Abstract
The actin cytoskeleton powers organelle movements, orchestrates responses to abiotic stresses, and generates an amazing array of cell shapes. Underpinning these diverse functions of the actin cytoskeleton are several dozen accessory proteins that coordinate actin filament dynamics and construct higher-order assemblies. Many actin-binding proteins from the plant kingdom have been characterized and their function is often surprisingly distinct from mammalian and fungal counterparts. The adenylyl cyclase-associated protein (CAP) has recently been shown to be an important regulator of actin dynamics in vivo and in vitro. The disruption of actin organization in cap mutant plants indicates defects in actin dynamics or the regulated assembly and disassembly of actin subunits into filaments. Current models for actin dynamics maintain that actin-depolymerizing factor (ADF)/cofilin removes ADP–actin subunits from filament ends and that profilin recharges these monomers with ATP by enhancing nucleotide exchange and delivery of subunits onto filament barbed ends. Plant profilins, however, lack the essential ability to stimulate nucleotide exchange on actin, suggesting that there might be a missing link yet to be discovered from plants. Here, we show that Arabidopsis thaliana CAP1 (AtCAP1) is an abundant cytoplasmic protein; it is present at a 1:3 M ratio with total actin in suspension cells. AtCAP1 has equivalent affinities for ADP– and ATP–monomeric actin (Kd ∼ 1.3 μM). Binding of AtCAP1 to ATP–actin monomers inhibits polymerization, consistent with AtCAP1 being an actin sequestering protein. However, we demonstrate that AtCAP1 is the first plant protein to increase the rate of nucleotide exchange on actin. Even in the presence of ADF/cofilin, AtCAP1 can recharge actin monomers and presumably provide a polymerizable pool of subunits to profilin for addition onto filament ends. In turnover assays, plant profilin, ADF, and CAP act cooperatively to promote flux of subunits through actin filament barbed ends. Collectively, these results and our understanding of other actin-binding proteins implicate CAP1 as a central player in regulating the pool of unpolymerized ATP–actin.
INTRODUCTION
Rapid actin filament turnover is necessary for many cellular processes such as intracellular transport, motility, cytokinesis, endocytosis and polarized cell growth. A variety of ubiquitous actin-binding proteins, including actin-depolymerizing factor (ADF)/cofilin, profilin, and capping protein cooperate in regulating these dynamic processes. Most current models for regulation of actin turnover agree that ADF/cofilin severs filaments and consequently increases the depolymerization rate; this provides a pool of ADP–G-actin that is available for binding to profilin (Didry et al., 1998; Balcer et al., 2003; Nicholson-Dykstra et al., 2005; Andrianantoandro and Pollard, 2006). The ability of profilin to catalyze nucleotide exchange on monomeric (G)-actin is also generally accepted and serves to “recharge” the subunit pool for highly polymerizable ATP–G-actin. The profilin–actin complex delivers monomers on to free barbed ends, whereas profilin has sequestering properties when barbed ends are blocked with capping proteins (Didry et al., 1998; Kang et al., 1999). Several studies show that adenylyl cyclase-associated protein (CAP), an actin monomer-binding protein (Gieselmann and Mann, 1992; Freeman et al., 1995), cooperates with ADF/cofilin to accelerate turnover (Moriyama and Yahara, 2002; Balcer et al., 2003; Bertling et al., 2004; Mattila et al., 2004). CAP (known as Srv2p in budding yeast) was identified originally as an adenylyl cyclase-interacting protein, facilitating cyclase activation by RAS (Fedor-Chaiken et al., 1990; Field et al., 1990), and recent studies link cAMP signaling, actin dynamics, and apoptosis in yeast (Gourlay and Ayscough, 2006). Biochemical and genetic analyses reveal how this protein can perform multiple functions. CAP has two distinct domains; the N terminus binds adenylyl cyclase and the C terminus binds G-actin (Gerst et al., 1991). Adenylyl cyclase binding, however, may not be conserved in animal and plant kingdoms (Hubberstey and Mottillo, 2002).
CAP binds actin monomers with a dissociation equilibrium constant of 0.5–5.0 μM (Gieselmann and Mann, 1992; Freeman et al., 1995; Gottwald et al., 1996; Zelicof et al., 1996) and Srv2p/CAP prefers ADP–G-actin (Kd = 0.02 μM) to ATP–G-actin (Kd = 1.2 μM) (Mattila et al., 2004). Recent studies expand our view beyond simple monomer binding by implicating CAP in regulating turnover of the actin cytoskeleton and revealing novel functions for the C-terminal domain. For example, Balcer et al., (2003) showed that Srv2p/CAP recycles ADF/cofilin and G-actin for new rounds of actin filament polymerization by “handing off” actin monomer from the ADF—ADP–G-actin complex to profilin, which then delivers subunits to filament barbed ends. Subsequently, it was reported that the C terminus of Srv2p/CAP competes with ADF/cofilin for ADP–G-actin binding, and specifically blocks addition of subunits onto filament barbed ends, which can be relieved by the addition of profilin (Mattila et al., 2004). A somewhat different picture emerges from studies with human CAP1, which has actin-binding activity on both N and C termini (Moriyama and Yahara, 2002). Human CAP1 also allows cofilin and actin to recycle during filament turnover and accelerates ADF/cofilin-induced depolymerization (Moriyama and Yahara, 2002). However, human CAP1 bound to actin monomers stimulates nucleotide exchange and allows elongation of filaments at their barbed ends in the presence of ADF/cofilin (Moriyama and Yahara, 2002).
The importance of Srv2p/CAP in regulating actin dynamics in vivo is emphasized by cytological and genetic studies. In yeast, Srv2p/CAP localizes to cortical actin patches through an interaction with the actin filament binding protein Abp1 (Lila and Drubin, 1997; Balcer et al., 2003). The srv2 mutant yeast have reduced actin patch turnover, lose their actin cables, and demonstrate genetic interactions with cof1 and pfy1 alleles (Gerst et al., 1991; Vojtek et al., 1991; Balcer et al., 2003; Bertling et al., 2007). In mammalian cells, CAP depletion leads to an accumulation of actin and ADF/cofilin in abnormal cytoplasmic aggregates and also decreases rates of actin turnover, indicating that CAP is required for proper subcellular localization of ADF/cofilin (Bertling et al., 2004). In contrast to these loss-of-function phenotypes that result in reduced actin polymerization or filament levels, Drosophila mutants for CAP (Act Up) show increased actin filament levels during eye differentiation and defects in eye imaginal disk formation (Benlali et al., 2000). Further studies on lamellipodia formation in Drosophila S2 cells show that Act Up (and profilin) RNA interference lines have diffuse actin filament arrays throughout the cytoplasm, which contrasts with abundant filament localization predominantly at the leading edge in wild-type cells (Rogers et al., 2003). These diverse results suggest that CAP function might be unique in different organisms and that it might depend on specific cellular conditions or the presence and activity of other actin-binding proteins, such as profilin and ADF/cofilin, for regulation of actin filament dynamics.
CAP has been identified from the plant kingdom, and initial data suggest that it, too, will be a regulator of actin dynamics (Kawai et al., 1998; Barrero et al., 2002; Barrero et al., 2003). The C terminus of Arabidopsis CAP1 interacts with bovine actin in a simple pull-down assay and complements the srv2 mutant of yeast (Barrero et al., 2002). Overexpression of AtCAP1 results in the loss of actin filaments from suspension culture cells, arrests cell division, and reduces cell expansion (Barrero et al., 2002, 2003), whereas cap1 knockout mutant Arabidopsis plants show disrupted actin organization in roothairs and trichomes, a reduced plant stature, and altered morphology of several different cell types (our unpublished data; Deeks et al., 2007). The marked differences in biochemical and in vivo activities for yeast, fly, and vertebrate CAP suggest that a detailed examination of the properties for CAP from diverse organisms may reveal both novel and conserved mechanisms for coordinating actin dynamics in eukaryotic cells.
Through biochemical and cell biological characterization of AtCAP1, we can gain new insight about the role of this protein in actin dynamics, and we can understand the molecular basis for the mutant phenotypes. Here, we present biochemical evidence showing that AtCAP1 is an abundant protein with a moderate affinity for monomeric actin. AtCAP1 acts as an actin-monomer–sequestering protein at steady state as well as during nucleation and elongation reactions. However, AtCAP1 enhances the rate of nucleotide exchange on G-actin directly, even in the presence of ADF/cofilin. This is significant because plant profilin has no effect or may actually inhibit nucleotide exchange on G-actin (Perelroizen et al., 1996; Kovar et al., 2000a, 2001), yet it can shuttle actin subunits onto filament barbed ends (Michelot et al., 2005). We develop a new model for plant actin dynamics in which CAP is centrally positioned between ADF/cofilin and profilin in regulating the activity of the unpolymerized actin pool.
MATERIALS AND METHODS
Protein Purification
The cDNA encoding A. thaliana cyclase-associated protein (AtCAP1) was amplified with polymerase chain reaction (PCR) from a size-fractionated root cDNA library (CD4-16; Arabidopsis Biological Resource Center, The Ohio State University, Columbus, OH). Oligonucleotide primers were synthesized based on the predicted cDNA sequences available at GenBank (accession no. NM_119614). The primers for amplification of the AtCAP1 coding region were 5′-GGGGAATTCTAATGGAAGAGGATTTGATT-3′ containing the initiation codon and an EcoRI site (underlined), and 5′-GGGCTCGAGTTAGGCACCTGAATGCGA-3′ containing the stop codon and an XhoI site (underlined). The amplified product was A-tailed, cloned into pGEM-T, and verified by sequence analysis. For expression of full-length AtCAP1 in Escherichia coli, the ∼1400-base pair PCR fragment was subcloned into pGEX-KG (Guan and Dixon, 1991) prepared with the aforementioned restriction enzyme sites. The pGEX-AtCAP1 construct was transformed into strain BL21 (DE3) Tuner (Novagen, Madison, WI) of E. coli. Protein expression in bacterial cells was induced for >24 h at 10°C after the addition of 0.4 mM isopropyl β-d-thiogalactopyranoside. Cells were collected by centrifugation, resuspended in phosphate-buffered saline with a 1:200 dilution of protease inhibitors from a stock solution (Ren et al., 1997), and sonicated. The fusion protein was purified as described by Kovar et al. (2000b). An extinction coefficient for AtCAP1, based on the approach of Gill and von Hippel (1989), was determined to be 33,671 M−1 cm−1. Actin was purified from rabbit skeletal muscle acetone powder (Spudich and Watt, 1971) and monomeric Ca-ATP–actin was purified by Sephacryl S-300 chromatography (MacLean-Fletcher and Pollard, 1980) in buffer G (5 mM Tris-HCl, pH 8, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM dithiothreitol [DTT], and 0.1 mM azide). Actin was labeled on Cys-374 with pyrene iodoacetamide (Kouyama and Mihashi, 1981; Pollard, 1984) or on Lys-372 with 7-chloro-4-nitrobenzeneno-2-oxa-1,3-diazole (NBD-Cl) (Detmers et al., 1981; Weeds et al., 1986). Maize pollen actin (Ren et al., 1997), Arabidopsis ADF1 (Carlier et al., 1997), Arabidopsis profilin 2 (PRF2; At4g29350), and profilin 4 (PRF4; At4g29340) were purified as described previously (Kovar et al., 2000a). The plasmid for expression of human plasma gelsolin was kindly provided by T. Pollard (Yale University, New Haven, CT), and recombinant protein was purified roughly according to Pope et al. (1997) with modifications (Huang et al., 2004).
Polyclonal Antibody Production
Purified AtCAP1 was used to elicit polyclonal antisera in New Zealand White rabbits according to standard procedures as described previously (Gibbon et al., 1999). Polyclonal antibody, at a 1:5000 dilution, was used for Western blotting. Affinity purification of serum was performed according to Huang et al. (2003), and the eluate was used at 1:100 dilution. Phosphoenol pyruvate (PEP) carboxylase antibody (Rockland Immunochemicals, Gilbertsville, PA) at a 1:2000 dilution was used to ensure equal loading of Arabidopsis tissue extracts.
Quantitative Immunoblotting
Intracellular protein concentrations for CAP, actin, ADF, and profilin from wild-type Arabidopsis leaf and suspension cell extracts were determined by quantitative immunoblotting (Wu and Pollard, 2005). A linear standard curve was generated using native maize pollen actin, recombinant AtCAP1, AtPRF2, and AtADF1. Leaf and suspension cell protein extracts were prepared in 2× grinding buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 5 mM DTT, and 1:200 phenylmethylsulfonyl fluoride/protease inhibitor cocktail). For determination of actin and CAP concentrations, 25 μg of total protein was loaded, whereas for profilin and ADF/cofilin, 50 μg of total protein was loaded on the same SDS-polyacrylamide gel electrophoresis (PAGE) as the standard curve samples. The primary polyclonal antibodies used were anti-AtCAP1, anti-maize pollen actin (Gibbon et al., 1999), anti-ZmPRO4 (Gibbon et al., 1998), and anti-AtADF2 (Ashworth and Staiger, unpublished data). AlexaFluor 800-labeled secondary antibody was used for infrared visualization of protein bands. Quantification of protein in tissue was done with the Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE).
Critical Concentration Determination
The critical concentration (Cc) for actin polymerization was determined as described by Brenner and Korn (1983). Increasing concentrations of actin (2% pyrene labeled) were polymerized in 1× KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole-HCl, pH 7.0) in the absence or presence of 0.5 or 1 μM AtCAP1 for 16 h at room temperature in the dark. Human plasma gelsolin at 1:200 stoichiometry was used to cap the barbed ends of actin filaments. Fluorescence measurements for this and additional pyrenyl-actin assays described below were performed at room temperature using a PTI QuantaMaster spectrofluorometer (QM-2000-SE; Photon Technology International, South Brunswick, NJ) with excitation at 365 nm and emission at 407 nm. Linear best fit of the data, plotted as arbitrary fluorescence units versus actin concentration, was used to determine the intercept with the x-axis to yield a Cc for actin assembly. The free [Ca2+] was calculated with “EGTA” software available at www.liv.ac.uk/luds/people/cds/bds/pms/cal.htm. The dissociation constant (Kd) was calculated using the following equation:
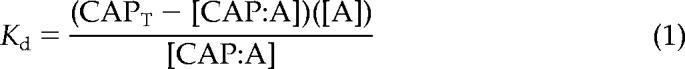 |
where CAPT is the total AtCAP1 concentration, A is the Cc for actin alone, and [CAP:A] is the concentration of the assumed 1:1 complex, which is determined as the difference between Cc values for actin in the presence and absence of AtCAP1.
Actin Monomer Binding Assay
The interaction of AtCAP1 with actin monomers was examined by measuring the fluorescence change of NBD-labeled G-actin (Detmers et al., 1981; Weeds et al., 1986). Monomeric, ATP-loaded, NBD–actin was prepared by adding 1 mM ATP to NBD–G-actin in Buffer G and incubating for 75 min at 4°C. Monomeric NBD–actin was converted to ADP-loaded form roughly as described by Pollard (1986). Then, 20 μM NBD–G-actin in buffer G was incubated with 20 U/ml hexokinase (Sigma-Aldrich, St. Louis, MO) and 1 mM glucose for 3–4 h at 4°C. The ATP– and ADP–G-actin (NBD labeled) were converted to Mg–actin by adding 0.1 volume of 10× ME (0.5 mM MgCl2 and 1 mM EGTA) for 2 min at 25°C before each measurement. Final binding reactions containing 0.2 μM NBD–G-actin in buffer G, with or without ATP, were brought to polymerization conditions by the addition of 1× KMEI. Various amounts of AtCAP1 or AtADF1 were titrated into these reactions and the fluorescence change monitored by fluorimetry. The normalized change in fluorescence, as determined by the equation
 |
was measured with the spectrofluorometer using 468-nm excitation and 520-nm emission wavelengths (Weeds et al., 1986). The data were modeled using Kaleidagraph version 3.6 software (Synergy Software, Reading, PA), and they were fitted with equations described by Mattila et al. (2004).
Assembly from Monomeric Actin
The polymerization of G-actin in the absence and presence of various amounts of AtCAP1 was followed by pyrene fluorescence. Actin monomers (3 μM; 5% pyrene labeled) were polymerized with the addition of 0.1 volume of 10× KMEI, and the change in fluorescence was followed for 30 min.
Seeded Elongation Assay
To assay the addition of monomeric actin onto the ends of preformed actin seeds, 5 μM actin was polymerized for 2 h at room temperature. For the elongation reaction, 1 μM (5% pyrene-labeled) Mg-ATP–G-actin was prepared in G-buffer-Mg (buffer G with 1 mM EGTA and 0.1 mM MgCl2) to which an appropriate amount of AtCAP1 and 0.4 μM F-actin seeds were added. Polymerization was initiated by adding KMEI to a final concentration of 1×, and assembly was monitored with the fluorometer as described above. For assembly at the pointed end of actin filaments, the procedure followed Higgs et al. (1999) with the use of gelsolin–actin seeds (80 nM final concentration). Briefly, 2 μM (5% pyrene-labeled) Mg-ATP–G-actin was prepared in G-buffer-Mg, and aliquots of the seeds and AtCAP1 were added as drops to the side of the tube. Elongation was initiated with 10× KMT (500 mM KCl, 10 mM MgCl2, and 100 mM Tris-HCl, pH 8.0) added to 1× final concentration. Determination of the apparent affinity of AtCAP1 for G-actin used the equations and approach of Higgs et al. (1999).
Nucleotide Exchange Analysis
The rate of nucleotide exchange on G-actin in the absence or presence of the indicated concentrations of AtCAP1, AtPRF4, HPRO1, and AtADF1 was determined by measuring the increase in fluorescence upon incorporation of 1-N6-ethenoadenosine 5′-triphosphate (ε-ATP; Sigma-Aldrich) (Goldschmidt-Clermont et al., 1992). The ε-ATP (50 μM) and AtCAP1, profilin, or AtADF1 were mixed with physiological buffer (2 mM Tris, pH 7.5, 0.5 mM DTT, 50 mM KCl, 1 mM EGTA, and 0.5 mM MgCl2) or low salt buffer (2 mM Tris, pH 7.5, and 0.5 mM DTT) for 5 s. Monomeric actin was added to a final concentration of 0.5 μM, mixed for 3 s, and the change in fluorescence was measured with excitation at 350 nm and emission at 410 nm for 600 s. The observed initial rate (kobs, s−1) of ε-ATP incorporation using pseudo first-order conditions was determined by fitting the data for the first 240 s to a single exponential function (Kovar et al., 2001):
where [PT] is the initial ε-ATP:G-actin reaction product, t is time, and k is the kobs, s−1 in arbitrary units per second. To determine the amount of G-actin bound at a given concentration of actin-binding protein, [PA], the following quadratic equation was used:
where, [A]T is total G-actin, [P]T is total actin-binding protein, and Kd is that determined at steady state (Perelroizen et al., 1995).
Actin Filament Turnover Assay
The turnover of filaments was estimated by measuring the rate at which fluorescently labeled ε-ADP–actin filaments, assembled from ε-ATP–actin subunits, became nonfluorescent ADP–actin filaments after the addition of ATP into the reaction mixture, according to Didry et al. (1998). The fluorescence of ε-ADP was monitored at 20°C in a Safas Xenius spectrofluorimeter (Safas, Monte-Carlo, Monaco) with excitation and emission wavelengths of 360 and 410 nm, respectively.
RESULTS
Cyclase-associated Protein Is an Abundant Cellular Protein in Arabidopsis
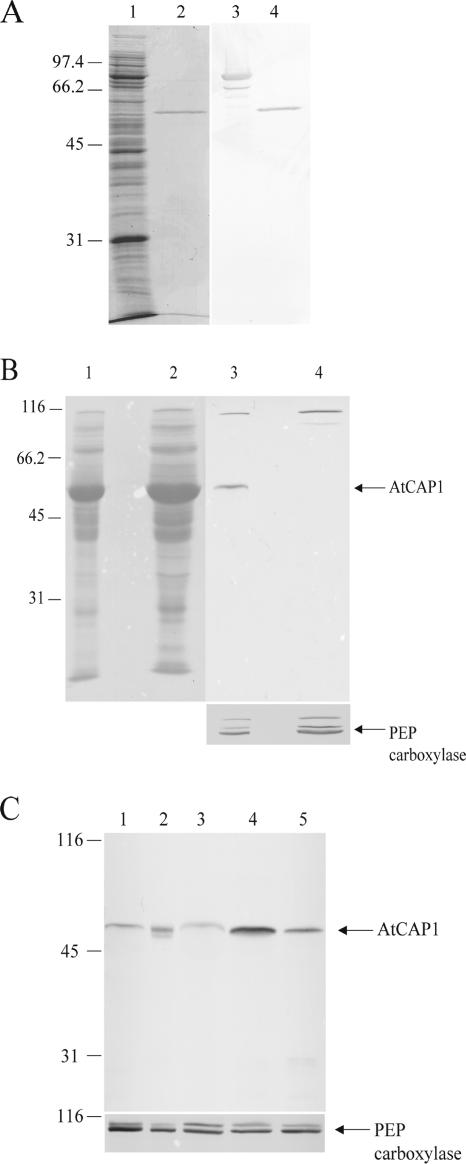
The AtCAP1 (NM_119614, At4g34490) present in current databases seems to be encoded by a single gene (Barrero et al., 2002). The full-length cDNA encodes a polypeptide of 477 amino acids with a predicted molecular mass of ∼50,900 and pI of 6.22. AtCAP1 shares 28–37% amino acid identity with CAP from vertebrates, yeast, and lower eukaryotes. To assess the biochemical and cell biological properties of plant CAP, a glutathione S-transferase (GST)-fusion protein with full-length AtCAP1 polypeptide was expressed in E. coli. Recombinant protein from bacterial extract (Figure 1A, lane 1) was purified by chromatography on glutathione-agarose. After digestion with thrombin to remove the GST moiety, the resulting AtCAP1 protein was 90–95% pure and migrated at 50–52 kDa on SDS-PAGE gels (Figure 1A, lane 2).
Figure 1.
AtCAP1 antibody recognizes recombinant and native protein from A. thaliana. (A) Coomassie-stained gel (lanes 1 and 2) and Western immunoblot (lanes 3 and 4) showing the purification of recombinant GST-AtCAP1. Lanes 1 and 3, 5 μg of total extract from bacterial cells; lanes 2 and 4, thrombin-cleaved protein eluate (1 μg). Lanes 3 and 4 were probed with affinity-purified anti-AtCAP1 antibody. Purified, recombinant AtCAP1 migrated at 50–52 kDa. (B) Twenty-five micrograms of A. thaliana wild-type (lanes 1 and 3) and homozygous mutant (lanes 2 and 4) leaf tissue extracts were separated by SDS-PAGE and Ponceau S-stained (lanes 1 and 2); lanes 3 and 4 were probed with polyclonal anti-AtCAP1 antibody (top) and subsequently with PEP carboxylase antibody as a loading control (bottom). Anti-CAP1 recognized an appropriate molecular weight polypeptide in wild-type but not in mutant tissue. (C) Twenty-five micrograms each of extracts from total plant (lane 1), roots (lane 2), leaves (lane 3), stems (lane 4), and flowers (lane 5) were probed with anti-AtCAP1 antibody (top). The same blot was reprobed with anti-PEP carboxylase antibody, which recognized a band of ∼116 kDa, to show loading controls (bottom). Migration of molecular weight standards is given at left in kilodaltons.
An affinity-purified polyclonal antibody that specifically recognized the recombinant protein was generated (Figure 1A, lanes 3 and 4). Immunoblots of Arabidopsis rosette leaf tissue from wild-type plants showed a band ∼50–52 kDa (Figure 1B, lane 3), whereas homozygous cap mutant plants (SALK_112802; Alonso et al., 2003; Deeks et al., 2007) were missing this polypeptide (Figure 1B, lane 4). A nonspecific band of ∼116 kDa was observed in both wild-type and homozygous mutant plants when crude polyclonal serum was used (Figure 1B). These results verified that the affinity-purified and polyclonal antibodies recognized recombinant and native AtCAP1. To examine expression patterns throughout the plant, blots with extracts from various plant tissues were probed with anti-AtCAP1. Figure 1C shows that the 51-kDa AtCAP1 protein was present in roots, leaves, flowers, and stems and that it appears to be most abundant in stems and flowers (Figure 1C, lanes 4 and 5) as well as in pollen (data not shown). Microarray data from GENEVESTIGATOR (Zimmermann et al., 2004) also indicate that AtCAP1 is ubiquitously expressed in A. thaliana, with relatively higher expression levels in node, pedicel, and pollen.
The relative abundance of AtCAP1 and two other actin-binding proteins important for actin dynamics, profilin and ADF/cofilin, were determined by quantitative immunoblotting (Table 1). Using Arabidopsis rosette leaves and leaf suspension-cultured cell extracts, molar ratios of total actin to CAP of 7:1 and 3:1 were determined. Profilin was in approximately a 3:1 molar ratio with total actin in both extracts. These findings are consistent with previous results for profilin and total actin levels in pollen (Vidali and Hepler, 1997; Gibbon et al., 1999; Snowman et al., 2002) and tobacco suspension cells (Wang et al., 2005). The total actin to ADF ratio was 3:1 and 1:1 for leaf and suspension cells, respectively. To our knowledge, cellular levels of ADF/cofilin have never been estimated for plant tissues. These results demonstrate that AtCAP1 is a fairly abundant protein in plant tissues and that profilin and ADF are present in ratios equal to or greater than total actin.
Table 1.
Intracellular protein amount for actin-binding proteins from A. thaliana
| Protein | Rosette leaves % of total protein | Suspension cells % of total protein | ABP:actin molar ratio (leaves/susp. cell) |
|---|---|---|---|
| Actin | 0.36 ± 0.03 (6) | 0.15 ± 0.03 (4) | — |
| CAP1 | 0.0593 ± 0.0106 (6) | 0.0554 ± 0.0113 (5) | 1:7/1:3 |
| Profilin | 0.32 ± 0.09 (4) | 0.14 ± 0.02 (4) | 3:1/3:1 |
| ADF | 0.048 ± 0.005 (4) | 0.07 ± 0.02 (4) | 1:3/1:1 |
Values represent mean percentage ± SD of a particular actin-binding protein with respect to total protein. Rosette leaves were collected at approximately principle growth stage 6 (Boyes et al., 2001). Molar ratios of each ABP to total actin were determined by multiplying the percentage of protein by the ratio of molecular weights and normalizing to actin concentration.
AtCAP1 Has a Moderate Affinity for Monomeric Actin
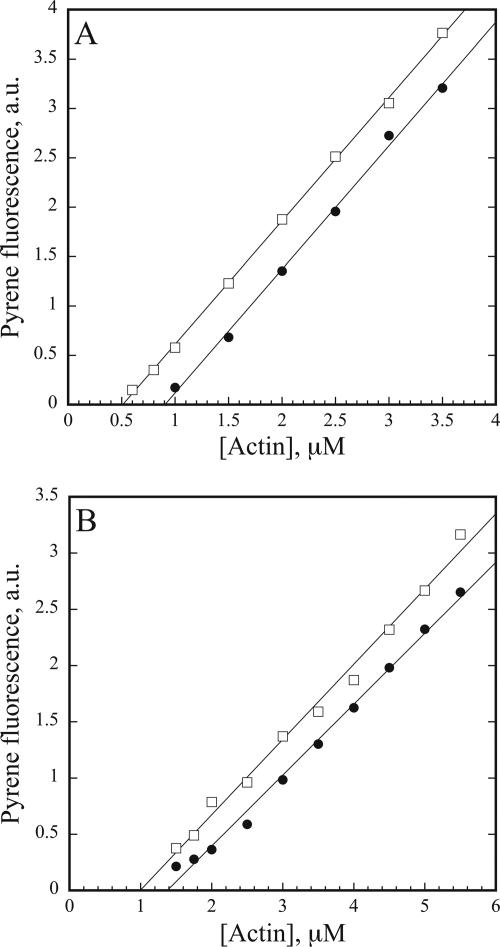
The dissociation constant (Kd) value for AtCAP1 binding to G-actin at steady state was estimated by measuring the shift in the Cc for the assembly of rabbit skeletal muscle actin and maize pollen actin. In these assays, a Cc is determined from the x-intercept of plots for fluorescence of pyrene–actin versus actin concentration. The difference in Cc values for actin alone or in the presence of AtCAP1 was used to determine the apparent Kd value for the assumed 1:1 interaction between AtCAP1 and G-actin. To compare directly AtCAP1 with profilin and to minimize the effects of barbed end addition, filaments were capped with 1:200 human plasma gelsolin. Experiments with maize profilin (ZmPRO5) and human profilin served as monomer-binding protein controls for Kd determination (Table 2). In a representative experiment with muscle actin shown in Figure 2A, the calculated Kd value for AtCAP1 was 0.8 μM. From several such experiments (n = 7), an average Kd value of 0.6 ± 0.2 μM was determined (Table 2). By comparison, an average Kd value of 0.5 ± 0.2 μM (n = 6) for pollen actin binding to AtCAP1 was obtained (see Figure 2B for a representative experiment). It should be noted that the Cc for assembly at the barbed end (0.36 μM; see also Ren et al., 1997 and Kovar et al., 2000a) and pointed end (1.2 μM; Table 2) of plant actin filaments is substantially higher than for muscle actin filaments. However, the apparent Kd values for AtCAP1 binding to plant and muscle actin were not statistically different (p > 0.05), indicating that AtCAP1 has equivalent ability to bind and sequester monomeric actin from both sources. For convenience, most biochemical characterizations were performed with rabbit muscle actin unless specified otherwise.
Table 2.
Apparent Kd values for steady-state binding to G-actin when filament ends are capped
| Plant actin | Muscle actin | |
|---|---|---|
| AtCAP1 | 0.5 ± 0.2 (6) | 0.6 ± 0.2 (7) |
| ZmPRO5 | 0.19 ± 0.08 (4) | 0.71 ± 0.14 (4) |
| HPRO1 | n.d. | 0.37 ± 0.13 (5) |
n.d., not determined.
The apparent Kdvalues (micromolar, mean ± SD [n]) for AtCAP1 binding to rabbit muscle actin and maize pollen actin (plant) with 1:200 gelsolin–actin were determined by the shift in Cc at steady state at 100 nM free Ca2+. The Cc for plant and muscle actin was 1.2 ± 0.2 (13) and 0.56 ± 0.07 (6), respectively, when filament ends were capped. The apparent Kd for human profilin (HPRO1) binding to plant actin could not be measured using this method.
Figure 2.
AtCAP1 changes the critical concentration (Cc) for actin assembly. (A) Representative experiment showed that AtCAP1 changes the steady state Cc for rabbit muscle actin assembly. Increasing concentrations of 1:200 gelsolin:actin (5% pyrene labeled) was polymerized in the absence (squares) and presence (circles) of 1 μM AtCAP1. The Cc values (intercept of line of best fit with the x-axis) were 0.5 μM for actin alone and 0.9 μM with AtCAP1. The resulting Kd value for this experiment was 0.8 μM. (B) Representative experiment performed with maize pollen actin, in the presence of gelsolin to cap filament barbed ends, gives Cc values of 1 μM for actin alone (squares) and 1.4 μM with AtCAP1 (circles), with a resulting Kd = 0.3 μM. a.u., arbitrary fluorescence units.
AtCAP1 Has Similar Kd Values for ATP– and ADP–G-actin
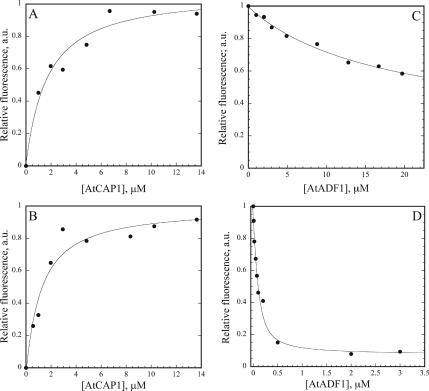
The C-terminal actin-binding domain from yeast Srv2p/CAP shows ∼100-fold higher affinity for ADP– versus ATP–G-actin (Mattila et al., 2004). The affinity of AtCAP1 for ATP– and ADP–G-actin was tested with NBD-labeled actin (Detmers et al., 1981; Weeds et al., 1986; Mattila et al., 2004) under polymerizing salt conditions (50 mM KCl and 1.25 mM MgCl2). A dose-dependent increase in fluorescence for 0.2 μM NBD–G-actin was observed in the presence of AtCAP1. As shown in Figure 3, the fluorescence change of NBD–G-actin was maximal in the 4–6 μM range of AtCAP1 for both ATP– and ADP–G-actin (Kd = 1.6 and 1.1 μM for the representative experiments in Figure 3, A and B, respectively). Calculated average Kd values (n = 5) of 1.3 ± 0.6 μM for ATP– and 1.3 ± 0.9 μM for ADP–G-actin were determined. Statistically, AtCAP1 did not have a marked preference for either nucleotide-loaded state of actin (two-tailed t test; p > 0.05), but rather it has a moderate affinity for both. Therefore, plant CAP and yeast Srv2p/CAP are different for this actin-binding property. Experiments with AtADF1, using the same batches of ATP– and ADP–actin as those for AtCAP1, served as controls to demonstrate that differences between ATP– and ADP–actin were measurable (e.g., Figure 3, C and D). AtADF1 gave average Kd values (n = 4) of 0.2 ± 0.1 μM for ADP–actin and 10 ± 6 μM for ATP–actin, in agreement with published data (Carlier et al., 1997).
Figure 3.
AtCAP1 binds to both ATP– and ADP–G-actin. (A) The increase in fluorescence of 0.2 μM Mg–ATP–G-actin (50% NBD labeled) was plotted as a function of [AtCAP1]. Dissociation equilibrium constants were determined by fitting the data as described in Materials and Methods. A single representative experiment which gave a Kd of 1.6 μM is shown. (B) The affinity of AtCAP1 for ADP–G-actin was determined as outlined above. A single representative experiment, which gave a Kd of 1.1 μM, is shown. (C) The affinity of AtADF1 for ATP–G-actin was followed by quenching of fluorescence. A single representative experiment, which gave a Kd of 17 μM, is shown. (D) The affinity of AtADF1 for ADP–G-actin, from a single representative experiment, gave a Kd of 0.05 μM.
AtCAP1 Inhibits Spontaneous Nucleation of Actin Filaments
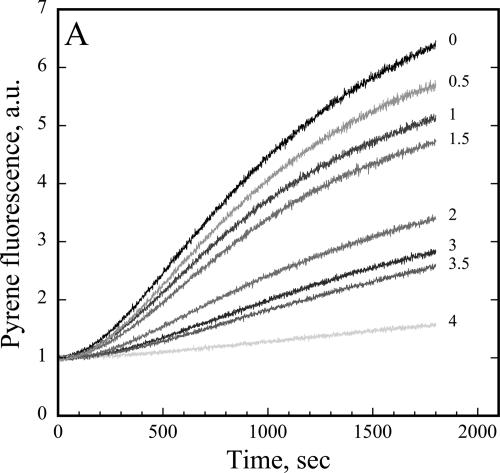
Actin nucleation involves the formation of a stable nucleus and is the rate-limiting step for actin polymerization, which is indicated by a distinctive lag period in a plot of filament mass versus time. Actin-monomer binding proteins have the ability to prevent or delay the nucleation step during assembly reactions (Pollard and Cooper, 1984; Korenbaum et al., 1998). We tested for this activity by adding various amounts of AtCAP1 to a constant amount of G-actin (5% pyrene labeled) and monitored filament assembly by the increase in pyrene fluorescence after addition of polymerization salts (Figure 4). AtCAP1 delayed polymerization and reduced the final extent of assembly compared with actin only (Figure 4, top curve). From these data, we conclude that AtCAP1 sequesters actin monomers and inhibits actin polymerization in a dose-dependent manner.
Figure 4.
AtCAP1 prevents spontaneous actin polymerization. Polymerization of actin monomers (3 μM; 5% pyrene labeled) was measured by fluorescence increase in the presence of various concentrations of AtCAP1. Pyrene fluorescence was plotted versus time after addition of polymerization salts. AtCAP1 concentrations were, from top to bottom: 0, 0.5, 1, 1.5, 2, 3, 3.5, and 4 μM. AtCAP1 suppressed actin nucleation in a dose-dependent manner and reduced the final extent of polymerization.
AtCAP1 Inhibits Actin Elongation at Both Ends of Actin Filaments
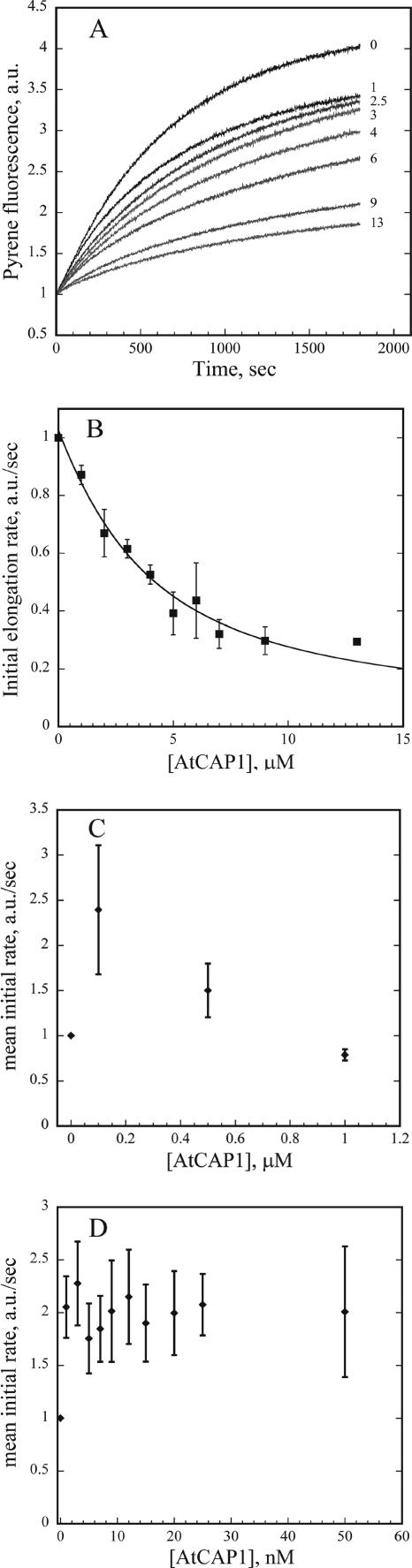
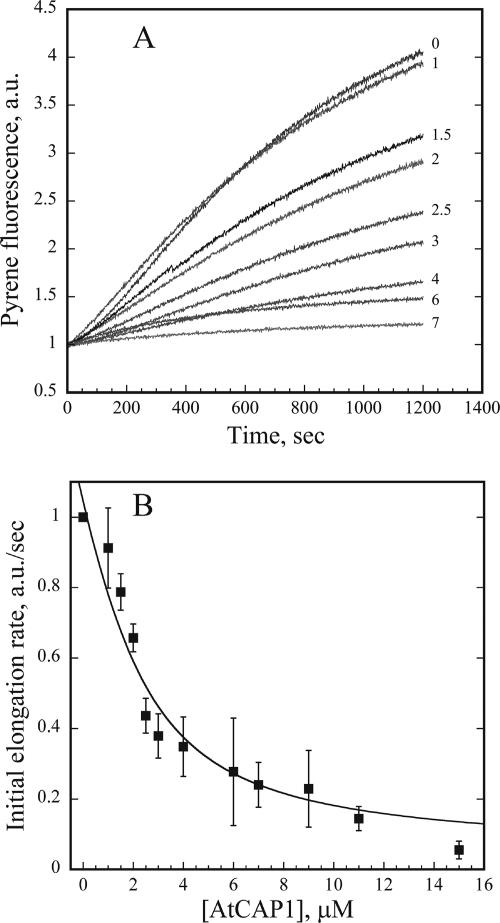
An elongation assay that monitors assembly of pyrene-actin onto preformed F-actin seeds was used to test the ability of AtCAP1 to regulate assembly at filament ends (Schafer et al., 1996; Huang et al., 2003). The initial polymerization rate depends on the ability of AtCAP1 to bind monomers and thereby prevent elongation. Various amounts of AtCAP1 and 0.4 μM actin filament seeds were added to 1 μM G-actin (5% pyrene labeled), and polymerization salts were used to initiate assembly. The concentration of filament ends was assumed to be constant, and it was calculated from the control curves. The initial elongation rate decreased in the presence of micromolar concentrations of AtCAP1, consistent with actin monomer-sequestering activity of AtCAP1 (Figure 5A). By fitting the data for the representative experiment shown in Figure 5A with the methods of Higgs et al. (1999), an apparent Kd value of 1.6 μM for actin monomer binding was calculated. From several such experiments (n = 4), an average Kd of 1.63 ± 0.13 μM was determined (Figure 5B). Interestingly, a modest enhancement of elongation rates was observed at submicromolar concentrations of AtCAP1 (0.01–0.5 μM), as shown in Figure 5D. This effect disappeared at molar ratios of CAP:G-actin greater than 1:1 (Figure 5C), and it is similar to the behavior of human CAP1 C-terminal domain on the elongation at filament barbed ends in the presence of cofilin (Moriyama and Yahara, 2002) We next tested whether the AtCAP1–G-actin complex can elongate filaments from pointed ends. Using gelsolin to cap the barbed ends of actin filament seeds, elongation from pointed ends was initiated with 2 μM G-actin (5% pyrene labeled). The initial elongation rate decreased as a function of increasing AtCAP1 concentration (Figure 6A). From several such experiments (n = 4), an apparent Kd value for actin monomers of 0.9 ± 0.1 μM was obtained (Figure 6B). Together, these results indicate that AtCAP1 prevents the assembly of monomeric actin at both barbed and pointed ends of actin filaments.
Figure 5.
AtCAP1 inhibits actin filament elongation in a dose-dependent manner. (A) Actin monomers (1 μM; 5% pyrene labeled) were supplemented with varying amounts of AtCAP1 and 0.4 μM actin seeds. Pyrene fluorescence was plotted as a function of time after addition of polymerization salts. AtCAP1 concentrations were, from top to bottom: 0, 1, 2.5, 3, 4, 6, 9, and 13 μM. AtCAP1 inhibited actin elongation from uncapped F-actin seeds in a dose-dependent manner. (B) Initial elongation rates were measured for the first 100 s and normalized to the rates in the absence of AtCAP1. The means ± SD from n = 4 experiments are shown. The data were fit as described in Materials and Methods to determine an apparent dissociation constant for AtCAP1 binding to actin monomer (Kd = 1.6 ± 0.1 μM). (C and D) AtCAP1–actin complex adds on to free barbed ends of actin filaments. Initial rates were determined as described for B using 0.01–0.05 μM (D) and 0.1–1 μM AtCAP1 (C). Mean values ± SD from four experiments are plotted.
Figure 6.
AtCAP1 inhibits pointed end elongation in a dose-dependent manner. (A) Actin monomers (2 μM; 5% pyrene labeled) were supplemented with different concentrations of AtCAP1 and 80 nM gelsolin-actin seeds. Pyrene fluorescence was plotted versus time after the addition of polymerization salts. AtCAP1 concentrations were, from top to bottom: 0, 1, 1.5, 2, 2.5, 3, 4, 6, and 7 μM. (B) Initial elongation rates at the pointed end were measured for the first 100 s, as in Figure 5. The means ± SD for n = 4 experiments were fit as described in Materials and Methods to determine an apparent Kd value of 0.9 ± 0.1 μM.
AtCAP1 Stimulates Nucleotide Exchange on Actin
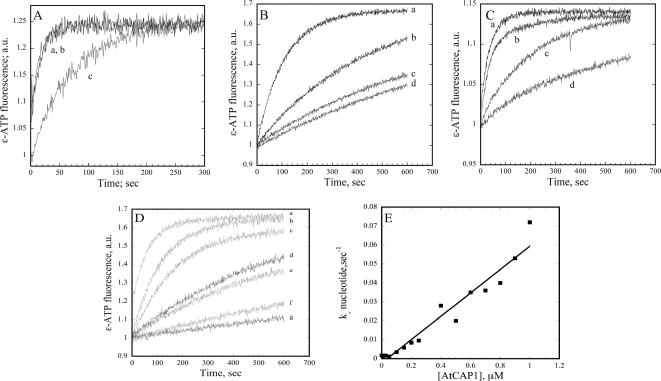
Yeast and human CAP differ in the ability to enhance nucleotide exchange on G-actin; the C-terminal domain of human CAP stimulates nucleotide exchange (Moriyama and Yahara, 2002), whereas Srv2p has no effect (Balcer et al., 2003). The rate of nucleotide exchange for 0.5 μM maize pollen actin alone (Figure 7A, curve c) and in the presence of AtCAP1 (Figure 7A, curve b) was determined by measuring the change in fluorescence upon the incorporation of the ATP analogue ε-ATP under low ionic strength buffer conditions (Goldschmidt-Clermont et al., 1992). This method allowed us to control the rate of nucleotide exchange on maize actin, which is intrinsically faster than vertebrate actin (Kovar et al., 2000a, 2001). An equimolar amount of AtCAP1 (0.5 μM) enhanced the rate of nucleotide exchange rather dramatically (Figure 7A, curve b). This is similar to the effect observed with 0.1 μM human profilin 1 (Figure 7A, curve a). For comparison, 0.5 μM AtPRF4, a plant profilin, slightly inhibited nucleotide exchange (data not shown) and 1 μM AtADF1 markedly inhibited nucleotide exchange (data not shown), as reported previously (Carlier et al., 1997). A similar pattern emerged when muscle actin was tested under the same conditions (Figure 7B). To evaluate these effects quantitatively, nucleotide exchange rates from several experiments (n = 4), in which >90% of actin subunits are calculated to be bound by actin-binding protein, were measured (Table 3). Human profilin I increased the nucleotide exchange rate by 35-fold and AtCAP1 showed 64-fold increase in rate when >90% of actin subunits were bound. Like other plant profilins (Perelroizen et al., 1996; Eads et al., 1998; Kovar et al., 2000a, 2001), AtPRF4 had very little or no effect on nucleotide exchange (Table 3). Similarly, AtADF1 inhibited exchange by fourfold compared with actin at only 50% bound G-actin (Table 3). A lower fold enhancement of nucleotide exchange for maize actin (data not shown) in the presence of AtCAP1 and HPRO1 may be due to the ∼10- to 20-fold higher endogenous rate of nucleotide exchange exhibited by plant actin (Kovar et al., 2001; this study).
Figure 7.
AtCAP1 increases nucleotide exchange on plant and muscle actin. (A) Representative experiment shows nucleotide exchange on 0.5 μM maize pollen actin (curve c). In the presence of 0.5 μM AtCAP, the rate was substantially faster (curve b) compared with actin alone, and it was similar to that observed in the presence of 0.1 μM human profilin (curve a). Not shown: a plant profilin, AtPRF4, and AtADF1 slightly or markedly inhibited nucleotide exchange under these conditions. These experiments were performed in a low ionic strength salt solution. (B) Representative experiment shows nucleotide exchange on 0.5 μM muscle actin at low ionic strength (curve c) and in the presence of 0.05 μM AtCAP1 (curve b). A more dramatic enhancement of nucleotide exchange rate was stimulated by the presence of 0.05 μM human profilin (curve a), whereas 1 μM AtADF1 inhibited nucleotide exchange (curve d). (C) Representative experiment shows nucleotide exchange on 0.5 μM muscle actin under physiological ionic conditions (curve c), 0.05 μM AtCAP1 (curve b), 0.05 μM human profilin (curve a), and 1 μM AtADF1 (curve d). (D) AtCAP1 relieves the inhibition of nucleotide exchange caused by AtADF1. Increasing concentrations of AtCAP1 were added to constant amounts of 0.5 μM AtADF1 and 0.5 μM actin. AtCAP1 concentrations were 0.01 μM (curve f), 0.025 μM (curve e), 0.15 μM (curve c), 0.2 μM (curve b), and 0.5 μM AtCAP1 (curve a). Control curves were actin + 0.5 μM AtADF1 (curve g) and 0.5 μM muscle actin alone (curve d). (E) The dose-dependent effect of the ability of increasing AtCAP1 to relieve nucleotide exchange inhibition by AtADF1 was plotted against nucleotide exchange rate and fit with a linear curve. For this experiment, the exchange rates for 0.5 μM actin alone, actin + 0.5 μM AtADF1, and actin + 1 μM AtCAP1 were 0.0017, 0.0003, and 0.07 a.u./s, respectively.
Table 3.
Nucleotide exchange rates for muscle actin at low ionic strength in the presence of four different actin-binding proteins
| Actin only | +HPRO1 | +AtCAP1 | +AtPRF4 | +AtADF1a |
|---|---|---|---|---|
| 0.005 ± 0.002 (17) | 0.174 ± 0.012 (4) | 0.32 ± 0.07 (4) | 0.0074 ± 0.0005 (3) | 0.00142 ± 0.00013 (3) |
Nucleotide exchange rates are reported as the change in fluorescence (arbitrary units) per second ± SD (n), as determined by fitting experimental data to a single exponential function. The amount of actin-binding protein added corresponds to an amount that would give near saturation of monomers or >90% bound, based on the Kd values in Table 2 and our unpublished data. For human profilin (HPRO1), this amount corresponded to 3.8 μM; for AtCAP1, it was 6.03 μM; and for Arabidopsis profilin 4 (AtPRF4), it was 6.5 μM.
a Because of limitations in the amount of protein that could be added to a reaction, AtADF1 was tested at 50% bound or 11.2 μM.
Similar nucleotide exchange experiments were performed with muscle actin in a physiological salt buffer (Figure 7C). A range of 0.01–0.15 μM AtCAP1 was tested in the presence of 0.5 μM actin. At 11% bound G-actin, HPRO1 enhanced the rate threefold, whereas AtCAP1 enhanced the rate ∼2.7-fold. At 28% bound G-actin, the highest measurable activity tested, AtCAP1 enhanced the overall rate by ∼4.7-fold. Again, the high endogenous rate of nucleotide exchange under these conditions prevented us from making measurements in the presence of near-saturating concentrations of actin-binding protein. Nevertheless, the sum of these results indicates that AtCAP1 can enhance nucleotide exchange on both plant and vertebrate actin.
The data are consistent with AtCAP1 being the physiologically relevant stimulator of nucleotide exchange on plant actin, because AtADF1 inhibits and plant profilin has no effect, or slightly inhibits exchange. Given that AtADF1 has a marked preference for ADP–G-actin, the form of monomer that disassembles from actin filaments, and because ADF is probably more abundant in plant cells than is AtCAP1 (Table 1), it was important to test the ability of AtCAP1 to stimulate nucleotide exchange in the presence of AtADF1. Using muscle actin at physiological ionic strength, we observed that substoichiometric amounts of AtCAP1 (e.g., 0.01 and 0.025 μM; Figure 7D, curves e and f) were able to partially or completely overcome the inhibition of nucleotide exchange caused by 0.5 μM AtADF1 (Figure 7D, curve g). Relative to actin alone (Figure 7D, curve d), concentrations of AtCAP1 greater than 0.1 μM led to increased nucleotide exchange rates (Figure 7D, curves a–c), even in the presence of AtADF1. By titrating increasing amounts of AtCAP1 in the presence of constant 0.5 μM AtADF1, we found a dose-dependent enhancement of nucleotide exchange stimulated by AtCAP1 (Figure 7E). These results demonstrate that AtCAP1 can promote nucleotide exchange even in the presence of an excess of AtADF1.
AtCAP and ADF Act Synergistically to Enhance Actin Filament Turnover
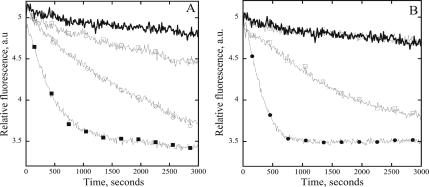
We used “single-turnover” experiments (Didry et al., 1998) to investigate the effect of AtCAP1 on the time course of actin filament turnover in the presence or absence of plant ADF (AtADF1) and/or plant profilin (AtPRF4). In these assays, turnover is measured by a decrease in fluorescence when “cold” ATP is added to a solution of ε-ADP–actin filaments at equilibrium. Because nucleotide exchange occurs only on monomers, rather than on subunits within filaments, the kinetics of turnover is affected by proteins that alter the rate of endogenous nucleotide exchange on monomers, the ability of actin and actin-binding protein complexes to assemble and disassemble at filament ends, or both. In the absence of any actin-binding protein, actin filament turnover was very slow (Figure 8, A and B, thick solid line), due to the slow rate of actin depolymerization from the pointed end. The presence of AtCAP or profilin had only a modest effect on the rate of actin filament turnover (3-fold increase; Figure 8A, open squares or Figure 8B, open circles), whereas ADF/cofilin increased the turnover rate up to 14-fold (Figure 8A, open circles). In the presence of both AtCAP and ADF, the turnover is further increased (34-fold; Figure 8A, closed squares), consistent with the ability of AtCAP1 to promote nucleotide exchange on monomeric actin even in the presence of ADF. In contrast to this, plant profilin and ADF increased turnover only 18.6-fold (Figure 8B, open squares). However, the presence of both profilin and AtCAP together with ADF increased the rate by up to 42-fold (Figure 8B, closed circles), demonstrating that these proteins function synergistically to increase actin turnover.
Figure 8.
AtCAP1 enhances the rate of actin filament turnover in the presence of ADF/cofilin. Shown is the exchange of fluorescent ε-ADP–actin in filaments for unlabeled ADP–actin, in the presence of various actin binding proteins, as a function of time. (A) Plotted are reactions containing 4 μM actin alone, thick black line; 4 μM actin and 1 μM AtCAP1, open squares; 4 μM actin and 3 μM AtADF1, open circles; and 4 μM actin and 3 μM AtADF1 and 1 μM AtCAP1, closed squares. (B) Plotted are 4 μM actin alone, thick black line; 4 μM actin and 4 μM AtPRF4, open circles; 4 μM actin with 3 μM AtADF1 and 4 μM AtPRF4, open squares; and 4 μM actin with 3 μM AtADF1 and 1 μM AtCAP1 and 4 μM AtPRF4, closed circles. The plots in A and B are from representative experiments performed on the same day. Turnover rates were estimated by fitting the data with a monoexponential function. Mean rates ± SD obtained from four to six experiments were as follows: actin alone = 0.00005 ± 0.00005 a.u./s, actin + ADF = 0.0007 ± 0.0003 a.u./s, actin + AtCAP = 0.00015 ± 0.0001 a.u./s, actin + PRF4 = 0.0009 ± 0.0009 a.u./s, actin + ADF + PRF4 = 0.0009 ± 0.0006 a.u./s, actin + ADF + AtCAP = 0.0017 ± 0.0011 a.u./s, and actin + ADF + CAP + PRF4 = 0.0021 ± 0.0015 a.u./s.
DISCUSSION
To accurately interpret mutant phenotypes and to gain insight about the in vivo function of diverse actin-binding proteins, it is imperative to fully understand their biochemical activities and cellular concentrations. This is especially pertinent for plants where many recent examples of unique and divergent properties for actin and actin-binding proteins have been reported (Staiger and Blanchoin, 2006). Here, we used multiple, complementary biochemical assays to examine the in vitro properties for a plant CAP. Several lines of evidence demonstrate unambiguously that AtCAP1 is an abundant cellular protein that binds to monomeric actin with moderate affinity (Kd of 0.5–1.6 μM). This plant protein shows no preference for the source of actin, binding with equal affinity to purified pollen and muscle actins. It also binds with identical affinity to ATP– and ADP–G-actin. At steady state, AtCAP1 reduces the amount of filamentous actin, consistent with formation of a 1:1 CAP–G-actin complex that does not assemble. During polymerization, AtCAP suppresses spontaneous nucleation of filaments from a pool of monomers and inhibits the addition of actin subunits at both barbed and pointed ends of filaments. These behaviors are consistent with an actin monomer-sequestering function, and they are somewhat reminiscent of vertebrate thymosin β4 (Safer et al., 1990; Carlier et al., 1993; Pantaloni and Carlier, 1993; Huff et al., 2001), a sequestering protein that seems to be absent from the plant kingdom (Hussey et al., 2002; Meagher and Fechheimer, 2003). In contrast to thymosin β4, however, AtCAP1 has the ability to act as a nucleotide exchange factor for monomeric actin, and it does not show a preference for ATP–G-actin. This is the first biochemical evidence for a plant protein that enhances the nucleotide exchange rate on G-actin, and it is quite relevant because plant profilins lack this ability or even inhibit nucleotide turnover (Perelroizen et al., 1996; Eads et al., 1998; Kovar et al., 2000a, 2001). Indeed, AtCAP1 stimulates nucleotide exchange on G-actin even in the presence of AtADF1. ADF/cofilin facilitates actin disassembly by severing filaments and increasing the loss of ADP–G-actin from filament pointed ends (Maciver et al., 1991; Carlier et al., 1997); however, this latter property was recently challenged with results obtained from total internal reflection fluorescence microscopy of single actin filaments (Andrianantoandro and Pollard, 2006). They also bind preferentially to ADP–G-actin and inhibit nucleotide exchange (Carlier et al., 1997; Blanchoin and Pollard, 1998; this study). We conclude that AtCAP1 is a central plant factor that regulates actin dynamics by serving as a key intermediate between ADF/cofilin-mediated disassembly and the profilin-based nucleation and elongation machinery.
The properties of plant CAP overlap but are distinct from either human CAP1 or yeast Srv2p/CAP. The C-terminal domain of Srv2p/CAP binds ADP–G-actin with high affinity (Kd = 0.02 μM) compared with ATP–G-actin (Kd = 1.2 μM); does not allow elongation at barbed or pointed ends of actin filaments; competes with ADF/cofilin for binding to ADP–G-actin; accelerates ADF/cofilin-induced depolymerization; binds to profilin (Bertling et al., 2007) and accelerates profilin-induced nucleotide exchange, but it does not enhance nucleotide exchange on ADP–actin on its own (Balcer et al., 2003; Mattila et al., 2004). By contrast, AtCAP1 stimulates nucleotide exchange on G-actin directly and binds equally well to ATP– and ADP–G-actin. Human CAP1 accelerates cofilin-induced depolymerization similar to Srv2p/CAP, but it differs in many other respects. Although its preference for ATP– versus ADP–G-actin is not known, full-length human CAP1 permits addition of subunits on to filament barbed ends but not on to pointed ends (Moriyama and Yahara, 2002). It also enhances nucleotide exchange on G-actin directly (Moriyama and Yahara, 2002), similar to AtCAP1. Interestingly, human CAP1 has actin-binding sites on both N- and C-terminal domains that may or may not overlap with the cofilin-binding site on actin monomers (Moriyama and Yahara, 2002). Plant CAP also facilitates filament elongation at barbed ends, even in the absence of ADF/cofilin (Figure 5D). Several possibilities can be considered, including modest filament-severing activity that increases the number of ends, binding and stabilization of filaments, and nucleotide exchange activity that converts any ADP–G-actin in these reactions to highly polymerizable ATP–G-actin.
Nucleotide exchange and the nucleotide-loaded state of G- and F-actin are generally held to be important for actin dynamics. The nucleotide status of subunits in the filament regulates ADF/cofilin-severing and depolymerization activity, and it acts as a timer for filament aging, with ADF/cofilin preferring the older ADP-bound filament (Maciver et al., 1991; Carlier et al., 1997). ADF/cofilin also prefers to bind ADP–G-actin and inhibits the turnover of nucleotide on monomers (Nishida, 1985; Lappalainen et al., 1997; Blanchoin and Pollard, 1998). Profilin, in contrast, enhances nucleotide exchange on G-actin (Nishida, 1985; Goldschmidt-Clermont et al., 1992; Perelroizen et al., 1996), prefers to bind ATP–G-actin over ADP–G-actin (Vinson et al., 1998), and delivers ATP–G-actin for assembly at filament barbed ends (Pantaloni and Carlier, 1993; Kang et al., 1999). The importance of nucleotide exchange by profilin has been demonstrated genetically in yeasts (Wolven et al., 2000; Lu and Pollard, 2001). In Schizosaccharomyces pombe, the ability of mutant profilins to complement profilin null and temperature-sensitive (ts) strains parallels the ability to enhance nucleotide exchange on actin. Profilin mutations that enhance nucleotide exchange complement both null and ts strains, whereas mutant profilins that are totally deficient for exchange activity cannot (Lu and Pollard, 2001). Similarly, in Saccharomyces cerevisiae, an actin mutant (act1-157) with high intrinsic nucleotide exchange activity rescues a profilin mutant (pfy1-4) deficient for nucleotide exchange activity (Wolven et al., 2000). Thus, nucleotide exchange activity of AtCAP1 fits into an emerging picture for plant actin turnover where profilin has no effect on nucleotide exchange (Perelroizen et al., 1996; Kovar et al., 2000a, 2001), and plant ADF/cofilins inhibit nucleotide exchange (Carlier et al., 1997; Didry et al., 1998). AtCAP1 may therefore be a key intermediate that couples ADF/cofilin-mediated depolymerization of actin filaments with assembly of ATP–G-actin by profilin at filament barbed ends.
Although plant cells are not motile, they use the actin cytoskeleton to power a plethora of dynamic intracellular processes (Staiger et al., 2000; Hussey et al., 2006; Staiger and Blanchoin, 2006). Surprisingly, these actin-based functions are accomplished with a relatively small proportion of the total actin protein assembled into actin filaments at any given time (Staiger and Hussey, 2004; Staiger and Blanchoin, 2006). In pollen grains and pollen tubes for example, filamentous actin has been estimated to be ∼5–10% of the total actin pool, or roughly 10–15 μM actin in polymer (Gibbon et al., 1999; Snowman et al., 2002), whereas in tobacco BY-2 cells this value is on the order of just 1–2% (Wang et al., 2005). These observations hint that actin subunits cycle through the polymer system with extremely high rates in plant cells. Consistent with this, the monomer-binding proteins are also highly abundant. Profilin has been shown previously to be in a 1:1 stoichiometry with total actin in pollen (Vidali and Hepler, 1997; Gibbon et al., 1999; Snowman et al., 2002) and in tobacco suspension cells (Wang et al., 2005). It is generally considered to be the main buffer of the actin monomer pool, suppressing filament nucleation and providing a large population of subunits for formin-mediated polymerization (Staiger and Hussey, 2004; Staiger and Blanchoin, 2006). In the first attempts to quantify actin and actin-binding protein levels from Arabidopsis, we estimated that profilin levels may actually exceed the total actin pool (Table 1). The use of maize pollen actin and the corresponding maize actin antibody for generation of a standard curve, however, may have led to underreporting the amount of actin in Arabidopsis extracts. Nevertheless, profilin appears to be quite abundant and is at least equimolar with total actin in leaf tissues and suspension cells. Not previously documented for plants, we found that the ratio of ADF/cofilin to total actin was 1:3 in leaf tissue and roughly 1:1 in suspension cell tissues (Table 1). Our results also show that AtCAP1 is present in a 1:7 and 1:3 ratio with total actin in leaf tissues and suspension cells, respectively (Table 1). This is similar to the 1:10 ratio reported for yeast (Balcer et al., 2003) and the 1:4 CAP1 to actin ratio in mammalian cells (Bertling et al., 2004). Based on its moderate affinity (Kd ∼ 1 μM) for G-actin, we estimate that roughly 10–30% of the cellular monomeric actin pool is bound by AtCAP1. The high concentration for three synergistic regulators of actin dynamics in plant cells is consistent with virtually all of the actin monomer bound to actin monomer-binding proteins and a prediction for high turnover rates of actin filaments.
Based on the results reported herein and on previous studies of ADF/cofilin and profilin function (Staiger and Hussey, 2004; Hussey et al., 2006; Staiger and Blanchoin, 2006), we propose a new model for plant actin filament turnover that incorporates the unique properties of AtCAP1. Central to this model, AtCAP1 serves as an intermediate between the severing/depolymerizing activity of ADF/cofilin and the assembly promoting function of profilin at filament barbed ends. This is superficially similar to actin dynamics fostered by yeast Srv2p/CAP (Balcer et al., 2003; Mattila et al., 2004); in plants, however, CAP rather than profilin stimulates nucleotide exchange on G-actin. Specifically, the nucleotide exchange activity of AtCAP1 will accelerate the shuttling of actin monomers between the ADF/cofilin- and profilin-bound pools, thereby fostering rapid actin dynamics. Flux of subunits between the filament and subunit pools is as follows: Older regions of actin filaments are made up of ADP-actin, which promotes ADF/cofilin binding, severing, and depolymerization. G-actin bound by ADF/cofilin remains ADP loaded, due to the inhibition of nucleotide exchange. AtCAP1 competes with ADF/cofilin for binding to G-actin, and it stimulates the exchange of ADP for ATP. Given the similar affinities for monomeric actin and the abundance of profilin (Table 1), ATP–G-actin is transferred from AtCAP1 to profilin. The pool of profilin–ATP–G-actin can then add to filament barbed ends and is also the likely mechanism for formin-mediated nucleation of new actin filaments (Michelot et al., 2005). The model assumes that nucleotide exchange is a limiting factor for actin polymerization and that plant profilin has low affinity for ADF–ADP–G-actin. Further biochemical evidence is necessary to elucidate whether plant profilin indeed has a preference for ATP–G-actin and can compete with AtCAP1 for binding monomers. Given the reported interactions between profilin, ADF/cofilin, and CAP in nonplant systems (Moriyama and Yahara, 2002; Bertling et al., 2007), models in which allosteric regulation of profilin activity by CAP and of CAP activity by ADF can also be considered. It should be noted, however, that the proline-rich domain in yeast CAP that is implicated in profilin binding (Bertling et al., 2007) is poorly conserved or absent in AtCAP1 (Barrero et al., 2002). Nevertheless, our simple model and the central role of AtCAP1 in actin dynamics are consistent with the severe disruption to actin filament arrays observed in atcap1 homozygous mutant plants (Deeks et al., 2007). Measurements of actin filament turnover rates and polymer levels in wild-type and atcap1 mutant plants will be necessary to further test the validity of the model in planta.
ACKNOWLEDGMENTS
We thank Shanjin Huang for helpful comments, Rabbiya Chaudhry for assistance with statistical analysis, Alan Weeds for advice and the NBD–actin protocol, and the Arabidopsis Biological Resources Center for supplying seed stocks and cDNA libraries. Patrick Hussey and Michael Deeks (Durham University) kindly shared unpublished data on cap mutant phenotypes. This research was supported by the National Research Initiative-Competitive Grants Program, U.S. Department of Agriculture, grant 2002-35304-12412 (to C.J.S.) and a Centre National de la Recherche Scientifique-Programmes Internationaux de Coopération Scientifique grant (to L.B.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-11-1041) on May 30, 2007.
REFERENCES
- Alonso J. M., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T. D. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Balcer H. I., Goodman A. L., Rodal A. A., Smith E., Kugler J., Heuser J. E., Goode B. L. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Barrero R. A., Umeda M., Yamamura S., Uchimiya H. Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division. Plant Cell. 2002;14:149–163. doi: 10.1105/tpc.010301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero R. A., Umeda M., Yamamura S., Uchimiya H. Over-expression of Arabidopsis CAP causes decreased cell expansion leading to organ size reduction in transgenic tobacco plants. Ann. Bot. 2003;91:599–603. doi: 10.1093/aob/mcg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlali A., Draskovic I., Hazelett D. J., Treisman J. E. Act up controls actin polymerization to alter cell shape and restrict hedgehog signaling in the Drosophila eye disc. Cell. 2000;101:271–281. doi: 10.1016/s0092-8674(00)80837-5. [DOI] [PubMed] [Google Scholar]
- Bertling E., Hotulainen P., Mattila P. K., Matilainen T., Salminen M., Lappalainen P. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell. 2004;15:2324–2334. doi: 10.1091/mbc.E04-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertling E., Quintero-Monzon O., Mattila P. K., Goode B. L., Lappalainen P. Mechanism and biological role of profilin-Srv2p/CAP interaction. J. Cell Sci. 2007;120:1225–1234. doi: 10.1242/jcs.000158. [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T. D. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J. Biol. Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Boyes D. C., Zayed A. M., Ascenzi R., McCaskill A. J., Hoffman N. E., Davis K. R., Görlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. L., Korn E. D. On the mechanism of actin monomer-polymer subunit exchange at steady state. J. Biol. Chem. 1983;258:5013–5020. [PubMed] [Google Scholar]
- Carlier M.-F., Jean C., Rieger K. J., Lenfant M., Pantaloni D. Modulation of the interaction between G-actin and thymosin β4 by the ATP/ADP ratio: possible implication in the regulation of actin dynamics. Proc. Natl. Acad. Sci. USA. 1993;90:5034–5038. doi: 10.1073/pnas.90.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.-F., Laurent V., Santolini J., Melki R., Didry D., Xia G.-X., Hong Y., Chua N.-H., Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks M. J., Rodrigues C., Dimmock S., Ketelaar T., Maciver S. K., Malhó R., Hussey P. J. Arabidopsis CAP1, a key regulator of actin organization and development. J. Cell Sci. 2007 doi: 10.1242/jcs.007302. (in press) [DOI] [PubMed] [Google Scholar]
- Detmers P., Weber A., Elzinga M., Stephens R. E. 7-Chloro-4-nitrobenzeno-2-oxa-1,3-diazole actin as a probe for actin polymerization. J. Biol. Chem. 1981;256:99–105. [PubMed] [Google Scholar]
- Didry D., Carlier M.-F., Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 1998;273:25602–25611. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Eads J. C., Mahoney N. M., Vorobiev S., Bresnick A. R., Wen K-K., Rubenstein P. A., Haarer B. K., Almo S. C. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Fedor-Chaiken M., Deschenes R. J., Broach J. R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990;61:329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- Field J., et al. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell. 1990;61:319–327. doi: 10.1016/0092-8674(90)90812-s. [DOI] [PubMed] [Google Scholar]
- Freeman N. L., Chen Z., Horenstein J., Weber A., Field J. An actin monomer ninisbinding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J. Biol. Chem. 1995;270:5680–5685. doi: 10.1074/jbc.270.10.5680. [DOI] [PubMed] [Google Scholar]
- Gerst J. E., Ferguson K., Vojtek A., Wigler M., Field J. CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol. Cell. Biol. 1991;11:1248–1257. doi: 10.1128/mcb.11.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B. C., Kovar D. R., Staiger C. J. Latrunculin B has different effects on pollen germination and tube growth. Plant Cell. 1999;11:2349–2363. doi: 10.1105/tpc.11.12.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B. C., Zonia L. E., Kovar D. R., Hussey P. J., Staiger C. J. Pollen profilin function depends on interaction with proline-rich motifs. Plant Cell. 1998;10:981–994. doi: 10.1105/tpc.10.6.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann R., Mann K. ASP-56, a new actin sequestering protein from pig platelets with homology to CAP, an adenylate cyclase-associated protein from yeast. FEBS Lett. 1992;298:149–153. doi: 10.1016/0014-5793(92)80043-g. [DOI] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont P. J., Furman M. I., Wachsstock D., Safer D., Nachmias V. T., Pollard T. D. The control of actin nucleotide exchange by thymosin β4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald U., Brokamp R., Karakesisoglou I., Schleicher M., Noegel A. A. Identification of a cyclase-associated protein (CAP) homologue in Dicytostelium discoideum and characterization of its interaction with actin. Mol. Biol. Cell. 1996;7:261–272. doi: 10.1091/mbc.7.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:6487–6501. doi: 10.1128/MCB.00117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Higgs H. N., Blanchoin L., Pollard T. D. Influence of the c terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Chaudhry F., Franklin-Tong V. E., Staiger C. J. A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J. Biol. Chem. 2004;279:23364–23375. doi: 10.1074/jbc.M312973200. [DOI] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D. R., Staiger C. J. Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 2003;278:44832–44842. doi: 10.1074/jbc.M306670200. [DOI] [PubMed] [Google Scholar]
- Hubberstey A. V., Mottillo E. P. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 2002;16:487–499. doi: 10.1096/fj.01-0659rev. [DOI] [PubMed] [Google Scholar]
- Huff T., Müller C.S.G., Otto A. M., Netzker R., Hannappel E. β-thymosins, small acidic peptides with multiple functions. Int. J. Biochem. Cell Biol. 2001;33:205–220. doi: 10.1016/s1357-2725(00)00087-x. [DOI] [PubMed] [Google Scholar]
- Hussey P. J., Allwood E. G., Smertenko A. P. Actin-binding proteins in the Arabidopsis genome database: properties of functionally distinct plant actin-depolymerizing factors/cofilins. Phil. Trans. R. Soc. Lond. B. 2002;357:791–798. doi: 10.1098/rstb.2002.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey P. J., Ketelaar T., Deeks M. J. Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 2006;57:109–125. doi: 10.1146/annurev.arplant.57.032905.105206. [DOI] [PubMed] [Google Scholar]
- Kang F., Purich D. L., Southwick F. S. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 1999;274:36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Kawai M., Aotsuka S., Uchimiya H. Isolation of a cotton CAP gene: a homologue of adenylyl cyclase-associated protein highly expressed during fiber elongation. Plant Cell Physiol. 1998;39:1380–1383. doi: 10.1093/oxfordjournals.pcp.a029346. [DOI] [PubMed] [Google Scholar]
- Korenbaum E., Nordberg P., Björkegren-Sjögren C., Schutt C. E., Lindberg U., Karlsson R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry. 1998;37:9274–9283. doi: 10.1021/bi9803675. [DOI] [PubMed] [Google Scholar]
- Kouyama T., Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labeled F-actin. Eur. J. Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Kovar D. R., Drøbak B. K., Staiger C. J. Maize profilin isoforms are functionally distinct. Plant Cell. 2000a;12:583–598. doi: 10.1105/tpc.12.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Staiger C. J., Weaver E. A., McCurdy D. W. AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 2000b;24:625–636. doi: 10.1046/j.1365-313x.2000.00907.x. [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Yang P., Sale W. S., Drøbak B. K., Staiger C. J. Chlamydomonas reinhardtii produces a profilin with unusual biochemical properties. J. Cell Sci. 2001;114:4293–4305. doi: 10.1242/jcs.114.23.4293. [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Fedorov E. V., Fedorov A. A., Almo S. C., Drubin D. G. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila T., Drubin D. G. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Pollard T. D. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver S. K., Zot H. G., Pollard T. D. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean-Fletcher S., Pollard T. D. Mechanism of action of cytochalasin B on actin. Cell. 1980;20:329–341. doi: 10.1016/0092-8674(80)90619-4. [DOI] [PubMed] [Google Scholar]
- Mattila P. K., Quintero-Monzon O., Kugler J., Moseley J. B., Almo S. C., Lappalainen P., Goode B. L. A high-affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein (CAP) Mol. Biol. Cell. 2004;15:5158–5171. doi: 10.1091/mbc.E04-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R. B., Fechheimer M. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2003. The Arabidopsis cytoskeletal genome; pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C. J., Blanchoin L. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell. 2005;17:2296–2313. doi: 10.1105/tpc.105.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K., Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 2002;115:1591–1601. doi: 10.1242/jcs.115.8.1591. [DOI] [PubMed] [Google Scholar]
- Nicholson-Dykstra S., Higgs H. N., Harris E. S. Actin dynamics: growth from dendritic branches. Curr. Biol. 2005;15:R346–R357. doi: 10.1016/j.cub.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Nishida E. Opposite effects of cofilin and profilin from porcine brain on rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1985;24:1160–1164. doi: 10.1021/bi00326a015. [DOI] [PubMed] [Google Scholar]
- Pantaloni D., Carlier M.-F. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Perelroizen I., Carlier M.-F., Pantaloni D. Binding of divalent cation and nucleotide to G-actin in the presence of profilin. J. Biol. Chem. 1995;270:1501–1508. doi: 10.1074/jbc.270.4.1501. [DOI] [PubMed] [Google Scholar]
- Perelroizen I., Didry D., Christensen H., Chua N.-H., Carlier M.-F. Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J. Biol. Chem. 1996;271:12302–12309. doi: 10.1074/jbc.271.21.12302. [DOI] [PubMed] [Google Scholar]
- Pollard T. D. Polymerization of ADP-actin. J. Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D., Cooper J. A. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pope B., Gooch J. T., Weeds A. G. Probing the effects of calcium on gelsolin. Biochemistry. 1997;36:15848–15855. doi: 10.1021/bi972192p. [DOI] [PubMed] [Google Scholar]
- Ren H., Gibbon B. C., Ashworth S. L., Sherman D. M., Yuan M., Staiger C. J. Actin purified from maize pollen functions in living plant cells. Plant Cell. 1997;9:1445–1457. doi: 10.1105/tpc.9.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safer D., Golla R., Nachmias V. T. Isolation of a 5-kilodalton actin-sequestering peptide from human blood platelets. Proc. Natl. Acad. Sci. USA. 1990;87:2536–2540. doi: 10.1073/pnas.87.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. A., Jennings P. B., Cooper J. A. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowman B. N., Kovar D. R., Shevchenko G., Franklin-Tong V. E., Staiger C. J. Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell. 2002;14:2613–2626. doi: 10.1105/tpc.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Staiger C. J., Baluska F., Volkmann D., Barlow P. Dordrecht, The Netherlands: Kluwer Academic Publisher; 2000. Actin: A Dynamic Framework for Multiple Plant Cell Functions. [Google Scholar]
- Staiger C. J., Blanchoin L. Actin dynamics: old friends with new stories. Curr. Opin. Plant Biol. 2006;9:554–562. doi: 10.1016/j.pbi.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Staiger C. J., Hussey P. J. Actin and actin-modulating proteins. In: Hussey P. J., editor. The Plant Cytoskeleton in Cell Differentiation and Development. Oxford, United Kingdom: Blackwell Publishers; 2004. pp. 32–80. [Google Scholar]
- Vidali L., Hepler P. K. Characterization and localization of profilin in pollen grains and tubes of Lilium longiflorum. Cell Motil. Cytoskeleton. 1997;36:323–338. doi: 10.1002/(SICI)1097-0169(1997)36:4<323::AID-CM3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Vinson V. K., De La Cruz E. M., Higgs H. N., Pollard T. D. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- Vojtek A., Haarer B., Field J., Gerst J., Pollard T. D., Brown S., Wigler M. Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell. 1991;66:497–505. doi: 10.1016/0092-8674(81)90013-1. [DOI] [PubMed] [Google Scholar]
- Wang H.-Y., Yu Y., Chen Z.-L., Xia G.-X. Functional characterization of Gossypium hirsutum profilin 1 gene (GhPFN1) in tobacco suspension cells. Planta. 2005;222:594–603. doi: 10.1007/s00425-005-0005-2. [DOI] [PubMed] [Google Scholar]
- Weeds A., Harris H., Gratzer W., Gooch J. Interactions of pig plasma gelsolin with G-actin. Eur. J. Biochem. 1986;161:77–84. doi: 10.1111/j.1432-1033.1986.tb10126.x. [DOI] [PubMed] [Google Scholar]
- Wolven A. K., Belmont L. D., Mahoney N. M., Almo S. C., Drubin D. G. In vivo importance of actin nucleotide exchange catalyzed by profilin. J. Cell Biol. 2000;150:895–903. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., Pollard T. D. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Zelicof A., Protopopov V., David D., Lin X.-Y., Lustgarten V., Gerst J. E. Two separate functions are encoded by the carboxyl-terminal domains of the yeast cyclase-associated protein and its mammalian homologs. J. Biol. Chem. 1996;271:18243–18252. doi: 10.1074/jbc.271.30.18243. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]