Abstract
The poleward flux of tubulin subunits through spindle microtubules is a striking and conserved phenomenon whose function and molecular components remain poorly understood. To screen for novel components of the flux machinery, we utilized RNA interference to deplete regulators of microtubule dynamics, individually and in various combinations, from S2 cells and examined the resulting impact on flux rate. This led to the identification of two previously unknown flux inhibitors, KLP59C and KLP67A, and a flux promoter, Mini-spindles. Furthermore, we find that flux rate is regulated by functional antagonism among microtubule stabilizers and destabilizers specifically at plus ends. Finally, by examining mitosis on spindles in which flux has been up- or down-regulated or restored after the codepletion of antagonistic flux regulators, we show that flux is an integral contributor to anaphase A but is not responsible for chromosome congression, interkinetochore tension, or the establishment of normal spindle length during prometaphase/metaphase.
INTRODUCTION
The microtubule array of the mitotic spindle is notable for its dynamicity; as a whole, the entire array assembles and morphs, and as individual components, spindle microtubules frequently polymerize or depolymerize at their ends. Dynamic microtubules are essential because they serve several critical mitotic functions including, for example, generating and positioning the spindle, searching for and docking with kinetochores, congressing and segregating chromosomes, and performing a central role in the spindle checkpoint (Wittmann et al., 2001; Kline-Smith and Walczak, 2004). A particularly dramatic manifestation of spindle dynamics is poleward flux: the minus-end–directed flow of tubulin subunits through spindle microtubules, driven by the disassembly of microtubules at their minus ends (oriented toward the spindle poles) and assembly at their plus ends (oriented toward the spindle equator; Rogers et al., 2004; Mitchison, 2005). Functionally, poleward flux has been proposed to provide a mechanism for moving chromosomes toward spindle poles and for controlling spindle length. However, the overall contribution of flux to these activities has remained a point of contention (Khodjakov and Kapoor, 2005; Ganem and Compton, 2006).
The flux of tubulin through microtubules of the steady-state metaphase spindle appears, on its surface, to be a wasteful and rather useless phenomenon. However, manipulation of the balance of polymerization/depolymerization that gives rise to flux may provide a rapid, flexible, and adaptive means of stimulating changes in microtubule length, which can be translated into movements of chromosomes or spindle poles at appropriate moments in the cell cycle. For example, the poleward movement of chromatids during anaphase A could be induced by the down-regulation of polymerization at kinetochore microtubule plus ends, thus allowing chromatids to be reeled into spindle poles by minus-end depolymerization. More generally, the position and movement of chromosomes on the spindle throughout mitosis could be directly determined by coordinated changes in the balance of polymerization and depolymerization at the ends of fluxing kinetochore microtubules. A similar principle could be applied to the regulation of spindle length, as well.
Unfortunately, though it is a conserved process among higher eukaryotes, the functional significance of flux and its regulation could not be directly assessed until recently, because the molecular components of flux were unknown and so could not be selectively inhibited. Studies in Drosophila have revealed two components of the “flux machinery” that function by altering microtubule end dynamics. The first to be identified was KLP10A, a microtubule-destabilizing kinesin-13 that targets to microtubule minus ends at spindle poles. In early embryos, the inhibition of KLP10A causes a near cessation of flux, significantly decreases the velocity of chromatid-to-pole motion and stimulates a rapid elongation of the spindle, consistent with the hypothesis that KLP10A removes tubulin subunits from microtubule minus ends (Rogers et al., 2004). Inhibition of Kif2A, the mammalian orthologue of KLP10A, was recently found to also significantly diminish flux in human cells, confirming the requirement for a kinesin-13 at spindle poles to drive flux (Ganem et al., 2005).
The second flux component to be identified is the CLASP protein, Mast/Orbit (Mast), which induces microtubule polymerization and can be found on kinetochore-associated plus ends (Inoue et al., 2000; Lemos et al., 2000). The inhibition of Mast also perturbs flux but causes spindles to shorten or collapse, consistent with the hypothesis that this protein actively incorporates tubulin subunits into the plus ends of kinetochore microtubules (Maiato et al., 2005). Coinhibition of Mast and KLP10A in S2 cells restores the spindle collapse frequency and the spindle length to control level (Laycock et al., 2006). Together these data outline a mechanism that involves a controlled balance of KLP10A and Mast activities, and suggest that these proteins affect spindle length (and perhaps other features of mitotic spindles) by their control of poleward flux.
Whether KLP10A and Mast function in isolation or work cooperatively with additional regulators of microtubule dynamics is unknown. There are multiple additional candidate proteins for the flux machinery present in Drosophila. For example, in addition to CLASP, microtubule polymerization is promoted by members of the XMAP215/TOG and +TIP families of proteins. Mini-spindles (Msps), the only known member of the Drosophila XMAP215/TOG family of microtubule-associated proteins, is recruited to centrosomes by D-TACC during mitosis (although it eventually disperses onto spindles) and promotes microtubule assembly, probably by stabilizing the minus ends of centrosome-associated microtubules (Cullen et al., 1999; Lee et al., 2001; Barros et al., 2005).
Microtubule polymerization and stability is also promoted by the plus-end tracking protein (+TIP), EB1 (Rogers et al., 2002; Tirnauer et al., 2002; Wen et al., 2004). The specific changes to microtubule dynamics vary between experimental systems, but generally EB1 stabilizes microtubules by increasing the rescue frequency and decreasing the time spent in a paused state. In mitotic Drosophila cells, loss of EB1 activity sharply increases the frequency of aberrant spindle phenotypes and significantly decreases spindle lengths (Rogers et al., 2002; Maiato et al., 2005). Microtubule depolymerization, on the other hand, is stimulated by several members of the kinesin superfamily (Wordeman, 2005). In Drosophila, these include KLP10A, KLP59C (both kinesin-13 members; Rogers et al., 2004), and KLP67A (a kinesin-8; Goshima and Vale, 2003; Gandhi et al., 2004). KLP59C and KLP67A localize to centromeres/kinetochores during mitosis, suggesting that these kinesins could stimulate depolymerization of kinetochore microtubule plus ends (Rogers et al., 2004; Savoian et al., 2004; Goshima and Vale, 2005). Consistent with this hypothesis, the inhibition of KLP59C in early embryos slows anaphase chromatid-to-pole motion (Rogers et al., 2004). KLP67A also appears to impact chromatid-to-pole motion in meiosis but not mitosis (Savoian et al., 2004; Goshima and Vale, 2005).
As a screen to identify new components of the molecular machinery driving poleward flux, we used double-strand RNA interference (dsRNAi) to deplete, individually and in combination, selected proteins known to regulate microtubule dynamics in S2 cells. The interactions of flux components were studied to reveal the existence of functional antagonism between microtubule-polymerizing and -depolymerizing proteins. Finally, the performance of important mitotic processes (establishment of spindle length, generation of interkinetochore tension, congression, and segregation of chromosomes) was evaluated under conditions of perturbed flux rates to test the functional link between flux and mitotic process. Overall, this study sought to identify new components of the mechanism driving poleward flux and to determine if certain mitotic processes (which have, in the past, been proposed to be flux-dependent) actually require flux or just the activities of regulators of microtubule dynamics.
MATERIALS AND METHODS
S2 Cell Culture
All experiments used Schneider S2 cells that had been stably transfected with enhanced green fluorescent protein (eGFP)-α-tubulin under control of a constitutive promoter (pAC5.1; Invitrogen). These S2 cells were a gift from the lab of R. Vale (UCSF). S2 cells were cultured at room temperature (23°C) in standard culture medium: Schneider's Drosophila medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen) and penicillin/streptomycin. For live cell microscopy, S2 cells were plated for at least 2 h in 35-mm glass-bottom microwell dishes (MatTek, Ashland, MA) coated with 10 μg concanavalin A (Sigma-Aldrich, St. Louis, WI) to promote cell spreading (Rogers et al., 2002). For immunofluorescence, S2 cells were similarly plated on concanavalin A–coated coverslips to induce cell spreading, after which the coverslips were fixed and immunostained.
dsRNAi
Regions of the target proteins' coding sequences were used to generate the double-stranded RNA oligonucleotides applied to the cultured S2 cells. Briefly, target-specific PCR primers (Supplementary Figure 1D) were used to amplify DNA templates from S2 cell cDNA or Drosophila expressed sequence tagged oligos (Drosophila Genomics Resource Center, Bloomington, IN). The 5′ end of each primer begins with T7 promoter sequence (TAATACGACTCACTATAGGG) to allow dsRNA synthesis using T7 reverse transcriptase. dsRNA was synthesized from the DNA templates using commercial T7 transcription kits (MEGAscript T7, Ambion, Austin, TX; T7 RiboMAX, Promega, Madison, WI).
To knockdown target proteins, S2 cells cultured in six-well plates were incubated in 0.75 ml of Schneider Drosophila cell medium (lacking FBS and antibiotics) with 20 μg of gene-specific dsRNA added for each protein to be knocked down. For controls, sufficient control dsRNA was added to equal the highest total dsRNA applied to any treatment in each experiment. After a 1-h incubation, 0.75 ml of Schneider medium containing 20% heat-inactivated FBS was added. This procedure was used to apply fresh dsRNA on days 0, 2, 4, and 6 of the treatment period. On day 7, cells were transferred to other culture dishes for live cell recording or immunofluorescence.
Knockdown was evaluated from Western blots of S2 cell lysates. Blots were first probed with mouse anti-α-tubulin monoclonal (DM1A, Sigma-Aldrich) to compare sample loadings and then were stripped and reprobed with rabbit polyclonal antibodies specific to the RNAi targets. Rogers et al. (2004) demonstrated that KLP59C was efficiently knocked down in S2 cells using the same dsRNA as was used here, but their anti-KLP59C antibody is no longer stable after affinity purification. Therefore, relative RT-PCR was used to demonstrate that the KLP59C transcript is diminished in S2 cells treated with KLP59C dsRNA (Supplementary Figure 1A). After first-strand cDNA production from RNA isolated from day 7 RNAi-treated S2 cells, PCR was used to amplify the DNA of KLP59C or the housekeeping gene, DmGAPDH1. During PCR, aliquots were taken at cycles 25 and 50 for analysis. In Supplementary Figure 1B, unpurified anti-KLP59C serum was used because affinity purification inactivates the antibody and because crude serum was found to recognize a single prominent band with the predicted molecular mass of KLP59C.
To quantitate extent of knockdown, integrated intensities of immunoblot bands were measured by densitometry of unsaturated blot images. Values for integrated intensities were then adjusted by background subtraction and normalized to compensate for loading volume differences (using the adjusted integrated intensities for α-tubulin bands). Percent knockdowns relative to controls are presented in Supplementary Figure 1C.
Measurement of Poleward Microtubule Flux
Poleward flux rates of preanaphase spindles of S2 cells were measured by tracking marks photobleached onto fluorescent spindles of preanaphase S2 cells stably expressing eGFP-α-tubulin at 22–24°C. Only cells with bipolar spindles oriented perpendicular to the optical axis were analyzed. Narrow rectangular regions were photobleached across each fluorescent half-spindle of a cell using a TCS SP2 confocal system (Leica, Heidelberg, Germany) on a Leica DMIRE2 inverted microscope (Plan Apo 63× objective, 1.4 NA). Typically, photobleached marks remained visible for 30–60 s and were most sharply defined when the bleached mark was created roughly two thirds of the distance from pole to equator (Supplementary Movie 1). The distance of the photobleach mark to the pole was manually measured using the MetaMorph software calipers tool (Molecular Devices, Sunnyvale, CA) on sequential images of single confocal sections captured at 1.8- or 3.6-s intervals. Flux rate was calculated from the distance progressed and time elapsed before the bleached zone disappeared as fluorescence recovered. For each spindle, flux rates were measured from each of the spindle's prominent fluorescent filaments (probably kinetochore microtubule bundles) because their bleach marks were more defined and persistent, and then a mean spindle flux rate was obtained by averaging the multiple flux measurements. The resulting “spindle average” constituted a single data point of the entire data set used for analysis.
Fluorescence Recovery after Photobleaching Measurement
Preanaphase, eGFP-α-tubulin–expressing S2 cells were photobleached (as described above) to create two bleached zones: one near the spindle equator (plus ends of kinetochore and nonkinetochore microtubules) and the second at the pole of the other half-spindle (which we refer to as consisting of minus ends, but may also contain plus ends of short microtubules). Experiments were performed at an ambient temperature of 22–24°C. The bleached regions were produced in different half-spindles to prevent their cross-interference during recovery. After photobleaching, images were captured at 3.6-s intervals for 2–3 min. ImageJ (NIH) was used to measure the fluorescence intensities of regions-of-interest positioned within the bleached zones in each image. The same size/shaped region-of-interest was used for all analyses. Loss of fluorescence due to photobleaching during image capture was measured as the decrease in total fluorescence of the entire cell; fluorescence recovery after photobleaching (FRAP) values were adjusted for this loss. The corrected fluorescence intensities were best-fit plotted to a hyperbolic or sigmoidal function using nonlinear regression (SigmaPlot, Systat Software, Evanston, IL). An asymptote was calculated from the parameters of the best-fit equation and was used to calculate the percent recovery and the time to reach 50% of the final recovered fluorescence (T1/2) for each data set. Spindles that shifted or collapsed were not analyzed, and spindles with <30% fluorescence recovery were not used to calculate T1/2.
Measurement of Anaphase Segregation Velocity
Immediately before recording, 300 ng/ml (final) Hoechst 33258 was added to the culture medium to vital stain chromosomes. At an ambient temperature of 22–24°C, live S2 cells expressing eGFP-α-tubulin were recorded as they progressed through anaphase using the Leica confocal microscope described above. Only cells with spindles oriented perpendicular to the optical axis were recorded. The chromatid-to-pole distances were measured manually on frames from recorded movies using the calipers tool of MetaMorph. Specifically, distances were measured from the leading edges of stained chromatids (which should correspond to the orientations of the kinetochores). As long as the leading edge of one chromatid did not superimpose with the stained region of a second chromatid, then leading edges were sharply defined and could be readily tracked. Chromatid-to-pole segregation rate was calculated as the distance moved as a function of time.
Measurement of Interkinetochore Distance and Congression
RNAi-treated S2 cells were fixed in −20°C methanol, rehydrated with 0.04% Tween-20 in phosphate-buffered saline (PBS/T), blocked with PBS/T containing 5% normal goat serum, and then incubated with 1.3 μg/ml of DM1A monoclonal (to label microtubules in any cells expressing low titers of eGFP-α-tubulin; Sigma Aldrich) and an antibody to the Drosophila CENP-A homolog, Cid, to label centromeres (Blower and Karpen, 2001) in blocking buffer, 1–2 h. After washing with PBS/T, cells were incubated with 1.5 μg/ml Cy2-conjugated anti-mouse IgG and 1.5 μg/ml rhodamine-conjugated anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) in blocking buffer for 1–2 h, washed, and then mounted in anti-fade solution (1% N-propyl-gallate, 100 mM Tris, pH 8, 50% glycerol). Images of mitotic cells were captured with confocal microscopy as z-stacks as described above. Only spindles whose pole-to-pole axes were perpendicular to the light path were analyzed.
To evaluate the tension on kinetochores, distances between centromeres of sister chromatids were measured using the calipers tool of MetaMorph. For each spindle, the interkinetochore distances of all pairs of sister chromatids whose anti-Cid stained centromeres lay within the same confocal z-section were measured. All distance measurements of an individual spindle were averaged. The resulting “spindle average” constituted a single data point of the entire data set used for analysis.
To analyze chromosome congression, the distances between centromeres and spindle equator were measured. All measurements from an individual spindle were averaged, and this “spindle average” was used later for analysis. Centromere displacement results are presented graphically in two ways, either as a percent of spindle length (Figure 4A′; Supplementary Figure 5A) or simply as a linear distance (Supplementary Figure 5A′). Evaluating displacement as a percent of spindle length avoids a potential artifact that might arise when comparing chromosome congression as linear displacements on short spindles (like those typical after Mast or Msps RNAi)—which have a narrow range of possible displacement distances—with those on long spindles (e.g., after KLP67A or KLP10A RNAi) with their large range of possible distances. Uncongressed chromosomes on short spindles will necessarily have smaller linear displacements than uncongressed chromosomes on long spindles, assuming that chromosomes are distributed throughout the spindles.
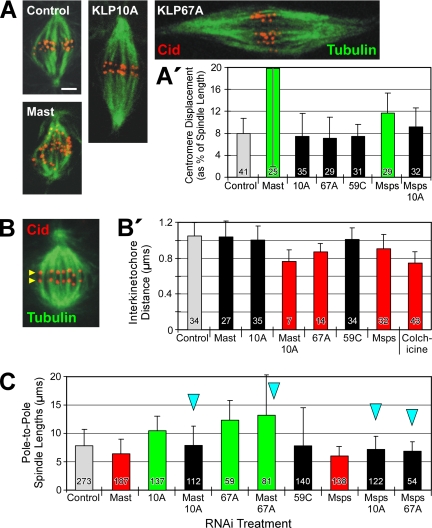
Figure 4.
Regulators of microtubule dynamics affect congression, tension, and spindle length, but do not exert their influence via flux. (A) Spindles of RNAi-treated cells were immunostained for the centromere marker, Cid (red), in order to measure chromosome congression on spindles. Microtubules, green. Scale bar, 2 μm. (A′) Congression was evaluated by the average centromere displacement from the spindle equator, expressed as a percent of the pole-to-pole spindle length. Notably, some treatments that perturb flux rate (RNAi of KLP10A or KLP67A, or co-RNAi of Msps/KLP10A) do not inhibit congression. (B) Interkinetochore tension was evaluated by measuring the distance between sister centromeres immunolabeled with Cid antibody. Yellow arrowheads mark one pair of Cid-immunostained sister centromeres on a control metaphase spindle. (B′) Average distance between sister centromeres after RNAi to knockdown the indicated target proteins. Note that some RNAi treatments (specifically, Mast or KLP10A) decrease flux but fail to significantly decrease centromere spacing. (C) Average pole-to-pole spindle lengths after RNAi to deplete the activities of the indicated regulators of microtubule dynamics. A number of treatments significantly decrease flux but have no significant effect on spindle length.
Spindle Length Measurement
After plating on concanavalin A–coated coverslips to promote cell spreading, cells were fixed as described above and then immunostained with the primary antibodies DM1A and anti-phosphohistone H3 (Ser10; 1.3 μg/ml; Upstate Biotechnology, Lake Placid, NY). Mitotic cells were identified by their positive phosphohistone immunostain, and then each mitotic cell was recorded as a z-stack with an Ultraview spinning disk confocal system (Perkin Elmer, Waltham, MA) on a Nikon TE200 inverted microscope (PlanApo 100× objective, 1.4 NA). Only bipolar spindles oriented perpendicular to the light path were used for analysis. Pole-to-pole spindle lengths were measured on projections of each spindle using the calipers tool of MetaMorph.
Statistical Analysis
A nonparametric statistical test, the Kurskal-Wallis one-way analysis of variance on ranks, was performed because not all datasets had a Gaussian distribution. Dunn's posttest analysis was used to test for significant differences between all treatments compared with control (SK RNAi). For pairwise comparisons of two treatments, the nonparametric Mann-Whitney rank sum test was used. Comparisons are considered significantly different if p < 0.05. Unless stated otherwise, data points used for analysis were “spindle averages”, which were obtained by averaging all the measurements for an individual spindle. Statistical analyses were performed with the software packages, SigmaStat 3 (Systat Software) or GraphPad Prism 4 (GraphPad, San Diego, CA).
RESULTS
Mitotic microtubules are more dynamic than their interphase counterparts (reviewed by Kline-Smith and Walczak, 2004), and the application of taxol or colchicine at low concentrations to suppress spindle microtubule dynamics blocks mitotic progression (reviewed by Jordan, 2002), indicating that one or more mitotic processes requires the dynamic behavior of microtubules. Indeed, microtubule dynamics could conceivably impact every aspect of spindle assembly and function. Do different mitotic processes share a common need for poleward flux, or are they more loosely connected by their requirements for the activities of proteins controlling microtubule dynamics?
To begin answering these questions, we examined the effects of knocking down three microtubule-depolymerizing kinesins, KLP10A, KLP59C, and KLP67A, and three putative microtubule-stabilizing proteins, Mast, Msps, and EB1, on several mitotic processes in Drosophila S2 cells. These proteins were chosen for their reported or putative effects on microtubule dynamics. (Their activities are generally important for proper completion of mitosis because RNAi of these proteins, either singly or in combination, usually impacts the mitotic index or the spindle morphology profile of S2 cells [data not shown].) The extent of knockdown ranged from 92 to 100%, including those treatments to knock down two targets by co-RNAi (Supplementary Figure 1C). Therefore, the following experiments were performed in conditions in which most, but not necessarily all, of the target proteins were knocked down.
Regulation of Poleward Flux by Mast and KLP10A
By its very nature, the generation of poleward flux is closely linked to the regulation of the dynamic behaviors of spindle microtubule ends. Recent work suggests that flux involves the active removal of tubulin subunits from microtubule minus ends at spindle poles by a kinesin-13 (KLP10A in Drosophila) and the addition of subunits to plus ends at kinetochores by a CLASP (Mast in Drosophila; Rogers et al., 2004; Ganem et al., 2005; Maiato et al., 2005). We sought to determine whether additional, previously unidentified molecular components of flux exist among the group of proteins controlling microtubule dynamics.
In agreement with previous studies (Maiato et al., 2003, 2005; Rogers et al., 2004), we found that the flux rate was significantly decreased after RNAi treatments to deplete either Mast or KLP10A (Figure 1, A and B; Supplementary Movie 2). The extent of flux rate decrease after Mast or KLP10A RNAi (decreased 74 or 65%, respectively, relative to control rate) was comparable to the decrease measured in cells treated with low concentrations of colchicine or taxol to suppress microtubule dynamics (Supplementary Figure 2).
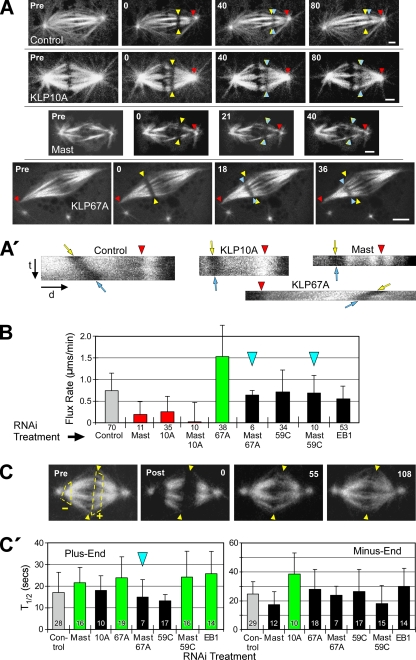
Figure 1.
Mast and multiple microtubule-destabilizing kinesins regulate the rate of poleward flux. (A) After RNAi of the indicated target proteins, flux rates were measured by photobleaching rectangular bars across fluorescent spindle microtubules and tracking their poleward motions. Yellow arrowheads mark the initial positions, blue arrowheads mark the current positions of the photobleached bars and red arrowheads mark the reference positions. Pre, prebleach spindle. Numbers are time (seconds) elapsed after the initial photobleached image. Scale bars, 2 μm. (A′) Kymographs generated from kinetochore fibers spanning the bleached regions of the spindles shown in A. Red arrowheads mark the same features as those in A; t and d indicate the time and distance axes, respectively. The angles of the bleached marks' tracks (between yellow and blue arrows) reflect the flux rates: the rate decreases as the track becomes vertical. These kymographs are illustrative only; flux rates were measured directly from movie frames like those in A. (B) Poleward flux rates after RNAi of targeted proteins. Statistically significant differences (p < 0.05) are indicated by green (greater than control), red (less than control), or black bars (not different from control). This scheme is used for all figures. Blue arrowheads mark co-RNAi treatments that are mentioned in the text as phenotype rescues. Error bars, +SD. Numbers below bars, N. (C) FRAP of microtubule ends was used to measure α-tubulin subunit turnover after RNAi. Pre, spindle before photobleaching. Arrowheads mark the spindle equator; dashed lines outline regions at plus (+) and minus ends (−) to be photobleached. Post, spindle immediately after photobleaching; numbers are elapsed time (seconds) after the first postbleach image. (C′) FRAP half-times (T1/2) of microtubule plus (left) and minus ends (right) after RNAi treatment. Error bars, +SD. Numbers within bars, N.
We used FRAP to measure the turnover of fluorescent α-tubulin subunits at both ends of spindle microtubules to determine the regions of spindles displaying altered microtubule dynamics after RNAi treatment (Figure 1C). Mast RNAi suppressed α-tubulin turnover (i.e., significantly increased the T1/2) specifically at microtubule plus ends (T1/2 increased 1.3 times relative to control), whereas KLP10A RNAi had the same effect specifically on minus ends (1.6× increase; Figure 1C′). The changes in microtubule end dynamics after knockdown of these proteins support the model that flux is driven by Mast-stimulated plus-end assembly and KLP10A-stimulated minus-end disassembly of spindle microtubules. This model predicts that simultaneous depletion of Mast and KLP10A by co-RNAi should not rescue flux, and as expected, flux rate was not restored when both Mast and KLP10A activities were simultaneously knocked down by co-RNAi (which decreased flux by 97%; Figure 1B). Therefore, Mast and KLP10A are components of the molecular machinery driving flux, and the respective sites of their effects on tubulin turnover agree with their proposed sites of activity. Is flux solely the manifestation of balanced Mast and KLP10A activities, or do other regulators of microtubule dynamics also control the flux rate?
Regulation of Flux at Plus Ends: KLP67A and KLP59C Antagonize Mast
To identify other proteins that could be controlling flux by virtue of their effects on plus-end dynamic behavior, we first examined the activities of two other members of the microtubule-destabilizing kinesin-13 family. In contrast to KLP10A, which appears to act predominantly on microtubule minus ends at spindle poles, the other two microtubule-destabilizing enzymes examined here, KLP67A and KLP59C, concentrate proximally to plus ends. KLP67A is positioned on outer kinetochores, whereas KLP59C concentrates at centromeres (Rogers et al., 2004; Goshima and Vale, 2005). Given that metaphase flux requires net polymerization of plus ends, we predicted that neither KLP67A nor KLP59C would contribute significantly to flux rates. Instead, we found that depletion of either protein did impact flux but in distinct ways.
KLP67A knockdown caused the average flux velocity to double (Figure 1B; Supplementary Movie 3), indicating that KLP67A is normally an active flux suppressor. FRAP analysis indicates that KLP67A activity specifically alters microtubule plus-end dynamics: KLP67A RNAi significantly increases T1/2 by 1.4 times for α-tubulin turnover at the spindle equator but not at poles (Figure 1C′; Supplementary Figure 3A). We propose that KLP67A's disassembly-promoting activity decreases the net polymerization rate of spindle microtubule plus ends, which our data suggest is the rate-limiting factor for flux velocity under normal conditions.
This result suggests that microtubule-stabilizing and -destabilizing activities at plus ends control the flux rate by functioning antagonistically to modulate the assembly state of plus ends. Consistent with the hypothesis of functional antagonism at plus ends, the codepletion of KLP67A and Mast restored both flux rate and the plus-end turnover time (T1/2) to levels statistically indistinguishable from controls (Figure 1, B and C′). The restoration of plus-end tubulin turnover after the double knockdown could result from a restoration of balance between residual Mast/KLP67A or from other plus-end factors affecting dynamics.
In contrast to KLP67A, the depletion of KLP59C alone did not significantly impact flux (rate was 98% vs. control), nor did it significantly alter α-tubulin turnover at plus or minus ends (T1/2 was 79 or 108%, respectively, vs. controls; Figure 1, B and C′). However, KLP59C functionally antagonizes Mast to regulate flux, because codepletion of KLP59C and Mast restores flux to control velocities (94% vs. control; Figure 1B). Thus, although KLP59C is not a flux suppressor like KLP67A, it shares with KLP67A the ability to interact antagonistically with Mast. In summary, both KLP59C and KLP67A oppose Mast-stimulated flux, but their specific mechanisms of action differ (see Discussion).
Finally, we examined another regulator of microtubule dynamics, EB1, a +TIP protein (Maiato et al., 2002; Rogers et al., 2002). Like Mast, EB1 activity alters the dynamics of plus ends: RNAi of EB1 causes microtubule plus ends to turnover more slowly (1.5 times slower than control; Figure 1C′). However, in contrast to Mast, knockdown of EB1 did not have a significant impact on flux rate (though the rate decreased 25% vs. control; Figure 1B). This is particularly interesting because depletion of EB1 slows plus-end turnover to a greater extent than Mast depletion (Figure 1C′) and suggests that Mast is the more dominant assembly-promoting activity for regulation of flux rate (but why this would be so is unclear).
Regulation of Flux at Microtubule Minus Ends: Mini-Spindles and KLP10A Are Essential Flux Components
Along with KLP10A, a second regulator of microtubule dynamics positioned at minus ends is Msps, the sole member of the Drosophila XMAP215 family. Msps has been reported to promote mitotic microtubule assembly and to be concentrated at centrosomes (Cullen et al., 1999; Barros et al., 2005). Therefore, our initial hypothesis was that Msps inhibits flux by stabilizing microtubule minus ends against KLP10A-induced disassembly; Msps RNAi should therefore increase flux velocity. Instead, we observed the opposite effect. Msps depletion sharply suppressed poleward flux (by 79%; Figure 2, A and B; Supplementary Movie 4).
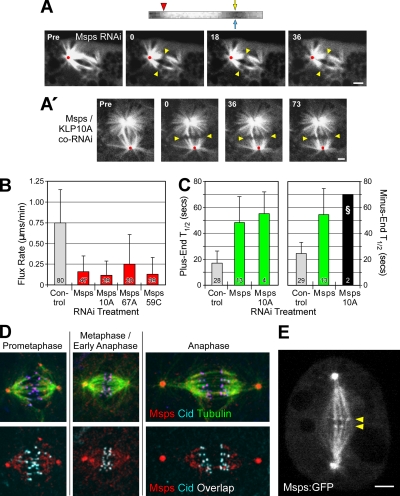
Figure 2.
Msps is required for poleward flux. (A) A photobleached mark on a spindle after Msps RNAi is stationary, indicating an absence of flux. Yellow arrowheads mark the initial photobleach position; red dots mark the spindle reference point. Scale bars, 2 μm. The kymograph (top) was generated from a kinetochore fiber spanning the bleached region; the markings are the same as in Figure 1A′. The vertical track of the photobleached region is a result of flux cessation. (A′) CoRNAi of Msps and KLP10A does not restore flux. (B) Average flux rates after RNAi of the indicated target proteins. (C) Average fluorescence recovery half-times (T1/2) after photobleaching. The Msps/KLP10A co-RNAi treatment (§) was not be statistically analyzed because too few measurements (N = 2; SD = 0) were obtained. (D) Immunolocalization of Msps and Cid (a kinetochore marker) in mitotic eGFP-α-tubulin–expressing S2 cells. Images are maximum intensity projections. In the bottom panels, Msps and Cid colocalization is indicated by areas of white overlap. (E) A single confocal z-section from a live, stably transfected S2 cell expressing Msps-eGFP. The small but clearly visible puncta aligned at the spindle equator (yellow arrowheads) presumably mark the positions of kinetochores. This image was obtained from a frame of Supplementary Movie 5. Scale bar, 2 μm.
We were also surprised to find that Msps activity affects the dynamics of both microtubule ends. FRAP analysis revealed that Msps RNAi sharply decreases α-tubulin subunit turnover at plus and minus ends (increasing T1/2 2.9× and 2.2×, respectively; Figure 2C; Supplementary Figure 3A). This conclusion is further supported by the observation that Msps is present not only at centrosomes, but also at kinetochores. In S2 cells, Msps concentrates on centrosomes, distributes in a punctate pattern throughout most of the spindle, and is present at kinetochores throughout mitosis (Figure 2D). The unexpected kinetochore localization was confirmed by analysis of stably transfected S2 cells expressing Msps-eGFP (Figure 2E; Supplementary Movies 5 and 6). Therefore, Msps is appropriately positioned to influence the dynamic behaviors of both microtubule ends.
Unlike Mast, the impact of Msps depletion on flux rate could not be rescued by co-RNAi with any microtubule-destabilizing kinesin tested (Figure 2B; Supplementary Movies 7 and 8). Likewise, the reduced tubulin turnover could not be rescued by codepletion of Msps with KLP10A: the bipolar spindles of these cells still exhibited very reduced microtubule dynamicity (Figure 2C). Indeed, the majority of these spindles recovered little eGFP-α-tubulin within the bleached zones (Supplementary Figure 3B). These results argue against the hypothesis that Msps stabilizes microtubule minus ends at poles and functions to counterbalance KLP10A-driven disassembly. We tested the possibility that Msps is needed to recruit or sequester KLP10A to poles by examining the immunolocalization of KLP10A and Msps in cells after either KLP10A or Msps RNAi. RNAi of KLP10A or Msps reduced the immunolocalization of the appropriate target protein, but did not displace or visibly reduce the immunostaining of the second protein (i.e., Msps or KLP10A, respectively; Supplementary Figure 4A).
Finally, we found that KLP10A, like Msps, is necessary for flux, since the flux rate could not be rescued by the co-RNAi of KLP10A with any of the putative microtubule-stabilizing proteins, Mast, EB1, or Msps (Figures 1B and 2B, Supplementary Figure 2). Thus, minus-end disassembly for flux is promoted by an essential factor (KLP10A), whereas plus-end assembly is controlled by several interacting, nonessential factors (Mast, KLP67A, and KLP59C). In addition, both ends share a requirement for Msps, which might actually function to enable dynamic behavior at microtubule ends rather than to strictly promote assembly (see Discussion).
Flux Generates Poleward Forces on Chromosomes during Anaphase
The identification of conditions in which flux can be up- or down-regulated or restored in the absence of specific regulatory proteins creates the opportunity to address the question: Is a specific mitotic process dependent on flux or rather on the regulatory proteins that coincidentally control flux? It has long been suggested that microtubule flux or treadmilling within spindles provides a driving force for chromosome motility, particularly during anaphase A (Margolis et al., 1978). However, this matter is currently controversial because the results of some recent studies provide support for this notion, whereas others downplay the importance of flux in chromatid-to-pole motion (Rogers et al., 2004; Ganem et al., 2005; Ganem and Compton, 2006). If flux contributes significantly to chromatid motility, then any change in the flux rate should correspond to a similar alteration of the velocity of anaphase A.
In control S2 cells, the average flux rate decreases slightly after the start of anaphase, from 0.75 (before anaphase; Figure 1B) to 0.70 μm/min after anaphase start (Figure 3B), whereas chromatids segregate at an average 1.2 μm/min (Figure 3C; Supplementary Movie 9). Thus, a complete elimination of flux should decrease the anaphase segregation velocity by ∼0.7 μm/min, a loss of 58%. The chromatid-to-pole velocity actually decreases 49% after KLP10A RNAi and by 63% after co-RNAi of Mast and KLP10A (Figure 3C). These velocity decreases are significant and suggest that flux contributes to the rate of chromosome segregation. Previous studies have also observed decreases in chromatid segregation rates after inhibition of pole-positioned kinesin-13 (Rogers et al., 2004; Ganem et al., 2005), but the question remains: To segregate chromatids, are the activities of regulatory proteins promoting a dynamic behavior of microtubules that is distinct from poleward flux, or is flux itself that dynamic process?
Figure 3.
Flux contributes to the anaphase A segregation rate of chromosomes. (A) Measurement of flux and segregation rates in an anaphase spindle. Pre, prebleached spindle; microtubules, green; chromatin, red. Numbers are elapsed time (seconds) after photobleaching. The blue arrow marks the progression of the photobleach mark toward the pole as a result of flux. (The dashed yellow line marks the end of a pole and, coincidentally, the center of a centrosome.) The yellow arrow marks the leading edge of a selected chromosome as it moves poleward. Scale bar, 2 μm. (B) Average anaphase flux rates after the indicated RNAi treatments. (C) Average anaphase chromatid-to-pole velocities after RNAi treatment.
If flux supplies part of the force responsible for segregating chromatids, then chromatids should segregate at an increased velocity if the flux rate were experimentally increased. To test this prediction, we measured the chromatid-to-pole rates in S2 cells treated with KLP67A RNAi since the anaphase A flux rate of these cells is 1.3 times greater than the control rate (Figure 3B). The chromatid-to-pole velocity does increase significantly (almost 1.3 times) in anaphase cells after KLP67A RNAi (Figure 3C), supporting the hypothesis that changes in flux rate have a direct impact on the anaphase A separation rate of chromosomes. This result differs from that of a previous study that found an insignificant effect on segregation rate after KLP67A RNAi (Goshima and Vale, 2005). However, chromatids in meiotic Drosophila spermatocytes with null mutations of the KLP67A gene segregate faster than controls (Savoian et al., 2004). The reasons underlying these discrepancies are not clear. Typically, some KLP67A remains on kinetochores during anaphase A, but at a diminished level (Supplementary Figure 4B; Goshima and Vale, 2005). Therefore, active KLP67A at kinetochores could continue to inhibit flux (and segregation) during anaphase. However, we note that KLP67A RNAi has a smaller affect on flux rate during anaphase A (∼1.3× increase over control rate) than before anaphase (∼2× increase), which could be a consequence of the diminished amount of KLP67A normally present on anaphase kinetochores.
Finally, if flux is a component of the anaphase A segregation machinery, then co-RNAi treatments that restore the flux rate to the control level should also restore the chromatid-to-pole rate. We measured the anaphase A chromatid-to-pole rates of cells treated by co-RNAi of Mast and KLP59C, which increases anaphase A flux to a rate equivalent to control (Figure 3B). In this condition, segregation rate was also increased to a level equivalent to control (Figure 3C). These data support the hypothesis that loss or gain of flux rate has a commensurate impact on anaphase A rate. Therefore, changes in flux rate result in similar changes in anaphase A segregation rate.
Microtubule Dynamics, But Not Flux per se, Regulates Congression
If the dynamic behavior of microtubules is harnessed to segregate chromosomes during anaphase A and if flux is at least one aspect of that required dynamic behavior, then flux might also contribute to the forces applied to chromosomes of preanaphase spindles. First, we examined the requirement of the six microtubule regulating proteins (singly and in combination) for the congression of chromosomes to the spindle equator. S2 cells were fixed after RNAi treatment and their centromeres visualized by immunostaining the centromeric protein, Cid (Figure 4A; Blower and Karpen, 2001). The displacement of each centromere from the equators of spindles was measured and expressed as a percent of the spindle length. Surprisingly, the chromosomes of cells treated by RNAi to either suppress flux (KLP10A RNAi and Msps/KLP10A co-RNAi) or accelerate flux (KLP67A RNAi) had congressed as well as those of controls (Figure 4A′). Previous studies have reported normal congression after flux inhibition (Ganem et al., 2005; Laycock et al., 2006), but our results show that even abnormally rapid flux will not prevent normal congression. (However, we note that when the displacement is expressed simply as the linear distance between centromeres and spindle equator, then KLP67A RNAi does inhibit congression [Supplementary Figure 5A′]. We believe that expressing displacement as a fraction of spindle length [Figure 4A′] is more appropriate for evaluating congression for the reason described in Materials and Methods.) Therefore, a normal flux rate is not required for chromosome congression.
Even though depletion of any of the microtubule-destabilizing kinesins does not interfere with congression, depletion of either Mast or Msps significantly increases the scatter of chromosomes throughout the spindle (Figure 4A′; Maiato et al., 2002). In fact, because the congression failure after Mast RNAi could not be rescued by co-RNAi with any depolymerization-promoting kinesin tested (including those co-RNAi treatments that rescue the flux rate; Supplementary Figure 5A), then these results indicate that Mast activity is required for congression independently of its effect on flux. Therefore, congression and flux appear to be separable mitotic processes, linked only by their requirement for specific regulators of microtubule dynamic behavior.
Microtubule Dynamics, But Not Flux per se, Regulates Interkinetochore Tension
Second, previous work has suggested that the dynamic behavior of spindle microtubules exerts a poleward force on each kinetochore and so generates an interkinetochore tension (Waters et al., 1996; Zhou et al., 2002), which is conventionally assessed by measuring the distance between sister kinetochores (Figure 4B). Thus, inhibition of regulators of microtubule dynamics should alter the tension on kinetochores, changing the sister kinetochore separation. RNAi of either Mast or KLP10A does not reduce centromere separation significantly, indicating that normal tension can be generated during conditions of significantly reduced flux rate (Figure 4B′), which has been shown previously in mammalian cells (Ganem et al., 2005). Nevertheless, we expected that a large increase in flux rate would impose some additional poleward strain on kinetochores. Surprisingly, even though treatment of cells with KLP67A RNAi doubles the flux rate, it significantly decreases centromere separation, indicating that tension has actually been relieved (Figure 4B′). One explanation for this result is that the attachment between kinetochore and microtubule does not resist the extension of the microtubule away from the kinetochore as a result of plus-end growth. Instead, the attachment might resist the escape of the plus-end from the kinetochore, which exerts tension on the kinetochore. As a result, flux per se would not generate tension unless plus-end growth had been suppressed (a condition probably present in the study of Waters et al. [1996]).
Other RNAi treatments also decrease centromere separation, indicating a loss of tension. These include several treatments that strongly inhibit poleward flux: Mast/KLP10A co-RNAi and all treatments involving Msps RNAi (Figure 4B′; Supplementary Figure 5B). Taken together, these data are consistent with a model of kinetochore tension development that does not depend on flux per se, but does require regulators of microtubule dynamics—some of which also control flux rate. Kinetochore tension and flux are coregulated but not codependent. Future experiments are needed to explore the specific role of microtubule end dynamics in tension generation.
Microtubule Dynamics, But Not Flux per se, Regulates Spindle Length
Finally, because spindle length is defined by the length of the microtubule array, then processes that drive microtubule dynamic behavior should contribute to spindle length control. To investigate the general importance of poleward flux to spindle length control, we measured the pole-to-pole spindle lengths of preanaphase cells after knockdown of target proteins. Only the lengths of bipolar spindles were measured.
As previously reported (Maiato et al., 2002; Goshima et al., 2005; Laycock et al., 2006), RNAi of Mast or Msps significantly reduces spindle lengths, whereas RNAi of KLP10A or KLP67A generates long spindles (Figure 4C). The effects of these proteins on spindle length agree with their putative effects on microtubule dynamics. In the cases of the two newly identified flux regulators, KLP67A promotes plus-end disassembly (which inhibits both flux and spindle microtubule growth), whereas Msps probably promotes microtubule dynamic behavior (which is required for flux and microtubule growth).
Next, co-RNAi was used to knockdown pairs of target proteins to determine if spindle lengths, like flux rates, are controlled by the balance of activities regulating microtubule dynamics. The prediction was confirmed: the short spindles created by RNAi of one of the three putative microtubule-stabilizing proteins, Mast, Msps, or EB1, can be rescued by simultaneous RNAi with one of the microtubule-destabilizing proteins, KLP10A or KLP67A (Figure 4C; Supplementary Figure 5C). (Co-RNAi with KLP59C is less effective at rescuing the short spindle phenotype [Supplementary Figure 5C].) A recent study also found that Mast/KLP10A co-RNAi restored a normal spindle length (Laycock et al., 2006).
Notably, some treatments suppress flux but do not affect spindle length. In fact, all of the co-RNAi treatments that produce normal spindle lengths in Figure 4C are ones that also inhibit flux. Therefore, poleward flux is not required to generate spindles of normal length. The separation between flux and spindle length is further demonstrated by the observation that some co-RNAi treatments restore flux to control rates but generate spindles that are either too long (Mast/KLP67A co-RNAi) or too short (EB1/59C co-RNAi; Figure 4C; Supplementary Figure 5C). In summary, variations in flux rate have no consistent, corresponding effect on spindle length. Both flux rate and spindle length depend on the activities of microtubule-dynamics regulators, but flux and spindle length apparently are not codependent.
DISCUSSION
In this study we sought to identify new flux-regulating proteins from a group of candidate proteins that regulate microtubule dynamics and found that the activities of KLP59C, KLP67A, and Msps affect the pace of poleward flux. In addition, our findings suggest a new model for flux regulation, involving a mixture of proteins at minus ends with indispensable activities and other proteins at plus ends whose impacts on flux are colored by their functional interactions (Figure 5).
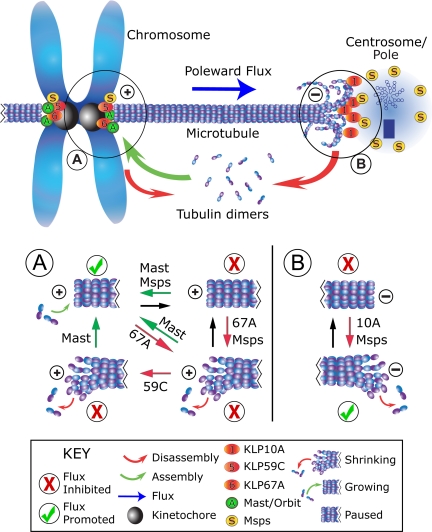
Figure 5.
Model of poleward flux regulation. In this representation of a metaphase half-spindle, flux rate is governed by the rates of gain (green arrow) or loss (red arrows) of tubulin subunits at microtubule ends, whose dynamic states are regulated by flux components (labeled spheres) positioned at either end. Plus-ends are switched between assembly states (i.e., either assembling, paused, or disassembling), subject to the activities of several proteins, but minus ends appear to be influenced by activities that promote disassembly. See figure key (bottom) for identification of markings. (A) At plus ends, flux rate is regulated by Mast and KLP67A, which antagonistically interact to determine the assembly state. KLP59C activity is not required for flux, but does functionally interact with Mast. This situation might arise if KLP59C does not induce catastrophes, but instead operates a step away by maintaining the disassembly state. Conditions that favor or inhibit flux are marked with a checkmark or ×, respectively. (B) At minus ends, flux requires both KLP10A and Msps. KLP10A drives minus-end disassembly. Msps may increase microtubule dynamicity by inhibiting the paused state at both microtubule ends.
We propose that the dynamic behavior of spindle microtubule ends is paramount, and factors that generally promote dynamics will also promote flux, perhaps by predisposing microtubule ends against pausing. Interestingly, Msps has been found to inhibit the paused state of microtubules in interphase S2 cells, by promoting switching from pause to growth and possibly to shrinkage (Brittle and Ohkura, 2005). In addition, the budding yeast orthologue of Msps/XMAP215, Stu2p, was found to increase the dynamics of kinetochore microtubules, by increasing both the rescue and catastrophe frequencies and reducing the pause duration of cytoplasmic microtubules (Kosco et al., 2001). If Msps increases microtubule dynamicity by forcing transitions from the paused state, then Msps could serve as a facilitator to other factors stimulating either growth or shrinkage (Figure 5, A and B).
Next, flux requires that microtubules disassemble at minus ends to allow the sustained, poleward flow of tubulin subunits. In Drosophila, KLP10A at spindle poles serves this necessary function (Figure 5B), and the loss of KLP10A activity cannot be compensated. On the other hand, the process of tubulin subunit incorporation at plus ends is governed by the collective activities of at least three kinetochore/centromeric proteins, Mast, KLP67A, and KLP59C (Figure 5A). Our model of flux regulation appears consistent with recent studies of kinetochore microtubule plus-end dynamics in PtK1 and Drosophila S2 cells, which demonstrated a heterogeneity of dynamic behaviors even in individual kinetochores (Maiato et al., 2006; VandenBeldt et al., 2006). In the context of flux regulation, inhibition of minus-end factors would therefore serve to arrest flux entirely, whereas changes in the relative activities of plus-end factors could generate incremental changes of flux rate.
This model explains the opposite effects of Mast versus KLP67A knockdown on flux rate, and the restoration of normal flux rate when both are simultaneously diminished. But KLP59C RNAi does not significantly affect flux, and yet rescues the flux rate when Mast has also been diminished. How does this occur? KLP59C RNAi has no apparent impact on the localization of Mast or KLP67A and vice versa (Supplementary Figure 4A), and thus regulation via a scheme of targeting codependence is unlikely. During interphase, KLP59C perpetuates microtubule disassembly by suppressing rescues; it does not initiate the process by promoting catastrophes (Mennella et al., 2005). Similarly, mitotic KLP59C may work downstream of the catastrophe-inducer KLP67A, to maintain the disassembly of a plus-end after a catastrophe (Figure 5A).
It merits noting that flux is restored to a near-normal rate after co-RNAi of Mast and either KLP59C or KLP67A. In a condition of depleted Mast, how is the rate of plus-end assembly sufficient to sustain a normal metaphase flux rate? One possibility is that, despite >90% knockdown by RNAi, sufficient residual Mast activity remains after RNAi that plus ends assemble at nearly normal rates if plus-end depolymerizing activity is also reduced. A second possibility is that another microtubule-stabilizing factor is present at plus ends, which restores plus-end assembly. EB1 might serve this purpose. For example, EB1 appears to share Mast's ability to functionally antagonize the activity of KLP67A: the fast-fluxing spindles of KLP67A RNAi-treated cells can be rescued to a near-normal rate by co-RNAi of EB1 (similar to Mast co-RNAi; Supplementary Figure 2). The functions of Mast and EB1 are not completely redundant, though, because we find that Mast RNAi and EB1 RNAi have different impacts on flux and several other mitotic processes.
It is also surprising that knockdown of KLP67A has two apparently incongruent results: slower tubulin turnover at plus ends and a near doubling of flux rate. How can plus-end turnover decrease even as flux is increased? One possibility is that dynamic switching between assembly/disassembly states at plus ends contributes more to turnover than does flux (for example, Gorbsky et al., 1990). If KLP67A increases the catastrophe frequency of plus ends, then a loss of KLP67A would decrease dynamic switching, causing tubulin turnover to decrease even though flux is moving more rapidly. Another possibility is that KLP67A actually stabilizes all or a subset of microtubule plus ends. Though we disfavor the last possibility because it would contradict most published studies of KLP67A activity and would fail to explain other results in this study (e.g., the functional antagonism between KLP67A and Mast), KLP67A has been reported to stabilize microtubules of anaphase central spindles (Gatt et al., 2005).
A final interesting question arising directly from our analysis of flux components is whether and how microtubule plus and minus ends “communicate” during the progression of flux. In particular, the doubling of flux rate after KLP67A RNAi is almost necessarily a consequence of a doubling of the KLP10A-catalyzed minus-end disassembly rate at poles. One simple explanation is that minus-end disassembly normally varies within a discrete range of rates. If the translocation of microtubule toward poles were increased (e.g., as a consequence of increased plus-end assembly and/or increased antiparallel microtubule sliding—though one study makes the latter seem unlikely; Goshima et al., 2005), then this change would be compensated by up to a twofold increase of minus-end disassembly (and flux rate). A decrease in polymer translocation to poles (e.g., decreased plus-end assembly after Mast RNAi) would slow minus-end disassembly and flux. Although this mechanism could buffer the spindle against limited fluctuations of plus-end assembly rate, extreme changes of assembly rate would eventually exceed the capacity of KLP10A to compensate, leading to significant spindle lengthening or collapse.
Finally, our findings suggest a complex and often indirect relationship between flux and several fundamental mitotic processes. Anaphase A appears to be directly linked to flux as any change in flux velocity results in a commensurate alteration of anaphase A. These data strongly support the decades old proposal that flux elicits a poleward pulling forces on anaphase chromosomes (Margolis et al., 1978). However, we find that while flux-regulating proteins also control chromosome congression, interkinetochore tension and spindle length, they do not appear do so directly via flux. This is particularly apparent in the case of chromosome congression. Alterations of flux velocity (or even the abolition of flux altogether) have no influence on congression. But congression does require Mast and Msps, which also promote flux. Perhaps this is because microtubule plus ends must be maintained in a growth state a large proportion of the time or because congression is particularly sensitive to treatments (like Mast depletion) that interfere with microtubule attachment to kinetochores (Maiato et al., 2002).
In conclusion, we hope that our findings can be adapted for analyses of the role of flux in other meiotic/mitotic processes, such as regulation of the spindle checkpoint and correction of errors in chromosome attachment. Proteins regulating microtubule dynamics apparently can have divergent influence on multiple mitotic processes, so flux can operate in parallel (rather than in series) with other mitotic processes. Therefore, future studies of the flux requirements of meiotic/mitotic processes should distinguish between a specific need for flux and the general need for regulated dynamic microtubules. This critical distinction can be made by evaluating mitotic/meiotic processes after the inhibition of various combinations of regulatory proteins to variably alter flux rates. This approach should prove a useful tool to further our understanding of flux and other mechanisms controlling spindle function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ron Vale (UCSF), Steve Rogers (University of North Carolina), and Gohta Goshima (UCSF) for the eGFP-α-tubulin–expressing S2 cells and the KLP67A and EB1 antibodies; Claudio Sunkel (Molecular and Cell Biology Institute, Portugal) for Mast antibody; Jordan Raff (Gurdon Institute, United Kingdom) for Msps antibody; G. Karpen (Salk Institute) for Cid antibody; and Greg Rogers (University North Carolina) for constructive criticism and artistic assistance. This work was supported by grants from the National Institutes of Health to D.J.S., and in part by Philip Morris USA. D.J.S. is a Scholar of the Leukemia and Lymphoma Society.
Abbreviations used:
- FRAP
fluorescence recovery after photobleaching.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-11-0994) on June 6, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Barros T. P., Kinoshita K., Hyman A. A., Raff J. W. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 2005;170:1039–1046. doi: 10.1083/jcb.200504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittle A. L., Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C. F., Deak P., Glover D. M., Ohkura H. Mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R., Bonaccorsi S., Wentworth D., Doxsey S., Gatti M., Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Compton D. A. Functional roles of poleward microtubule flux during mitosis. Cell Cycle. 2006;5:481–485. doi: 10.4161/cc.5.5.2519. [DOI] [PubMed] [Google Scholar]
- Ganem N. J., Upton K., Compton D. A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Gatt M. K., Savoian M. S., Riparbelli M. G., Massarelli C., Callaini G., Glover D. M. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J. Cell Sci. 2005;118:2671–2682. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- Gorbsky G. J., Simerly C., Schatten G., Borisy G. G. Microtubules in the metaphase-arrested mouse oocyte turn over rapidly. Proc. Natl. Acad. Sci. USA. 1990;87:6049–6053. doi: 10.1073/pnas.87.16.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J. M., Vale R. D. Length control of the metaphase spindle. Curr. Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Inoue Y. H., do Carmo Avides M., Shiraki M., Deak P., Yamaguchi M., Nishimoto Y., Matsukage A., Glover D. M. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J. Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. A. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr. Med. Chem. Anticancer Agents. 2002;2:1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Kapoor T. Microtubule flux: what is it good for? Curr. Biol. 2005;15:R966–R968. doi: 10.1016/j.cub.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Kosco K. A., Pearson C. G., Maddox P. S., Wang P. J., Adams I. R., Salmon E. D., Bloom K., Huffaker T. C. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laycock J. E., Savoian M. S., Glover D. M. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J. Cell Sci. 2006;119:2354–2361. doi: 10.1242/jcs.02957. [DOI] [PubMed] [Google Scholar]
- Lee M. J., Gergely F., Jeffers K., Peak-Chew S. Y., Raff J. W. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- Lemos C. L., Sampaio P., Maiato H., Costa M., Omel'yanchuk L. V., Liberal V., Sunkel C. E. Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO J. 2000;19:3668–3682. doi: 10.1093/emboj/19.14.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Fairley E. A., Rieder C. L., Swedlow J. R., Sunkel C. E., Earnshaw W. C. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- Maiato H., Hergert P. J., Moutinho-Pereira S., Dong Y., Vandenbeldt K. J., Rieder C. L., McEwen B. F. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 2006;115:469–480. doi: 10.1007/s00412-006-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Khodjakov A., Rieder C. L. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Sampaio P., Lemos C. L., Findlay J., Carmena M., Earnshaw W. C., Sunkel C. E. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J. Cell Biol. 2002;157:749–760. doi: 10.1083/jcb.200201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis R. L., Wilson L., Kiefer B. I. Mitotic mechanism based on intrinsic microtubule behaviour. Nature. 1978;272:450–452. doi: 10.1038/272450a0. [DOI] [PubMed] [Google Scholar]
- Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J. Mechanism and function of poleward flux in Xenopus extract meiotic spindles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:623–629. doi: 10.1098/rstb.2004.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian M. S., Gatt M. K., Riparbelli M. G., Callaini G., Glover D. M. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J. Cell Sci. 2004;117:3669–3677. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- Tirnauer J. S., Grego S., Salmon E. D., Mitchison T. J. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBeldt K. J., Barnard R. M., Hergert P. J., Meng X., Maiato H., McEwen B. F. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr. Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Mitchison T. J., Rieder C. L., Salmon E. D. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell. 1996;7:1547–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Wittmann T., Hyman A., Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 2001;3:E28–E34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Zhou J., Panda D., Landen J. W., Wilson L., Joshi H. C. Minor alteration of microtubule dynamics causes loss of tension across kinetochore pairs and activates the spindle checkpoint. J. Biol. Chem. 2002;277:17200–17208. doi: 10.1074/jbc.M110369200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.