Abstract
The kinetochore, a protein complex that links chromosomes to microtubules (MTs), is required to prevent spindle expansion during S phase in budding yeast, but the mechanism of how the kinetochore maintains integrity of the bipolar spindle before mitosis is not well understood. Here, we demonstrate that a mutation of Spc24, a component of the conserved Ndc80 kinetochore complex, causes lethality when cells are exposed to the DNA replication inhibitor hydroxyurea (HU) due to premature spindle expansion and segregation of incompletely replicated DNA. Overexpression of Stu1, a CLASP-related MT-associated protein or a truncated form of the XMAP215 orthologue Stu2 rescues spc24-9 HU lethality and prevents spindle expansion. Truncated Stu2 likely acts in a dominant-negative manner, because overexpression of full-length STU2 does not rescue spc24-9 HU lethality, and spindle expansion in spc24-9 HU-treated cells requires active Stu2. Stu1 and Stu2 localize to the kinetochore early in the cell cycle and Stu2 kinetochore localization depends on Spc24. We propose that mislocalization of Stu2 results in premature spindle expansion in S phase stalled spc24-9 mutants. Identifying factors that restrain spindle expansion upon inhibition of DNA replication is likely applicable to the mechanism by which spindle elongation is regulated during a normal cell cycle.
INTRODUCTION
Preserving the integrity of the genome is a fundamental requirement for eukaryotic cell viability. DNA replication must be completed before segregation of the chromosomes to prevent the transmission of partially replicated chromosomes to daughter cells. In the budding yeast Saccharomyces cerevisiae, cells undergo a closed mitosis and the microtubule (MT) organizing centers, or spindle pole bodies (SPBs), are imbedded in the nuclear envelope. SPB duplication begins at the end of anaphase and spindle formation begins during S phase when duplicated SPBs separate from each other (Adams and Kilmartin, 1999; Jaspersen and Winey, 2004). Because chromosomes remain attached to kinetochore MTs throughout the cell cycle, spindle expansion must be restrained until all 16 chromosomes have duplicated and kinetochores on sister chromatids have formed bipolar MT attachments. When DNA replication is stalled by hydroxyurea (HU) treatment, cells arrest with a large bud, an undivided nucleus positioned at the mother-bud neck and a short bipolar spindle (Allen et al., 1994). Maintaining a short spindle is crucial for cell survival during HU-induced arrest, which activates the DNA replication checkpoint effectors Mec1 and Rad53 (Kolodner et al., 2002). When mec1 and rad53 mutants are treated with HU, the DNA replication checkpoint is not activated, and, as a result, replication forks are not stabilized, the spindle expands, and unequal division of incompletely replicated nuclear material occurs—all of these events contribute to cell lethality (Allen et al., 1994; Weinert et al., 1994; Lopes et al., 2001).
In human cells, stalled replication forks activate the ataxia telangiectasia mutated and Chk2 kinases, which are Mec1 and Rad53 homologues, respectively, and arrest the cell cycle by inhibiting mitotic entry (Canman, 2001). Until recently, it was presumed that mec1 and rad53 mutants enter mitosis prematurely upon HU treatment. However, two recent studies have shown that this is not the case, suggesting that spindle expansion is actively restrained when DNA replication is stalled (Krishnan et al., 2004; Bachant et al., 2005; Krishnan and Surana, 2005). Two mechanisms, which are not mutually exclusive, have been proposed for how spindle expansion is prevented during the DNA replication checkpoint. One mechanism suggests that spindle-associated proteins are regulated in a Mec1/Rad53-dependent manner (Krishnan and Surana, 2005). Spindle expansion and nuclear division of mec1-1 mutants is reduced in cells carrying mutations of the kinesin-5/BimC orthologue Cin8 and XMAP215 orthologue Stu2 (Krishnan et al., 2004). The second mechanism proposes that tension imposed by the bipolar attachment of kinetochores to MTs emanating from opposite SPBs is responsible for maintaining a short spindle upon inhibition of DNA replication (Bachant et al., 2005).
In budding yeast, each kinetochore, a multiprotein complex that resides on centromere (CEN) DNA, attaches to a single MT (McAinsh et al., 2003). After chromosome replication, kinetochores on sister chromatids must attach to MTs emanating from opposite SPBs to achieve bipolar attachment. The SPB pulling force opposes the cohesion holding sister chromatids together, and it creates tension that physically separates CEN regions during metaphase (Goshima and Yanagida, 2000; He et al., 2000; Pearson et al., 2001). The kinetochore not only attaches to spindle MTs but is also capable of regulating MT dynamics and spindle stability before and during mitosis. First, MT-associated proteins such as Stu2 and kinesin-related motor proteins localize to and function at kinetochores (He et al., 2001; McAinsh et al., 2003; Tanaka et al., 2005; Tytell and Sorger, 2006). Second, the Dam1 outer kinetochore complex encircles MTs, and mutations in Dam1 components severely affect MT dynamics (Cheeseman et al., 2001; Miranda et al., 2005; Westermann et al., 2005; Shimogawa et al., 2006). Third, a group of kinetochore proteins called chromosome passenger proteins relocalize from kinetochores to the spindle midzone during anaphase, and they regulate spindle stability and cytokinesis (Bouck and Bloom, 2005).
We have identified an HU-sensitive spc24-9 kinetochore mutant that prematurely expands its spindle upon HU treatment. By performing a high copy suppressor (HCS) screen, we have identified 10 genes that when overexpressed rescue the HU sensitivity and spindle expansion defect of the spc24-9 mutant strain. We characterized the rescue function of two of these genes—Stu1, a MT-associated protein that shares a region of similarity to the CLASP/Mast/Orbit subfamily of MT plus-end tracking proteins, and Stu2, a member of the conserved Dis1/XMAP215 family of MT plus-end binding proteins (Inoue et al., 2000; Yin et al., 2002; Gard et al., 2004). We demonstrate that both Stu1 and Stu2 are localized to the kinetochore early in the cell cycle and that Stu2 kinetochore binding depends on Spc24. By performing quantitative and time-lapse analysis of Stu2 fluorescence on spindles during HU treatment, we show that spindle expansion in spc24-9 cells correlates with mislocalization of Stu2. We propose that localization of Stu2 to the kinetochore in cells when DNA replication is stalled is imperative for maintaining a short spindle and preventing separation of incompletely replicated DNA.
MATERIALS AND METHODS
Strain Construction
Standard methods for yeast culture and transformation were followed (Guthrie and Fink, 1991). Rich medium (YPD) and supplemental minimal medium (SC) were used (Kaiser et al., 1994) as well as FPM (minimal media supplemented with adenine and 6.5g/l sodium citrate) for microscopy analysis (Pot et al., 2005). Yeast strains used in this study are described in Supplemental Table S1. Genes were deleted or epitope tagged using standard yeast methods (Longtine et al., 1998).
HCS Screen
We transformed a 2μ yeast genomic DNA library carrying 6- to 8-kb genomic DNA fragments (Connelly and Hieter, 1996) into the spc24-9 strain, and we plated 40,000 colonies onto SC-URA plates to select for the presence of the library plasmid. We then replica plated the colonies to 0.1 M HU (Sigma-Aldrich, Oakville, ON, Canada) SC-URA plates and incubated at 30°C to identify colonies that could rescue the HU lethality of spc24-9 mutants. Library plasmids were rescued from colonies growing on the 0.1 M HU SC-URA plates and transformed back into spc24-9 mutants to confirm the HU rescue phenotype. Plasmids were then sequenced using T3 and T7 primers to identify the flanking sequences of the genomic insert.
Plasmid Construction: Subcloning of HCS Genes
The coordinates of the genomic DNA identified in the HCS screen rescue plasmids and their subsequent subclones to confirm identity of the gene are as follows: STU1ΔN: Chr.II, 151363-158219; STU1ΔN subclone: Chr.II, 153753-158219; STU2ΔN: Chr.XII, 230442-237078; STU2ΔN subclone: Chr.XII 233569-237078; KIP2: Chr.XVI, 252390-259933; KIP2 subclone: Chr.XVI, 257172-259933; GIC1: Chr.VIII, 220246-228530; GIC1 subclone: Chr.VIII, 220246-222771; RCK2: Chr.XII, 634230-640397; RCK2 subclone: Chr.XII, 634230-636611; HCM1: Chr.III, 224203-231351; MCK1: Chr. XIV, 52447-58767; DMA1; Chr.VIII, 337353-344031; DMA1 subclone: Chr. VIII, 337353-342008. DMA1 was confirmed as the gene responsible for rescue by digesting the DMA1 subclone with EcoRI followed by Klenow treatment to create a frameshift in the DMA1 gene. Full-length MCK1 was a gift from Dr. Phil Hieter (University of British Columbia, Vancouver, BC, Canada) (Shero and Hieter, 1991), full-length HCM1 was a gift from Dr. Trisha Davis (University of Washington, Seattle, WA) (Zhu et al., 1993), and full-length STU1 and STU2 were gifts from Dr. Tim Huffaker (Cornell University, Ithaca, NY) (Pasqualone and Huffaker, 1994; Wang and Huffaker, 1997).
Microscopic Analyses
Immunofluorescence.
Cells shown or described in Figures 1, 3, 4, and 5 were imaged using a Zeiss Axioplan 2 microscope equipped with a CoolSNAP HQ camera (Photometrics, Tucson, AZ) and MetaMorph (Molecular Devices, Sunnyvale, CA) software. The indirect immunofluorescence microscopy studies in Figures 1 and 4 were performed as described previously (Hyland et al., 1999) with the following modifications. Cells were synchronized in G1 at 25°C by using 5 μg/ml α-mating factor (BioVectra, Charlottetown, Prince Edward Island, Canada), released into 0.2 M HU for 3 h at 30°C, and fixed with a final concentration of 3.7% formaldehyde for 1 h. Spindles were visualized by staining with Yol 1-34 rat anti-tubulin antibody (1:50) (Serotec, Oxford, United Kingdom) followed by fluorescein-conjugated goat anti-rat secondary antibody (1:2000). Single focal plane images were acquired with a 100× objective.
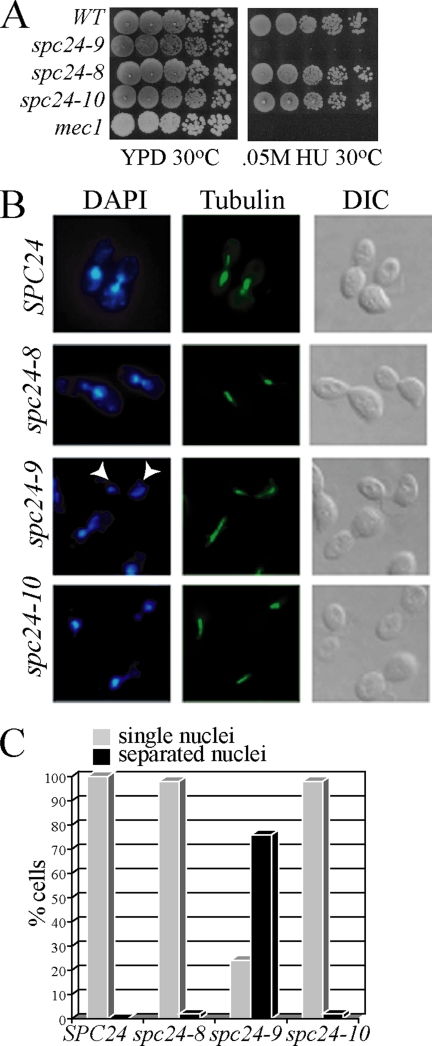
Figure 1.
spc24-9 mutants are sensitive to HU due to inappropriate spindle expansion. (A) Cell dilution assay of indicated strains grown on YPD and 0.05 M HU at 30°C for 3 d. (B) Immunofluorescence analysis of wild-type (SPC24), spc24-8, spc24-9, and spc24-10 cells synchronized in G1 phase with α-factor and then released into 0.2 M HU for 3 h at 30°C. Shown are representative cells after 3-h HU treatment imaged for DNA (4,6-diamidino-2-phenylindole [DAPI]), MTs (tubulin) and cell morphology (differential interference contrast [DIC]). White arrowheads point to separated nuclei in the spc24-9 mutant. (C) Percentage of cells (100 cells counted) described in B displaying single (gray bars) or separated (black bars) nuclei.
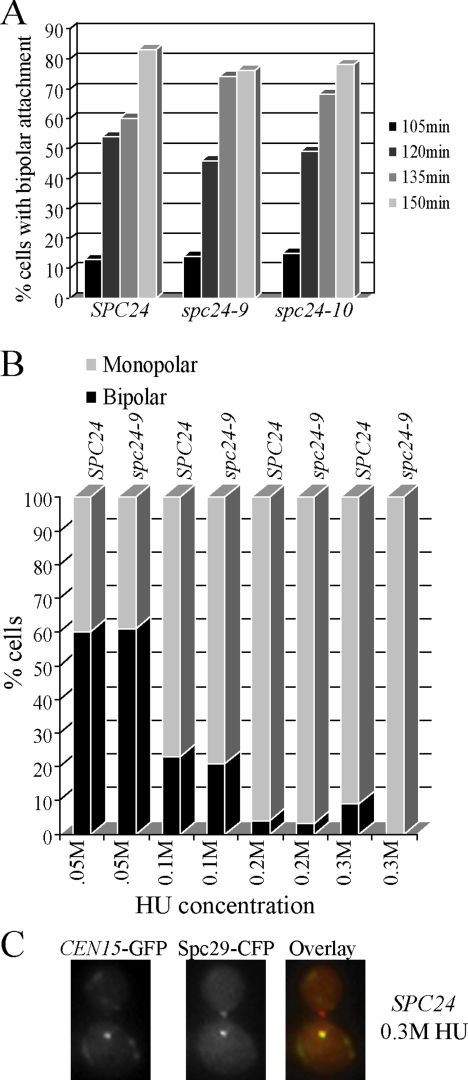
Figure 3.
spc24-9 mutants are capable of establishing bipolar attachment. (A) Wild-type (SPC24), spc24-9, and spc24-10 strains carrying LacO repeats integrated 1.8 kb from CEN15, LacI-GFP, and Spc29-CFP were synchronized in G1 phase with α-factor and released at 30°C. Samples were taken every 15 min and imaged using fluorescence microscopy for the presence of the CEN15-GFP and Spc29-CFP signal. Shown are the time points (105 min onward) at which the cells began to display bipolar attachment (separation of CEN15-GFP signals). Duplicate experiments were performed with similar results. Shown is the result of one experiment in which 100 cells containing a spindle of 0.5 μm or larger were counted for each time point. (B) Wild-type (SPC24) and spc24-9 CEN15-GFP Spc29-CFP strains were synchronized in G1 phase with α-factor, split into four cultures, and indicated concentrations of HU were added for 3 h at 30°C. Similar results were seen with duplicate experiments; thus, data from one experiment are shown (100 cells counted). (C) Example of a wild-type (SPC24) cell from B after 3 h of 0.3 M HU treatment that displays monopolar CEN15 attachment. In the overlay, CEN15-GFP is green and Spc29-CFP is red.
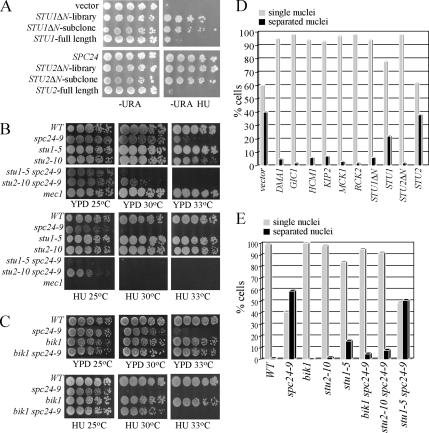
Figure 4.
Spindle expansion in spc24-9 mutants depends on active Stu2. (A) Cell dilution assay of spc24-9 mutants carrying the 2μ plasmid vector (pRS202), STU1ΔN-library (HCS clone identified in screen), STU1ΔN-subclone (subclone of STU1ΔN-library containing only the STU1 gene), STU1 full-length (gift from T. Huffaker), full-length SPC24, STU2ΔN-library (HCS clone identified in screen), STU2ΔN-subclone (subclone of STU2ΔN-library containing only the STU2 gene), and STU2 full-length (gift from T. Huffaker) were grown on −URA plates at 30°C for 4 d or −URA 0.05 M HU at 30°C for 5 d. (B and C) Cell dilution assay of indicated strains grown on YPD at 25°C (2 d), 30 and 33°C (3 d), and 0.05 M HU at 25, 30, and 33°C (3 d). (D) Immunofluorescence analysis of spc24-9 mutants carrying the indicated HCS plasmids synchronized in G1 phase with α-factor and then released into 0.2 M HU for 3 h at 30°C. Cells were counted (100 per sample) for single nuclei (gray bars) or separated nuclei (black bars) by DAPI staining. Duplicate experiments were performed with similar data, and shown is the result of one experiment. (E) Immunofluorescence analysis of indicated strains treated and analyzed as described in D.
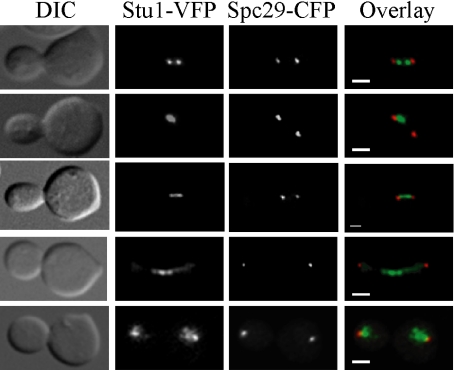
Figure 5.
Stu1 localizes to kinetochores and the spindle midzone. Wild-type Stu1-VFP Spc29-CFP cells were synchronized in G1 phase with α-factor and released into the cell cycle at 30°C. Cells were fixed in 70% ethanol every 15 min for 90 min, and they were imaged as described in the Materials and Methods. In the overlay, green is VFP signal and red is CFP. Bar, 2 μm for all images.
Analysis of Green Fluorescent Protein (GFP)-Centromeres, Stu1-VFP, and Stu2-VFP Localization in Fixed Cells.
CEN15-GFP–tagged (Figure 3; Goshima and Yanagida, 2000) cells were synchronized with α-mating factor, and LacI-GFPHIS LacO::URA3-CEN15(1.8) was activated with 30 mM 3-aminotriazole in SC–HIS media. Cells were released into indicated concentrations of HU in FPM media for 3 h. Cells were washed and fixed in a total concentration of 4% paraformaldehyde for 15 min. Image stacks were acquired with a 100× objective at a step of 0.2 μm to span the entire cell. Stu1-VFP fluorescence (Figure 5) was imaged as described above with the following alterations. Cells were grown in FPM media at 30°C, synchronized with α-mating factor, and released to 30°C. After 30 min., samples were taken every 15 min and fixed in 70% ethanol. Stu2-VFP in fixed cells (Figure 7) was imaged with a WaveFX spinning disk confocal microscope (Quorum Technologies, Guelph, Ontario, Canada) as described previously (Cuschieri et al., 2006) without agar pads. Optical sections (0.5 μm) were acquired through a ±2.5-μm z-plane (total of 5.0-μm stack) by using Volocity 3DM acquisition software (Improvision, Conventry, United Kingdom).
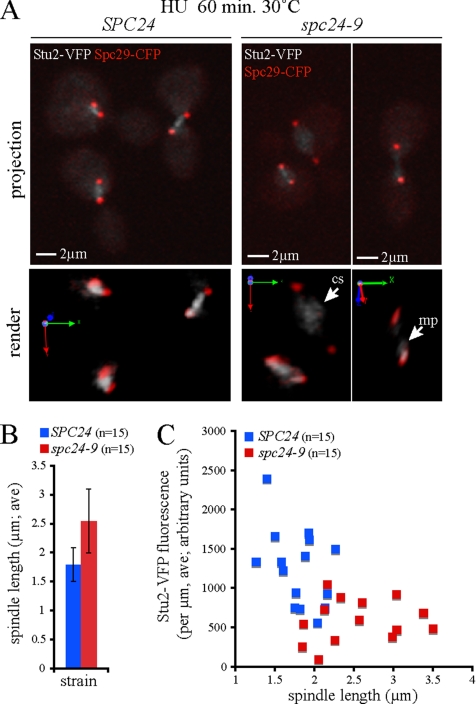
Figure 7.
Increased spindle length correlates with Stu2 mislocalization and reduction. Wild-type (SPC24) and spc24-9 cells expressing Stu2-VFP Spc29-CFP were synchronized in G1 phase with pheromone, released into 0.2 M HU at 25°C for 1.5 h, and shifted to 30°C in HU for 60 min, and then fixed. (A) Representative images (extended focus and three-dimensional render) of Stu2-VFP (gray scale) and Spc29-CFP (red) fluorescence in wild-type and spc24-9 cells at 60 min after shift to 30°C are shown; mp indicates monopolar localization (near the SPB), whereas cs indicates localization to the central spindle. (B) Average (ave) spindle length of wild-type (SPC24) and spc24-9 cells after a 60-min incubation in 0.2 M HU at 30°C (n = 15). (C) Quantitative analysis of Stu2-VFP fluorescence per micrometer plotted as a function of spindle length.
Live Cell Analysis
For live-cell imaging of Stu2-VFP fluorescence intensities and spindle length measurements (Figure 8 and Supplemental Figure 3), overnight cultures were grown in YPD (containing 2× adenine sulfate) at 25°C to a cell density of ∼0.3-0.4 OD600 units ml−1. Cultures were then diluted to 0.2 OD600 units ml−1 and grown for an additional generation. Cells were arrested with 5 μg/ml α factor for 1.5 h at 25°C, washed, and then released into media containing 0.2 M HU for 1.5 h at 25°C. One-milliliter samples of each strain were taken and resuspended in ∼50 μl of 30°C prewarmed media containing 0.2 M HU. Cells were mounted on a prewarmed 30°C heated stage and allowed to equilibrate for 15 min before imaging.
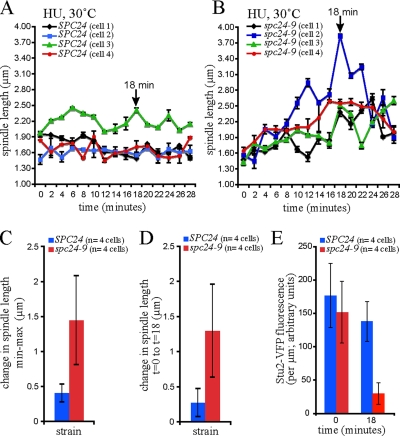
Figure 8.
Decreased Stu2 in the spc24-9 mutant results in oscillation of spindle length. Wild-type (SPC24) and spc24-9 cells carrying Stu2-VFP Spc29-CFP were synchronized in G1 phase with pheromone, released into 0.2 M HU at 25°C for 1.5 h, mounted in FP supplemented with HU, shifted to 30°C on a heated stage, and time-lapse microscopy was performed. Time zero is time in HU at 30°C after equilibration on the stage at 30°C for 15 min. Spindle length is plotted for four cells of each strain (A, SPC24; B, spc24-9) as a function of time, each depicted with a different color, and shows oscillation with a net increase in length observed in all four spc24-9 cells at 18 min. (C) In contrast with wild-type cells, spindle length increases in spc24-9 cells. (D) Spindle length is significantly increased in all four spc24-9 cells at 18 min relative to length at t = 0. (E) At 18 min, Stu2-VFP fluorescence on the spindle is significantly decreased in all spc24-9 mutant cells relative to wild-type cells.
Multichannel four-dimensional imaging of Spc29-CFP and Stu2-VFP fluorescent fusion proteins was performed using a WaveFX spinning disk confocal system (Quorum Technologies) as described previously (Cuschieri et al., 2006). A Tokai Hit stage warmer was used to shift cells from 25 to 30°C, image acquisition commenced 15 min after the stage reached 30°C. Optical sections (0.5 μm) were acquired through a ±2.5-μm z-plane (total of 5.0-μm stack) at 2-min intervals for 30 min by using Volocity 3DM acquisition software (Improvision).
Spindle Length Measurements
Calculation of spindle lengths in fixed and live cell analyses shown in Figures 7 and 8 was performed using Volocity Classification (Improvision). Spindle lengths (in micrometers) were measured in triplicate for each time point, and the average value and SE of the mean determined. Lengths were determined by measuring the linear distance (micrometers; in x, y, z) between the midpoint of one SPB (Spc29-CFP channel) to the midpoint of the opposite SPB. All spindle lengths were measured in the XYZ plane view by using the line length measurement tool. Average spindle lengths and SEs were calculated using Excel software (Microsoft, Redmond, WA).
Stu2-VFP Fluorescence Measurements
For Stu2-VFP fluorescence measurements shown in Figures 7 and 8, image stacks were acquired using an exposure of 91 ms/frame (providing an unsaturated image). For both fixed and live cell analyses, fluorescence intensity was measured by rastering a 4 × 4 voxel volume along the length of the long axis of the spindle (4 × 4 voxel: spindle fluorescent unit), including both SPBs. Background subtraction as performed as follows: the fluorescence in a 4 × 4 voxel volume positioned in the cytoplasm was measured, and subtracted from each fluorescence unit acquired along the spindle, resulting in corrected spindle fluorescence units (arbitrary units). For live cell analyses, background fluorescence was determined for each time point. The corrected fluorescence per unit length (fluorescence/micrometer) was calculated. Background subtractions, corrected fluorescence values, and standard deviations were calculated using Excel software.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP experiments and primers used for polymerase chain reaction (PCR) analysis were performed as described previously (Measday et al., 2002; Pot et al., 2003). The linear range for PCR analysis was determined, and dilutions used for Figure 2A were total chromatin (T; 1:200), immunoprecipitation (IP; 1:1); for Figure 2B, T (1:200), IP (5:1); for and Figure 2C, T (1:200), IP (5:1). Dilutions used for Figure 6A were T (1:780), IP (1:6); for Figure 6B, T (1:780), IP (1:2.5); and for Figure 6, C and D, T (1:125), IP (1:1).
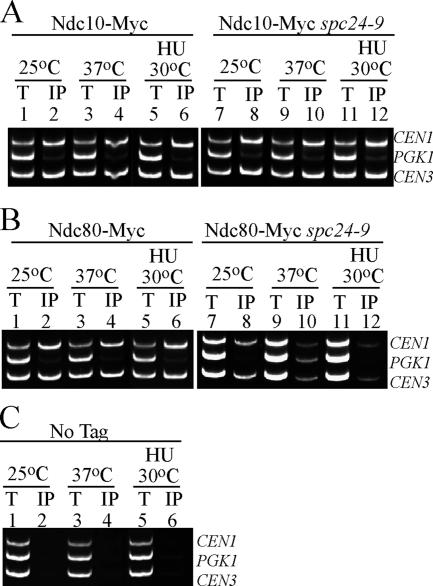
Figure 2.
Ndc80 CEN association is disrupted in spc24-9 mutants. Multiplex PCR analysis of CEN1, PGK1, and CEN3 loci was performed with total chromatin (T) or immunoprecipitate (IP) as PCR templates. Strains were grown at 25°C to log phase and then either shifted to 37°C for 3 h or incubated in 0.2 M HU at 30°C for 3 h. (A) Ndc10-Myc wild type (lanes 1–6) or spc24-9 cells (lanes 7–12). (B) Ndc80-Myc wild-type (lanes 1–6) or spc24-9 cells (lanes 7–12). (C) Wild-type strain carrying no epitope tag (No Tag, lanes 1–6) shown as a control. An untagged spc24-9 mutant was also used as a control and showed similar results (data not shown).
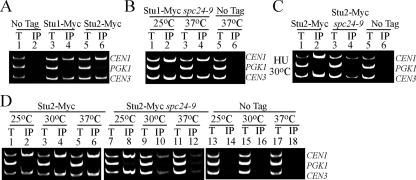
Figure 6.
Stu2 CEN binding is abolished in spc24-9 mutants, whereas Stu1 is still able to associate with CEN DNA. Multiplex PCR analysis of CEN1, PGK1, and CEN3 loci was performed with total chromatin (T) or immunoprecipitate (IP) as PCR templates. Strains were grown at 25°C to log phase and then either kept at 25°C or incubated at 30 or 37°C for 3 h. (A) Wild-type log phase cells grown at 30°C and carrying no epitope tag (No Tag; lanes 1 and 2), Stu1-Myc (lanes 3 and 4), and Stu2-Myc (lanes 5 and 6). (B) spc24-9 mutants carrying Stu1-Myc (lanes 1–4) and a wild-type strain with no tag at 37°C (lanes 5 and 6). (C) Stu2-Myc in a wild-type (lanes 1 and 2), spc24-9 (lanes 3 and 4), and an untagged wild-type strain (No Tag; lanes 5 and 6). Strains were grown to log phase at 25°C, HU was added to a final concentration of 0.2 M HU, and cells were shifted to 30°C for 3 h. (D) Stu2-Myc in a wild-type (lanes 1–6) and spc24-9 (lanes 7–12) strains. No Tag strain (lanes 13–18) is an untagged wild-type strain. For all ChIP assays where the spc24-9 mutant was used, we included both a wild-type and spc24-9 untagged control, and we saw similar results; thus, only the wild-type untagged control is presented.
RESULTS
Spc24 Is Required for Viability and for Preventing Spindle Expansion during HU Arrest
The budding yeast Ndc80 central kinetochore complex, which is composed of the four coiled-coil proteins, Ndc80, Nuf2, Spc24, and Spc25, is required for proper attachment of chromosomes to spindle MTs and activation of the spindle checkpoint in the presence of defects in attachment (Janke et al., 2001; Wigge and Kilmartin, 2001; Le Masson et al., 2002; Montpetit et al., 2005; Pinsky et al., 2006). It has recently been shown that the kinetochore has a role in maintaining a short (1.5–2 μm) spindle when DNA replication is stalled by treatment of cells with HU (Bachant et al., 2005). Previously, we created two mutants in the coiled-coil region of Spc24 (spc24-8 and spc24-10) and one mutant in the C terminus of Spc24 (spc24-9) (Montpetit et al., 2005). We tested these alleles for viability in the presence of HU at semipermissive temperature (30°C), and we found that the growth of the spc24-9 mutant is sensitive to levels of HU that do not inhibit the growth of spc24-8 and spc24-10 mutants and the wild-type strain (Figure 1A). Next, we monitored the spindle length and bulk segregation of DNA in spc24 mutants arrested in G1 and released into 0.2 M HU media. This analysis revealed that 100% of wild type and 98% of spc24-8 and spc24-10 mutants maintain a short spindle and undivided nuclei after 3-h exposure to HU; however, the majority (76%) of spc24-9 mutants displayed elongated spindles and segregated nuclei (Figure 1, B and C). Our data suggest that the spc24-9 mutation results in a defect in the function of the Ndc80 complex in preventing expansion of the spindle in cells with partially replicated DNA.
Characterization of the Kinetochore in spc24-9 Mutants
The Ndc80 complex is composed of two subcomplexes, Nuf2/Ndc80 and Spc24/Spc25, that are linked via their coiled-coil domains (Wei et al., 2005). The C-terminal mutation in spc24-9 lies within the Spc24 globular domain (Montpetit et al., 2005; Wei et al., 2005). To determine the state of the kinetochore and the Ndc80 complex in spc24-9 mutants, we performed ChIP assays by using a member of the inner kinetochore CBF3 complex, Ndc10, and the Ndc80 protein. We found that Ndc10 was able to interact with centromere (CEN) DNA in spc24-9 cells at both restrictive temperature (37°C) and after 3 h of 0.2 M HU treatment at 30°C, suggesting that the core kinetochore is still intact in spc24-9 mutants (Figure 2A, lanes 10 and 12). Ndc80, however, showed a clear defect in its ability to associate with CEN DNA in spc24-9 cells both at 37°C and after 3 h of 0.2 M HU treatment at 30°C (Figure 2B, lanes 10 and 12). In corroboration with our ChIP data, we found that Ndc80-VFP localization was also perturbed in spc24-9 mutants at 37°C and that it is even more affected in HU-arrested cells (Supplemental Figure 1). Eighty percent of HU-treated spc24-9 mutants showed diffuse and weak Ndc80-VFP staining, suggesting that the Ndc80 complex is disrupted when spc24-9 cells are exposed to 0.2 M HU (Supplemental Figure 1).
Bipolar Attachment Is Not a Requirement for Maintaining a Short Spindle in HU-treated Cells
Previous studies have suggested that specific kinetochore mutants display inappropriate spindle expansion during HU exposure due to their inability to establish kinetochore–MT bipolar attachment and thus appropriate tension on the spindle (Bachant et al., 2005). Using a CEN15-GFP–marked strain, we found that a similar percentage of wild-type, spc24-9, and spc24-10 cells displayed bipolar attachment in cells released from a G1 block to 30°C (Figure 3A). Thus, spc24-9 kinetochores are capable of bipolar attachment to spindle poles during a normal cell cycle at the same temperature (30°C) that results in spc24-9 HU lethality. Whether kinetochores attain bipolar attachment in a wild-type strain in the presence of partially duplicated DNA (as a result of treatment with HU) is unclear. We reasoned that exposing cells to increasing concentrations of HU would increase replication fork stalling (.05M-0.3M HU) and impact the number of CENs that were replicated (Clarke et al., 2001). We monitored CEN15 separation as a sign of bipolar attachment using CEN15-GFP and Spc29-CFP–tagged wild-type and spc24-9 mutant strains. Wild-type cells treated with the lowest concentration of HU (0.05 M HU) exhibited CEN15 bipolar attachment in 60% of cells after 3 h (Figure 3B). Importantly, spc24-9 mutants displayed the same percentage of separated CEN15 foci, suggesting once again, that spc24-9 mutants are capable of bipolar attachment (Figure 3B). Similar to previous studies, we found that separated CEN15 foci were detected in 23% of wild-type cells exposed to 0.1 M HU, yet only 4% of separated CEN15 foci were seen in 0.2 M HU-treated wild-type cells (Goshima and Yanagida, 2000; Krishnan et al., 2004). The majority of wild-type cells treated with the highest concentration of HU (0.3 M HU) contained one CEN15 foci that colocalized with one SPB, suggesting either that CEN15 had not yet replicated or that it had replicated but it had not yet established bipolar attachment (Figure 3C). Previous work has demonstrated that unreplicated monocentric minichromosomes also remain in the vicinity of one SPB (Dewar et al., 2004). Thus, the ability of CEN15 to attain bipolar attachment is not correlated with maintaining a short spindle during the DNA replication checkpoint.
Identification of HCS Genes That Rescue spc24-9 HU Lethality
To understand why Spc24 is required for viability when DNA replication is stalled, we performed an HCS screen to identify genes that, when overexpressed, could suppress the lethality of spc24-9 cells exposed to HU. Ten genes were identified in our HCS screen (Table 1). Multiple isolates of SPC24 and its interacting partner SPC25 were recovered (Janke et al., 2001; Wigge and Kilmartin, 2001). We also identified four genes encoding proteins that regulate spindle dynamics—MT-associated proteins Stu1 and Stu2, the kinesin-related motor protein Kip2, and a protein involved in spindle positioning called Dma1 (Roof et al., 1992; Pasqualone and Huffaker, 1994; Wang and Huffaker, 1997; Fraschini et al., 2004). Two protein kinases were isolated—Mck1, which has a role in chromosome segregation, and Rck2, which has a role in the osmotic stress response pathway (Neigeborn and Mitchell, 1991; Shero and Hieter, 1991; Bilsland-Marchesan et al., 2000). We identified Gic1, which has roles in cell polarity and mitotic exit (Brown et al., 1997; Chen et al., 1997; Hofken and Schiebel, 2004). Finally, we identified the Hcm1 transcription factor, which has also been isolated in a variety of synthetic lethal screens pertaining to the cell division cycle and chromosome segregation (Zhu and Davis, 1998; Horak et al., 2002; Sarin et al., 2004; Montpetit et al., 2005; Daniel et al., 2006). Interestingly, Hcm1 has recently been shown to activate expression of spindle and chromosome segregation proteins specifically in S phase (Pramila et al., 2006). None of the HCS genes were able to rescue the inviability of a mec1 mutant on HU plates (data not shown), suggesting that they were suppressing the specific defect of spc24-9 mutants.
Table 1.
HCS screen of spc24-9 HU lethality
| HU rescuea | Gene name | Open reading frame | Biological processb |
|---|---|---|---|
| + | DMA1 | YHR115C | Spindle position and orientation |
| + | RCK2 | YLR248W | Oxidative and osmotic stress signaling |
| + | STU1 truncatedc | YBL034C | MT dynamics |
| ++ | GIC1 | YHR061C | Cell polarity |
| +++ | KIP2 | YPL155C | Mitotic spindle positioning |
| +++ | MCK1 | YNL307C | Mitotic and meiotic chromosome segregation |
| ++++ | HCM1 | YCR065W | Transcription |
| +++++ | STU2 truncatedd | YLR045C | MT dynamics |
| +++++ | SPC24 | YMR117C | Chromosome segregation |
| +++++ | SPC25 | YER018C | Chromosome segregation |
a Growth of spc24-9 mutant carrying rescue clone struck on 0.05 M HU plates at 30°C from weak (+) to strong (+++++) growth.
b GO Annotation from Saccharomyces Genome Database.
c The STU1 rescue clone is missing the N-terminal 97 amino acids.
d The STU2 rescue clone is missing the N-terminal 252 amino acids.
HCS Rescue Occurs through Restraining Spindle Expansion
We next determined whether the HCS genes rescued spc24-9 HU lethality by restraining spindle expansion and thus premature chromosome segregation. spc24-9 mutants carrying the HCS rescue plasmids were synchronized in G1 phase, released into HU for 3 h, and immunofluorescence was performed to analyze chromosome segregation and spindle morphology. All of the HCS genes were able to restore a single nuclei phenotype to spc24-9 HU-treated cells (Figure 4D). The STU1 and STU2 rescue clones that we identified in our screen lacked the N-terminal 97 and 252 amino acids of Stu1 and Stu2, respectively (herein referred to as STU1ΔN and STU2ΔN). We tested whether high copy full-length STU1 or STU2 expression plasmids were capable of rescuing spc24-9 HU lethality. Expression of full-length STU2 was clearly not able to rescue either the HU lethality or chromosome segregation defects of spc24-9 mutants, suggesting that the N-terminal truncation is an important feature of the STU2ΔN rescue activity (Figure 4, A and D). Expression of full-length STU1 was able to rescue spc24-9 HU lethality and chromosome separation at levels above vector alone, but not as well as the STU1ΔN clone (Figure 4, A and D).
We reasoned that expression of STU2ΔN might be rescuing the spindle expansion defect in HU-treated spc24-9 mutants by destabilizing MTs. Consistent with this hypothesis, the STU2ΔN clone lacks the TOG1 domain of Stu2, which binds tubulin heterodimers (Al-Bassam et al., 2006, 2007). Previous work has shown that expression of Stu2 lacking its TOG1 domain, which still binds MTs, results in decreased mitotic spindle length and slows down anaphase spindle elongation (Al-Bassam et al., 2006). We asked whether Stu2ΔN localization is similar to endogenous Stu2 by tagging Stu2ΔN with venus fluorescent protein (VFP) (which still retains its spc24-9 HU rescue activity) and endogenous Stu2 with cyan fluorescent protein (CFP). Indeed, we found that Stu2ΔN-VFP localization overlapped with Stu2-CFP in both wild-type and spc24-9 mutants in log phase or HU-treated cells, consistent with previous data, demonstrating that Stu2ΔTOG1-GFP still binds MT plus ends (Supplemental Figure 2; Al-Bassam et al., 2006). To test whether depletion of Stu2 activity by using another mutant form of Stu2 is also capable of preventing spindle expansion in spc24-9 cells, we combined spc24-9 with the stu2-10 temperature-sensitive (Ts) mutation (Severin et al., 2001) and tested the double mutant for separation of nuclei upon HU treatment. Only 8% of stu2-10 spc24-9 mutants segregated nuclei after 3 h of HU treatment compared with 59% of spc24-9 mutants (Figure 4E). Therefore, functional Stu2 is required for the spindle expansion defect in spc24-9 mutants. Although stu2-10 spc24-9 double mutants maintained a short spindle during HU treatment, they were lethal on HU plates at 30°C (Figure 4B), suggesting that the defects in both Stu2 and Spc24 prevent cell cycle recovery after HU exposure.
Stu2 interacts with two other MT plus-end tracking proteins, Bim1 and Bik1 (Chen et al., 1998; Lin et al., 2001; Wolyniak et al., 2006). Because we had previously shown that bim1 spc24–9 mutants have a synthetic growth defect (Montpetit et al., 2005), we deleted BIK1 in spc24-9 cells and analyzed growth phenotypes. The bik1 spc24-9 double mutant rescued the nuclei separation defect of HU treated spc24-9 mutants and both the HU (at 30°C) and Ts (at 33°C) lethality of spc24-9 mutants (Figure 4, C and E). Therefore, the activity of Stu2 and Bik1 is responsible for the spindle expansion and subsequent nuclei separation and lethality of spc24-9 cells upon HU exposure.
We also determined whether Stu1 is required for the spindle expansion activity in spc24-9 HU-treated cells by creating a stu1-5 spc24-9 double mutant (Yin et al., 2002). The stu1-5 spc24-9 mutant behaved in a similar manner to spc24-9 mutants and elongated their spindles when treated with HU, suggesting that, unlike Stu2 and Bik1, Stu1 activity is not required for spindle expansion in HU exposed spc24-9 mutants. Although both spc24-9 and stu1-5 individual mutants grow well at 30°C on rich media (YPD), the double mutant is synthetically lethal at 30°C (Figure 4D). In addition, the spc24-9 stu1-5 double mutant is viable at 25°C in rich media but inviable when grown on HU plates (Figure 4D). The sensitivity of spc24-9 stu1-5 double mutants to HU and the synthetic lethal interaction between spc24-9 and stu1-5 mutants suggests that Stu1 and Spc24 have a joint or parallel role in restraining spindle expansion during the DNA replication checkpoint.
Stu1 Localizes to Kinetochores before Anaphase
Stu1, which was originally isolated as a suppressor of a tub2 (β-tubulin) mutation, interacts with Tub2 and localizes to the spindle midzone in anaphase spindles (Pasqualone and Huffaker, 1994; Yin et al., 2002). However, the localization of Stu1 in relation to a SPB marker has not been assessed. We imaged Stu1 fused to VFP in relation to the Spc29-CFP SPB protein by synchronizing cells in G1 phase with mating pheromone and then releasing them into the cell cycle and fixing cells every 30 min. The budding yeast spindle reaches a length of 1.5–2 μm before entering anaphase (Pearson et al., 2001). Before anaphase, we detected three evenly distributed patterns of Stu1-VFP localization in fixed cells: a bilobed distribution pattern in between the Spc29-CFP foci, which is a hallmark localization pattern for a kinetochore protein (Figure 5, top row) (He et al., 2001; Measday et al., 2002); a single foci located closer to one of the SPBs (Figure 5, second row); and a continuous signal in between SPBs (Figure 5, third row). In agreement with previous results, we also found that Stu1-VFP localized to the midzone of anaphase spindles (Figure 5, fourth row) (Yin et al., 2002). Finally, in telophase, we observed a dispersed Stu1 signal near the SPBs (Figure 5, bottom row).
Our localization data suggest that Stu1 may interact with the kinetochore. We tested whether Stu1 localizes to the kinetochore by performing Stu1-Myc ChIP assays from logarithmically growing cells. Stu1-Myc specifically associated with CEN1 and CEN3 DNA but not with a non-CEN loci, PGK1 (Figure 6A, lane 4). We performed a Stu1-Myc ChIP assay in a spc24-9 mutant strain at both permissive (25°C) and restrictive (37°C) temperature (Figure 6B). Stu1-Myc is still able to associate with CEN DNA at restrictive temperature, suggesting that Stu1 does not require Spc24 to bind kinetochores (Figure 6B, lane 4). In summary, our localization and ChIP data demonstrate that Stu1 localizes to kinetochores early in the cell cycle in an Spc24-independent manner and that it relocalizes to the spindle around the time of the metaphase to anaphase transition.
Stu2 Localizes to Kinetochores Early in the Cell Cycle
In anaphase cells, Stu2-GFP localizes to the cytoplasmic side of the SPB and along the spindle MTs as determined by immunoelectron microscopy (Kosco et al., 2001). In addition, Stu2 has been shown to colocalize with kinetochores and SPBs and to bind CEN DNA in metaphase cells (He et al., 2001). We analyzed Stu2-VFP localization in short (<1.5-μm) spindles (acquired at the time of bud emergence) and during normal spindle expansion by using time-lapse microscopy (Supplemental Figure 3). We observed that Stu2-VFP signal displayed a bilobed pattern between Spc29-CFP foci in short spindles (Supplemental Figure 3, image 10b). Time-lapse microscopy using longer exposures (which saturate Stu2-VFP spindle fluorescence) revealed that Stu2-VFP also tracks on astral microtubules and transiently associates with SPBs as astral MT shorten (data not shown). Our imaging data are consistent with Stu2 localizing primarily to kinetochores and/or the nuclear spindle before metaphase. Once spindles had lengthened (>2 μm), we detected colocalization of Stu2-VFP with Spc29-CFP as well as Stu2-VFP at the spindle midzone as described previously (Supplemental Figure 3, image 0c) (He et al., 2001; Kosco et al., 2001). The localization of Stu2 to kinetochores early in the cell cycle suggests that defects in kinetochore function when DNA replication is stalled by HU treatment may significantly affect Stu2 activity.
Stu2 Is Mislocalized in HU-treated spc24-9 Cells
We tested whether Stu2 localization to the kinetochore depends on functional Spc24 by using both ChIP and microscopy analyses. Stu2-Myc displayed a decreased ability to interact with CEN DNA in spc24-9 cells as we increased the temperature from permissive (25°C) to semipermissive (30°C) conditions (Figure 6D, lanes 8 and 10). Stu2-Myc did not coprecipitate with CEN DNA in spc24-9 mutants shifted to restrictive temperature (37°C), suggesting that Stu2 requires Spc24 to interact with the kinetochore (Figure 6D, lane 12). Although Stu2 requires Spc24 for proper CEN localization in logarithmically growing cells, this does not necessarily reflect the situation when cells are exposed to HU. We performed a Stu2-Myc ChIP assay in wild-type and spc24-9 cells after treating cells with HU for 3 h at 30°C. Stu2 interaction with CEN DNA was highly reduced in spc24-9 mutants compared with wild-type cells (Figure 6C, compare lanes 2 and 4). Thus, Spc24 is required for Stu2 to efficiently interact with CEN DNA in both log phase and HU-treated cells. To test whether Stu2 localization is perturbed in spc24-9 cells, we performed a quantitative analysis of Stu2-VFP fluorescence during HU exposure. Cells were released from a G1 pheromone block into HU at 25°C, shifted to 30°C, and fixed after 60 min of incubation. At this time point, spindle expansion had clearly begun in spc24-9 cells as spindle lengths averaged 2.5 μm in mutant cells compared with 1.8 μm in wild-type cells (Figure 7B). Analysis of individual cells revealed that Stu2 remained as bilobed foci in wild-type cells, whereas Stu2 signal was clearly mislocalized along the spindle midzone (cs) or next to one pole (monopolar) in spc24-9 cells (Figure 7A, three-dimensional render, rotated). Analysis of Stu2-VFP fluorescence on the spindle indicated that Stu2-VFP fluorescence intensity decreased significantly in the spc24-9 mutant relative to wild type (Figure 7C). Stu2-VFP also redistributed from discrete foci to diffuse fluorescence along the length of the spindle (Figure 7A); thus, Stu2-VFP fluorescence on the spindle per unit length (micrometers; see Materials and Methods for details) was used for the comparison of Stu2-VFP in wild-type versus spc24-9 cells. In general, this analysis revealed that spindles in the spc24-9 mutant had decreased Stu2-VFP fluorescence and were longer, suggesting that spindle expansion correlates with mislocalization of Stu2 (Figure 7C). However, we noticed that low levels of Stu2-VFP fluorescence were found on both long and short spindles in spc24-9 cells, suggesting that the relationship between spindle length and Stu2 levels was not absolute. More specifically, we wondered whether the observed spindle expansion observed in spc24-9 cells was permanent or represented oscillations in spindle length.
To further explore the dynamics of Stu2 interaction with the spindle and spindle expansion, we analyzed dynamic changes in Stu2-VFP fluorescence and spindle length in living cells using time-lapse microscopy. For this analysis, HU-arrested wild-type and spc24-9 mutant cells were shifted to 30°C on the microscope stage, and the HU arrest was maintained throughout the time lapse by mounting the cells in FP medium supplemented with HU. This analysis revealed that spindle length remains relatively static in wild-type cells, with an average net change (either shrinking or elongating) in length of 0.40 ± 0.125 μm during the time lapse for all cells (n = 4) analyzed (Figure 8, A and C). Spindle length at the start of the time lapse was not significantly different between wild-type and spc24-9 cells, and it was similarly static in spc24-9 cells (n = 4 cells/strain) during the first 5 min of the time lapse (Figure 8B). In contrast with wild-type cells, spindle length increased significantly (1.45 ± 0.638 μm) in the spc24-9 mutant over time (Figure 8, B and C). At 18 min after the shift to 30°C, a net increase in spindle length occurred in all four spc24-9 cells; in each cell spindle length had increased (0.9–2.3 μm; mean length increase of 1.3 ± 0.66 μm) relative to the length at the start of the time lapse and relative to all four wild-type cells (Figure 8, B and D). Thus, we chose to compare the intensity of Stu2-VFP fluorescence on the spindle at time 0 and at 18 min. Stu2-VFP fluorescence was significantly decreased in spc24-9 cells compared with wild-type cells (Figure 8E), suggesting that mislocalization of Stu2 is correlated with the spindle expansion observed (Figure 8D). Finally, we detected significant oscillation of spindle length between 10 and 28 min in spc24-9 cells (Figure 8B), suggesting that mislocalization of Stu2 results in transient spindle expansion. The transient nature of this defect is consistent with our observation of a subpopulation of spc24-9 cells with short spindles and mislocalized or low Stu2-VFP levels.
DISCUSSION
The kinetochore is required to restrain spindle expansion in budding yeast when DNA replication is stalled; however, the mechanism by which kinetochores maintain spindle length in this state is not well understood. We identified genes that when overexpressed, rescue the lethality and spindle expansion defects of the spc24-9 kinetochore mutant when exposed to HU. Two MT plus-end binding proteins, the CLASP-related protein Stu1 and XMAP215 homologue Stu2, were identified, and their interactions with the kinetochore were explored further. We find that Stu1 localizes to kinetochores early in the cell cycle and relocalizes to the spindle midzone after metaphase and that Stu2 binds to the kinetochore in a Spc24-dependent manner. In addition, inappropriate spindle expansion in HU treated spc24-9 cells can be prevented by inhibiting Stu2 activity. We propose that mislocalization of Stu2 in spc24-9 cells enables spindle expansion during the DNA replication checkpoint.
Kinetochore–MT Bipolar Attachment and the DNA Replication Checkpoint
DNA microarray studies have suggested that most CENs are replicated upon exposure to HU (Yabuki et al., 2002; Feng et al., 2006). Thus, budding yeast CENs may be capable of attaining bipolar attachment during HU treatment. However, our data suggest that bipolar attachment may not be the only mechanism by which kinetochores maintain short spindles during HU arrest. First, wild-type and spc24-9 cells display similar percentages of CEN15-GFP bipolar foci when treated with different concentrations of HU, yet only spc24-9 spindles expand (Figure 3B). Second, when wild-type cells are treated with high concentrations of HU, we detect a single CEN15-GFP focus that clearly colocalizes with one SPB (Figure 3C). These data are similar to previous studies demonstrating that a GFP-marked, unreplicated minichromosome colocalizes with one SPB after SPB separation (Dewar et al., 2004). Although our data do not distinguish between replicated and unreplicated CEN15, CEN15 is clearly attached to one pole, yet the spindle remains short. Thus, we propose that the ability to attain bipolar attachment during HU treatment is not correlated with restraining spindle expansion.
Stu2 Activity Enables Spindle Expansion in spc24-9 HU-treated Cells
We isolated a truncated version of the XMAP215 homologue STU2 (STU2ΔN), in our spc24-9 HCS screen that lacks the N-terminal 252 amino acids of Stu2 (Table 1). We propose that overexpression of STU2ΔN rescues spc24-9 HU lethality by restraining MT dynamics induced by mislocalization of Stu2. STU2ΔN lacks the N-terminal TOG1 domain that binds tubulin heterodimers but retains the TOG2 domain that binds MT plus ends (Al-Bassam et al., 2006). Previous studies have shown that Stu2 lacking its TOG1 domain binds MT plus ends but cannot promote plus-end MT growth, suggesting that STU2ΔN inhibits spc24-9 HU spindle expansion via the same mechanism (Al-Bassam et al., 2006). The spc24-9 mutant and the resultant mislocalization of Stu2 (see next section) is an important feature of STU2ΔN's rescue function, because overexpression of STU2ΔN does not inhibit spindle expansion in a wild-type cell cycle (Supplemental Figure 4). Another possible Stu2ΔN rescue mechanism, which is not mutually exclusive with the previous mechanism, is that overexpression of STU2ΔN is titrating out a Stu2-interacting protein that is mediating spindle expansion in spc24-9 HU cells. Stu2 interacts with the CLIP-170 orthologue Bik1 at the C terminus of Stu2 (Wolyniak et al., 2006). We find that deletion of the Stu2-interacting protein Bik1 rescues the spc24-9 spindle expansion defects and HU lethality at 30°C and that the bik1 spc24-9 double mutant grows at a higher temperature than the spc24-9 mutant alone (Figure 4, C and E). Thus, inhibition of Stu2 or Bik1 plus-end MT activity prevents spindle expansion when spc24-9 mutants are under HU arrest.
Stu2 Retention at the Kinetochore is Important for Maintaining a Short Spindle when DNA Replication is Stalled
Our data suggest that Stu2 activity is required for spindle expansion in spc24-9 HU-treated cells. The stu2-10 spc24-9 double mutant no longer displays inappropriate spindle expansion when exposed to HU (Figure 4E). Why is Stu2 able to promote spindle expansion in HU-treated spc24-9 cells but not wild-type cells? We propose that the inability to recruit and retain Stu2 at the kinetochore in spc24-9 mutants enables Stu2 to promote MT dynamics. Our ChIP data suggest that the interaction of Stu2 with CEN DNA is perturbed in spc24-9 mutants both in log phase cells and during HU treatment (Figure 6, C and D). In agreement with our studies, Stu2 does not associate with CEN DNA at restrictive temperature in an ndc80-1 Ts mutant (He et al., 2001). Thus, the Ndc80 complex is required for recruitment of Stu2 to the kinetochore. We performed a detailed analysis of Stu2-VFP fluorescence and spindle length in both live and fixed spc24-9 HU-treated cells. Shortly after shift to restrictive temperature (30°C for spc24-9 cells exposed to HU), we detected spindle expansion and mislocalization of Stu2-VFP as well as its diffusion along the axis of the spindle (Figure 7). Stu2-VFP also displayed variable localization patterns including movement to one pole and to the spindle midzone in these cells (Figure 7A). Our time-lapse analysis revealed that HU treatment induces spc24-9 mutants to undergo oscillations in spindle length, unlike in wild-type cells where relatively little change in spindle length is detected (Figure 8). We identified a window of time when the spindle was at an average maximum length in these analyses, and we quantitated Stu2-VFP fluorescence levels at this time. Stu2-VFP fluorescence was significantly reduced compared with wild-type cells, suggesting a correlation between reduction in Stu2-VFP fluorescence and spindle expansion (Figure 8E). The oscillations observed also explain why short spindles with decreased Stu2 are observed in populations of spc24-9 cells. Finally, oscillations in spindle length were also detected in HU-exposed rad53 mutants, suggesting that activation of the DNA replication checkpoint regulates spindle dynamics, and in the absence of the checkpoint, this restraint is compromised (Bachant et al., 2005). Our studies have uncovered the role of an effector of the checkpoint, Stu2, in restraining spindle dynamics while localized at the kinetochore.
The Role of Stu2 during the DNA Replication Checkpoint
Does Spc24 participate in regulating spindle expansion only during the DNA replication checkpoint or also during an unperturbed cell cycle? To address this question, we measured spindle length in a wild-type versus spc24-9 mutant after release from a G1 block to restrictive temperature (Supplemental Figure 5). We found that early in the cell cycle, spc24-9 mutants had longer spindles than wild-type cells consistent with a defect in the S phase checkpoint during a normal cell cycle. As cells progressed, spc24-9 spindle expansion lagged behind wild-type cells, suggesting a delay in anaphase. These data are consistent with our analysis of DNA content during a synchronous cell cycle at restrictive temperature, which demonstrated that spc24-9 cells progress more rapidly through S phase than wild-type cells (compare 60-min time point between wild-type and spc24-9; Supplemental Figure 6). However, once DNA has replicated in spc24-9 cells, a 2N content of DNA is maintained for 2 h before 1N DNA content is once again detected (Supplemental Figure 6; Montpetit et al., 2005). Thus spc24-9 cells accelerate through S phase, but are delayed in anaphase.
Mechanism of Spindle Expansion in rad53 and mec1 Mutants
The results shown here suggest that the kinetochore regulates spindle integrity during an HU-induced DNA replication checkpoint by sequestering proteins such as Stu2 that regulate MT dynamics. Why then do mec1 and rad53 mutants elongate their spindles during the DNA replication checkpoint? A previous study used a CEN transcription readthrough assay to demonstrate that the kinetochore is still capable of blocking access to the transcription machinery in a rad53-21 strain, suggesting that the inner CBF3 kinetochore complex that binds DNA is still intact (Bachant et al., 2005). Ndc10, a CBF3 component, is also present on CEN DNA in spc24-9 cells, suggesting that the spindle expansion is not due to defects in inner kinetochore assembly (Figure 2A). Not all central kinetochore mutants display CEN transcription readthrough; thus, the central kinetochore may be compromised in rad53 or mec1 mutant strains (Doheny et al., 1993). mec1-1 HU-treated cells display up-regulation of STU2 and CIN8 mRNA and protein levels, suggesting that increased levels of MT regulatory proteins may contribute to spindle expansion in mec1-1 cells (Krishnan et al., 2004). Our data suggest that mislocalization of Stu2 by disruption of a central kinetochore complex also causes spindle expansion during the DNA replication checkpoint.
By using HU as a method to stall cells in the process of DNA replication, we have uncovered a role for the kinetochore in regulating spindle dynamics in S phase. We have discovered a role for Spc24 in recruiting Stu2 to the kinetochore to mediate MT dynamics before metaphase. Mutation of Spc24 results in mislocalization of Stu2 and deregulation of spindle dynamics when DNA replication is stalled and likely during an unperturbed S phase as well. We propose that the kinetochore regulates spindle integrity during an HU-induced DNA replication checkpoint by sequestering proteins such as Stu2 that play central roles in controlling spindle MT dynamics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julien Guizetti (Vogel laboratory) for shared discussions of data. We acknowledge Ravi Jadusingh for technical assistance with cell dilution assays and Ken Thorne for creating the bik1 deletion strain. We thank Dr. Tim Huffaker for gifts of STU1 and STU2 overexpression plasmids, and stu1–5 and stu2–10 mutants; Dr. Trisha Davis for the gift of an HCM1 overexpression plasmid; and Dr. Phil Hieter for the gift of an MCK1 overexpression plasmid. Finally, we thank Dr. Kristin Baetz, Dr. Munira Basrai, Dr. Ben Montpetit, James Knockleby, Nina Piggott, and Ken Thorne for critical comments on the manuscript. This research was supported by operating grant MOP-67224 from the Canadian Institutes of Health Research (CIHR) and Michael Smith Foundation for Health Research (MSFHR) establishment grant CI-SCH-022(03–1)BIOM (to V.M.) and by CIHR operating grant MOP-64404 and infrastructure grants CFI 7395 from the Canada Foundation for Innovation (to J.V.) and to the Developmental Biology Research Initiative of McGill University (CFI 8298). V.M. is a Canada Research Chair in Enology and Yeast Genomics and an MSFHR Scholar. J.V. is supported by New Investigator award MSH 69117 from the CIHR.
Abbreviations used:
- CEN
centromere
- ChIP
chromatin immunoprecipitation
- CFP
cyan fluorescent protein
- GFP
green fluorescent protein
- HCS
high copy suppressor
- HU
hydroxyurea
- MT
microtubule
- SPB
spindle pole body
- Ts
temperature-sensitive
- VFP
venus fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0882) on May 16, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Larsen N. A., Hyman A. A., Harrison S. C. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J., van Breugel M., Harrison S. C., Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. B., Zhou Z., Siede W., Friedberg E. C., Elledge S. J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- Bachant J., Jessen S. R., Kavanaugh S. E., Fielding C. S. The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J. Cell Biol. 2005;168:999–1012. doi: 10.1083/jcb.200412076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsland-Marchesan E., Arino J., Saito H., Sunnerhagen P., Posas F. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol. Cell Biol. 2000;20:3887–3895. doi: 10.1128/mcb.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D., Bloom K. The role of centromere-binding factor 3 (CBF3) in spindle stability, cytokinesis, and kinetochore attachment. Biochem. Cell Biol. 2005;83:696–702. doi: 10.1139/o05-161. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Jaquenoud M., Gulli M. P., Chant J., Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman C. E. Replication checkpoint: preventing mitotic catastrophe. Curr. Biol. 2001;11:R121–R124. doi: 10.1016/s0960-9822(01)00057-4. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Enquist-Newman M., Muller-Reichert T., Drubin D. G., Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. C., Kim Y. J., Chan C. S. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. P., Yin H., Huffaker T. C. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J. Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D. J., Segal M., Jensen S., Reed S. I. Mec1p regulates Pds1p levels in S phase: complex coordination of DNA replication and mitosis. Nat. Cell Biol. 2001;3:619–627. doi: 10.1038/35083009. [DOI] [PubMed] [Google Scholar]
- Connelly C., Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuschieri L., Miller R., Vogel J. γ-Tubulin is required for proper recruitment and assembly of Kar9-Bim1 complexes in budding yeast. Mol. Biol. Cell. 2006;17:4420–4434. doi: 10.1091/mbc.E06-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. A., Keyes B. E., Ng Y. P., Freeman C. O., Burke D. J. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 2006;172:53–65. doi: 10.1534/genetics.105.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H., Tanaka K., Nasmyth K., Tanaka T. U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Doheny K. F., Sorger P. K., Hyman A. A., Tugendreich S., Spencer F., Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Collingwood D., Boeck M. E., Fox L. A., Alvino G. M., Fangman W. L., Raghuraman M. K., Brewer B. J. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Bilotta D., Lucchini G., Piatti S. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol. Biol. Cell. 2004;15:3796–3810. doi: 10.1091/mbc.E04-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard D. L., Becker B. E., Josh Romney S. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int. Rev. Cytol. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. New York: Academic Press; 1991. [Google Scholar]
- Haase S. B., Lew D. J. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 1997;283:322–332. doi: 10.1016/s0076-6879(97)83026-1. [DOI] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P. K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- He X., Rines D. R., Espelin C. W., Sorger P. K. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hofken T., Schiebel E. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 2004;164:219–231. doi: 10.1083/jcb.200309080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., Gerstein M., Snyder M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland K. M., Kingsbury J., Koshland D., Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y. H., do Carmo Avides M., Shiraki M., Deak P., Yamaguchi M., Nishimoto Y., Matsukage A., Glover D. M. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J. Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz J., Lechner J., Shevchenko A., Shevchenko A., Magiera M. M., Schramm C., Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kolodner R. D., Putnam C. D., Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- Kosco K. A., Pearson C. G., Maddox P. S., Wang P. J., Adams I. R., Salmon E. D., Bloom K., Huffaker T. C. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Nirantar S., Crasta K., Cheng A. Y., Surana U. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell. 2004;16:687–700. doi: 10.1016/j.molcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Krishnan V., Surana U. Taming the spindle for containing the chromosomes. Cell Cycle. 2005;4:376–379. doi: 10.4161/cc.4.3.1537. [DOI] [PubMed] [Google Scholar]
- Le Masson I., Saveanu C., Chevalier A., Namane A., Gobin R., Fromont-Racine M., Jacquier A., Mann C. Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol. Microbiol. 2002;43:1431–1443. doi: 10.1046/j.1365-2958.2002.02844.x. [DOI] [PubMed] [Google Scholar]
- Lin H., de Carvalho P., Kho D., Tai C. Y., Pierre P., Fink G. R., Pellman D. Polyploids require Bik1 for kinetochore-microtubule attachment. J. Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C. S., Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- McAinsh A. D., Tytell J. D., Sorger P. K. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- Measday V., Hailey D. W., Pot I., Givan S. A., Hyland K. M., Cagney G., Fields S., Davis T. N., Hieter P. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 2002;16:101–113. doi: 10.1101/gad.949302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Montpetit B., Thorne K., Barrett I., Andrews K., Jadusingh R., Hieter P., Measday V. Genome-wide synthetic lethal screens identify an interaction between the nuclear envelope protein, Apq12p, and the kinetochore in Saccharomyces cerevisiae. Genetics. 2005;171:489–501. doi: 10.1534/genetics.105.045799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Mitchell A. P. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- Pasqualone D., Huffaker T. C. STU1, a suppressor of a beta-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J. Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Salmon E. D., Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Pot I., Knockleby J., Aneliunas V., Nguyen T., Ah-Kye S., Liszt G., Snyder M., Hieter P., Vogel J. Spindle checkpoint maintenance requires Ame1 and Okp1. Cell Cycle. 2005;4:1448–1456. doi: 10.4161/cc.4.10.2106. [DOI] [PubMed] [Google Scholar]
- Pot I., Measday V., Snydsman B., Cagney G., Fields S., Davis T. N., Muller E. G., Hieter P. Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell. 2003;14:460–476. doi: 10.1091/mbc.E02-08-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T., Wu W., Miles S., Noble W. S., Breeden L. L. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof D. M., Meluh P. B., Rose M. D. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Ross K. E., Boucher L., Green Y., Tyers M., Cohen-Fix O. Uncovering novel cell cycle players through the inactivation of securin in budding yeast. Genetics. 2004;168:1763–1771. doi: 10.1534/genetics.104.029033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Habermann B., Huffaker T., Hyman T. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 2001;153:435–442. doi: 10.1083/jcb.153.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shero J. H., Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1) Genes Dev. 1991;5:549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- Shimogawa M. M., et al. Mps1 phosphorylation of dam1 couples kinetochores to microtubule plus ends at metaphase. Curr. Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C., Tanaka T. U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tytell J. D., Sorger P. K. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. J., Huffaker T. C. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R. R., Sorger P. K., Harrison S. C. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H. W., Niederstrasser H., Wong J., Drubin D. G., Nogales E., Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Wigge P. A., Kilmartin J. V. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak M. J., Blake-Hodek K., Kosco K., Hwang E., You L., Huffaker T. C. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N., Terashima H., Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Yin H., You L., Pasqualone D., Kopski K. M., Huffaker T. C. Stu1p is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol. Biol. Cell. 2002;13:1881–1892. doi: 10.1091/mbc.01-09-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Davis T. N. The fork head transcription factor Hcm1p participates in the regulation of SPC110, which encodes the calmodulin-binding protein in the yeast spindle pole body. Biochim. Biophys. Acta. 1998;1448:236–244. doi: 10.1016/s0167-4889(98)00135-9. [DOI] [PubMed] [Google Scholar]
- Zhu G., Muller E. G., Amacher S. L., Northrop J. L., Davis T. N. A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork head family of DNA-binding proteins. Mol. Cell Biol. 1993;13:1779–1787. doi: 10.1128/mcb.13.3.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.