Abstract
Human PROTEIN ARGININE METHYLTRANSFERASE5 (PRMT5) encodes a type II protein arginine (Arg) methyltransferase and its homologs in animals and yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe) are known to regulate RNA processing, signal transduction, and gene expression. However, PRMT5 homologs in higher plants have not yet been reported and the biological roles of these proteins in plant development remain elusive. Here, using conventional biochemical approaches, we purified a plant histone Arg methyltransferase from cauliflower (Brassica oleracea) that was nearly identical to AtPRMT5, an Arabidopsis (Arabidopsis thaliana) homolog of human PRMT5. AtPRMT5 methylated histone H4, H2A, and myelin basic protein in vitro. Western blot using symmetric dimethyl histone H4 Arg 3-specific antibody and thin-layer chromatography analysis demonstrated that AtPRMT5 is a type II enzyme. Mutations in AtPRMT5 caused pleiotropic developmental defects, including growth retardation, dark green and curled leaves, and FlOWERING LOCUS C (FLC)-dependent delayed flowering. Therefore, the type II protein Arg methyltransferase AtPRMT5 is involved in promotion of vegetative growth and FLC-dependent flowering time regulation in Arabidopsis.
Protein Arg methylation, a posttranslational modification event, plays essential roles in modulating transcription, RNA processing, DNA repair, and signal transduction (Bedford and Richard, 2005; Lee et al., 2005a). According to the biochemical characteristics, protein Arg methyltransferases (PRMTs) are classified into type I and type II enzymes. Type I enzymes form monomethyl-Arg (MMA) and asymmetric dimethyl-Arg (ADMA) residues, whereas type II enzymes catalyze the formation of MMA and symmetric dimethyl-Arg (SDMA) residues. To date, nine human PRMTs (PRMT1–PRMT9) have been identified. Among them, PRMT1, PRMT3, PRMT4/CARM1 (coactivator-associated Arg methyltransferase 1), PRMT6, and PRMT8 are type I enzymes (Bedford and Richard, 2005; Lee et al., 2005b), whereas PRMT5 (Branscombe et al., 2001), PRMT7 (Lee et al., 2005c), and PRMT9 (Cook et al., 2006) belong to the type II group. Human PRMT5 represents the major type II enzyme and methylates histone H2A, H3, H4, and nonhistone proteins, including myelin basic protein (MBP), small nuclear ribonucleoproteins (snRNPs; SmD1 and SmD3), methyl-DNA-binding domain proteins (MBDs), and transcription elongation factor SPT5 (suppressor of Ty 5) (Branscombe et al., 2001; Meister et al., 2001; Kwak et al., 2003; Pal et al., 2004; Ancelin et al., 2006; Tan and Nakielny, 2006). PRMT5 has been implicated in modulating RNA processing, signal transduction, and gene expression (Pollack et al., 1999; Branscombe et al., 2001; Friesen et al., 2001; Fabbrizio et al., 2002; Friesen et al., 2002; Meister and Fischer, 2002; Pal et al., 2004; Ancelin et al., 2006). Unlike PRMT1 and PRMT4, two important type I PRMTs that are associated with gene activation, PRMT5 usually functions in repressing target gene expression (Fabbrizio et al., 2002; Pal et al., 2004; Ancelin et al., 2006).
PRMT5 homologs have been identified in yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe), fruitfly (Drosophila melanogaster), mouse, and filamentous fungus (Gilbreth et al., 1996; Trojer et al., 2004; Ancelin et al., 2006; Gonsalvez et al., 2006). For example, in yeast, null mutants of Skb1 (Shk1 kinase-binding protein1), the yeast homolog of PRMT5, are less elongated in morphology and slower in cell growth rate, whereas cells overexpressing Skb1 are hyperelongated (Gilbreth et al., 1996; Bao et al., 2001). In addition, Skb1 is more important in regulating cell growth and polarity under hyperosmotic stress condition (Bao et al., 2001). DROSOPHILA ARGININE METHYLTRANSFERASE5, the Drosophila ortholog of PRMT5, methylates Sm proteins for snRNP biogenesis and is essential for germ cell specification and maintenance (Gonsalvez et al., 2006). In mouse, PRMT5 has been shown to repress the expression of Dhx38, a DEAH box containing RNA helicase in mouse germ cells (Ancelin et al., 2006).
However, the importance of protein Arg methylation in Arabidopsis (Arabidopsis thaliana) has just emerged and the roles of the PRMT5 counterpart in Arabidopsis remain unclear. Using a biochemical approach, we have previously purified a PLANT HISTONE ARGININE METHYLTRANSFERASE10 (PHRMT10) from cauliflower (Brassica oleracea; L.F. Niu, F.L. Lu, Y.X. Pei, C.Y. Liu, and X.F. Cao, unpublished data). Simultaneously, we successfully purified another histone H4 Arg methyltransferase, PHRMT5, from cauliflower. PHRMT5 and its Arabidopsis homolog AtPRMT5 symmetrically methylate histone H4 at Arg-3. Loss-of-function mutations in AtPRMT5 cause pleiotropic phenotypes. Most interestingly, AtPRMT5 is found to play important roles in promoting seedling growth and the transition from vegetative to reproductive development. These data provide the experimental evidence supporting that the type II PRMT AtPRMT5 is involved in regulating plant vegetative growth and control of flowering time in Arabidopsis.
RESULTS
Purification and Characterization of a Histone H4 Methyltransferase
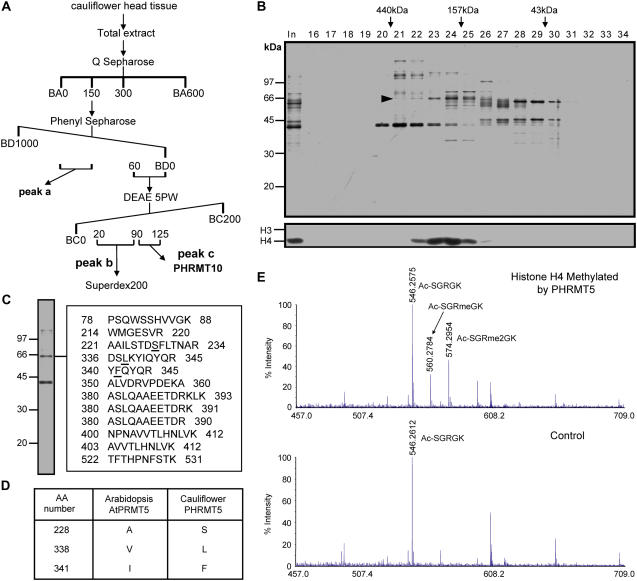
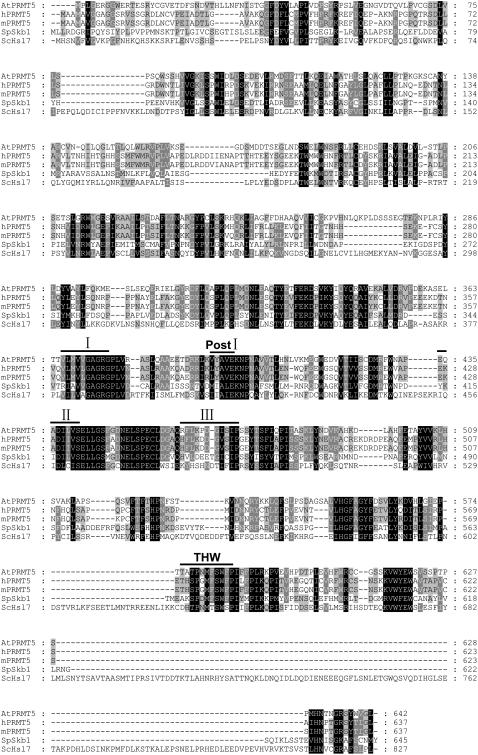
By following histone methyltransferase activity, we have previously purified a histone H4 Arg 3 (H4R3) methyltransferase, PHRMT10, from cauliflower (L.F. Niu, F.L. Lu, Y.X. Pei, C.Y. Liu, and X.F. Cao, unpublished data). During the purification procedure, multiple histone methyltransferase activity peaks were observed. The strongest activity against histone H4 was in peak b (Fig. 1A; data not shown). This activity peak was pooled and further fractionated on a Superdex200 gel filtration column. Silver staining of an SDS-PAGE gel containing the column fractions revealed that a polypeptide band migrating at around 67 kD coeluted with the histone methyltransferase activity (Fig. 1B). This band was excised and subjected to mass spectrometry analysis. Peptides derived from liquid chromatography-tandem mass spectrometry (MSMS) showed the highest similarity to the Arabidopsis PRMT (AtPRMT) encoded by the gene At4g31120 (Fig. 1, C and D). We named this protein PHRMT5 for PLANT HISTONE ARGININE METHYLTRANSFERASE5. Mass spectrometry analysis of histone H4 methylated by PHRMT5 revealed that methylation occurred at histone H4R3 in both mono- and dimethylated manners based on the peptide corresponding to the first five amino acids of histone H4 (Fig. 1E). Compared with human PRMT proteins, AtPRMT coded by At4g31120 showed the highest similarity (47% identical at the amino acid level) to human PRMT5 and we named it AtPRMT5. AtPRMT5 contains 23 exons and encodes a 642-residue protein with conserved catalytic core including methyltransferase region I, posts I, II, III, and THW loop of the PRMT family (Fig. 2).
Figure 1.
Purification of a histone H4 methyltransferase from cauliflower. A, Purification scheme of the histone H4 methyltransferase. Numbers represent salt concentration (mm). BA, BC, and BD refer to buffer A, buffer C, and buffer D, respectively. Details are described in “Materials and Methods.” B, Silver-stained gel (top section) and histone methyltransferase activity against core histone (bottom section) of fractions derived from Superdex200 column. Proteins cofractionated with H4 methyltransferase activity are indicated by an arrowhead. Elution profile of Mr standard is indicated at the top. Numbers on the left indicate Mr standard of SDS-PAGE. In represents input. Numbers above the gel lanes indicate fraction number. C and D, Protein that cofractionated with histone H4 methyltransferase activity, PHRMT5, is nearly identical to AtPRMT5 by mass spectrometry analysis. Peptides derived from MSMS analysis are listed. Numbers represent amino acids number of AtPRMT5. Underlined amino acids are amino acids different between PHRMT5 in cauliflower and AtPRMT5 in Arabidopsis, which are also summarized in D. E, Mass spectrometry analysis of histone H4 methylated by PHRMT5 (top section). Histone H4 in a mock-performed histone methyltransferase activity assay where enzyme was omitted is used as a control (bottom section). Methylated peptides and corresponding unmodified peptide peaks are indicated. Ac-SGRGK, Ac-SGRmeGK, and Ac-SGRme2GK are unmethylated, R3 monomethylated, and R3 dimethylated peptides corresponding to amino acids 1 to 5 of histone H4. Peptide sequences are confirmed by tandem MS analysis (data not shown). [See online article for color version of this figure.]
Figure 2.
Sequence alignment of PRMT5 homologs in Arabidopsis, human, mouse, and yeast. Conserved methyltransferase region I, posts I, II, III, and THW loop are labeled by lines.
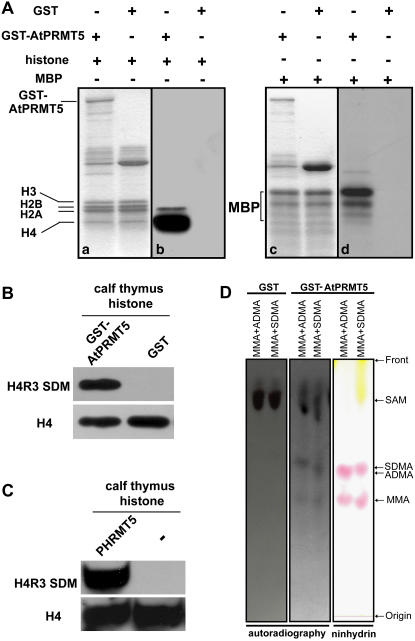
To substantiate the methyltransferase activity of AtPRMT5, recombinant glutathione S-transferase (GST)-AtPRMT5 fusion protein was analyzed. Figure 3A showed both Coomassie blue-stained gels (a and c) and autoradiographs (b and d) of the proteins methylated in the reactions. Among the four core histones (H2A, H2B, H3, and H4) assayed, H4 was preferentially methylated and H2A was also methylated in vitro (Fig. 3A, b). In addition to histones, nonhistone protein MBP was also the substrate of AtPRMT5 (Fig. 3A, d). PHRMT5 mono- and dimethylated histone H4R3 and PRMT5 in animals could symmetrically dimethylate histone H4R3. Therefore, to determine whether AtPRMT5 also showed the same type II methyltransferase specificity, western blotting was performed and calf thymus histone methylated by AtPRMT5 was recognized by the antibody against symmetric dimethyl histone H4R3 (Fig. 3B), as was calf thymus histone methylated by PHRMT5 (Fig. 3C). To further confirm that AtPRMT5 was a type II methyltransferase, a thin-layer chromatography (TLC) assay was performed after acidic hydrolysis of histone H4 that was methylated by AtPRMT5 in the presence of 3H-labeled S-adenosyl-l-Met (SAM). Compared with MMA and either ADMA or SDMA standards, we observed that the 3H-labeled methylated Args comigrated with both MMA and SDMA, but not ADMA (Fig. 3D). Hence, the mass spectrometry analysis, western blot, and TLC analysis support the conclusion that both AtPRMT5 and PHRMT5 mono- and symmetrically dimethylate histone H4 at R3 and that AtPRMT5 is a bona fide type II PRMT.
Figure 3.
Methyltransferase activity and specificity of recombinant AtPRMT5 in vitro. A, Recombinant proteins were incubated with indicated substrates in the presence of the methyl donor S-adenosyl-l-[methyl-3H]. Recombinant GST-AtPRMT5 methylates histone (a and b) and MBP (c and d). Coomassie blue-stained gel is shown on the left (a and c) and the autoradiograph of the gel is shown on the right (b and d). Positions of GST-AtPRMT5, histones, and MBP are indicated. B, Calf thymus histone methylated by GST-AtPRMT5 is probed with antibody against symmetric dimetylated histone H4R3 (Abcam, ab5823). GST is used as a negative control. The membrane is stripped and reprobed with antibody against histone H4 to serve as a loading control. C, Calf thymus histone methylated by PHRMT5 is probed with antibody against symmetric dimethyl histone H4R3 (Abcam, ab5823). Reaction performed in the absence of PHRMT5, which is indicated as “−” is used as a negative control. The membrane is stripped and reprobed with antibody against histone H4 to serve as a loading control. D, TLC analysis of H4R3 methylated by AtPRMT5. The positions of MMA, ADMA, and SDMA are visualized by ninhydrin staining (right section). Autoradiography of this plate shows that the radio activities comigrated with ninhydrin-stained MMA and SDMA but not ADMA (middle section). Autoradiography of GST alone used as negative control is shown on the left. Free 3H-labeled SAM is indicated. Separation starting site is indicated as Origin and solvent front is indicated as Front. [See online article for color version of this figure.]
Isolation and Identification of atprmt5 Mutants
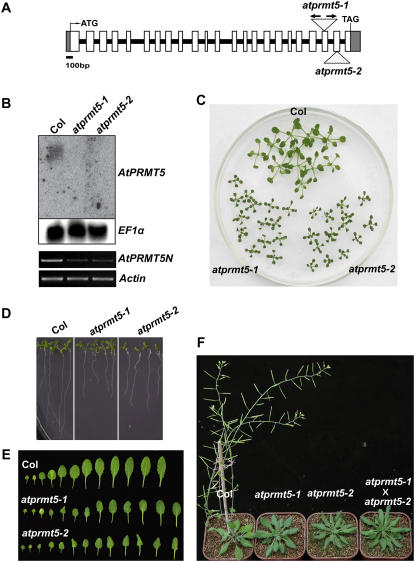
To investigate the functions of AtPRMT5 in growth and development in Arabidopsis, we searched for atprmt5 T-DNA insertion mutant alleles and obtained atprmt5-1 and atprmt5-2. The atprmt5-1 mutant contains two inverted T-DNA insertions in the 21st exon, while atprmt5-2 has an insertion in the 22nd exon (Fig. 4A). Each insertion was confirmed by PCR and sequencing analysis. In contrast to the RNA-blot analysis in which neither full-length nor truncated form of AtPRMT5 transcript was detectable in atprmt5 mutants (Fig. 4B, top sections), low levels of an N-terminally truncated form of AtPRMT5 were detected (Fig. 4B, bottom sections), indicating that atprmt5-1 and atprmt5-2 might not be null alleles.
Figure 4.
AtPRMT5 gene structure, expression, and pleiotropic phenotypes in atprmt5-1 and atprmt5-2 mutants. A, AtPRMT5 gene structure and the T-DNA insertion sites of SALK lines. White boxes and gray boxes represent exons and 5′ or 3′ untranslated region, respectively. Lines mean introns. The T-DNA insertion sites in each mutant are indicated by triangles. In atprmt5-1 mutant, arrows represent two inverted T-DNA insertions. B, AtPRMT5 full-length but not the N-terminal transcript is absent in the atprmt5 mutants. Top sections: The total RNA from seedlings with four to five rosette leaves of atprmt5 mutants and wild-type Col is probed by RNA blot with full-length coding sequence of AtPRMT5. EF1α gene is used as a control for constitutive expression. Bottom sections: RT-PCR analyses were performed for the N-terminally expressed AtPRMT5 (AtPRMT5N) in wild-type Col and atprmt5-1 and atprmt5-2 mutants. Equal amounts of cDNAs were determined by RT-PCR at the Actin locus. C to F, Pleiotropic phenotypes of atprmt5 mutants. Growth retardation of young seedling leaves at 12 d (C) and primary roots at 9 d (D). E, Comparison of rosette leaves between wild-type Col and atprmt5 plants. F, atprmt5 mutants are late flowering. Plants shown here are 8 weeks old grown at 23°C under LD. aprmt5-1 × aprmt5-2 means F1 progeny from the crosses between aprmt5-1 and aprmt5-2.
Lesions in AtPRMT5 Cause Pleiotropic Developmental Defects in Arabidopsis
Compared with the wild-type accession Columbia (Col), atprmt5 mutants showed pleiotropic phenotypes. The atprmt5 mutants displayed growth retardation such that both the size of cotyledons and rosette leaves and the length of primary roots in atprmt5-1 and atprmt5-2 mutants were strikingly smaller and shorter than those of wild-type Col during early vegetative development (Fig. 4, C and D). The leaves of atprmt5 mutants continued to expand and eventually reached the same size as Col leaves. In addition to growth retardation, some of the early juvenile leaves were curled in atprmt5 mutants as compared with wild-type Col (Fig. 4E). At later developmental stages, when the leaves of wild-type Col had senesced, atprmt5 leaves were more curled and dark green in color. Most obviously, mutations in AtPRMT5 led to dramatically delayed flowering time and increased number of rosette and cauline leaves, indicating that AtPRMT5 was involved in regulating the transition from vegetative to reproductive development (Fig. 4F).
To characterize the atprmt5 mutants genetically, F1 progenies from crosses between wild-type Col and atprmt5 mutants were generated. All F1 plants displayed normal developmental phenotypes. In the F2 generation, plants exhibited a 1:3 segregation ratio of mutant to wild-type phenotypes and all plants showing the mutant phenotypes were atprmt5 homozygous. These genetic analyses revealed that atprmt5 mutants were recessive and controlled by a single locus. F1 progenies (atpmrmt5-1 × atprmt5-2) from reciprocal crosses between atprmt5-1 and atprmt5-2 displayed the same phenotype as their parents at both early and late developmental stages (Fig. 4F; data not shown), indicating that atprmt5-1and atprmt5-2 mutants are allelic. An extra copy of genomic DNA from AtPRMT5 rescued atprmt5 mutant phenotypes, indicating that the above pleiotropic phenotypes indeed were caused by disruption of AtPRMT5 (data not shown).
AtPRMT5 Regulates Flowering Time through the Autonomous Pathway
The floral transition is one of the most important plant developmental processes that influence reproductive success. In Arabidopsis, flowering time is regulated by four major pathways, including photoperiod, gibberellin (GA), vernalization, and autonomous pathways (Simpson et al., 1999; Reeves and Coupland, 2000; Sheldon et al., 2000a; Mouradov et al., 2002; Finnegan et al., 2004; Komeda, 2004; Amasino, 2005; Dennis et al., 2006). To investigate the role of AtPRMT5 in the transition from vegetative to reproductive development, we first compared the flowering time of wild type with that of atprmt5 mutants grown under both long days (LDs; 16-h light and 8-h dark) and short days (SDs; 8-h light and 16-h dark). Numbers of rosette and cauline leaves were counted after plants had flowered. Compared with Col, atprmt5-1 and atprmt5-2 mutants showed delayed flowering under both LDs and SDs (Fig. 5A), indicating that the late-flowering phenotype of atprmt5 is not mediated by the photoperiod pathway. To examine the effect of exogenous GA on the flowering time of atprmt5 mutants under SDs, atprmt5 mutants and wild-type Col were sprayed with a solution of 20 μm GA twice a week until they flowered. The atprmt5 plants displayed a normal response to GA treatment. To explore the effect of vernalization on flowering time of atprmt5 mutants, atprmt5 mutants and wild-type Col were grown under LD after vernalization for 6 weeks at 2°C to 6°C and leaf numbers at flowering were counted. Vernalization treatment promoted flowering in both wild-type and atprmt5 plants, indicating that AtPRMT5 affected flowering time in a vernalization-independent manner.
Figure 5.
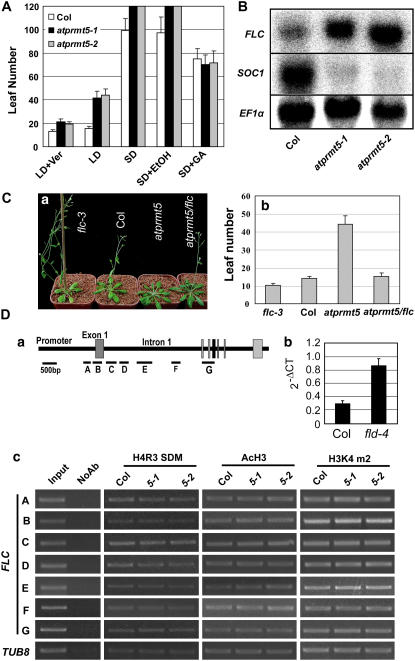
Late flowering of atprmt5 is FLC dependent. A, Flowering time of atprmt5 mutants is assessed by total leaf number after plants stop producing new leaves under different conditions or treatments. LD + Ver (vernalization) treatment means plants grown at 4°C under SDs for 6 weeks before transferred to 23°C under LD. SD + GA indicates GA treatment under SDs. SD + ethanol is as a control. Bars represent sd. Under SD or SD + ethanol conditions, atprmt5 plants did not yet flower after producing 120 leaves and the experiment was terminated at this point. Bars represent sd. B, RNA-blot analysis of mRNA level of FLC and SOC1 in seedlings with four to five rosette leaves of wild-type Col and atprmt5 plants. EF1α is used as an internal control for constitutive expression. C, Phenotype (a) and flowering time (b) of atprmt5/flc-3 double mutant grown at 23°C under LD. Bars represent sd. D, Chromatin modifications at FLC locus. a, Diagram of FLC genomic DNA. A to G regions are amplified in ChIP assay (Bastow et al., 2004). b, Positive control ChIP assay. ChIP-PCRs of FLC-C are shown from immunoprecipitated by anti-AcH3 antibody using seedlings of Col and fld-4 mutants. The quantitative PCR results were shown in the right. c, Representative ChIP-PCR results are shown. Input is chromatin before immunoprecipitation that is diluted 25 times for PCR amplification. NoAb refers to precipitation performed without antibody. AcH3, H3K4 m2, and H4R3 SDM refer to the corresponding immunoprecipitations using antibodies against acetylated histone H3, dimethylated histone H3 at K4, and symmetric dimethylation at histone H4R3, respectively. FLC-A to FLC-G represent different regions of FLC chromatin (Bastow et al., 2004) and the diagram is shown at the top. TUB8 is used as an internal control. [See online article for color version of this figure.]
All above physiological characters of atprmt5 mutants were similar to those late-flowering mutants in the autonomous pathway. Autonomous pathway genes control flowering time mainly through repressing the major flowering repressor FLOWERING LOCUS C (FLC), a MADS-box transcription factor that quantitatively blocks the floral transition in Arabidopsis (Michaels and Amasino, 1999; Sheldon et al., 1999, 2000b, 2002, 2006; Rouse et al., 2002; Liu et al., 2004; Jean Finnegan et al., 2005). Therefore, to investigate whether expression of FLC was controlled by AtPRMT5, we examined mRNA levels of FLC in wild-type Col and atprmt5 plants. Expression of FLC was dramatically increased due to mutations in AtPRMT5 (Fig. 5B, top section). As a result, expression of SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1)/AGAMOUS-LIKE20, a floral integrator downstream of FLC, was greatly reduced (Fig. 5B, middle section). We also generated double mutants between atprmt5 and flc-3, a null allele mutant of FLC (Michaels and Amasino, 1999). Homozygous progeny of atprmt5/flc double mutants flowered significantly earlier than atprmt5 (Fig. 5C, a and b). Taken together, we concluded that AtPRMT5 regulates flowering time through the autonomous pathway and the late-flowering phenotype of atprmt5 is FLC dependent.
In humans, PRMT5 has been implicated in transcriptional repression of the cyclin E promoter by symmetric dimethylation of histone H4R3 (Fabbrizio et al., 2002). Since AtPRMT5 possesses similar enzymatic features to PRMT5 and the expression of FLC was increased in atprmt5 mutants, we wondered whether AtPRMT5 affected the histone modifications of FLC chromatin. Therefore, chromatin immunoprecipitation (ChIP) assays were performed to analyze chromatin modifications of FLC in Col and atprmt5 plants (Fig. 5D). The chromatin fragments from wild-type and atprmt5 mutants were immunoprecipitated with antibodies against symmetrical dimethyl H4R3, acetyl-histone H3 (AcH3), and dimethyl histone H3K4, respectively. Following isolation of precipitated DNA, seven distinct regions of the FLC-chromatin were amplified (Fig. 5D, a; Bastow et al., 2004). FLC-C region in fld was enriched by AcH3 antibody, indicating that ChIP procedures were appropriate (Fig. 5D, b). When compared with wild-type control, all histone modifications tested were not significantly changed in atprmt5 mutants at these regions (Fig. 5D, c).
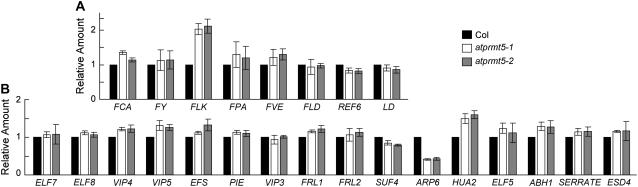
In addition to FLC, it was possible that upstream regulators of FLC were affected by AtPRMT5. To test this possibility, real-time PCR was performed to evaluate transcript levels of FLC modulators. Among eight repressors and 16 activators of FLC, only the mRNA level of FLOWERING LOCUS K (FLK), a component of autonomous pathway, was increased by 2-fold, and ARP6 (ACTIN-RELATED PROTEIN6), an activator of FLC, was decreased by 2-fold in atprmt5 mutants as compared to wild-type Col (Fig. 6).
Figure 6.
Expression levels of upstream regulators of FLC in atprmt5 plants. mRNA expression levels of FLC repressors (A) and FLC activators (B) are analyzed by real-time PCR. Relative amount of RNA levels normalized to UBQ (A) or Actin (B) is shown. Bars represent sd. For FCA, δ and γ forms are detected.
DISCUSSION
In this study, we purified PHRMT5 from cauliflower, a histone H4 methyltransferase that confers the strongest histone H4 methyltransferase activity. Peptide sequences obtained by mass spectrometry showed that PHRMT5 is an ortholog of Arabidopsis AtPRMT5. In vitro methyltransferase activity assays showed that recombinant GST-AtPRMT5 methylates histone H4, H2A, and MBP, similar to PRMT5 in animals. In addition, recombinant AtPRMT5 and purified PHRMT5 catalyze symmetric dimethylation at histone H4R3, indicating that AtPRMT5 and PHRMT5 are type II PRMTs. Mutations in AtPRMT5 cause pleiotropic developmental defects, including seedlings growth retardation, darker green color, curled leaves, and delayed flowering, which indicates that AtPRMT5 may be involved in regulating fundamental cellular processes and development in Arabidopsis.
At the young juvenile stage, a striking defect of atprmt5 plants is the growth retardation, which indicates that wild-type AtPRMT5 enhances vegetative development. Consistent with this phenotype, yeast cells carrying the mutation in Skb1 are less elongated in morphology and show a slower rate of cell growth (Gilbreth et al., 1996; Bao et al., 2001). Furthermore, the in vitro methyltransferase activity and specificity of AtPRMT5 are similar to those of PRMT5 in animals, suggesting a conserved function.
The regulation of flowering time by developmental signals and external environmental clues is a complex biological process and can be regulated at transcriptional and posttranscriptional levels (Henderson and Dean, 2004; Simpson et al., 2004; Fischle et al., 2005). In this study, we demonstrate that lesions in AtPRMT5, the major enzyme responsible for symmetrically dimethylated histone H4R3, display features of autonomous pathway mutants. Arabidopsis is a LD plant and LD promotes flowering in wild-type plants. atprmt5 mutants flower later than wild-type Col under both LD and SD, indicating that atprmt5 responds normally to photoperiod. Moreover, plant hormone GA is essential to flowering under SD and atprmt5 plants also display a normal response to GA treatment. In addition, this late-flowering phenotype of atprmt5 can be rescued by a prolonged cold exposure (vernalization). All the above-mentioned characteristics of atprmt5 mutants resemble those of autonomous pathway mutants in Arabidopsis. We further observe that the mRNA level of FLC is up-regulated and loss of function of FLC suppresses the late-flowering phenotype of atprmt5 plants.
Although FLC mRNA levels were up-regulated in atprmt5 plants, we did not observe any significant difference in histone modifications between wild-type and atprmt5 plants at FLC locus under our conditions (12-d-old seedlings grown at 23°C under LD condition; Fig. 5, B and D). A similar situation was observed in loss of function of a histone acetyltransferase (Deng et al., 2007; Han et al., 2007). In Arabidopsis, mutations in HAC1 cause pleiotropic developmental defects, including shortened primary roots, partially reduced fertility, and an FLC-dependent late flowering (Deng et al., 2007). However, chromatin modifications of FLC are not altered in hac1 as compared to wild type, although mRNA level of FLC is increased greatly in hac1 mutants. One possibility to explain why no changes of chromatin modifications were detected between the wild-type Col and atprmt5 mutants is that atprmt5-1 and atprmt5-2 are not null alleles. Another possibility is that the mark of symmetric dimethyl H4R3 at FLC only occurs at certain developmental stages but not the stage we examined. Alternatively, FLC is not the direct target of AtPRMT5. In this case, AtPRMT5 could modulate FLC expression through a histone Arg methylation-independent mechanism. In animals, in addition to histones, PRMT5 also methylates MBP, snRNPs SmD1 and SmD3, MBD, and SPT5 (Branscombe et al., 2001; Meister et al., 2001; Kwak et al., 2003; Pal et al., 2004; Ancelin et al., 2006; Tan and Nakielny, 2006), many of which are involved in RNA metabolism. Therefore, AtPRMT5 may also function at posttranslational level by methylating proteins other than histones, which may affect FLK and ARP6 at posttranscriptional level to modulate FLC expression. In combination of our biochemical and genetic data, we demonstrate that type II PRMTs are involved in regulation of flowering time in an FLC-dependent manner. Therefore, Arg methylation confers an extra layer of complexity to flowering time regulation in Arabidopsis. Further studies aimed at dissecting the molecular mechanism of AtPRMT5 in the FLC-dependent pathway will be of interest.
MATERIALS AND METHODS
Purification of the Histone H4 Methyltransferase
One kilogram upper layer of cauliflower (Brassica oleracea) was homogenized in a waring blender in liquid nitrogen and further in 1 L buffer A [50 mm Tris-HCl, 0.1 mm EDTA, 20 mm (NH4)2SO4, 25% (v/v) glycerol, 2 mm dithiothreitol (DTT), and 0.2 mm phenylmethylsulfonyl fluoride (PMSF), pH 8.0] supplemented with protease inhibitor cocktail (Roche) for three times of 10 s bursts (Chamovitz et al., 1996). Insoluble substance was removed by filtering through four layers of cheesecloth followed by centrifuging at 18,800g for 90 min. After conductivity and pH adjusted to buffer A using Tris base and distilled, deionized water, the supernatant was filtered through a 0.45 μm filter and applied to a 270 mL Q Sepharose column (Amersham Biosciences) equilibrated in buffer A. Bound protein was step eluted with BA150 (buffer A supplied with 150 mm NaCl), BA300, and BA600. Solid ammonium sulfate was gradually added to BA150 fraction to a final concentration of 1 m. The mixture was stirred for 1 h at 4°C and cleared by centrifuging at 15,000g for 15 min. The supernatant was loaded on to a 53 mL phenyl Sepharose column equilibrated in BD1000 (buffer D: 50 mm Tris-HCl, 0.1 mm EDTA, 10% [v/v] glycerol, pH 8.0, 2 mm DTT, 0.2 mm PMSF) with 1 m ammonium sulfate and eluted by a linear gradient to BD0. The second H4 histone methyltransferase activity peak was pooled, diluted with 1 volume of buffer C, and loaded onto a 3.3 mL DEAE-5PW column (TOSOH) equilibrated in buffer C (50 mm Tris-HCl, 0.1 mm EDTA, 10% [v/v] glycerol, pH 8.0, 2 mm DTT, 0.2 mm PMSF). Bound protein was eluted by a linear gradient from BC30 to BC200 followed by linear gradient to BC1000. The first H4 methyltransferase activity peak was pooled, concentrated, and loaded onto a 24 mL Superdex200 (Amersham Biosciences) column equilibrated in buffer BC150. Fractions were used for silver staining and activity analysis. Samples were concentrated and loaded onto a preparative gel for mass spectrometry analysis.
Protein Identification
To identify the 65 kD band that coeluted with H4 histone methyltransferase, the Coomassie-stained 65 kD band was excised, digested with trypsin, and subjected to liquid chromatography-MSMS analysis according to standard procedure. DTA files for MSMS spectra were generated by Bioworks software (ThermoFinnigan) and searched against Arabidopsis (Arabidopsis thaliana) database using the Global Proteome Machine search engine (Craig et al., 2004). Point mutations were allowed during model refinement. The identified protein of interest was further manually confirmed.
Methyltransferase Activity Assay
Full-length AtPRMT5 coding sequence was amplified from cDNA made from the inflorescence of wild-type Col Arabidopsis. Primers used for AtPRMT5 cloning were cx1108 (5′-CATGCCATGGATATGCCGCTCGGAGAGAGAGGAGGATGGG-3′) and cx0927 (5′-TCCCCCGGGAAAAGTCTCTAATCCTAAAGGC-3′). Recombinant GST-AtPRMT5 was expressed in Escherichia coli strain BL21 (RIL) and purified using glutathione Sepharose beads (Amersham Biosciences). Purified recombinant protein or column fractions were incubated with indicated substrates in the presence of the methyl donor, S-adenosyl-l-[methyl-3H]Met (Amersham Biosciences), in HMT buffer (20 mm Tris-HCl, pH 8.0, 4 mm EDTA, 1 mm PMSF, 0.5 mm DTT) for 1 to 3 h at 30°C. Proteins were separated by 15% SDS-PAGE and visualized by Coomassie staining. Gels were then treated with Amplifier (Amersham Biosciences) for 15 to 30 min, dried, and exposed to Kodak Biomax MS film at −80°C for the appropriate time.
Western-Blot Analysis
Proteins were separated by 10% or 15% SDS-PAGE and transferred to Hybond ECL membrane (Amersham Biosciences) or Immobilon P SQ polyvinylidene difluoride membrane (Millipore) for histones. Supersignal West Dura substrate (Pierce) was used to detect the HRP-conjugated secondary antibodies. Antibodies used for western blot are antisymmetric dimethyl-H4R3 (H3R3 SDM; Abcam, ab5823) and anti-histone H4 (Upstate, 07-108).
TLC Analysis
In vitro histone methylation reactions (50 μL) were performed with 3H-labeled SAM (Amersham biosciences), calf thymus histone H4 (Roche), and appropriate enzymes. Methylation reaction mixtures were hydrolyzed in ampule tubes by adding 5 volumes 6 n HCl (250 μL). The tubes were heat sealed and incubated at 110°C for 24 h. The reaction mixtures were then dried completely on a heat block set at 90°C. Twenty microliters of water was added into each tube and the tubes were let stand for about 30 min. Twenty-five percent hydrolyzed amino acids (5 μL) were mixed with 30 nmol Arg standards (either SDMA plus MMA or ADMA plus MMA) and spotted onto TLC plate (silica gel 60 F254 with concentrating zone, Merck). The TLC plate was run with ammonium hydroxide:chloroform:methanol:water (2:0.5:4.5:1) as previously reported (Friesen et al., 2001). Standards were visualized by ninhydrin (0.1% ninhydrin in ethanol) staining and 3H labeled Args were visualized by exposing dried plate to Biomax film with Biomax transcreen LE intensifying screen (Kodak) for the appropriate time.
Plant Materials and Growth Conditions
All the Arabidopsis lines used in this study were the Col ecotype. flc-3 and fld-4 seeds are the kind gifts of Dr. R. Amasino. For flowering time measurements, plants were grown side by side in Versatile Environmental test chamber (MLR-350H, SANYO) at 23°C under 16 h of light followed by 8 h of darkness for LD or 8 h of light followed by 16 h of darkness for SD. For vernalization treatment, atprmt5 mutants and wild-type plants were grown in soil side by side at 2°C to 6°C for 6 weeks under SD before transferred to 23°C under LD condition. Nonvernalized control plants were grown under SD at 2°C to 6°C for 3 d before being transferred to 23°C under LD condition. For GA treatment, 20 μm GA3 solution was sprayed twice a week under SD until flowering. GA3 was dissolved in ethanol, therefore, equal amounts of ethanol was sprayed as untreated control. Flowering time was measured by counting the total numbers of rosette and cauline leaves at flowering.
Identification of T-DNA Insertion Mutants
atprmt5-1 (SALK_065814) and atprmt5-2 (SALK_095085) mutants were obtained from the SALK collection (http://signal.salk.edu/). T-DNA border primer cx101 LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) or cx102 LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) was used to amplify DNA from mutants. Gene-specific primers were cx166 (5′-TCTTGTGACAAAAATACAGCACAA-3′) and cx167 (5′-CATCCATTTGGCAGGTTAAGGC-3′) for atprmt5-1 and cx168 (5′-ATGTTTTGTCAAACGGCCGAG-3′) and cx169 (5′-TCTACCAAGCGATGCTGGCTC-3′) for atprmt5-2. The flc-3 seed was a gift from Dr. R. Amasino. Primers used for flc-3 genotyping were cx743 (5′-TTGCATCACTCTCGTTTACCC-3′) and cx744 (5′-GCGTCACAGAGAACAGAAAGC-3′). T-DNA insertion fragments from PCR were further confirmed by sequencing.
Analysis of Transcript Levels
mRNA expression levels were measured either by real-time PCR or by RNA gel blot. Seedlings with four to five rosette leaves of Col and atprmt5 plants grown in Murashige and Skoog plates with 3% Suc were used to extract total RNA (Liu et al., 2005). About 20 μg total RNA of each sample was separated in 1% agarose/formaldehyde gels and transferred onto N+ nylon membrane (Hybond-N+, Amersham-Pharmacia). EF1α was used as a constitutive expression control. The primers for the probe template amplification were as follows: cx1454 (5′-ACGCGTCGACATGCCGCTCGGAGAGAGAGGAGGATGGG-3′) and cx1456 (5′-ACGCGTCGACCTAAAGGCCAACCCAGTACGAACG-3′) for AtPRMT5; cx461 (5′-CGCGGATCCATGGGAAGAAAAAAACTAGA-3′) and cx462 (5′-CCGGAATTCCTAATTAAGTAGTGGGAGAGT-3′) for FLC; cx992 (5′-CGCGGATCCATGGTGAGGGGCAAAACTCAGATG-3′) and cx993 (5′-CGCGGATCCCATTCTAGAGAGGCAAGTGTAAGAA-3′) for SOC1; cx1561 (5′-ATGCCCCAGGACATCGTGATTTCAT-3′) and cx1562 (5′-TTGGCGGCACCCTTAGCTGGATCA-3′) for EF1α. Reverse transcription (RT)-PCR analyses were performed for the N-terminally expressed AtPRMT5 (AtPRMT5N) with the following primers: cx1454 (5′-ACGCGTCGACATGCCGCTCGGAGAGAGAGGAGGATGGG-3′) and cx2421 (5′-CCTGGGCAGCATGATCAAAA-3′). For the internal control of Actin2/7 gene, the following primer pairs were used: cx415 (5′-CTCAGCACCTTCCAACAGATGTGGA-3′) and cx416 (5′-CCAAAAAAATGAACCAAGGACCAAA-3′). Real-time PCR was performed using the Chromo4 Real Time PCR instrument (MJ) and SYBR Green I (Invitrogen, S-7567). Primers for real-time PCR of autonomous pathway genes were described in Deng et al. (2007). Cx0773 (5′-GAACTGGACAGCAGCAAGGCTGTTG-3′) and cx1691 (5′-AGGGTGCCTATGCGTTCTCTCTCC-3′) were used for FCA δ and the functional form FCA γ detection. Primers for real-time PCR of FLC activator genes were as follows: cx2217 (5′-CGGCTCCCAGATCCAACTAAAGTG-3′) and cx2218 (5′-CTCCATGTGAGGCCATATCTCGCT-3′) for ABH1; cx2219 (5′-GTACCTGAGACGTTATTCCAGCCTGC-3′) and cx2220 (5′-CTCCATACACCTAGTATGGGGTCCTC-3′) for ARP6; cx2221 (5′-CCTCCTTTTGCAGGGATGGAAAGGTT-3′) and cx2222 (5′-TGGTTCTGCAGGTCTTGGGGACTGAT-3′) for EFS; cx2223 (5′-CACACCTGAGCTTACATCCATGGTCCC-3′) and cx2224 (5′-GTGCGCCAAGAGCTTTCATGTCCTCTA-3′) for ELF5; cx2225 (5′-CCTGGCACATACCTGGTATCATTTGAC-3′) and cx2226 (5′-ACCAACCCTAGAAGAGTAAACCCCTGA-3′) for ELF7; cx2227 (5′-GAGGACGATGAAGAAGAAGCTGCCACTA-3′) and cx2228 (5′-CGCCTTACACCCGAAGTAGGTACCTCAT-3′) for ELF8; cx2229 (5′-GCTCTGGCAAAATACATGGGTGATGAAG-3′) and cx2230 (5′-GAAGTATGGCATGTGTTCCTGGCTGAA-3′) for ESD4; cx2231 (5′-TCACAAGTTCCAGTTCCGAGTCAGCA-3′) and cx2232 (5′-TTGAGGCGGCAACACATATCCAGTTT-3′) for FRL1; cx2233 (5′-TACTTCCTGGTGTTGCTGTCCCATAT-3′) and cx2234 (5′-TAGAGAGCATAGACCTGTTCGAGCAG-3′) for FRL2; cx2235 (5′-ACACAAGAGCTCCAATAGGTCAAGGA-3′) and cx2236 (5′-AGTTAACGTAAGGCCATGTCTCTTGC-3′) for VIP5; cx2250 (5′-CTTACCCTTTCCCACCTCAACGTGAT-3′) and cx2238 (5′-ATCTGGGGAGCAGTATGACCTGGAAT-3′) for HUA2; cx2239 (5′-TGCTGCAGAGAACCGTTACAGGAATG-3′) and cx2240 (5′-GAGATTCGGATCATTTGGCCTAACCC-3′) for PIE1; cx2241 (5′-CGGATATAAACCCACCACCAATGCTG-3′) and cx2242 (5′-AGCATCTAGGTCTTGGTAGCTGCGCA-3′) for SERRATE; cx2243 (5′-CATTCTTATGCCTCTGGCCCAAACACT-3′) and cx2244 (5′-CTGCATTTATCGAGTTCATCTGGCTGG-3′) for SUF4; cx2245 (5′-TGGGGAATGCAATTTGAACCTAAGGGT-3′) and cx2246 (5′-TCGTACCATCCATTGAACCACAAGCAA-3′) for VIP3; cx2247 (5′-GCCAGACCGTCAAGACGTCAAATGGAG-3′) and cx2248 (5′-ATCCTTCTCTGCCCTACCTCCCGCAA-3′) for VIP4; cx1730 (5′-GGGCACTCAAGTATCTTGTTAGC-3′) and cx1580 (5′-TGCTGCCCAACATCAGGTT-3′) for Ubiquitin (UBQ); and cx1722 (5′-GGAACTGGAATGGTGAAGGCTG-3′) and cx1723 (5′-CGATTGGATACTTCAGAGTGAGGA-3′) for Actin-specific gene expression was normalized to the housekeeping gene UBQ or Actin.
ChIP
Twelve- or 14-d-old plants with four to five true leaves were harvested from Murashige and Skoog plates containing 3% Suc grown under LD at 23°C. ChIP assay was performed as previously described (Johnson et al., 2002; Gendrel et al., 2005). Immunoprecipitations were performed with antisymmetric dimethyl-H4R3 (Abcam, ab5823), anti-AcH3 (Upstate; 06-599), and antidimethyl-histone H3K4 (Upstate, 07-030) antibodies. The PCR analysis was performed using specific primers corresponding to different regions of FLC chromatin (Bastow et al., 2004). TUB was used as an internal control (Mathieu et al., 2005). Input samples were diluted 25 times before PCR amplification. The cycle numbers of PCR were 30 to 32 for AcH3 and dimethyl-H3K4, and 33 to 35 for dimethyl H4R3 of FLC chromatin.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtPRMT5: NP_194841; hPRMT5: NP_006100; mPRMT5: NP_038796; SpSkb1: P78963; and ScHs17: NP_009691.
Note Added in Proof
Wang et al. (Wang X, Zhang Y, Ma Q, Zhang Z, Xue Y, Bao S, Chong K [2007] SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J 26: 1934–1941) have also found that SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis.
Acknowledgments
We thank Dr. Haiyan Zheng (Robert Wood Johnson Medical School, Piscataway, NJ) for conducting the mass spectrometric analysis, Dr. R. Amasino (University of Wisconsin, Madison, WI) for providing flc-3 and fld-4 seeds, Dr. Ian Henderson (University of California, Los Angeles) for English editing, and the Arabidopsis Biological Resources Center at Ohio State for providing SALK T-DNA-insertion lines.
This work was supported by the National Basic Research Program of China (grant no. 2005CB522400 to X.C.), by the National Natural Science Foundation of China (grant nos. 30430410, 30621001, and 30325015 to X.C. and 30571032 to C.L.), and by the Chinese Academy of Sciences (grant no. CXTD–S2005–2).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xiaofeng Cao (xfcao@genetics.ac.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Amasino RM (2005) Vernalization and flowering time. Curr Opin Biotechnol 16 154–158 [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8 623–630 [DOI] [PubMed] [Google Scholar]
- Bao S, Qyang Y, Yang P, Kim H, Du H, Bartholomeusz G, Henkel J, Pimental R, Verde F, Marcus S (2001) The highly conserved protein methyltransferase, Skb1, is a mediator of hyperosmotic stress response in the fission yeast Schizosaccharomyces pombe. J Biol Chem 276 14549–14552 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18 263–272 [DOI] [PubMed] [Google Scholar]
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S (2001) PRMT5 (janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem 276 32971–32976 [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121 [DOI] [PubMed] [Google Scholar]
- Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S (2006) FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun 342 472–481 [DOI] [PubMed] [Google Scholar]
- Craig R, Cortens JP, Beavis RC (2004) Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 3 1234–1242 [DOI] [PubMed] [Google Scholar]
- Deng W, Liu C, Pei Y, Deng X, Niu L, Cao X (2007) Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLC in Arabidopsis. Plant Physiol 143 1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Helliwell CA, Peacock WJ (2006) Vernalization: spring into flowering. Dev Cell 11 1–2 [DOI] [PubMed] [Google Scholar]
- Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, et al (2002) Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep 3 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Sheldon CC, Jardinaud F, Peacock WJ, Dennis ES (2004) A cluster of Arabidopsis genes with a coordinate response to an environmental stimulus. Curr Biol 14 911–916 [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438 1116–1122 [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G (2001) The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 21 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G (2002) A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem 277 8243–8247 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2 213–218 [DOI] [PubMed] [Google Scholar]
- Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb MH, Marcus S (1996) The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc Natl Acad Sci USA 93 13802–13807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG (2006) The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol 16 1077–1089 [DOI] [PubMed] [Google Scholar]
- Han SK, Song JD, Noh YS, Noh B (2007) Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J 49 103–114 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Dean C (2004) Control of Arabidopsis flowering: the chill before the bloom. Development 131 3829–3838 [DOI] [PubMed] [Google Scholar]
- Jean Finnegan E, Kovac KA, Jaligot E, Sheldon CC, James Peacock W, Dennis ES (2005) The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J 44 420–432 [DOI] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S (2002) Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12 1360–1367 [DOI] [PubMed] [Google Scholar]
- Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55 521–535 [DOI] [PubMed] [Google Scholar]
- Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor RB (2003) Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell 11 1055–1066 [DOI] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR (2005. a) Role of protein methylation in regulation of transcription. Endocr Rev 26 147–170 [DOI] [PubMed] [Google Scholar]
- Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT (2005. b) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem 280 32890–32896 [DOI] [PubMed] [Google Scholar]
- Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S (2005. c) PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem 280 3656–3664 [DOI] [PubMed] [Google Scholar]
- Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X (2005) Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol 139 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He Y, Amasino R, Chen X (2004) siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev 18 2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J (2005) Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J 24 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U (2001) Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 11 1990–1994 [DOI] [PubMed] [Google Scholar]
- Meister G, Fischer U (2002) Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J 21 5853–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell (Suppl) 14 S111–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem 274 31531–31542 [DOI] [PubMed] [Google Scholar]
- Reeves PH, Coupland G (2000) Response of plant development to environment: control of flowering by daylength and temperature. Curr Opin Plant Biol 3 37–42 [DOI] [PubMed] [Google Scholar]
- Rouse DT, Sheldon CC, Bagnall DJ, Peacock WJ, Dennis ES (2002) FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J 29 183–191 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Finnegan EJ, Dennis ES, Peacock WJ (2006) Quantitative effects of vernalization on FLC and SOC1 expression. Plant J 45 871–883 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Finnegan EJ, Rouse DT, Tadege M, Bagnall DJ, Helliwell CA, Peacock WJ, Dennis ES (2000. a) The control of flowering by vernalization. Curr Opin Plant Biol 3 418–422 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000. b) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C (1999) When to switch to flowering. Annu Rev Cell Dev Biol 15 519–550 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Quesada V, Henderson IR, Dijkwel PP, Macknight R, Dean C (2004) RNA processing and Arabidopsis flowering time control. Biochem Soc Trans 32 565–566 [DOI] [PubMed] [Google Scholar]
- Tan CP, Nakielny S (2006) Control of the DNA methylation system component MBD2 by protein arginine methylation. Mol Cell Biol 26 7224–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Dangl M, Bauer I, Graessle S, Loidl P, Brosch G (2004) Histone methyltransferases in Aspergillus nidulans: evidence for a novel enzyme with a unique substrate specificity. Biochemistry 43 10834–10843 [DOI] [PubMed] [Google Scholar]