switch
It is of prime interest to understand why the cell operates such a change between α and γ isoforms. The present study provides compelling evidence that HP1α plays a specific and nonredundant role in cell cycle gene silencing during NDT. Individual functions of HP1 proteins are so far poorly defined. A recent study by Cammas et al (2007) demonstrated the isoform-specific implication of HP1β proteins in differentiation of embryonic carcinoma F9 cells. Our findings also support the notion that despite structural and biochemical similarities, HP1 proteins may exert different functions. We show that HP1α and HP1γ act both as silencers on E2F-dependent transcription in a neuronal context. Interestingly, e2f-dependent promoters are less accessible to DNaseI treatment on HP1α binding. Our interpretation is that HP1γ-induced repression of e2f-targeted genes in neuroblasts would occur within euchromatin during the regular cell cycle and silencing by HP1α would be associated to a higher-order heterochromatin structure. Recently, Smallwood et al (2007) reported that HP1α/β silencing of the survivin gene occurred through p53 recruitment of G9a and dnmt1, thus translating methylation information from histone to DNA and it will be of great interest to characterize other p130/E2F4-associated factors during NTD. Thus, HP1α binding may be necessary to maintain E2F1 silenced throughout the next differentiation steps.
Our results clearly implicate HP1s in regulating cell cycle processes. Strikingly, time-lapse experiments revealed that the three HP1 isoforms induced completely different transcriptional dynamics. Rb was shown to bind HP1 on promoters of cell cycle regulators such as cyclin E (Nielsen et al, 2001b) and deregulated cell cycle kinetics and gene expression are hallmarks of Rb deficient cells (Herrera et al, 1996). Our data showing that p130 binds HP1 on E2F-dependent promoters may thus underlie a mechanism by which HP1 exerts its ability to change cell cycle rate. A recent report showed that HP1 depletion by RNAi induced a drastic cell cycle progression defect (De Lucia et al, 2005), yet the different HP1 isoforms were not investigated. More recently, Auth et al (2006) showed that stable depletion of HP1alpha in NIH3T3 led to a change in cell proliferation rate, as well as an aberrant cell cycle progression. Our siRNA experiments performed in neuronal cells clearly demonstrated that if HP1α is not there in the right time frame, not only the cell cycle is accelerated and e2f1 gene transcription is re-activated, but also neurons do not differentiate properly and do not express typical neuronal markers, such as MAP2. This demonstration of the role of HP1α and γ in regulating E2F-dependent transcription during NTD provides new insights in understanding HP1s function, showing that they also act as important cell cycle regulators.
In conclusion, we present evidence that HP1 isoforms are capable of timing repressions observed at e2f-dependent promoters in differentiating neurons. During NTD, likely because HP1α levels increase, the association of HP1γ on E2F sites progressively switches to a mixed HP1α/HP1γ configuration, an event playing a major role in timing the last cell cycle of granule progenitors (see model in Figure 7C). Nevertheless, inappropriate reactivation of such E2F-dependent genes (cell cycle genes, pro-apoptotic genes) is a hallmark of neuronal death during neurodegenerative diseases and the displaceability of HP1α binding may underlie molecular events implicated in these mechanisms. Overall, this report is the first evidence that HP1s dynamically orchestrate the proper timing of a physiological process, as important as neuronal differentiation.
Materials and methods
Drugs and antibodies
The HDAC inhibitor TSA, poly-ornithin, BrdU, proteinase K and the rabbit polyclonal anti-MAP2 were purchased from Sigma (St-Louis, MO, USA). Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG, rabbit polyclonal E2F4 (A-20), Sp4 and p130 (C-20) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Monoclonal anti-dimethyl histone H3K9, HP1α, HP1γ and rabbit polyclonal anti-acetyl histone H3K9, HP1β antibodies were obtained from Upstate biotechnology (Lake Placid, NY, USA). HRP-conjugated secondary goat anti-rabbit IgG and Cy3-coupled sheep anti-mouse IgG, FITC-coupled goat anti-rabbit IgG were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
Cell cultures
Cerebellar granule neuron (CGN) cultures were obtained from dissociated cerebelli of 7-day-old mice as described previously (Boutillier et al, 2003). P19 embryonic carcinoma cells were cultured in 7.5% fetal calf serum (Harlan Seralab,UK) and 2.5% fetal bovine serum (Gibco)-supplemented MEM medium. Neuronal differentiation was induced by 1 μM ‘all-trans'-RA (Sigma) as described previously (Martins et al, 2005). Differentiation starts with the formation of non-adherent embryonic bodies 2 days after RA treatment. Neuronal maturation is complete in 8 days. All cells were maintained in a humidified incubator at 5% CO2 and 37°C.
Isolation of E2F-RE-binding proteins
We used the microMACS Streptavidin Kit (Miltenyi Biotec GmbH, Germany) for binding assays. Briefly, 400 μg of CGN nuclear protein extracts (Dignam et al, 1983) were mixed with microMACS Streptavidin MicroBeads and applied to a microColumn staying in a magnetic field. Nonspecifically bound molecules were removed by four steps of stringent washes and bound proteins were eluted. All fractions were analyzed by western blot. See details of the protocol in Supplementary Data S5.
Chromatin immunoprecipitation
In vitro ChIP experiments (CGN cultures) were performed as described previously (Boutillier et al 2003). In vivo ChIP experiments on whole cerebelli were performed identically, except for the first steps (see Supplementary Data S5).
Immunocytochemistry
CGN. Cells were previously incubated with 50 μM of BrdU for 4 h when required, fixed with 4% paraformaldehyde (15 min) and permeabilized with 0.1% Triton X-100 plus 5% goat serum in PBS. Cells were then treated with 2 N HCl, 10 min, 37°C and subsequently incubated overnight with the primary antibody and 1 h with secondary antibodies. Staining was visualized with a Leitz microscope. Photographs were taken with Nikon Digital Camera Dxm 1200.
P19 cells. For suspension of embryonic bodies (1 ml), all the steps described below were preceded by a 3 min spin at 5000 r.p.m. to pellet the cells. Cells were rinsed twice with PBS and subsequently fixed with 4% paraformaldehyde (15 min). Cells were blocked (1% BSA, 0.1% Triton X-100 in PBS), and primary antibody was incubated for 2 h (HP1α, euromedex; MAP2, AbCam). After three washes, secondary Cy3-conjugated anti-mouse IgG antibody (Sigma) was incubated for 1 h. Cells in suspension were transferred in glass-bottom Iwaki dishes for microscope observation (LSM510; Carl Zeiss MicroImaging Inc.) with a plan-apochromat × 63/1.4 NA oil immersion objective.
Immunohistochemistry
Brains of Math1-GFP transgenic mice (Lumpkin et al, 2003) were dissected and directly frozen in isopentane cooled to −40°C in dry ice. Cryostat sections (10 μM) of cerebelli were postfixed with 3% paraformaldehyde for 5 min and permeabilized with 0.5% Triton X-100 in PBS for 4 min. Slides were blocked in 5% horse serum and 5% fetal calf serum in PBS. Primary antibodies were incubated overnight (HP1α, euromedex, Calbindin, Chemicon). Sections were then washed three times with PBS and incubated for 45 min with appropriate secondary antibodies and the Hoechst dye 33342 (1 μg/ml).
Plasmids and gene transfer
Plasmids encoding mouse HP1 variants and mutation were a kind gift from Dr R. Losson (Strasbourg, France). −176 to +63/E2F-luc was obtained from Dr P. Farnham (Wisconsin, USA). pEGFP was from Clonetech. P19 cells were transfected using Fugene-6 (Roche Applied Sciences, UK) according the manufacturer's instructions.
Live cell imaging of the luminescent reporter gene
After induction of the differentiation and luciferin addition (0.5 μM, Biosynth AG, Switzerland), 2 ml of cells were plated onto 35 mm glass coverslip culture dishes (Iwaki), and incubated on the microscope stage at 37°C, 5% CO2. Imaging was carried out using a Zeiss Axiovert 100 microscope with a Fluor × 10 0.5 NA objective. The photons emitted were collected using a Hamamatsu VIM intensified CCD camera (Hamamatsu Photonics Ltd, UK). A series of images were acquired using a 30-min integration time over a period of 24 h. AQM advanced 6 Software (Kinetic Imaging, UK) was used for image analysis with background correction. All these experiments were performed at least three times and in each experiment at least 30 cells were recorded and analyzed.
Flow cytometric analysis
At 24 h before induction of differentiation, P19 cells were co-transfected with HP1α and EGFP-expressing plasmids. At indicated time points, 1 ml of cell suspension was taken, centrifuged, PBS washed and fixed with 0.5% paraformaldehyde for 10 min. After two subsequent PBS washes, the cells were resuspended in 1 ml of PBS and stained by addition of 250 μl of 50 μg/ml propidium iodide before analysis in a flow cytometer (Ortho Cytoron Absolute). When co-transfection is performed, most of the cells are coexpressing both plasmids. We therefore used the presence of EGFP to sort the transfected cell population (also expressing HP1α) from the nontransfected cells. The same experiment was carried out with the control empty plasmid (pSG5).
siRNA transfection
P19 cells where co-transfected in 60-mm dishes with 90 pmoles of siRNA-HP1α (5′-AATTCAGGACAATCCAAGTTC-3′) or siRNA-Cyclophilin A (Upstate Biotechology) as negative control along with 1.5 μg of pEGFP-expressing vector using 7.5 μl of lipofectamine 2000 (Invitrogen). At 70 h after transfection, 5 × 105 cells/ml were used to induce differentiation in 10 cm dishes as described above. A 1 ml portion of cell suspension was used for FACS analysis at described time points or to perform immunocytochemistry.
DNase I-sensitivity assays
Nuclei were isolated from cultured neurons using the first steps of Dignam et al (1983), resuspended in digestion buffer (see Supplementary Data S5) and 20 U of DNase I (Promega) were added to 200 μl aliquots for 0–15 min at 25°C. Reactions were stopped with 5 μl of 0.5 M EDTA, pH 8.0. After proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation, the purified DNA was dissolved in 50 μl and PCR were performed with primers described in Supplementary Data S5.
For western blot, immunoprecipitation and RT–PCR analyses, quantifications and statistics see Supplementary Data S5.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Data S5
Acknowledgments
IP has an INSERM fellowship (2006-2007). We thank the French Ministry and ‘Association Française pour les Myopathies, AFM' for supporting her in 2003–2004 and 2005, respectively. SB was a recipient of the French association ‘France Alzheimer' (2003 and 2004). HP1 variants and E2F plasmids were generously provided by R Losson (Strasbourg, France) and P Farnham (Wisconsin, USA), respectively. We thank L Dupuis (Strasbourg, France) and R Losson for critical reading of an earlier version of this manuscript. We also thank J Johnson for helpful comments on Math1-GFP mice. The laboratory is supported by grants from ARC, FRC, ARS and AREMANE. We thank funding of the UK BBSRC (David Phillips fellowship) to VS and Ligue Nationale contre le Cancer, PIC Rétinoblastome, Cancéropôle Idf, ANR (2005-005396) to GA.
References
- Auth T, Kunkel E, Grummt F (2006) Interaction between HP1alpha and replication proteins in mammalian cells. Exp Cell Res 312: 3349–3359 [DOI] [PubMed] [Google Scholar]
- Badugu R, Yoo Y, Singh PB, Kellum R (2005) Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution. Chromosoma 113: 370–384 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Boutillier AL, Trinh E, Loeffler JP (2002) Constitutive repression of E2F1 transcriptional activity through HDAC proteins is essential for neuronal survival. Ann N Y Acad Sci 973: 438–442 [DOI] [PubMed] [Google Scholar]
- Boutillier AL, Trinh E, Loeffler JP (2003) Selective E2F-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. J Neurochem 84: 814–828 [DOI] [PubMed] [Google Scholar]
- Cammas F, Herzog M, Lerouge T, Chambon P, Losson R (2004) Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev 18: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas F, Janoshazi A, Lerouge T, Losson R (2007) Dynamic and selective interactions of the transcriptional corepressor TIF1beta with the heterochromatin protein HP1 isotypes during cell differentiation. Differentiation, Mar 23 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- De Lucia F, Ni JQ, Vaillant C, Sun FL (2005) HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res 33: 2852–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W (2006) Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle 5: 2842–2851 [DOI] [PubMed] [Google Scholar]
- Eskeland R, Eberharter A, Imhof A (2007) HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol Cell Biol 27: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ (2004) Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117: 2173–2181 [DOI] [PubMed] [Google Scholar]
- Ghosh MK, Harter ML (2003) A viral mechanism for remodeling chromatin structure in G0 cells. Mol Cell 12: 255–260 [DOI] [PubMed] [Google Scholar]
- Greil F, van der Kraan I, Delrow J, Smothers JF, de Wit E, Bussemaker HJ, van Driel R, Henikoff S, van Steensel B (2003) Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev 17: 2825–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera RE, Makela TP, Weinberg RA (1996) TGF beta-induced growth inhibition in primary fibroblasts requires the retinoblastoma protein. Mol Biol Cell 7: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K, Neve R, Ackerman SL, Copani A (2004) Divide and die: cell cycle events as triggers of nerve cell death. J Neurosci 24: 9232–9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiragami K, Festenstein R (2005) Heterochromatin protein 1: a pervasive controlling influence. Cell Mol Life Sci 62: 2711–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DO, Mattei MG, Horsley D, Cowell IG, Singh PB (2001) The gene and pseudogenes of Cbx3/mHP1 gamma. DNA Seq 12: 147–160 [DOI] [PubMed] [Google Scholar]
- Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnins VI (1982) RA induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol 94: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Liu DX, Nath N, Chellappan SP, Greene LA (2005) Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev 19: 719–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE (2003) Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns 3: 389–395 [DOI] [PubMed] [Google Scholar]
- Martins AH, Resende RR, Majumder P, Faria M, Casarini DE, Tarnok A, Colli W, Pesquero JB, Ulrich H (2005) Neuronal differentiation of P19 embryonal carcinoma cells modulates kinin B2 receptor gene expression and function. J Biol Chem 280: 19576–19586 [DOI] [PubMed] [Google Scholar]
- Maison C, Almouzni G (2004) HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304 [DOI] [PubMed] [Google Scholar]
- Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R (2006) Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol 8: 407–415 [DOI] [PubMed] [Google Scholar]
- Nicolas E, Roumillac C, Trouche D (2003) Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol Cell Biol 23: 1614–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J 18: 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R (2001a) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell 7: 729–739 [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T (2001b) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 25: 338–342 [DOI] [PubMed] [Google Scholar]
- Smallwood A, Esteve PO, Pradhan S, Carey M (2007) Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev 21: 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA (2005) Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 19: 381–391 [DOI] [PubMed] [Google Scholar]
- Wang VY, Zoghbi HY (2001) Genetic regulation of cerebellar development. Nat Rev Neurosci 2: 484–491 [DOI] [PubMed] [Google Scholar]
- Zhao T, Heyduk T, Eissenberg JC (2001) Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J Biol Chem 276: 9512–9518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Data S5