Abstract
We have identified 24 members of the DnaK subfamily of heat shock 70 proteins (Hsp70s) in the complete genomes of 5 diverse photosynthetic eukaryotes. The Hsp70s are a ubiquitous protein family that is highly conserved across all domains of life. Eukaryotic Hsp70s are found in a number of subcellular compartments in the cell: cytoplasm, mitochondrion (MT), chloroplast (CP), and endoplasmic reticulum (ER). Although the Hsp70s have been the subject of intense study in model organisms, very little is known of the Hsp70s from early diverging photosynthetic lineages. The sequencing of the complete genomes of Thalassiosira pseudonana (a diatom), Cyanidioschyzon merolae (a red alga), and 3 green algae (Chlamydomonas reinhardtii, Ostreococcus lucimarinus, Ostreococcus tauri) allow us to conduct comparative genomics of the Hsp70s present in these diverse photosynthetic eukaryotes. We have found that the distinct lineages of Hsp70s (MT, CP, ER, and cytoplasmic) each have different evolutionary histories. In general, evolutionary patterns of the mitochondrial and endoplasmic reticulum Hsp70s are relatively stable even among very distantly related organisms. This is not true of the chloroplast Hsp70s and we discuss the distinct evolutionary patterns between “green” and “red” plastids. Finally, we find that, in contrast to the angiosperms Arabidopsis thaliana and Oryza sativa that have numerous cytoplasmic Hsp70, the 5 algal species have only 1 cytoplasmic Hsp70 each. The evolutionary and functional implications of these differences are discussed.
INTRODUCTION
The heat shock 70 proteins (Hsp70s) are a ubiquitous protein family that is highly conserved across all domains of life (Gupta and Golding 1993; Karlin and Brocchieri 1998). The Hsp70s are chaperones and are crucial housekeeping proteins. They have roles in the transport of proteins across membranes into organelles, the folding of newly translated proteins, and the repair of misfolded proteins (Bukau and Horwich 1998; Hartl and Hayer-Hartl 2002; Mayer and Bukau 2005). During times of heat stress, certain Hsp70s are upregulated and participate in the refolding of denatured proteins (Bukau and Horowich 1998; Hartl and Hayer-Hartl 2002; Mayer and Bukau 2005). All Hsp70s possess 3 distinct domains: an N-terminal adenosine triphosphatase (ATPase) domain of approximately 400 amino acids, a substrate-binding domain of approximately 200 amino acids, and a highly variable C-terminal domain.
Eukaryotes possess at least 3 types of Hsp70s, each of which localizes to a different cellular compartment: cytoplasm, mitochondrion (MT), and endoplasmic reticulum (ER). In addition, photosynthetic eukaryotes also possess chloroplast (CP) localized Hsp70s. The Hsp70s targeted to specific subcellular compartments share a close evolutionary history (Boorstein et al 1994; Rensing and Maier 1994; Karlin and Brocchieri 1998; Nikolaidis and Nei 2004). Evolutionary analysis of the Hsp70s reveals that they have evolved via 2 different pathways: gene duplication with subsequent divergence (in the case of the ER and cytoplasmic Hsp70s) and endosymbiosis with lateral gene transfer to the nucleus (the MT and CP Hsp70s) (Boorstein et al 1994; Gupta and Golding 1993; Karlin and Brocchieri 1998). Although the evolutionary history of the Hsp70s has been of considerable interest, the taxonomic sampling in previous studies has been uneven, primarily due to a lack of complete genome sequence data. For instance, Lin et al (2001) compared the Hsp70s in the complete genome of Arabidopsis thaliana (an angiosperm) to those found in yeast. The great evolutionary distance in this comparison was due to the lack of any complete genome datasets for any other photosynthetic eukaryotes. The recent sequencing of the complete genomes of a diatom Thalassiosira pseudonana, a red alga Cyanidioschyzon merolae, and 3 green algae (Chlamydomonas reinhardtii, Ostreococcus lucimarinus and O. tauri), now allow us to conduct comparative genomics studies of the Hsp70s present in diverse photosynthetic eukaryotic lineages. The purpose of this study was to identify Hsp70 homologs, analyze trends of Hsp70 evolution, and examine hypotheses concerning the diversity of Hsp70s. We hope this work will facilitate future studies of Hsp70s in these and related species.
MATERIALS AND METHODS
Identification of algal Hsp70 homologs
We use the term algae or algal to refer to aquatic photosynthetic eukaryotes. Algae are a diverse group of organisms that all share plastids. Algae are not a monophyletic group and we cannot assume that the organisms themselves have close evolutionary relationships.
The Hsp70 sequences were obtained from the Joint Genome Institute (JGI) genome sites: Thalassiosira pseudonana v3.0 (http://genome.jgi-psf.org/thaps3/thaps3.home.html), Chlamydomonas reinhardtii v3.0 (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), Ostreococcus lucimarinus v2.0 (http://genome.jgi-psf.org/Ost9901_3/Ost9901_3.home.html), and Ostreococcus tauri v.2.0 (http://genome.jgi-psf.org/Ostta4/Ostta4.home.html). The C. merolae genome site can be found at (http://merolae.biol.s.u-tokyo.ac.jp/). The databases were queried by both keywords (Hsp70 and heat shock protein 70) and sequence similarity using BLAST (Altschul et al 1997) searches with A. thaliana Hsp70 sequences (Lin et al 2001). We used an E-value cut-off of less than 0.001. The genome databases had differing levels of annotation and, in some genomes, multiple gene models for the same chromosomal locations were found during the searches. The most complete gene model for each chromosomal location was chosen for study. These gene models were compared to known expressed sequence tag (EST) sequences (see EST database searches section for details). The estimated molecular weights for each protein were determined by using the ProtParam program (Wilkins et al 1999).
Hsp70 protein nomenclature
To easily refer to the proteins discovered in the genome databases examined, we have designated the following naming system: for those HSP70 proteins from Thalassiosira pseudonana, Tphsp70-x; C. merolae, Cmhsp70-x; C. reinhardtii, Crhsp70-x; O. lucimarinus Olhsp70-x; O. tauri; Othsp70-x. The letter x denotes the protein number. This number is given so that the many Hsp70s in each genome can be identified individually. The list of the Hsp70s used in the phylogenetic analysis along with their gene accessions numbers is available in online
.
Phylogenetic analysis
In order to understand the origins and evolution of the Hsp70s in the diverse species studied here, the Hsp70 protein sequences were imported into the BioEdit Sequence Alignment Editor program (v7.0.5; Hall 1999) and aligned with ClustalW (Thompson et al 1994). Further refinement of the alignment was performed by hand. In this alignment, we included the Hsp70s identified in the 5 genomes mentioned above. In addition, we included Hsp70 homologs from other eukaryotes for which complete genome sequences are available, including Saccharoymces cerevisiae, Plasmodium falciparum, Plasmodium yoelii, Arabidopsis thaliana, and Oryza sativa. Our choice of Hsp70s included in this alignment was guided by the availability of complete genome data and the desire to include taxa that are closely related to the photosynthetic eukaryotes. For example, the evolutionary jump from green algae to angiosperms (A. thaliana and O. sativa) is large, but this is due to the lack of available genome datasets. P. falciparum and P. yoelii were included because they are both apicoplexans, they have relictual plastids that are of red algal origin, and they represent an important early diverging eukaryotic lineage (Baldauf et al 2000; Keeling 2004a, 2004b). Additional Plasmodium and other parasitic protist genomes exist; however, addition of these genomes may unnecessarily include additional divergent or long branches in our analysis. The need to clarify the evolutionary relationships of the CP Hsp70s led us to include CP-genome encoded Hsp70s from 5 red algae (Cyanidium caldarium, Gracilaria tenuistipitata, Porphyra haitanensis, Poryphyra purpurea, and Porphyra yezoensis), 1 cyroptophyte (Guillardia theta), and 1 diatom (Odontella sinensis). The DnaK proteins from the cyanobacteria Synechocystis sp strain PCC6803 and Escherichia coli also were included. Eighty-one sequences were in the final alignment. The full list of species and accession numbers is available in online
.
For the phylogenetic analysis, we excluded the variable and difficult to align C-terminal domain. In addition, the variable N-terminal region containing transit or leader sequences also was excluded. The alignment was then of the highly conserved ATPase and peptide-binding domains. However, some amino acid insertions were present in just single or a few proteins; these regions were removed from the alignment. The final alignment used for phylogenetic analysis is available as online
.
The phylogenetic relationships of the Hsp70s were analyzed with 2 different phylogenetic tree construction methods: neighbor-joining (NJ) in MEGA v.4.0 (Kumar et al 2004) and Bayesian in MrBayes v.3.1.2 (Ronquist and Huelsenbeck 2003). In MEGA, distance matrices were generated using the pairwise deletion option with the Dayhoff amino acid matrix. One thousand bootstrap replicates were created and trees were generated using NJ for each replicate. The bootstrap values reported for each branch reflect the percentage of the 1000 trees that contained that branch.
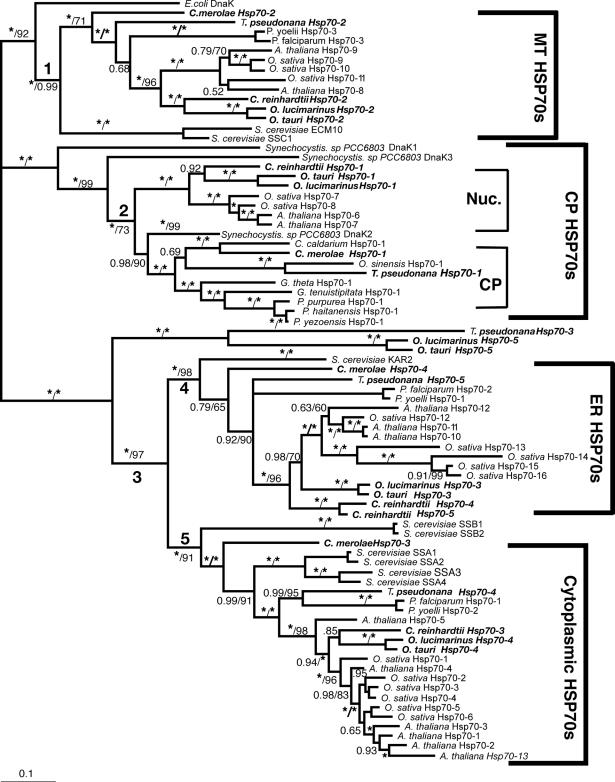
In MrBayes, we first performed an initial analysis using the mixed amino acid model. This analysis was conducted as described in the program manual (section 4.2.2 in MrBayes v.3.1; Ronquist et al 2005) and determined that the best fixed-rate model of protein evolution for the alignment was the WAG model (Whelan and Goldman 2001). This model (WAG) then was used in our subsequent phylogenetic analysis. Metropolis-coupled Markov chain Monte Carlo (MCMCMC) from a random starting tree was initiated in the Bayesian inference and run 2 000 000 generations with a sample frequency of 1000, print frequency of 100, and 4 chains. Three of the 4 chains run were heated and 1 was cold. All other settings or priors were set to the default used in MrBayes. We determined that the chains converged (the average standard deviation of the split frequencies was below 0.01) after 250 000 generations; this was used as our “burnin” and the first 250 trees were discarded. A consensus was created from the remaining trees (1750) and is presented in Figure 2. The topology of the NJ tree was highly congruent with the Bayesian tree and therefore only the bootstrap values are reported for the NJ analysis.
Fig 2.
EST database searches
Sequences obtained from the genome databases were used as queries in BLAST searches of available EST databases to determine if the genes are expressed. These EST databases can be found at the C. merolae genome site, a diatom site (http://avesthagen.sznbowler.com/), the Chlamy Center website (http://www.Chlamy.org/cgi-bin/webblast.pl), and an O. tauri EST site (http://bioinformatics.psb.ugent.be/blast/public/?project=ostreococcus).
The JGI T. pseudonana and C. merolae and O. tauri EST databases do not specify the conditions in which their ESTs were compiled. Therefore, for these species, there is no information on differential expression of Hsp70 EST sequences. However, the Chlamy Center database does list 7 different cDNA libraries from which ESTs were collected: core (normalized), S1D2 (normalized), deflagellation (pH shock and flagellum regrowth), gamete and zygote (nitrogen-deficient medium and collection during gametogenesis), and stress I, stress II, stress III. It is important to note that stress I and II cDNA libraries did not involve heat shock, but instead were grown in varying light conditions and TAP mediums with NO3, NH4, H2O2, and sorbitol. In addition, the stress III cDNA library was made from Chlamydomonas reinhardtii that had been exposed to different levels of copper (Shrager et al 2003). In the EST searches, only sequence matches of 95% sequence identity or higher were considered hits. A higher level of stringency could unnecessarily exclude true matches due to minor sequencing errors. A lower stringency could reflect a match to a closely related but still distinct homolog.
Subcellular predictions
The newly identified Hsp70 sequences were submitted to the prediction programs Psort, Predotar, and TargetP (Nakai and Horton 1999; Emanuelsson et al 2000; Small et al 2004) to determine their possible subcellular localization. Subcellular predictions also were based on the phylogenetic affinity or relationship of the proteins to other proteins with experimentally determined cellular locations (Heazlewood et al 2004).
RESULTS
The green algae
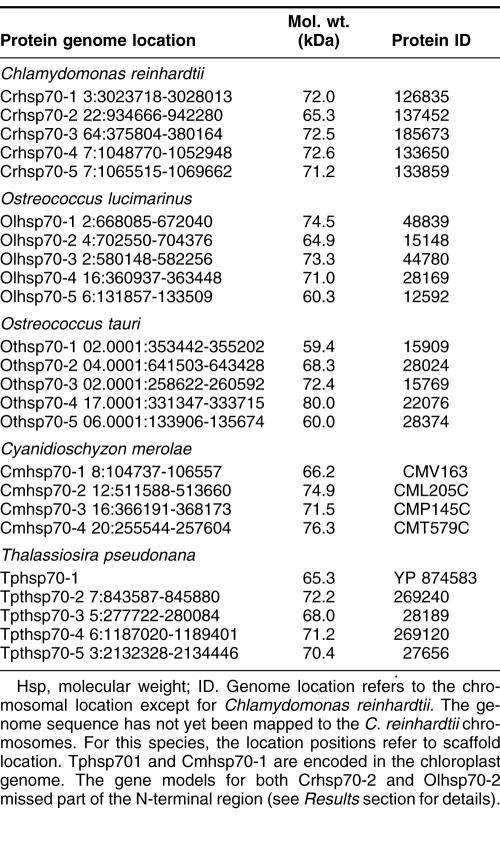
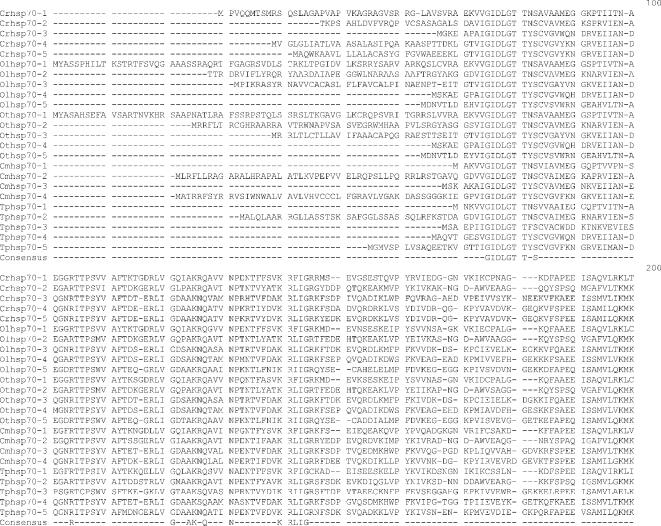
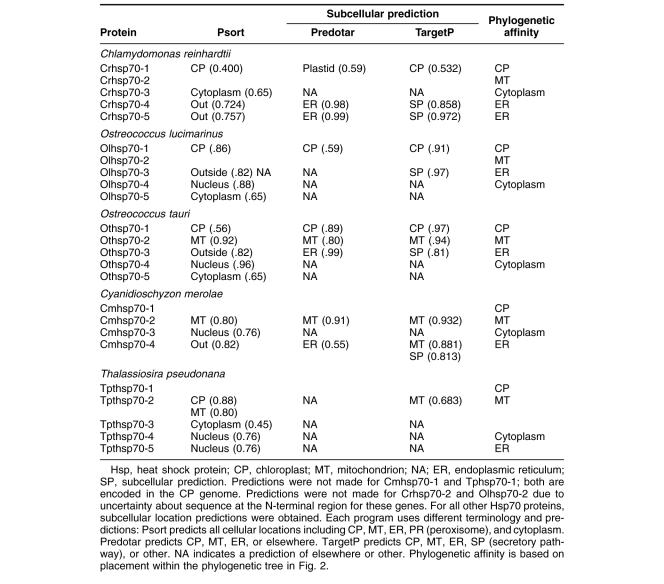
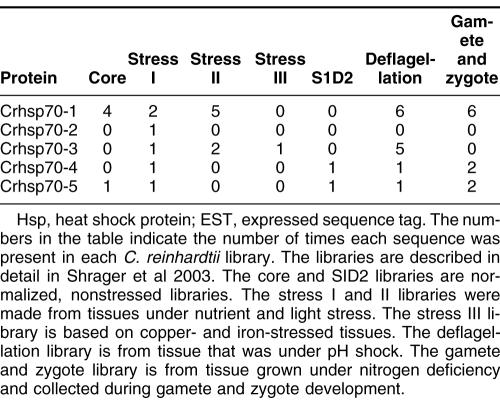
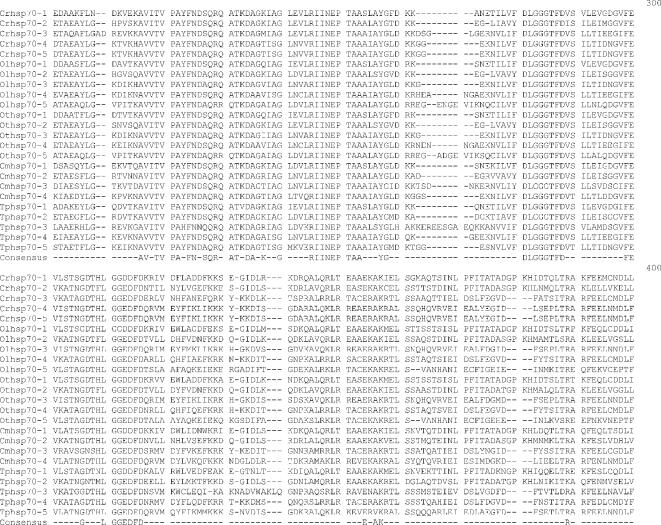
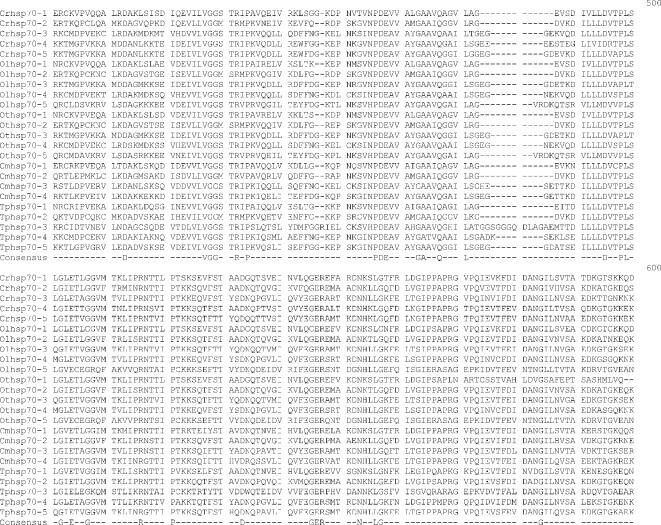
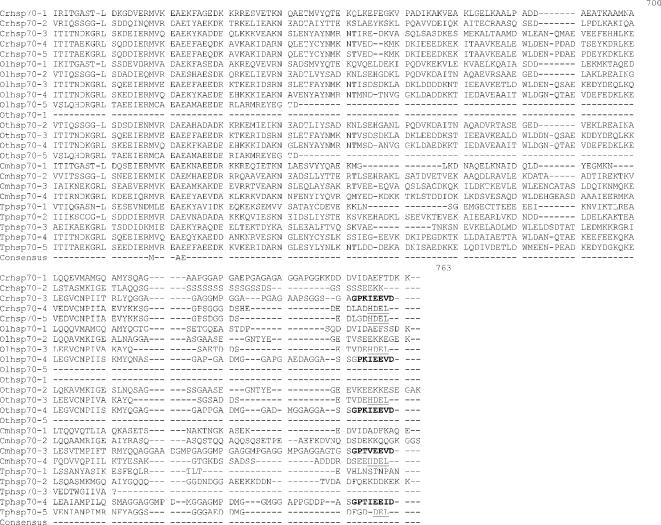
Five full-length Hsp70s from the DnaK subfamily were found in the Chlamydomonas reinhardtii nuclear genome (Table 1). Analysis of version 1 of the C. reinhardtii genome reported 7 Hsp70s (Schroda 2004); however, an analysis of version 2 and 3 data revealed that these additional 2 Hsp70 sequences are not complete with significant portions of usually conserved regions missing. Of the 5 Chlamydomonas reinhardtii Hsp70s, only 1, Crhsp70-3, is clearly a cytosolic protein. It has been established that cytosolic HSP70s have a conserved amino acid motif GP(T/K)(V/I)EEVD at their C-terminus (Boorstein et al 1994; Sung et al 2001). Crhsp70-3 contains the conserved cytosolic C-terminal sequence of GPKIEEVD and lacks any N-terminal signal or transit sequence (Fig 1). Crhsp70-3 is also clearly a member of the cytoplasmic Hsp70 family or lineage (Fig 2).
Table 1.
Algal HSP70 proteins of the DnaK subfamily
Fig 1.
Alignment of heat shock 70 proteins (HSP70s) amino acid sequences from Chlamydomonas reinhardtii (Cr), Ostreococcus lucimarinus (Ol), Ostreococcus tauri (Ot), Cyandioschyzon merolae (Cm), and Thalassiosira pseudonana (Tp). Amino acid residues 1–70 in the alignment include the variable N-terminal region. This region is absent in cytoplasmically localized HSP70s and contains the transit sequences for mitochondrion (MT), chloroplast (CP), and endoplasmic reticulum (ER) HSP70s. The much more highly conserved adenosine triphosphatase (ATPase) domain includes residues 70–475. This region displays considerable sequence conservation but also has regions of insertion or deletion of 1 to a few residues. The peptide-binding domain (residues 490–645) is extremely well conserved. The variable C-terminal region (645–760) is absent in some proteins and highly variable in others, and its function is not well established. It also contains ER and cytoplasmic consensus sequences. The cytoplasmic consensus sequence GP(T/K)(V/I)EEVD at residues 762–769 is in bold. The ER consensus sequence HDEL at residues 765–769 is underlined
All of the other C. reinhardtii Hsp70 proteins possess some kind of transit sequence. Crhsp70-4 and Crhsp70-5 both possess N-terminal transit sequences and the C-terminal ER retention signal HDEL, suggesting that these are ER proteins. The subcellular prediction programs indicate that these are targeted to the ER (Table 2), and both of these proteins are members of the ER Hsp70 lineage (Fig 2). Crhsp70-1 also possesses an N-terminal transit sequences (Fig 1) and all of the subcellular prediction programs indicate that this is a CP protein (Table 2). The phylogenetic placement of Crhsp70-1 within the lineage containing the CP-localized Hsp70s from A. thaliana and O. sativa (Fig 2) is consistent with this cellular location. The gene findings programs at the C. reinhardtii web site start Cmhsp70-2 at the MEG at positions 77–79 on the alignment (Fig 1). Our examination of the sequences 5′ of this ATG found addition sequence including a region coding for the conserved GIDLGTT region at resides 75–81 in Figure 1. With the addition of these amino acid residues, this protein appears to have a transit sequence but the true start of this protein is not known. EST clones that are an exact match to this gene are present in the EST databases but they are incomplete and do not include the start methionine. However, the phylogenetic placement of this protein clearly indicates that it is an MT-localized protein. Analysis of the EST data for all of the C. reinhardtii Hsp70s indicates that all the Hsp70 genes are expressed but that there is differential expression (Table 3). For instance, Crhsp70-2, Crhsp70-3, and Crhsp70-4 are not found in the core library but are found in other libraries.
Table 2.
Algal HSP70 predicted subcellular locations
Table 3.
Number of matches to Chlamydomonas reinhardtii Hsp70s in C. reinhardtii EST libraries
Each Ostreococcus genome (O. lucimarinus and O. tauri) contains 5 Hsp70s. One is a nuclear-encoded CP Hsp70 (Olhsp70-1, Othsp70-1); another is an MT Hsp70 (Olhsp70-2, Othsp70-2). One ER Hsp70 (Olhsp70-3 and Othsp70-3), 1 cytoplasmic Hsp70 (Olhsp70-4, Othsp70-4), and finally 1 Hsp70 of uncertain location (Olhsp70-5 and Othsp70-5; Tables 1 and 2, Figs 1 and 2) exist. The cytoplasmic Hsp70s contain the conserved consensus motif, and the organelle-localized proteins have the required N-terminal transit sequences (Fig 1). Although all the O. tauri Hsp70s were represented in the EST database, the cytoplasmic Othsp70-4 was the most highly represented at 132 matches compared to between 1 and 8 matches for the other Hsp70s. The O. tauri CPHsp70, Othsp70-1, is shorter than most other CPHsp70 and Olhsp70-1. It is likely that this gene model is correct. No sequence homologous to the C-terminal region of Olhsp70-1 was found in the O. tauri genome. In addition, this short gene model is supported by EST data. The 2 proteins of uncertain location (Olhsp70-5 and Othsp70-5) are closely related to Tphsp70-3. Both Olhsp70-5 and Othsp70-5 are shorter than the other HSP70s and are lacking the variable C-terminal region. These gene models are consistent with EST data, indicating that these proteins are expressed. The phylogenetic placement, outside of the ER+cytoplasmic lineage, of these proteins is not affected by their lack of a C-terminal domain because, due to its high level of variability, this region was excluded from the phylogenetic analysis. We also performed phylogenetic analyses with an even shorter alignment (with less gaps for these proteins) and it had the same topology as the tree in Figure 2. Due to their placement outside of the other Hsp70 lineages, it is not possible to predict where they are found in the cell.
Based on analysis of the EST clone sequences it is clear that both the O. lucimarinus (Olhsp70-2) and O. tauri (Othsp70-2) MT Hsp70s have N-terminal extensions not reflected in the gene models found at the JGI genome sites. A full-length EST sequence with a 100% match to the DNA sequence of Othsp70-2 (clone ot04g04210) was identified in the Ostreococcus EST database. This EST clone contains a clear MT-target sequence. DNA sequence encoding this N-terminal region is present in the genome sequence for OtHsp70-2 and a very similar region was identified in Olhsp70-2; however, the exact start Met residue for Olhsp70-2 is still uncertain. The additional N-terminal sequence protein sequence is presented in Figure 1. Both Othsp70-2 and Olhsp70-2 are clearly members of the MT family of Hsp70s (Fig 2). This is very similar to the situation with the missing N-terminal region of the C. reinhardtii MT Hsp70.
C. merolae: a red alga
Three Hsp70s from the DnaK subfamily have been found in the C. merolae nuclear genome (Table 1). Also an Hsp70 is in the C. merolae CP genome (Table 1). All of these Hsp70s are represented in the C. merolae EST database. Of the 4 C. merolae Hsp70s, only 1, Cmhsp70-3 (71.5 kDa), has a cytoplasmic Hsp70 sequence motif: GPTVEEVD (Fig 1). Cmhsp70-4 has a N-terminal transit sequence and ends in HDEL, suggesting that this could be an ER protein. This is consistent with its phylogenetic placement (Fig 2) and with the subcellular predictions (Table 2). Cmhsp70-2 also possesses an N-terminal transit sequence (Fig 1). The results of the subcellular predictions (Table 2) and the phylogenetic analysis indicate that Cmhsp70-2 is targeted to the mitochondria. The CP Hsp70, CmHsp70-1, is found within the larger plastid Hsp70s but is within the subfamily of CP-encoded Hsp70s, including Tphsp70-1 and other red algal and diatom CP Hsp70s (Fig 1).
Thalassiosira pseudonana: a diatom
Thalassiosira pseudonana has 5 Hsp70s. One Hsp70 (Tphsp70-1) is encoded in the CP genome (Table 1; Fig 2). An additional 4 genes for Hsp70s were identified in the JGI Thalassiosira pseudonana v3.0 nuclear genome (Table 1). All of these Hsp70s are represented in the Diatom EST database. Analysis of the alignment in Figure 2 reveals that Tphsp70-2 possesses an N-terminal leader sequence, suggesting either CP or MT localization (see also Table 2). Based on phylogenetic relationship (Fig 2) and subcellular prediction (Table 2) it is clear that Tphsp70-2 is MT protein. Tphsp70-4 contains the cytoplasmic amino acid motif (GPTIEEID). This evidence plus the placement of this protein within the cytoplasmic lineage suggests that it is cytoplasmically localized. Tphsp70-5 possesses a short N-terminal signal sequence and ends in DDEL (Fig 1), which indicates that this is ER localized. The ER location is consistent with Tphsp70-5 placement in the ER family in the phylogenetic tree (Fig 2). Tphsp70-3 lacks any N-terminal signal or transit sequence, suggesting a cytoplasmic location (Table 2; Fig 1). However, the C-terminal region is shorter than the cytoplasmic protein, and Tphsp70-3 lacks the cytoplasmic consensus region. Further, its placement outside of both the ER and cytoplasmic lineages (Fig 2) makes a prediction based on phylogenetic relationships problematic. At this time, the cellular location of this protein is unknown.
Gene family evolution
Analysis of the phylogenetic tree of Hsp70s in Figure 2 indicates that there are 5 well-supported lineages of Hsp70s. The first lineage (the branch leading to this lineage is labeled 1) includes all the MT-located Hsp70s (Fig 2), which are all nuclear encoded. The second major lineage includes the plastid Hsp70s. This lineage is closely related to Synechocystis DnaK1 and DnaK3, and it includes Synechocystis DnaK2. Within this CP lineage there are 2 distinct subfamilies. One subfamily includes the “green” plastid Hsp70s (the angiosperm and green algal CP Hsp70s). These proteins are all nuclear encoded. The other CP Hsp70 subfamily or the “red” plastid lineage includes the C. merolae and T. pseudonana CP-encoded Hsp70s, along with other CP-encoded Hsp70s. Another major branch in the Hsp70 tree (branch 3) includes both the ER and cytoplasmic Hsp70s. Within this lineage there are the well-supported ER (branch 4) and cytoplasmic (branch 5) Hsp70 lineages. Three Hsp70s fall outside of the ER+cytoplasmic lineage: Tphsp70-3, Olhsp70-5, and Othsp70-5.
DISCUSSION
In this study, we identified Hsp70 homologs in 5 complete genomes: C. reinhardtii, O. lucimarinus, O. tauri, T. pseudonana, and C. merolae. We found that each species had between 4 and 5 Hsp70s, with at least 1 each belonging to CP, MT, ER, and cytoplasmic lineages. This is considerably less than the 14 Hsp70s present in the A. thaliana genome and the 18 Hsp70s in O. sativa. As we describe the relationships and evolutionary history of the Hsp70s, it is useful to review the features and evolutionary relationships of the species examined in this study.
C. merolae, a red alga, is a single-celled organism that lives in acidic hot springs (Matsuzaki et al 2004). Therefore, it is well adapted to high temperatures. C. merolae has a small and compact genome (Matsuzaki et al 2004). The marine diatom T. pseudonana is also single-celled, has a worldwide distribution, and like other diatoms has silicified cell walls (Armbrust et al 2004). It also has a relatively small genome. C. reinhardtii is a chlorophyte green alga and, as such, is more closely related to land plants than are the diatoms and red algae (Baldauf 2000; Yoon et al 2004). Chlamydomonas is also single-celled and is not known to be adapted to extreme temperatures or other extreme conditions. It does not possess a streamlined genome. The Ostreococcus isolates are very interesting. They are Prasinophytes and members of the green algal lineage. O. tauri and lucimarinus are extremely small single-celled organisms; in fact, it has been reported that they are the smallest known free-living eukaryotes. They, like C. merolae, have very small and highly dense genomes (Derelle et al 2006). Both are found in marine environments. O. lucimarinus, usually isolated from surface waters, is adapted to high light intensities. O. tauri most often is found deeper in the water column.
C. merolae, C. reinhardtii, O. lucimarinus, O. tauri, and T. pseudonana all have 1 nuclear-encoded MT Hsp70 protein. This is comparable to the number of MT Hsp70s found in other organisms. Each Plasmodium genome has 1, S. cerevisiae and A. thaliana have 2, and O. sativa has 3 MT Hsp70s. The mitochondrion evolved once, very early in eukaryote evolution, prior to the divergence of the major eukaryotic lineages (Embley 2006). The transfer to the nucleus of many MT-endosymbiont genes occurred soon after the establishment of this endosymbiont (Embley and Martin 2006). In fact, the presence of Hsp70 (and Hsp60) genes in the nucleus of eukaryotes that now lack MT has provided the key evidence that MT were gained once in evolution, with multiple subsequent losses (Embley 2006; Embley and Martin 2006). The relationships of the early diverging eukaryote lineages still are uncertain and a single protein phylogeny is not expected to resolve these relationships (Embley and Martin 2006). However, the relationships of the MT Hsp70s (Fig 2, branch 1) in this study mostly follow organismal relationships. The green plant lineage (green algae plus plants or Chlorobiota) forms a well-supported lineage. The red alga and diatom fall outside of this lineage with the Plasmodium species. The relative lack of resolution among the red alga, diatom, and Plasmodium species is not unexpected because they represent early diverging lineages. The phylogenetic patterns among the MT Hsp70s indicate a fairly consistent evolutionary pattern for this protein across organismal lineages. However, it is clear that 1 duplication of the MT Hsp70s occurred prior to the monocot-dicot divergence and an additional duplication occurred within the lineage leading to rice.
The chloroplasts also are derived from bacterial endosymbionts, but the CP Hsp70s (Fig 2, branch 2) have a very different evolutionary history compared to the MT Hsp70s. All of the algal species examined here have 1 CP Hsp70. Again we see evidence of gene duplication within the angiosperms because A. thaliana and O. sativa each have 2 CP Hsp70s. What is most notable concerning the CP Hsp70s is that the green algae and plants all have nuclear-encoded CP Hsp70s, and the red algae and diatoms have CP-encoded Hsp70s. Reith and Munholland (1991) were the first to report that a red alga, Porphyra umbilicalis, had a CP-encoded Hsp70. Now, with the complete genome of C. merolae, we know that red algae do not have also a nuclear-encoded CP Hsp70.
A short discussion of plastid evolution is useful here in our evaluation of the CP Hsp70s. It is now clear that there was a single origin of primary plastids. However, the primary green, red, and glaucocystophyte plastid lineages diverged very early in plastid evolution (Keeling 2004a). The primary plastids are the product of a single endosymbiotic event in which a nonphotosynthetic eukaryote engulfed a cyanobacterium (Keeling 2004a). The primary plastids in turn have been involved in numerous secondary endosymbiotic events (Keeling 2004a, 2004b). In secondary endosymbiosis, nonphotosynthetic eukaryotes engulf a photosynthetic eukaryote (usually either a green or red alga) with a plastid (Armbrust et al 2004; Keeling 2004b). Much of the red or green alga then disappears after becoming an endosymbiont, leaving a plastid with multiple membranes. The plastids in diatoms are a product of a secondary endosymbiosis of a red alga. Therefore, the CP genomes of red algae and diatoms are much more closely related than the red algal and diatom nuclear genomes. From this we might expect more similarity between the C. merolae and T. pseudonana CP Hsp70s than we see in the other nuclear-encoded Hsp70 homologs in these 2 species.
From our phylogenetic analysis (Fig 2) it is clear that, although the CP Hsp70s in all the species studied are derived from the cyanobacterial endosymbiont, the green and red algal Hsp70s form 2 distinct lineages. Although it has been reported that much of the endosymbiont genome was transferred to the nucleus prior to the split of the plastid lineages (green and red) (Martin et al 1998; Martin 2003; Keeling 2004a, 2004b), it is apparent that the Hsp70 gene was not in 1 of these early transfer events. Rather, there was a transfer to the nucleus from the green plastid after the green and red plastid lineages diverged. No transfer to the nucleus of the CP Hsp70 gene occurred in the red plastid lineage.
We know that extant cyanobacteria have multiple DnaK proteins (Nimura et al 2001). This suggests that the CP endosymbiont also had multiple DnaKs. From our phylogeny, it appears that the red CP Hsp70s are more closely related to Synechocystis DnaK2 than are the green CP Hsp70s. It is then possible that the green and red algal CP Hsp70s are derived from different cyanobacterial DnaKs. We do not yet have a complete understanding of the functional differences among the DnaK homologs in cyanobacteria, but there is evidence that differences do exist (Kovacs et al 2001; Varvasovszki et al 2003). Many functional and biochemical studies have been done of green algal CP Hsp70s (mostly studies of the C. reinhardtii CP Hsp70s). In contrast, very little is known of the red algal CP Hsp70s. The different evolutionary histories of the green and red algal CP Hsp70s suggest functional differences between these CP Hsp70s. Comparative studies of the functional differences among the Synechocystis DnaKs, and red and green CP Hsp70s clearly are needed to address this interesting question.
The other major lineage of Hsp70s includes the ER and cytoplasmic Hsp70s (Fig 2, branch 3). This branching pattern is consistent with an early gene duplication generating the ER and cytoplasmic lineages. It is interesting that the TpHsp70-3, OlHsp70-5, and Othsp70-5 proteins fall outside this lineage. It is unlikely that they represent a family of Hsp70s that have been lost in other eukaryotic lineages because these species are not closely related. The branch uniting these proteins was well supported in both the NJ and Bayesian analysis. However, it is possible that they are divergent cytoplasmic Hsp70s whose placement outside of the cytoplasmic lineage is due to long-branch effects. Examination of additional homologs from other species will be needed to fully understand the evolution and function of these 3 Hsp70s. Like the MT Hsp70s, the ER Hsp70s (Fig 2, branch 4) display a stable or consistent evolutionary history across organismal lineages. Within the ER Hsp70s, phylogenetic relationships generally reflect organismal relationships. Of the algal species examined here, only C. reinhardtii has more than 1 ER Hsp70; by comparison A. thaliana has 3 and O. sativahas 5.
The evolution of the cytoplasmic Hsp70s deserves considerable attention. Though there is strong support for a cytoplasmic Hsp70 lineage (Fig 2, branch 5), within this lineage the relationships of the cytoplasmic Hsp70s do not reflect organismal relationships. This indicates a complex history of gene duplication, possible gene loss, and gene conversion. One A. thaliana cytoplasmic Hsp70, Athsp70-4, is found in a more basal position than the green algal cytoplasmic Hsp70s. However, most of the cytoplasmic A. thaliana and O. sativa Hsp70s form species-specific groups. These groupings suggest either recent gene duplication or gene conversion. It is also possible that both of these forces are acting on the cytoplasmic Hsp70s. Gene conversion among the angiosperm cytoplasmic Hsp70s quite likely is as rapid gene conversion among cytoplasmic Hsp70s as has been reported in Drosophila (Bettencourt et al 2002). In addition, in a study of Caenorhabditis elegans and C. briggsae Hsp70s, Nikolaidis and Nei (2004) also reported gene conversion among cytoplasmic Hsp70 genes. If gene conversion is active among the angiosperm cytoplasmic Hsp70s, then these duplicates could be quite old, and gene conversion, not recent duplication, is responsible for the high sequence similarity within species.
The complex evolutionary history of the cytoplasmic Hsp70s is also evident when the number of proteins across species is examined. Although some gene duplication has occurred within the organelle-localized Hsp70 lineages, it is evident from Figure 2 that these protein lineages have remained relatively stable over long periods of evolutionary time. In contrast, it appears that there has been the evolution of considerable diversity in the cytoplasmic Hsp70s since the last common ancestor between C. reinhardtii and the angiosperms. The angiosperms A. thaliana and O. sativa have 7 and 8 cytoplasmic Hsp70s, respectively, and the algal species studied have 1 each. In their study of the A. thaliana Hsp70s, Lin et al 2001 suggest that the reason that A. thaliana has so many more Hsp70s than other eukaryotes is the presence of the plastid. However, all 5 of our study species are photosynthetic and have plastids. Therefore, there must be another explanation for the differences in diversity of Hsp70s between the angiosperms (represented by A. thaliana and O. sativa) and these photosynthetic eukaryotes. An important distinction between the algae studied here and the flowering plants is that all the algae examined are single-celled. Is it possible that multiple cytoplasmic Hsp70s are associated with multicellularity? However, it has been noted that considerable differences in the numbers of cytoplasmic Hsp70s exist among other lineages. For example, the ascidian Ciona intestinalis (a multicellular animal) has only 2 cytoplasmic Hsp70s, although humans have 8 (Wada et al 2006). Other lineages that have multiple cytoplasmic Hsp70s include single-celled yeast and multicellular Drosophila. From these comparisons, it appears that there is no clear relationship between multicellularity and the number of cytoplasmic Hsp70s.
However, it has been well established that the cytoplasmic Hsp70s are an important part of the heat shock response and that these proteins can confer thermal tolerance. The importance of multiple copies of cytoplasmic Hsp70s in the ability of Drosophila to withstand high temperature stress has been shown in a number of studies (Feder and Krebs 1998; Krebs and Feder 1998; Garbuz et al 2003; Lerman and Feder 2004). These findings suggest that the multiple cytoplasmic Hsp70s in angiosperms may be related to an increased thermal tolerance in angiosperms compared to algae that have only 1 cytoplasmic Hsp70. However, the red alga C. merolae is adapted to extreme conditions, and the lack of additional Hsp70 homologs in this species indicates this adaptation was not gained by the evolution of diverse Hsp70s, suggesting that not all thermotolerant organisms have multiple cytoplasmic Hsp70s. The plant cytoplasmic Hsp70s have not been studied to the extent that their animal homologs have (Sung et al 2001) and further functional analysis of these proteins clearly is needed.
It is known that there have been numerous polyploidy events within the land plant lineages and that these events have played an important role in gene family diversification within plants. It is possible that the additional cytoplasmic Hsp70s in angiosperms are a product of multiple polyploid events. A fascinating study showed that duplications of cytoplasmically localized proteins are more likely to be retained than duplications of organelle-localized proteins (Blanc and Wolfe 2004). Other studies have found gene family expansions when angiosperms are compared to algae. For example, in a study of kinesins, it was found that C. merolae has 5, T. pseudonana has 22, C. reinhardtii has 23, and A. thaliana has 61 kinesins. The large increase in angiosperm kinesins compared to the C. reinhardtii is due to expansion in only 2 families of kinesins (Richardson et al 2006). It is also noteworthy that within the plant lineage there also has been a lineage-specific amplification of the small heat shock proteins or Hsp20s (Waters 2003). However, early diverging land plants also have a diversity of small heat shock proteins and so this expansion was not directly related to polyploidy in vascular plants. It has been suggested that the stresses of moving onto land, which included increased desiccation, increased ultraviolet exposure, and increased temperature extremes may have been a selective pressure that favored or drove an increase in the types and numbers of molecular chaperones (Waters 2003). Distinguishing between duplication due to the selective pressures of life on land and the effects of polyploidy for the Hsp70s and other gene families will require considerable additional information including the complete genome sequences of a number of plants representing early diverging land plant lineages. When this data is available, it will be possible to determine if multiple cytoplasmic Hsp70s are found in all land plants, suggesting that selection pressure for thermal tolerance early in land plant evolution drove the duplication of plant cytoplasmic Hsp70s or if only those land plant lineages that have undergone multiple polyploidy events have a diversity of cytoplasmic Hsp70s.
SUMMARY
From the complete genome sequences of 5 distantly related photosynthetic eukaryotes or algae we identified 24 Hsp70s of the DnaK subfamily. Analysis of these 24 proteins indicates that all are expressed and that they all possess the highly conserved ATPase and substrate-binding domains. Some, but not all, also possess the transit sequences for targeting to particular organelles and a few lack the highly variable C-terminal domain. The MT and ER Hsp70s have relatively stable evolutionary histories and the protein phylogenies approximate the organismal relationships. The CP Hsp70s have a very interesting evolutionary history that suggests the possibility of functional differences between red and green CP Hsp70. The green CP Hsp70s are all nuclear encoded. The red CP Hsp70s are all encoded in the CP genome. Our analysis also indicates that the red and green CP Hsp70 may be derived from distinct cyanobacterial DnaK homologs. Finally, the cytoplasmic Hsp70s have a much more complex evolutionary history than the MT and ER Hsp70s. We describe the absence of diverse cytoplasmic Hsp70s in the algal species. Each species appears to have only 1 Hsp70 that is clearly within the cytoplasmic Hsp70 lineage. This is quite distinct from the diverse number of cytoplasmic Hsp70s found in angiosperms or flowering plants.
Fig 1.
Continued
Fig 1.
Continued
Fig 1.
Continued
Acknowledgments
The authors thank 3 anonymous reviewers for their thoughtful comments on earlier versions of this manuscript. This work was supported by grant IBN:0313900 from the National Science Foundation (E.R.W.). An earlier version of this work was submitted to the Department of Biology for partial fulfillment of an Undergraduate Honors Thesis at San Diego State University (T.R.)
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389.0305-1048(1997)025[3389:GBAPAN]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156.0193-4511(2004)306[0079:TGOTDT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972.0193-4511(2000)290[0972:AKPOEB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bettencourt BR, Kim I, Hoffmann AA, Feder ME. Response to natural and laboratory selection at the Drosophila hsp70 genes. Int J Org Evol. 2002;56:569–586. doi: 10.1111/j.0014-3820.2002.tb00193.x. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410.1040-4651(2004)016[1679:FDODGF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the hsp70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490.0022-2844(1994)038[0001:MEOTHM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp90 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9.0092-8674(1998)092[0351:THAHCM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103.1091-6490(2006)103[11647:GAOTSF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijine G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903.0022-2836(2000)300[1005:PSLOPB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Embley TM. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos Trans Roy Soc Lond B Biol Sci. 2006;361:1055–1067. doi: 10.1098/rstb.2006.1844.1471-2970(2006)361[1055:MSOOTA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546.1476-4687(2006)440[0623:EECAC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Feder ME, Krebs RA. Natural and genetic engineering of thermotolerance in Drosphila melanogaster. Am Zool. 1998;38:503–517.0003-1569(1998)038[0503:NAGEOT]2.0.CO;2 [Google Scholar]
- Garbuz D, Evgenev MB, Feder ME, Zatsepina OG. Evolution of thermotolerance and the heat shock response: evidence from inter/intraspecific comparison and interspecific hybridization in the virilis species group of Drosophila. I. Thermal phenotype. J Exp Biol. 2003;206:2399–2408. doi: 10.1242/jeb.00429.0022-0949(2003)206[2399:EOTATH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gething M, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0.1476-4687(1992)355[0033:PFITC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gupta R, Golding G. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743.0022-2844(1993)037[0573:EOHGAI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.0261-3166(1999)041[0095:BAUBSA]2.0.CO;2 [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408.0193-4511(2002)295[1852:MCITCF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055.1040-4651(2004)016[0241:EAOTAM]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielson R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889.0193-4511(2001)294[2310:BIOPAI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution. J Mol Evol. 1998;47:565–577. doi: 10.1007/pl00006413.0022-2844(1998)047[0565:HSPFMS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Keeling PJ. A brief history of plastids and their hosts. Protist. 2004a;155:3–7. doi: 10.1078/1434461000156.1434-4610(2004)155[0003:ABHOPA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Diversity and evolutionary history of plastids and their hosts. Amer Jour Botany. 2004b;91:1481–1493. doi: 10.3732/ajb.91.10.1481.0002-9122(2004)091[1481:DAEHOP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kovacs E, van der Vies SM, Glatz A, Torok Z, Varvasovszki V, Horvath I, Vigh L. The chaperonins of Synechocystis PCC 6803 differ in heat inducibility and chaperone activity. Biochem Biophys Res Commun. 2001;289:908–915. doi: 10.1006/bbrc.2001.6083.0006-291X(2001)289[0908:TCOSPD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Krebs RA, Feder ME. Hsp70 and larval thermotolerance in Drosophila melanogaster: how much is enough and when is more too much? J Insect Physiol. 1998;44:1091–1101. doi: 10.1016/s0022-1910(98)00059-6.0022-1910(1998)044[1091:HALTID]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150.1467-5463(2004)005[0150:MISFME]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lerman DN, Feder ME. Naturally occurring transposable elements disrupt hsp70 promotor function in Drosophila melanogaster. Mol Biol and Evol. 2004;22:776–783. doi: 10.1093/molbev/msi063.0737-4038(2004)022[0776:NOTEDH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lerman DN, Michalak P, Helin AB, Bettencourt BR, Feder ME. Modification of heat shock gene expression in Drosophila melanogaster populations via transposable elements. Mol Biol Evol. 2003;20:135–144. doi: 10.1093/molbev/msg015.0737-4038(2003)020[0135:MOHSGE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress and Chaperones. 2001;6:201–208. doi: 10.1379/1466-1268(2001)006<0201:gaoths>2.0.co;2.1466-1268(2001)006[0201:GAOTHS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat shock proteins. Ann Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215.0066-4197(1988)022[0631:THSP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Martin W. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci U S A. 2003;100:8612–8614. doi: 10.1073/pnas.1633606100.1091-6490(2003)100[8612:GTFOTT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hansmann S, Hadegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234.1476-4687(1998)393[0162:GTTTNA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-i T. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae. Nature. 2004;428:653–657. doi: 10.1038/nature02398.1476-4687(2004)428[0653:GSOTUU]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell and Molec Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6.1420-682X(2005)062[0670:HCCFAM]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. Psort: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x.0376-5067(1999)024[0034:PAPFDS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas H, Deerfield D. GeneDoc: analysis and visualization of genetic variation. EMBNEW NEWS. 1997;4:14.1396-6073(1997)004[0014:GAAVOG]2.0.CO;2 [Google Scholar]
- Nikolaidis N, Nei M. Concerted and nonconcerted evolution of the hsp70 gene superfamily in two sibling species of nematodes. Mol Biol Evol. 2004;21:498–505. doi: 10.1093/molbev/msh041.0737-4038(2004)021[0498:CANEOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nimura K, Takahashi H, Yoshikawa H. Characterization of the dnaK multigene family in the cyanobacterium Synechococcus sp. strain PCC7942. J Bacteriol. 2001;183:1320–1328. doi: 10.1128/JB.183.4.1320-1328.2001.0021-9193(2001)183[1320:COTDMF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. Treeview: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357.1460-2059(1996)012[0357:TAATDP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Peeters N, Small I. Dual targeting to mitochondria and chloroplasts. Biochemica et Biophyscis Acta. 2001;1541:54–63. doi: 10.1016/s0167-4889(01)00146-x.0006-3002(2001)1541[0054:DTTMAC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Reith M, Munholland J. An hsp70 homolog is encoded on the plastid genome of the red alga, Porphyra umbilicalis. FEBS Lett. 1991;294:116–120. doi: 10.1016/0014-5793(91)81355-c.0014-5793(1991)294[0116:AHHIEO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Maier UG. Phylogenetic analysis of the stress-70 protein family. J Mol Evol. 1993;39:80–86. doi: 10.1007/BF00178252.0022-2844(1993)039[0080:PAOTSP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Richardson DN, Simmons MP, Reddy AS. Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics. 2006;7:18. doi: 10.1186/1471-2164-7-18.1471-2164(2006)007[0018:CCAOKI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180.1367-4803(2003)019[1572:MBPIUM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP, and van der Mark P 2005 MrBayes 3.1 Manual. http://mrbayes.csit.fsu.edu/manual.php. [Google Scholar]
- Scharf K-D, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins) Cell Stress and Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2.1466-1268(2001)006[0225:TEFOAT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M. The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynthesis Research. 2004;82:221–240. doi: 10.1007/s11120-004-2216-y.0166-8595(2004)082[0221:TCGRIS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Shrager J, Hauser C, Chang C-W. Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 2003;131:401–408. doi: 10.1104/pp.016899.0032-0889(2003)131[0401:CRGPAG]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776.1615-9861(2004)004[1581:PATFRS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sung DY, Kaplan F, Guy CL. Plant Hsp70 molecular chaperones: protein structure, gene family, expression, and function. Physiologia Plantarum. 2001;113:443–451.0031-9317(2001)113[0443:PHMCPS]2.0.CO;2 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673.0305-1048(1994)022[4673:CWITSO]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varvasovszki V, Glatz A, Shigapova N, Josvay K, Vigh L, Horvath I. Only one dnaK homolog, dnaK2, is active transcriptionally and is essential in synechocystis. Biochem Biophys Res Commun. 2003;305:641–648. doi: 10.1016/s0006-291x(03)00822-2.0006-291X(2003)305[0641:OODHDI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wada S, Hamada M, Satoh N. A genomewide analysis of gene for the heat shock protein 70 chaperone system in the ascidian Ciona intestinalis. Cell Stress and Chaperones. 2006;11:23–33. doi: 10.1379/CSC-137R.1.1466-1268(2006)011[0023:AGAOGF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E. Molecular adaptation and the origin of land plants. Mol Phy Evol. 2003;29:456–463. doi: 10.1016/j.ympev.2003.07.018.1095-9513(2003)029[0456:MAATOO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851.0737-4038(2001)018[0691:AGEMOP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools on the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531.1064-3745(1999)112[0531:PIAATO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075.0737-4038(2004)021[0809:AMTFTO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]