Abstract
Starch synthase IIIa (SSIIIa)-deficient rice (Oryza sativa) mutants were generated using retrotransposon insertion and chemical mutagenesis. The lowest migrating SS activity bands on glycogen-containing native polyacrylamide gel, which were identified to be those for SSIIIa, were completely absent in these mutants, indicating that they are SSIIIa null mutants. The amylopectin B2 to B4 chains with degree of polymerization (DP) ≥ 30 and the Mr of amylopectin in the mutant were reduced to about 60% and 70% of the wild-type values, respectively, suggesting that SSIIIa plays an important part in the elongation of amylopectin B2 to B4 chains. Chains with DP 6 to 9 and DP 16 to 19 decreased while chains with DP 10 to 15 and DP 20 to 25 increased in the mutants amylopectin. These changes in the SSIIIa mutants are almost opposite images of those of SSI-deficient rice mutant and were caused by 1.3- to 1.7-fold increase of the amount of SSI in the mutants endosperm. Furthermore, the amylose content and the extralong chains (DP ≥ 500) of amylopectin were increased by 1.3- and 12-fold, respectively. These changes in the composition in the mutants starch were caused by 1.4- to 1.7-fold increase in amounts of granules-bound starch synthase (GBSSI). The starch granules of the mutants were smaller with round shape, and were less crystalline. Thus, deficiency in SSIIIa, the second major SS isozyme in developing rice endosperm affected the structure of amylopectin, amylase content, and physicochemical properties of starch granules in two ways: directly by the SSIIIa deficiency itself and indirectly by the enhancement of both SSI and GBSSI gene transcripts.
Starch consists of two kinds of homopolymers of α-d-glucosyl units: the basically linear amylose and the highly branched amylopectin containing α-1,6 branch linkages. Most of the storage starch in higher plants is composed of 20% to 30% amylose and 70% to 80% amylopectin. Amylose is synthesized by the granules-bound starch synthase I (GBSSI) encoded by the Waxy gene in plants (Tsai, 1974; Sano, 1984), whereas amylopectin biosynthesis is catalyzed by the soluble starch synthases (SSs), starch branching enzymes (BEs), and starch debranching enzymes (DBEs; Smith et al., 1997; Myers et al., 2000; Nakamura, 2002; Ball and Morell, 2003). Many isoforms of these enzymes are implicated in regulation of starch biosynthesis in higher plants, and the analyses of several mutants (SSI, SSIIa, GBSSI, BEI, BEIIa, BEIIb, and isoamylse1 [ISA1]) and the manipulation of the genes coding for SSIIa, GBSSI, BEIIb, and ISA1 have given important clues to understanding the function of each isoform and the starch biosynthesis mechanisms (for review, see Nakamura, 2002). Furthermore, some of these mutants and transformants have produced novel starches having great potential as new foodstuffs and/or for industrial applications.
SS (EC 2.4.1.21) elongates α-glucans by adding Glc residues from ADP-Glc to the glucan nonreducing ends through α-1,4 glucosidic linkages. Among the enzymes responsible for starch biosynthesis, SS is the most difficult to be characterized because of its instability and the diversity of its isoform types. In rice (Oryza sativa), for example, there are 10 SS isoforms: SSI, SSIIa (SSII-3), SSIIb (SSII-2), SSIIc (SSII-1), SSIIIa (SSIII-2), SSIIIb (SSIII-1), SSIVa (SSIV-1), SSIVb (SSIV-2), GBSSI, and GBSSII. Among them, substantial expressions of SSI, SSIIa, SSIIc, SSIIIa, SSIVb, and GBSSI genes were observed in the endosperm (Hirose and Terao, 2004; Ohdan et al., 2005). SSI (Fujita et al., 2006), SSIIa (Umemoto et al., 2002; Nakamura et al., 2005), and GBSSI (Sano, 1984) have been characterized from biochemical studies using their mutants or transformants.
SSIII(a) is the second major SS isozyme in activity levels next to SSI in developing maize (Zea mays; Cao et al., 1999) or rice (Fujita et al., 2006) endosperm. There are two SSIII genes (SSIIIa and SSIIIb) in the rice genome. SSIIIa and SSIIIb are specifically expressed in the developing rice endosperm and leaf, respectively (Hirose and Terao, 2004; Dian et al., 2005; Ohdan et al., 2005). A 230 kD protein band on SDS-PAGE from the developing rice endosperm was detected by immunoblotting using an antiserum raised against maize SSIII (Dian et al., 2005). A putative N-terminal transit peptide, a C-terminal catalytic domain, and a central SSIII-specific domain containing repeat amino acid motifs were identified in the SSIII(a) genes of rice, wheat (Triticum aestivum), and maize (Gao et al., 1998; Li et al., 2000; Dian et al., 2005).
The maize dull-1 (du1) mutation results in mature kernels with a tarnished, glassy, and somewhat dull appearance, referred to as the dull phenotype (Mangelsdorf, 1947; Davis et al., 1955). Numerous prior reports have characterized the endosperm starch of the du1 mutants. However, using gene-tagging method, it was reported that the Du1 gene is identical to the SSIII gene in maize (Gao et al., 1998), and, by anion-exchange chromatography, the ‘SSII’ activity peak was accounted for by the zSSIII/DU1 product (Cao et al., 2000). The apparent amylose content in the endosperm starch of the maize du1 mutant is elevated compared with that of the wild type (Yeh et al., 1981; Inouchi et al., 1983; Boyer and Liu, 1985; Wang et al., 1993a, 1993b) and the long amylopectin chains are greatly reduced (Inouchi et al., 1983; Wang et al., 1993a, 1993b). Interestingly, total soluble SS activity in the du1 mutant is found to be increased (Singletary et al., 1997; Cao et al., 1999) because of the specific enhancement of SSI due to SSIII deficiency (Cao et al., 1999), although the SSII (SSIII) activity peak is significantly reduced (Boyer and Preiss, 1981). Partially purified SSIII from the developing maize endosperm has a lower Km for amylose compared to those for amylopectin and glycogen (Cao et al., 2000). These reports indicate that zSSIII/DU1 may have a specific function during amylopectin biosynthesis in the elongation of long glucan chains.
Very recently, rice SSIIIa mutant lines were generated by T-DNA insertion and isolated, although the reduction of SSIIIa activity of these mutants and pleiotropic effects on other isoforms were not confirmed (Ryoo et al., 2007). Scanning electron microscope (SEM) observation revealed that the starch granules in these mutants were smaller and round in shapes compared with wild type and crystallinity of the starch granules based on their x-ray diffraction patterns was decreased. The content of long chains of amylopectin with degree of polymerization (DP) ≥ 30 was reduced in these mutants compared with wild type, suggesting that SSIIIa plays an important role in generating relatively long chains in rice endosperm. In addition, the amylopectin chains with DP 6 to 8 and DP 16 to 20 appeared to be reduced, whereas the chains with DP 9 to 15 and 22 to 29 were increased. The gelatinization temperatures of endosperm starch were found to be 1°C to 5°C lower than those of wild type.
The reduction of potato (Solanum tuberosum) SSIII in antisense plants did not lead to any detectable changes in starch or amylose content, while starch granules with small subgranules, often with T-shaped cracks centered on the hilum, were observed (Abel et al., 1996; Marshall et al., 1996). When both of the main SS isoforms (SSII and SSIII) responsible for amylopectin synthesis in the tuber were reduced, chains with DP ≤ 15 and extralong chains (ELCs) in amylopectin were increased (Edwards et al., 1999; Lloyd et al., 1999).
The pattern of SS activity bands on native-PAGE/SS activity staining gel from the soluble fraction of leaves in Arabidopsis (Arabidopsis thaliana) was similar to that of the developing maize (Cao et al., 1999) and rice (Fujita et al., 2006) endosperm, indicating that SSI and SSIII account for the majority of soluble SS activity in Arabidopsis leaves. Recently, SSIII mutants of Arabidopsis were isolated and characterized (Zhang et al., 2005). The mutants had a starch excess phenotype in leaves due to an apparent increase in the rate of starch synthesis, indicating that SSIII has a negative regulatory function in the biosynthesis of transient starch in Arabidopsis (Zhang et al., 2005).
In Chlamydomonas (Chlamydomonas reinhardtii), sta3 mutant was isolated as a SSII-deficient mutant (Maddelein et al., 1994), however, they recently proposed to change the name of the Chlamydomonas SSII to SSIII according to the nomenclature of higher plant SS types (Ral et al., 2006). The chains with DP > 90, including ELCs and short amylopectin chains where DP < 20 is elevated, whereas chains of 20 ≤ DP ≤ 90 are reduced in sta3 mutant compared with those of wild type (Maddelein et al., 1994; Ral et al., 2006). The pleiotropic effects of SSIII deficiency on GBSSI are initially proposed in Chlamydomonas; the transcriptional regulation ensures full compensation of the absence of SSIII by GBSSI in sta3 mutant (Ral et al., 2006). Furthermore, based on the analysis of sta2/sta3 double mutant, they hypothesized that the both GBSSI and SSIII are responsible for the synthesis of the long B chains of amylopectin that in turn are required for the integrity of high mass amylopectin in Chlamydomonas.
In this study, two allelic SSIIIa mutant lines of rice generated by retrotransposon Tos17 insertion and N-methyl-N-nitrosourea (MNU) mutagenesis were isolated. We confirmed the defect of SSIIIa activity band on native-PAGE/SS activity staining and measured the total SS activity in vitro assays in these mutants. This article also describes detailed pleiotropic effects of SSIIIa deficiency on SSI and GBSSI for estimation of their amount of proteins and the activities of other enzymes related to the starch biosynthesis in these mutants although Ryoo et al. (2007) did not examine these effects. In this article, the analysis of the structure and physicochemical properties of the endosperm starch of these mutants clarified the function of rice SSIIIa on starch biosynthesis. Furthermore, the dramatic changes of structure of amylopectin and amylose content by the pleiotropic effects of the absence of SSIIIa on other SS isozymes were clearly interpreted.
RESULTS
Production of the OsSSIIIa Gene-Tagging Mutant Line
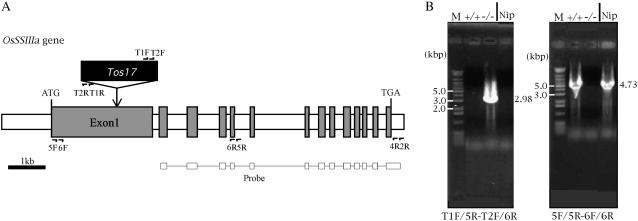
Only one line containing a Tos17 insertion in the rice SSIIIa gene (OsSSIIIa) was isolated by PCR screening of a Tos17 knockout rice population of approximately 40,000 lines (see “Materials and Methods”). The OsSSIIIa gene is composed of 14 exons—exon 1 being extremely long (2,695 bp), and 13 introns (Fig. 1A). Tos17 was inserted into exon 1 in the line used in this study (Fig. 1A). The genotype of the line was determined as either homozygous for Tos17 (−/−) or wild homozygous (+/+) using nested PCR (see “Materials and Methods”; Fig. 1B). In the line +/+, the PCR reaction using the T1F/5R and T2F/6R primer pairs had no product (Fig. 1B, left section, lane +/+), while 5F/5R and 6F/6R primer pairs generated an approximately 4.73 kb band (Fig. 1B, right section, lane +/+). In the line −/−, the PCR product of the T1F/5R and T2F/6R primer pairs was a 2.98 kb band (Fig. 1B, left section, lane −/−), while the 5F/5R and 6F/6R primer pairs had no product (Fig. 1B, right section, lane −/−). The −/− line was used as the SSIIIa mutant (ss3a-1) and the +/+ line (SS3a-1+/+) or ‘Nipponbare’ the wild-type parent (‘Nip’), as its control.
Figure 1.
Site of Tos17 insertion in the OsSSIIIa gene and determination of rice mutant line genotype by PCR. A, Structure of the OsSSIIIa gene. The exons and introns are depicted as gray and white boxes, respectively. ATG and TGA indicate the translation initiation and stop codons, respectively. The insertion site of Tos17 in a mutant line is indicated with a vertical arrow. Horizontal half arrows show the sites of primers for PCR for genotype determination (T1R, T2R, 5F, 6F, 5R, and 6R) and mutant line screening (T1F, T2F, T1R, T2R, 5F, 6F, 2R, 4R, 5R, and 6R). The primers T1F, T2F, T1R, and T2R were designed from the Tos17 sequence, while 5F, 6F, 2R, 4R, 5R, and 6R were designed from the OsSSIIIa gene sequence. The region used as a probe for Southern blotting to screen mutant line is indicated. B, Determination of genotype [homozygous for Tos17 insertion (−/−, left section) or wild homozygous (+/+ right section)] in a mutant line by nested PCR. Primer pairs are indicated below the photographs. T1F/5R-T2F/6R means that the primer pair T1F/5R was used for the first PCR and T2F/6R for the second PCR. M, Molecular markers.
Pleiotropic Effects on SS Isozymes in the SSIIIa Mutant Lines
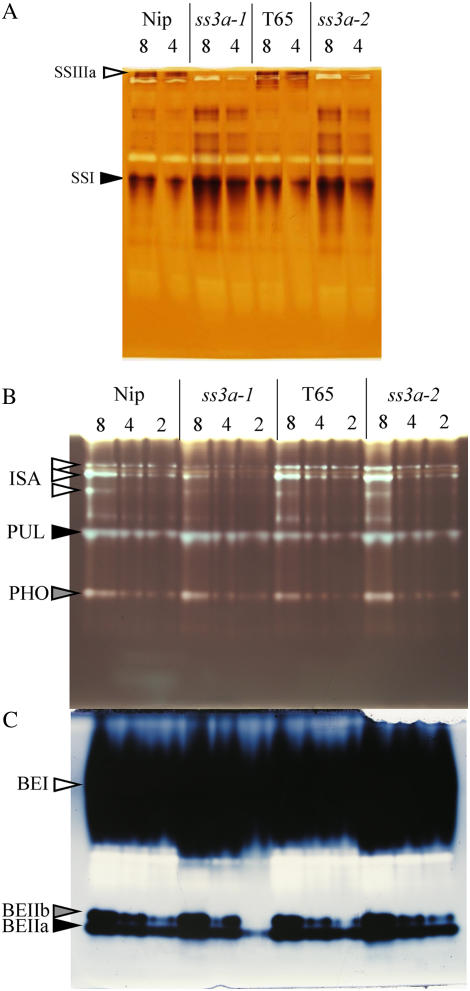
To evaluate the effect of the insertion of Tos17 into the OsSSIIIa gene, the activities of SS isoforms of the soluble fraction from the day after flowering (DAF) 12 developing endosperm were estimated by native-PAGE/SS activity staining using a gel containing oyster glycogen (Fig. 2A, lanes ‘Nip’ and ss3a-1). The brown bands on the gel were dependent on the addition of ADP-Glc in the incubation buffer (data not shown), meaning that these are due to the SS activities. The intermediate migrating bands on the gel have been identified to be the SSI isoform using the SSI mutants of rice (Fujita et al., 2006). The slowest migrating band was completely lacking in the SSIIIa mutant line (ss3a-1), indicating that it is a null SSIIIa mutant.
Figure 2.
Native-PAGE/activity staining of developing endosperm in rice SSIIIa mutant lines and the wild type. The numbers above the lanes are the volumes (μL) of the crude enzyme extract applied onto each lane. A, Native-PAGE/SS activity staining. The SSIIIa and SSI activity bands are indicated by arrowheads. B, Native-PAGE/DBE activity staining. The ISA (isoamylase), PUL (pullulanase), and PHO (phosphorylase) activity bands are indicated by arrowheads. C, Native-PAGE/BE activity staining. The BEI, BEIIa, and BEIIb activity bands are indicated by arrowheads. ‘Nip’, Wild type of ss3a-1, ss3a-1, SSIIIa mutant by Tos17 insertion; ‘T65’, Wild type parent of ss3a-2, ss3a-2, SSIIIa mutant induced by chemical mutagenesis. [See online article for color version of this figure.]
Another SSIIIa mutant line (ss3a-2) lacking the SSIIIa activity band was isolated from a population of the MNU-treated rice ‘Taichung 65’ (‘T65’; see “Materials and Methods”; Fig. 2A, lane ss3a-2). Analysis of genomic DNA sequence of SSIIIa gene in ss3a-2 revealed that GT at the 5′ splice donor site of intron 1 was replaced by AT (data not shown), and the point mutation results in the aberrant translation and stop codon in the intron 1. Therefore, lacking the SSIIIa band on native-PAGE/SS activity staining gel in ss3a-2 was thought to be caused by the point mutation in the SSIIIa gene by MNU mutagenesis. In both ss3a-1 and ss3a-2 mutants, the SSI activity and the minor activity bands between SSI and SSIIIa bands appeared to be enhanced relative to the wild type (Fig. 2A), consistent with the observation in the SSIII mutant of Arabidopsis (Zhang et al., 2005).
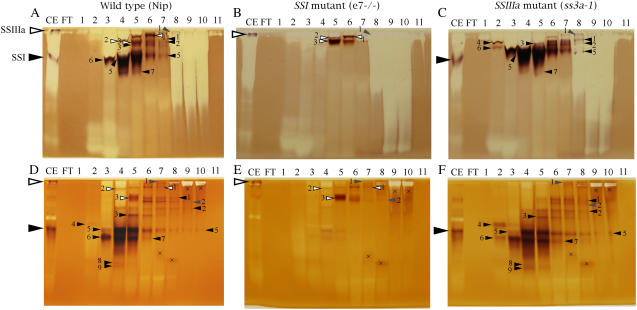
To examine the SS isozyme components in the rice developing endosperm in more details, soluble fractions from the developing endosperm of ‘Nip’ (wild type), e7−/− (SSI-deficient mutant; Fujita et al., 2006), and ss3a-1 were fractionated by anion-exchange (HiTrapQ, Pharmacia) chromatography with a linear gradient of 0 to 0.5 m NaCl, and native-PAGE/SS activity staining of each fraction was performed in a gel containing 0.1% rice amylopectin (Fig. 3, A–C) or 0.8% oyster glycogen as a substrate (Fig. 3, D–F). Fourteen SS activity bands were detected in the wild type, nine of which were missing in the SSI mutant (black arrowheads in Fig. 3, A, B, D, and E). These results suggest that SSI is divided into several bands having different mobilities and retention times probably due to protein modification by phosphorylation, polymerization, or unknown posttranscriptional modifications of SSI protein.
Figure 3.
Native-PAGE/SS activity staining. SP from 10 g of developing endosperm in the wild type (‘Nip’; A and D), the SSI mutant (e7−/−; B and E), and the SSIIIa mutant (ss3a-1; C and F) was fractionated by anion-exchange chromatography (HiTrapQ). Proteins in the fractions were separated by native-PAGE in gels containing 0.1% rice amylopectin (A, B, and C) or 0.8% oyster glycogen (D, E, and F). Gels were incubated overnight in a SS reaction buffer containing 0.5 m citrate (D, E, and F) or under a citrate-free condition (A, B, and C). The large white and black arrowheads indicate the positions of SSIIIa and SSI in the crude extract (CE), respectively. The small white, black, and gray arrowheads with numbers show that SS activity bands were detected in the ‘Nip’ and SSI mutant, in the ‘Nip’ and SSIIIa mutant, and in all lines, respectively. These activity bands were dependent on the addition of ADP-Glc in the incubation buffer, whereas the bands with X marks are not SS activity bands because they are also detected in the ADP-Glc-free incubation buffer (data not shown). FT, Flow through fraction; numbers 1 to 11, fraction numbers. [See online article for color version of this figure.]
The measurement of the SS activity of each fraction by HiTrapQ chromatography showed that SSIIIa activity is detected at around 0.35 m NaCl (Fujita et al., 2006). In this study, however, the SS activity bands equivalent to those of SSIIIa were not detected at 0.35 m NaCl (lane 8 in Fig. 3, C and F); SS activity bands numbers 1, 2, or 5 of black arrowheads on lanes 7 and 8 were detected in ‘Nip’ and ss3a-1, and the band number 1 of gray arrowhead on lanes 8 to 10 were detected in all lines, indicating that these bands are not SSIIIa. Unfortunately, the polyglucan chains elongated by SSIIIa might be digested by the hydrolytic enzymes during the SS reaction, since a strong hydrolytic activity band (white smear bands), presumably due to amylase or isoamylase, was contaminated on lanes 8 to 10. On the other hand, SS activity bands having different mobility (1–3 of white arrowheads) from that of SSIIIa (lane CE) were detected in ‘Nip’ and SSI mutant, but not in ss3a-1 on lanes 5 and 6 (Fig. 3, A, B, D, and E). The band number 3 of white arrowhead was enhanced in SSI mutant relative to that in ‘Nip’. It is unclear whether these three bands are SSIIIa or not because they were eluted at a lower concentration of NaCl than 0.35 m. Western blotting using antiserum raised against SSIIa (Nakamura et al., 2005) of HiTrapQ fractions showed that SSIIa was detected on lanes 8 to 10 in every line (data not shown), indicating that the bands detected in lanes 5 and 6 are not SSIIa. Unfortunately, the SS activity band of the SSIIa could not be detected on the native-PAGE/SS activity staining gel either because of its low activity in ‘Japonica’ (Nakamura et al., 2005) or the contamination of hydrolytic enzymes. One or two bands (gray arrowheads in Fig. 3, A–F) that are not thought to be SSI or SSIIIa were detected in every line.
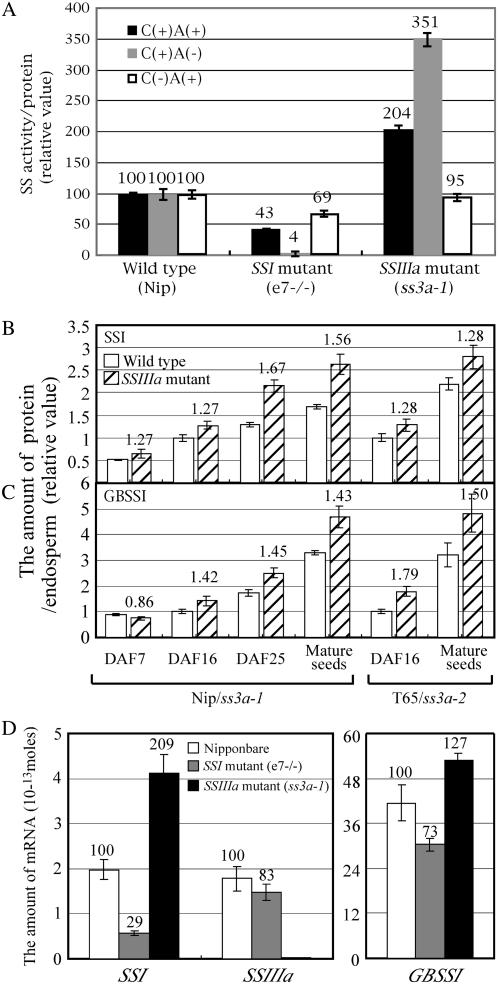
Total soluble SS activities increased in the du1 mutant in maize (Singletary et al., 1997; Cao et al., 1999). To clarify the case of rice, the total soluble SS activity of developing rice endosperm in ‘Nip’, SSI mutant, and ss3a-1 was measured in the presence or absence of 0.5 m citrate (C) and exogenous primers (rice amylopectin, A, Fig. 4A). Both SSI and SSIIIa activities have been detected in the presence of exogenous primers, whereas SSIIIa has not been detected in the absence of exogenous primers (Fujita et al., 2006). The total soluble SS activity of SSI mutant was 43% of that of ‘Nip’ in the presence of citrate and exogenous primer [C(+)A(+)], meaning that at least 57% of soluble SS activity of ‘Nip’ under this condition is accounted for by SSI (Fig. 4A). On the other hand, the soluble SS activity in ss3a-1 was increased approximately 2-fold relative to that of ‘Nip’. Only 4% of the SS activity of ‘Nip’ was detected in SSI mutant in the presence of citrate and the absence of exogenous primers [C(+)A(−)], indicating that most of the soluble SS activity of ‘Nip’ (96%) is due to SSI under this condition where the soluble SS (probably SSI) activity in ss3a-1 was increased approximately 3.5-fold relative to that of ‘Nip’ (Fig. 4A). These results suggest that the SSI activity was enhanced in ss3a-1 in the presence of citrate, especially under the C(+)A(−) condition. The total soluble SS activity of SSI mutant was 69% of that of ‘Nip’ when exogenous primer [C(−)A(+)], suggesting that in addition to SSI, another SS, probably SSIIIa, is active under this condition. Thus, these results and Figure 2A clarified the SSI enhancement in the SSIIIa-deficient mutant in rice and are consistent with the results obtained from the du-1 mutant in maize (Singletary et al., 1997; Cao et al., 1999).
Figure 4.
A, Total SS activity of the crude extract from developing endosperm in wild type ‘Nip’, the SSI mutant (e7−/−), and the SSIIIa mutant (ss3a-1). The SS activity was assayed in the presence (+) or absence (−) of 0.5 m citrate (C) and exogenous primers (2 mg/mL rice amylopectin; A), as indicated. The numbers on the graph are the percent of the activity in the SSI and SSIIIa mutants under each condition when the activity of ‘Nip’ was defined as 100%. The data are the mean ± se of three seeds. B and C, Amount of SSI (B) or GBSSI (C) protein in developing rice endosperm from DAF 7 through to the mature endosperm of the wild type ‘Nip’ and ‘T65’ and the SSIIIa mutant (ss3a-1 and ss3a-2). The total amount of three fractions (SP, LBP, and TBP; see “Materials and Methods”) of SSI or GBSSI protein was quantified by immunoblotting using antiserum raised against SSI or GBSSI (Fujita et al., 2006). The numbers on the graph are the rate of the amount of protein in SSIIIa mutants to that of the total SSI or GBSSI protein in the wild type. The data are the mean ± se of three seeds. D, Amount of mRNA of SSI, SSIIIa, and GBSSI genes in developing rice endosperm (DAF 10) of the SSI (e7) and SSIIIa (ss3a-1) mutants and the wild type (‘Nip’). The numbers on the graph are the percent of the amount of mRNA in the SSI and SSIIIa mutants when ‘Nip’ was defined as 100%. The data are the mean ± se of three replications.
In summary, SSI accounts for at least more than half of the total SS activity in the soluble fraction in the rice developing endosperm and the second major SS activity isozyme is SSIIIa. We cannot exclude the possibility that a small number of other isoforms expressed in the endosperm would be present in the soluble fraction of the developing rice endosperm and especially these isoforms might be enhanced when one of any SS was deficient (Fig. 3). However, such activity, if it exists, would be very minor in the developing endosperm of wild type in ‘Japonica’ rice.
To quantitatively examine the effect of SSIIIa deficiency, the amounts of SSI, GBSSI, and SSIIa proteins in SSIIIa mutants and the wild type at different stages of seed development (DAF 7, 16, 25, and mature) were estimated by western blotting (Fig. 4, B and C; data not shown). SSI exists not only in the soluble fraction but also binds to starch granules of developing rice endosperm (Fujita et al., 2006). The SSI proteins were separated into soluble protein (SP), loosely bound protein (LBP), and tightly bound protein (TBP) fractions, and total amounts of SSI were estimated (Fig. 4B). SSI in the SP was high at the early stages (DAF 7–16) but gradually decreased and was very low at the mature stage and the relative amounts of SSI in LBP and TBP gradually increased during seed development (data not shown; Fujita et al., 2006). Total amounts of SSI in three fractions of the SSIIIa mutants were 1.27 to 1.67 times higher than those of the wild type from DAF 7 through maturity (Fig. 4B). Almost GBSSI was detected in TBP (Fujita et al., 2006). The amount of the GBSSI protein of the SSIIIa mutant at DAF 7 was not significantly different from that of the wild type, whereas, from DAF 16 through maturity, those of the SSIIIa mutants were 1.42 to 1.79 times higher than those of the wild type (Fig. 4C). These results indicate that the amount of SSI and GBSSI protein were consistently higher in both SSIIIa mutants (ss3a-1 and ss3a-2) than in the wild type. SSIIa of ‘Japonica’ was detected in SP and LBP (Nakamura et al., 2005). The amounts of SSIIa in the SSIIIa mutants were not significantly different from those of the wild type (data not shown).
To clarify whether the increase of the amount of SSI and GBSSI proteins in the mutants is caused by transcriptional or posttranscriptional regulations, the amount of mRNA of SSI, SSIIIa, and GBSSI gene of developing endosperm (DAF 10) of SSI and SSIIIa mutant and wild type were estimated by reverse transcription (RT)-PCR method (Fig. 4D). In ss3a-1 mutant, the mRNA of SSIIIa gene was reduced to 1/100 of that of ‘Nip’, indicating that SSIIIa gene expression was strictly limited by Tos17 insertion into exon 1 of the gene. The amount of mRNA of SSI and GBSSI was about 2 and 1.27 times higher than that of the wild type, respectively (the values are significantly different at P < 0.05; Fig. 4D). These results suggested that the enhancement of the amount of SSI and GBSSI protein in SSIIIa mutant, if not the whole, could be explained by the transcriptional regulation of their genes derived from the SSIIIa deficiency. In contrast, there was no statistically significant difference between SSI mutant and wild type of the amount of mRNA of SSIIIa and GBSSI (Fig. 4D).
Pleiotropic Effects on Other Enzymes Related to Starch Biosynthesis in the SSIIIa Mutant Lines
To test whether the deficiency in SSIIIa activity has pleiotropic effects, the activities of other enzymes involved in the starch biosynthesis were measured in the SSIIIa mutant lines. As detected by native-PAGE/DBE activity staining, the ISA activity of ss3a-1 was slightly lower than that of the wild type, although that of ss3a-2 showed no obvious differences with that of the wild type (Fig. 2B). Pullulanase and phosphorylase activities detected by native-PAGE/DBE activity staining of SSIIIa mutants were not different from those of the wild type (Fig. 2B). Native-PAGE/BE activity staining showed that the BEI activity of ss3a-1 and ss3a-2 was slightly higher than that of the wild type; however, BEIIa and BEIIb activities showed no obvious differences (Fig. 2C). In contrast, BEIIa activity of the du-1 mutant in maize was lower than that of the wild type (Boyer and Preiss, 1981). AGPase activity in ss3a-1 was approximately 1.2 times (10.24 ± 0.17 mmol min−1 endosperm−1) higher than that of its control (8.52 ± 0.58) and similar observation is shown in the maize du-1 mutant (5 times higher than the wild type; Singletary et al., 1997) and rice SSI mutant (e7−/−; 1.6 times; Fujita et al., 2006).
Seed Morphology and Morphology and Crystallinity of Starch Granules of the SSIIIa Mutant Lines
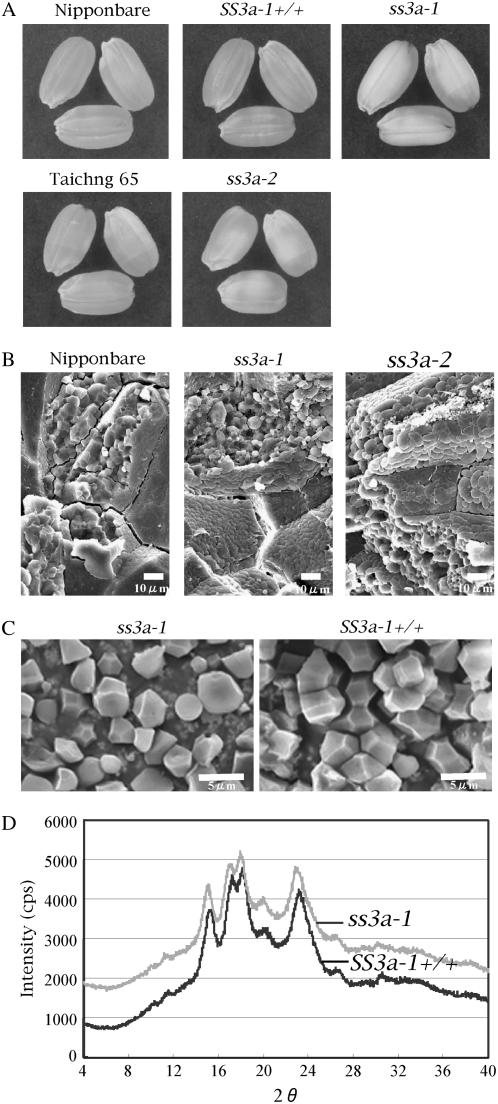
The seeds of the SSIIIa mutant lines, ss3a-1 and ss3a-2, had a chalky interior appearance (Fig. 5A). Their dehulled grain weight and starch content were slightly lower (93%–96% and 96%–98%, respectively) than those of the wild type (Table I), but there were no statistically significant differences in the dehulled grain weight and starch content between SSIIIa mutants and their control or the wild type.
Figure 5.
Characterization of the SSIIIa mutant lines ss3a-1 and ss3a-2, control line SS3a-1+/+, and the wild type, ‘Nip’ and ‘T65’. A, Seed morphology. B, SEM of the cross sections of mature endosperm. Bar = 10 μm. C, SEM observations of starch granules. Bar = 5 μm. D, X-ray diffraction patterns of endosperm starch.
Table I.
Dehulled grain weight and starch content (%) of rice SSIIIa mutant lines (ss3a-1, ss3a-2), its control (SS3a-1+/+), and wild-type rice ‘Nip’ and ‘T65’
| Lines | Dehulled Grain Weight | Starch Content | |
|---|---|---|---|
| mg | % | ||
| ‘Nip’ | a21.6 ± 0.3 | b(100) | c82.3 ± 2.6 |
| SS3a-1+/+ | 21.5 ± 0.3 | (99.5) | 85.6 ± 1.9 |
| ss3a-1 | 20.7 ± 0.3 | (95.8) | 80.9 ± 1.0 |
| ‘T65’ | 23.0 ± 0.4 | (100) | 81.1 ± 2.1 |
| ss3a-2 | 21.5 ± 0.3 | (93.5) | 78.1 ± 1.4 |
Mean ± se of 20 seeds.
Percent of wild type.
Mean ± se of three seeds.
To test whether the reduction of SSIIIa activity affects the distinct granular structure of starch in the endosperm, SEM observations of a cross section of rice seeds were conducted (Fig. 5B). In the wild type, the endosperm starch granules formed similarly sized polygonal granules with sharp edges. Several starch granules were tightly packed into the amyloplasts, which were very abundant in the endosperm cells. In contrast, the amyloplasts of SSIIIa mutants were round shaped and relatively loosely packed into endosperm cell (Fig. 5B). The isolated starch granules from the ss3a-1 endosperm contained slightly smaller and more rounded granules compared to those of the control (Fig. 5C).
The starch granules of ss3a-1 and SS3a-1+/+ displayed the typical A-type x-ray diffraction pattern (Fig. 5D). However, the height and sharpness of major peaks in ss3a-1 starch were 80% to 90% of those of the respective control line SS3a-1+/+, or the wild type ‘Nip’ (data not shown), indicating that the degree of crystallinity of the starch granules of the SSIIIa mutant was reduced.
Analysis of the Starch Component and Amylopectin Fine Structure of the SSIIIa Mutant Lines
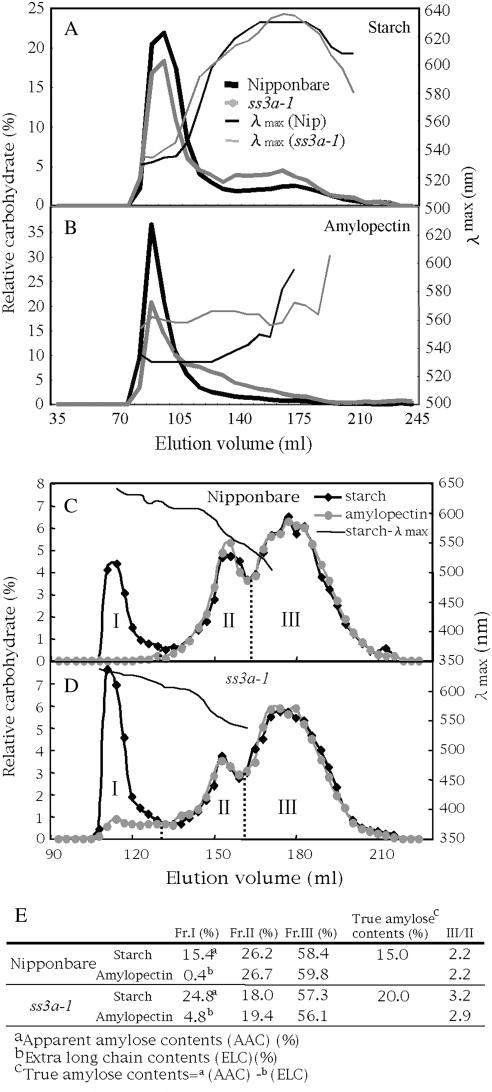
To test whether the reduction of SSIIIa activity affects the structure of endosperm starch, endosperm starch (Fig. 6A) and purified amylopectin (Fig. 6B) of SSIIIa mutants and the wild type were subjected to size-exclusion chromatography using Sephacryl S-1000SF. Judging from the λmax values of the polyglucan-iodine complex, fractions containing most, if not all, of the amylopectin and amylose were eluted at 70 to 120 mL and 130 to 220 mL, respectively (Fig. 6A). In the endosperm starch of ss3a-1 and ss3a-2, the amount of amylopectin decreased, while the amount of amylose increased (Fig. 6A; data not shown). It is also observed that the λmax values of the amylopectin fraction of ss3a-1 were higher than those of ‘Nip’ (Fig. 6A). In particular, the λmax values of purified amylopectin from ss3a-1 endosperm starch were about 30 nm higher than those of ‘Nip’ (Fig. 6B), suggesting that the amylopectin of ss3a-1 contains many more long chains than the ‘Nip’. The peak of purified amylopectin from ss3a-1 and ss3a-2 endosperm starch was lower in height and broader than that from the wild-type endosperm, indicating that the Mr of amylopectin of ss3a-1 and ss3a-2 is lesser than that of the wild type (Fig. 6B; data not shown). The weight-average Mr of amylopectin in ss3a-1 endosperm starch predicted by the HPSEC-MALLS-RI method was 74.6% of that of ‘Nip’ (data not shown).
Figure 6.
Size separation of endosperm starch and purified amylopectin from the SSIIIa mutant, ss3a-1, and the wild type, ‘Nip’. A and B, Elution profiles by gel filtration chromatography through Sephacryl S-1000SF of starch (A) and purified amylopectin (B) from ‘Nip’ (black lines) and ss3a-1 (gray lines). C and D, Elution profiles of isoamylase-debranched starch (black lines) and purified amylopectin (gray lines) by gel filtration chromatography through Toyopearl HW55S-HW50S columns from ‘Nip’ (C) and ss3a-1 (D). Each fraction was divided according to the following range of λmax values of the glucan-iodine complex: Fr. I, λmax ≥ 620 nm; Fr. II, 540 nm ≤ λmax < 620 nm; Fr. III, λmax < 540 nm. E, Percentage comparisons of each fraction separated by gel filtration (C and D) in the total carbohydrate of endosperm starch and purified amylopectin from ‘Nip’ and ss3a-1.
For further analysis of the structure of ss3a-1 endosperm starch, the isoamylolysates of endosperm starch and purified amylopectin of ‘Nip’ (Fig. 6C) and ss3a-1 (Fig. 6D) were subjected to size-exclusion chromatography using Toyopearl HW55S and HW50S. Judging from the λmax values of the polyglucan-iodine complex, fraction I (Fr. I) eluted at 100 to 130 mL and Fr. II and III that eluted at 130 to 220 mL contain most, if not all, of the amylose and amylopectin, respectively. Fr. II and III included long and short chains of amylopectin, respectively. A small amount of fraction I was detected in the purified amylopectin from ‘Nip’ endosperm starch (Fig. 6C) and was called a super long chain or ELC (DP ≥ 500) of amylopectin (Takeda et al., 1987; Horibata et al., 2004). Therefore, Fr. I from endosperm starch includes both the true amylose (TAM) and ELC. The value-subtracted ELC content from the apparent amylose content of starch is equivalent to the TAM of starch (Horibata et al., 2004). Based on Figure 6, C and D, each component of the starch of ss3a-1 and ‘Nip’ was calculated and is shown in Figure 6E. The Fr. I contained in ss3a-1 (24.8%) was 1.6 times larger than that of ‘Nip’ (15.4%). This result was derived from the increase in TAM (20.0%) and ELC (4.8%) in ss3a-1 endosperm starch compared with ‘Nip’ (15.0% and 0.4%, respectively). The higher λmax values of the amylopectin-iodine complex of ss3a-1 (Fig. 6, A and B) must have been caused by the increase in ELC content in ss3a-1 amylopectin, but not in amylase content. The ratio of Fr. III to Fr. II of the amylopectin chains from ss3a-1 endosperm starch (2.9) was higher than that from ‘Nip’ endosperm starch (2.2), indicating that the long amylopectin chains in ss3a-1 were fewer than those in ‘Nip’ (Fig. 6E).
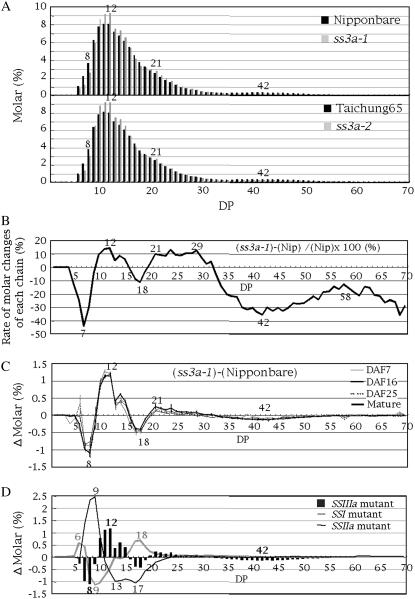
To evaluate the deficiency of the SSIIIa activity on the fine structure of the endosperm amylopectin, the chain-length distributions of isoamylolysate of endosperm amylopectin in the two SSIIIa mutant lines were determined using capillary electrophoresis, although this method cannot analyze the ELC of amylopectin. The chain-length distribution patterns of amylopectin chains with DP ≤ 70 in ss3a-1 and SS3a-2 were very similar (Fig. 7A); there were more chains with DP 10 to 15 and DP 20 to 25 and fewer chains with DP 6 to 9, DP 16 to 19, and DP 30 to 60 than in the wild type (Fig. 7A, and mature in Fig. 7C). These chain-length distribution patterns of SSIIIa mutants amylopectin are specific relative to those of other SS (SSI, SSIIa) mutant amylopectins analyzed so far (Fig. 7D). It is stressed that the pattern of changes in SSIIIa mutants in the range of DP ≤ 20 chains was almost an opposite image of that of SSI mutants (Fig. 7D). The rate of the molar change of each chain (Δmolar %/molar % × 100) in DP 5 to 60 of the rice SSIIIa mutant ss3a-1 was calculated from the chain-length distribution of endosperm amylopectin in ‘Nip’ (molar %; Fig. 7A) and the differences between the chain-length distribution of ss3a-1 and ‘Nip’ (Δmolar %; mature in Fig. 7C), as shown in Figure 7B. In this pattern, the decrease at DP 7 and DP 42 reached −40% (rate of molar changes), meaning that the amounts of long chains with DP 42 and 74 (data not shown) of ss3a-1 amylopectin decreased to about 60% of those of ‘Nip’. These results suggest that SSIIIa specifically elongates B2-B4 long chains of amylopectin.
Figure 7.
A, Chain-length distribution patterns of endosperm amylopectin in the mature endosperm of SSIIIa mutant lines (ss3a-1 and ss3a-2) and the wild-type parent ‘Nip’ and ‘T65’. B, Rate of molar changes of each chain relative to the amount of its chain (Δmolar %/molar % × 100), as calculated from A for DP 5 to 60 amylopectin chains of the SSIIIa mutant (ss3a-1). C, Differences in the chain-length distribution patterns of amylopectin in developing endosperm at DAF 7, 16, and 25 and the mature endosperm of the SSIIIa mutant line ss3a-1 and wild-type ‘Nip’. Vertical bars indicate ses. D, Comparison of differences in the chain-length distribution pattern (Δ molar %) among SS mutant lines (SSIIIa, SSI, and SSIIa). Values for the molar % in A and Δ molar % in B, C, and D for each DP are averages of three seeds arbitrarily chosen from a single homozygous plant. The numbers on the plots are the DP values.
Starch accumulation in rice endosperm becomes evident after DAF 5 (Sato, 1984; Hirose and Terao, 2004). Prior to this event, the expression of the OsSSIIIa gene starts at DAF 5 and is maintained at high levels until DAF 15 in endosperm (Hirose and Terao, 2004; Ohdan et al., 2005). The differences in the amylopectin chain-length distribution pattern between ss3a-1 and ‘Nip’ were analyzed during endosperm development (DAF 7 to maturity; Fig. 7C). The facts that the alteration in the chain-length distribution pattern in ss3a-1 was already apparent at DAF 7, and that the patterns at DAF 7 through seed maturity were almost the same (Fig. 7C) indicate that SSIIIa is functional from the very early stage of rice endosperm development through seed maturity and that effect does not vary during these periods.
Analysis of the Physicochemical Properties of Endosperm Starch in SSIIIa Mutant Lines
To evaluate the physicochemical properties of endosperm starch in SSIIIa mutant lines, the gelatinization temperature of endosperm starch was analyzed by differential scanning calorimetry (DSC). The temperatures for the onset (To) and peak (Tp) of gelatinization of endosperm starch in ss3a-1 and ss3a-2 were 5.2°C and 7.7°C and 1.7°C and 3.1°C, respectively, which were lower than those of control lines. There were no significant differences in the conclusion temperatures (Tc; Table II). The gelatinization enthalpy of endosperm starch in SSIIIa mutants was 1 to 2 mJ mg−1 lower than those of the wild type (Table II). In contrast, the To, Tp, and Tc of du-1 mutant starch in maize reported to be 2°C to 3°C higher than those of the wild type (Inouchi et al., 1991).
Table II.
Thermal properties of endosperm starch as determined by DSC
| Lines | Toa | Tpb | Tcc | ΔHd |
|---|---|---|---|---|
| °C | °C | °C | mJ mg−1 | |
| ‘Nip’ | 55.7 ± 0.4e | 62.2 ± 0.3 | 68.7 ± 0.5 | 8.3 ± 0.4 |
| SS3a-1+/+ | 53.7 ± 0.2 | 61.4 ± 0.2 | 67.7 ± 0.2 | 8.4 ± 0.2 |
| ss3a-1 | 50.5 ± 0.6 | 60.5 ± 0.1 | 68.1 ± 0.1 | 7.6 ± 0.5 |
| ‘T65’ | 56.1 ± 0.6 | 63.5 ± 0.4 | 69.9 ± 0.3 | 9.1 ± 0.1 |
| ss3a-2 | 48.4 ± 0.9 | 60.4 ± 0.1 | 69.9 ± 0.6 | 6.8 ± 0.4 |
Onset temperature.
Peak temperature.
Conclusion temperature.
Gelatinization enthalpy of starch.
The values are the averages of at least three replications (means ± se).
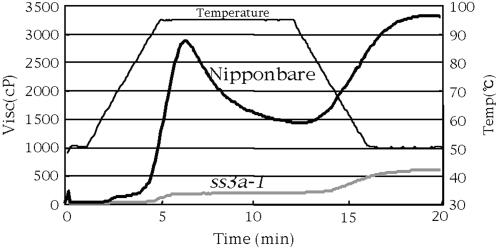
The pasting properties of the endosperm starch were analyzed using a rapid visco analyzer (RVA; Fig. 8). The viscosity pattern of the pasting of starch of ss3a-1 was dramatically different from that of ‘Nip’. The pasting starch of ss3a-1 did not show distinct parameters demonstrating peak viscosity and breakdown while the temperature was increased, and the viscosity was maintained at low level (only 6% of the peak viscosity of ‘Nip’). While the temperature was again lowered, the viscosity of the pasting of ss3a-1 starch increased slightly, and its final viscosity was 18% of that of ‘Nip’.
Figure 8.
Pasting properties of endosperm starch of the SSIIIa mutant line ss3a-1 (gray line) and the wild type ‘Nip’ (black line). The viscosity value at each temperature point is the average of three replications. The thin line indicates the change in temperature during measurement with a RVA.
DISCUSSION
Isolation of Rice SSIIIa Mutant Lines
The SSIII mutants of Arabidopsis (Zhang et al., 2005) and Chlamydomonas sta3 mutants (Maddelein et al., 1994; Ral et al., 2006), and the potato plants with antisense-induced SSIII reduction (Abel et al., 1996; Edwards et al., 1999; Lloyd et al., 1999; Fulton et al., 2002) have been reported, while in cereals, maize du1 mutants were reported as SSIII mutants (Mangelsdorf, 1947; Davis et al., 1955; Gao et al., 1998). In this study, the SSIIIa-deficient mutant of rice (ss3a-1) was isolated through reverse genetics by retrotransposon Tos17 insertion. SSIIIa is one of two SSIIIs (SSIIIa and SSIIIb) in the rice genome, and is specifically expressed in the endosperm. Tos17 was inserted into exon 1 of the SSIIIa gene in this mutant, and transcription of mRNA was inhibited (Fig. 4D). Moreover, the slowest migrating SS activity band in native-PAGE/SS activity staining gel was completely absent (Fig. 2A), indicating that this SS activity band is equivalent to the SSIIIa activity band. This band was also lacking in another SSIIIa mutant line ss3a-2 in which the SSIIIa gene was modified by a nucleotide replacement in the start of the intron 1 isolated from the rice ‘T65’ population of the MNU-treated plants (data not shown). The seed morphology (Fig. 5A), characteristics of starch granules (Fig. 5, B and C), the chain-length distribution pattern of endosperm amylopectin (Fig. 7A), and increase of amylose content in the endosperm starch (data not shown) of ss3a-2 were almost the same as those of ss3a-1, indicating that ss3a-2 is also an SSIIIa-deficient mutant line. Moreover, the traits of seed morphology, crystallinity of starch granules, chain-length distribution of amylopectin, thermal properties of starch, and enrichment of amylose content of SSIIIa mutants generated by T-DNA (Ryoo et al., 2007) were almost identical to those described in our results, indicating that these traits were caused by defect of SSIIIa gene in rice.
Characterization of Structure of Endosperm Starch in Rice SSIIIa Mutant Lines
In rice SSI mutants, the λmax of the starch-iodine complex and the crystallinity of endosperm starch were similar to those of the controls, although the chain-length distribution of their amylopectin and peak viscosity of their pasting starch changed (Fujita et al., 2006). In contrast, significant changes were observed in the starch component and structure in SSIIIa mutants of rice studied in this study. The traits of starch structure of rice SSIIIa mutant lines can be summarized as follows: (1) SSIIIa mutants are defective for the synthesis of long B chains (DP ≥ 30) of amylopectin (Figs. 6, B–E and Fig. 7). (2) SSIIIa mutants display a reduced weight-average molecular weight of amylopectin (data not shown). (3) SSIIIa mutants are enriched in amylose (Fig. 6, A, C, D, and E). (4) SSIIIa mutants are enriched in ELC of amylopectin (Fig. 6, C–E). (5) The proportion of short chains (DP < 30) of amylopectin of SSIIIa mutants changed (Fig. 7); chains with DP 6 to 9 and DP 16 to 19 decreased while chains with DP 10 to 15 and DP 20 to 25 increased.
The trait 1 seems to be a universal feature of all SSIII mutants and transformants with an exception of Arabidopsis (Zhang et al., 2005); e.g. maize du1 mutant (Yeh et al., 1981; Inouchi et al., 1983; Boyer and Liu, 1985; Wang et al., 1993a, 1993b), antisense-SSIII potato transformants (Fulton et al., 2002), and Chlamydomonas sta3 mutant (Ral et al., 2006). These reports and the results in this study support a consistent view that the major function of SSIII(a) is to elongate B2 and B3 chains and the longer chains of amylopectin. This idea is in accord with the observation that the partially purified SSIII(a) from developing maize and rice endosperm preferentially elongates long glucans (amylose) and longer chains of DP ≥ 12 broadly on native-PAGE gel, respectively, in vitro (Cao et al., 2000; Fujita et al., 2006).
The trait 2 is closely related to the trait 1, since the reduction by 74.6% in the Mr of SSIIIa mutant amylopectin molecules (data not shown) could be due to defect in the elongation of the long B chains connecting the clusters of amylopectin. The decrease of Mr of amylopectin was initially reported in sta3 mutant in Chlamydomonas (Maddelein et al., 1994).
The trait 3 was also shown in maize (Yeh et al., 1981; Inouchi et al., 1983; Boyer and Liu, 1985; Wang et al., 1993a, 1993b), but not shown in potato (Abel et al., 1996; Lloyd et al., 1999) and Arabidopsis (Zhang et al., 2005). The enrichment of amylose in endosperm starch is caused by two reasons as follows: one is caused by decrease of amylopectin derived from the SSIIIa deficiency responsible for the synthesis of amylopectin. However, the second reason was substantiated in this study; the increase of GBSSI protein (Fig. 4C) derived from transcriptional regulation of GBSSI gene (Fig. 4D).
GBSSI has been reported to be responsible for the biosynthesis of ELC (DP ≥ 500) of amylopectin as well as the amylose in rice (Takeda et al., 1987; Hizukuri, 1995), potato (Flipse et al., 1996), and Chlamydomonas (Delrue et al., 1992; Maddelein et al., 1994), indicating that trait 4 is also caused by the increase of GBSSI protein. The RT-PCR experiments in Chlamydomonas strongly suggest that the absence of SSIII in sta3 mutant, which is enriched in ELC, triggers a signal that is relayed to increase dramatically GBSSI mRNA and its activity abundance (Ral et al., 2006). On the other hand, the amylopectin with enriched ELC (DP700-1300) in antisense-SSIII potato transformants could be explained by the increase of GBSSI activity in vivo by the 5 times increase of ADP-Glc level compared with that of wild type, although amylose content and the GBSSI activity in vitro are not changed (Fulton et al., 2002). The increases in AGPase activity in developing endosperm in SSIII(a) mutant of rice (data not shown) and maize (Singletary et al., 1997) indicate the possibility that SSIII(a) deficiency leads to the increase of the ADP-Glc levels like as potato and maintains the amount of starch in the endosperm through the increase of the activities of other SS isoforms (especially those having high Km values like a GBSSI) in vivo, although it is very difficult to measure the level of ADP-Glc in amyloplast in rice because ADP-Glc in cereal endosperm is synthesized primarily in cytosol rather than in the plastid (Denyer et al., 1996; Beckles et al., 2001).
The trait 5 was shown in rice (Fig. 7) and maize (Jane et al., 1999). These changes in the range of DP < 30 of amylopectin in the rice SSIIIa mutants are almost opposite images of those of rice SSI mutants (Fig. 7D; Fujita et al., 2006), indicating that they are caused by the enhancement of SSI activity due to SSIIIa deficiency. The total soluble SS activity in the maize du1 mutant increased (Singletary et al., 1997; Cao et al., 1999) because of the specific enhancement of SSI activity based on the results of the immunodepletion of SS activity experiments (Cao et al., 1999). The soluble SS activity in the rice SSIIIa mutant also increased by approximately 2- and 3.5-fold more than that of ‘Nip’ under the condition of C(+)A(+) and C(+)A(−), respectively (Fig. 4A), and a marked enhancement of SSI activity was also shown by native-PAGE/SS activity staining (Fig. 2). These results show that up-regulation of SSI by a loss of the synthesis of SSIII(a) protein is a common compensatory phenomenon found in maize and rice SSIII(a) mutants. In developing rice SSIIIa mutants endosperm, it is most likely that the enhanced SSI due to SSIIIa deficiency elongates DP 10 to 15 chains from short DP 6 to 9 chains of A chains and DP 20 to 30 chains from DP 16 to 19 chains of B1 chains of amylopectin (Fig. 7, B and D). Moreover, the SSI mRNA abundance in SSIIIa rice mutant indicated that SSIIIa deficiency triggers a signal that is relayed to increase dramatically SSI activity by transcriptional regulation. In contrast, the short (DP < 30) proportion of chains of amylopectin of rice and maize mutants shows the quite different patterns in Chlamydomonas (Ral et al., 2006), potato (Lloyd et al., 1999; Fulton et al., 2002), and Arabidopsis (Zhang et al., 2005). The apparent discrepancy could be explained by the difference in the component of SS isoforms between Chlamydomonas, potato, Arabidopsis, and cereals.
In summary, though the literatures are limited in a few plants, the results with SSIII-deficient materials in green plants to date and those in this study show that the function of SSIII(a), elongation of long B chains connecting the cluster of amylopectin, is seemed to be widely conserved in green plant (traits 1 and 2), whereas pleiotropic effects of defect of SSIII(a) on other isozymes is not seemed to be conserved (traits 3, 4, and 5) and hence the starch structure and/or amylose content vary depending on green plant species. However, what are the triggers of indirect effects by SSIIIa deficiency remain to be resolved and further studies of transcriptional factors derived from SSIIIa deficiency will be required.
The Other Traits of Endosperm Starch in Rice SSIIIa Mutant Lines
The morphology and crystallinity of the starch granules of the SSI-deficient mutant were almost the same as those of wild type in rice plant (Fujita et al., 2006). In contrast, the crystallinity of starch granules in the SSIIIa mutant was reduced (Fig. 5D) and smaller, round-shaped starch granules accumulated in the amyloplasts of endosperm cells, in contrast to the sharp-edged polygonal granules in the wild type (Fig. 5, B and C). These phenomena could be caused by the enrichment of amylose content (Fig. 6) and the loose packing of starch granules in the amyloplast, respectively. These results suggest that the impact of SSIIIa deficiency on these characters is greater than that of the SSI deficiency in the rice endosperm. The starch granules of the maize du1 mutant (Yeh et al., 1981; Wang et al., 1993a) and antisense-SSIII potato (Edwards et al., 1999; Lloyd et al., 1999; Fulton et al., 2002) were smaller or abnormal in shape when compared to the controls. It is concluded that SSIII(a) of these plants has an indispensable function in the formation of normal starch granule morphology.
It is known that the gelatinization temperature of starch could be regulated by the ratio of DP ≤ 12 to 16 to DP > 12 to 16 chains in the A chains and the exterior part of the B chains of amylopectin, which compose the crystalline domains of starch granules (Hizukuri, 1986; Fujita et al., 2006). The decrease in short chains with DP 6 to 8 and the increase in chains with DP 14 to 15 elevate gelatinization temperature, while the increase in chains with DP 10 to 13 and decrease in chains with DP 16 to 19 (Fig. 7C) lower it in rice SSIIIa mutants. The total molar percent of the former (DP 6–8 and DP 14–15) is slightly smaller than that of the latter (DP 10–13 and DP 16–19), indicating that it is possible to predict that the gelatinization temperatures (To and Tp) of SSIIIa mutants will be lower than those of the wild type (Table II). In contrast, the To, Tp, and Tc of du1 mutant starch in maize are 2°C to 3°C higher than those of the wild type (Inouchi et al., 1991), although the chain-length distribution of amylopectin of maize is similar to that of rice. This inconsistency could be explained by the fact that the decrease of the chains with DP 17 to 18 of maize du1 mutant relative to the wild type is less than that obtained from the rice SSIIIa mutant, such that gelatinization temperature is higher in the maize du1 mutant compared to its wild-type parent.
The viscosity of the pasting starch of ss3a-1 at 95°C accounted for only 6% of the peak viscosity of that of ‘Nip’ (Fig. 8). The dramatic reduction in viscosity was also reproduced even when the starch granules were heated at a much slower rate of 1.5°C/min (data not shown). The viscosity of the pasting starch of ss3a-2 was also reduced to 40% of that of the wild type (Okuda et al., 2005), while that of the maize du1/wx mutant is also reduced to 53% of that of the maize wx mutant (Jane et al., 1999). These results suggest that the decrease of the viscosity of the pasting starch is closely related to the changes in the structure and components of endosperm starch due to the SSIII(a) deficiency in these plants. In contrast, that of antisense-SSII and SSII/SSIII potato is reduced about 50% of that of control, although that of antisense-SSIII potato is increased (Edwards et al., 1999).
The TAM contents of rice cultivars with diverse amylose contents are highly negatively correlated (R = −0.88) to the peak viscosity measured by RVA (Horibata et al., 2004). On the other hand, the peak viscosity of potato starch is known to be very high and seems to be related to its high phosphate monoester content (Jane et al., 1999). Other factors that might influence peak viscosity are the size of the starch granule and the swelling power of the starch. Furthermore, a significant reduction of peak viscosity occurs in some starch mutants of maize, such as ae and sug-1 (Jane et al., 1999) and chemically modified starches (N. Fujita, unpublished data). These reports and the results of this study show a higher possibility that the peak viscosity of pasting starch is caused by combined multiple factors including the size of starch granules, phosphate monoester content, the amylose content, swelling power of starch, as well as the structure of amylopectin. Further studies of several kinds of starches will be required to evaluate each contribution.
CONCLUSION
In summary, the changes in the physicochemical properties of SSIIIa mutants of rice are caused by changes in the structure and components of the endosperm starch derived directly from SSIIIa deficiency itself and indirectly from the transcriptional enhancement of SSI and GBSSI, respectively.
There was statistically no significant difference in the dehulled grain weight and starch content between SSIIIa mutants and the wild type (Table I), although the mutant starch is quite unique, indicating that the utilization of this endosperm starch could be expected for new functional foodstuffs such as resistant starch and gruel rice for people who have problems of deglutition and/or for industrial applications.
MATERIALS AND METHODS
Plant Materials
Two SSIIIa mutant lines were used in this study: the mutant line (ss3a-1) containing Tos17 insertion at the OsSSIIIa gene and ss3a-2 (EM790, Okuda et al., 2005), a product of NMU mutagenesis of rice ‘T65’ (Satoh and Omura, 1979). The SSIIIa activity band in native-PAGE/SS activity staining assay was lacking in these mutants (Fig. 2A). As control plants, SS3a-1+/+ (has no Tos17 insertion in the SSIIIa gene but had a genetic background common to the mutant line ss3a-1) and the parental ‘Nip’ and ‘T65’ were used. Rice plants were grown during the summer months in an experimental paddy field at Akita Prefectural University under natural environmental conditions.
Tos17 and MNU Mutagenesis and Screening for an SSIIIa-Deficient Mutant Line (ss3a-1 and ss3a-2)
Mutagenesis with Tos17 and pool sampling were performed as described by Hirochika (2001) and Kumar and Hirochika (2001). To screen for SSIIIa-deficient mutant lines, DNA fragments carrying the Tos17 transposon from DNA pools constructed using the three-dimensional sampling method from approximately 40,000 Tos17-containing plants were subjected to nested PCR using the transposon-specific primers T1F (for first PCR), T2F (for second PCR), T1R (for first PCR), and T2R (for second PCR, Fig. 1A) and the OsSSIIIa-specific primers 2R (for first PCR), 4R (for second PCR), 5R (for first PCR), and 6R (for second PCR, Fig. 1A) combinations. The PCR products were hybridized with the 2.2 kb OsSSIIIa cDNA SalI and NotI fragment probe (Fig. 1A), and positive products were gel purified and sequenced to identify those containing the real OsSSIIIa sequence and the location of Tos17 in the OsSSIIIa gene.
The first screening of SSIIIa mutants by MNU mutagenesis were carried out by the detection of deletion in the corresponding protein band in the native-PAGE/CBB staining analysis from 1,270 mutant lines induced from MNU treatment of fertilized egg cells in a rice ‘T65’. Out of 1,270 mutant lines, two mutants derived from the independent MNU treatment were lacking in the SSIIIa bands on native-PAGE/SS activity staining. One mutant line (EM790, ss3a-2) was used in this study.
Native-PAGE/Activity Staining and Enzyme Assay
Native-PAGE/activity staining of DBE and BE was performed using the methods of Fujita et al. (1999) and Yamanouchi and Nakamura (1992), respectively. SS activity staining was performed on 7.5% and 6.0% (w/v) acrylamide slab gel containing 0.8% (w/v) oyster glycogen (G8751, Sigma) and 0.1% rice amylopectin purified from rice waxy mutant line (EM-21), respectively, according to Nishi et al. (2001) with the modification that 0.5 m citrate was included in the reaction mixture for oyster gel but not for the rice amylopectin gel. The assay for AGPase was performed using the methods of Nakamura et al. (1989).
Preparation of Enzyme from Developing Rice Endosperm and Fractionation of Crude Enzyme Extract
Preparation of enzyme from developing rice grains of ‘Nip’, SSI mutant (e7−/−), and SSIIIa mutant (ss3a-1) at the midmilky stage (10 g fresh weight), and anion-exchange chromatography using HiTrapQ column was according to the method of Fujita et al. (2006).
SS Activity Analysis
One developing endosperm from around DAF 16 of each ‘Nip’, the SSI mutant (e7−/−), and the SSIIIa mutant (ss3a-1) were individually homogenized using a plastic pestle in 10 volumes of a cold grinding solution for SS (50 mm tris-HCl [pH 7.4], 2 mm EDTA, 5 mm dithiothreitol [DTT], 0.4 mm phenylmethylsulfonyl fluoride, and 12.5% [v/v] glycerol). The homogenate was centrifuged at 20,000g at 4°C for 10 min, and the supernatant was centrifuged once more under the same conditions. The supernatant was used for SS assays and the determination of the protein concentration as the crude enzyme extract.
The assay was conducted under three different conditions: C(+)A(+), 0.1 m Bicine-NaOH (pH 7.4), 0.5 m Citrate-Na (pH 7.4), 20 mm DTT, 2 mg/mL rice amylopectin, 2 mm ADP-[14C]Glc (100 dpm/nmoles; CFB144, Amersham), and the crude enzyme extract in a reaction volume of 100 μL; C(+)A(−), 0.1 m Bicine-NaOH (pH 7.4), 0.5 m Citrate-Na (pH 7.4), 20 mm DTT, 2 mm ADP-[14C]Glc (150 dpm/nmoles), and the crude enzyme extract in a reaction volume of 100 μL; or C(−)A(+), 0.1 m Bicine-NaOH (pH 7.4), 20 mm DTT, 2 mg/mL rice amylopectin, 2 mm ADP-[14C]Glc (150 dpm/nmoles), and the three time-diluted crude enzyme extract in a reaction volume of 100 μL. Reactions were done by incubation for 20 min at 30°C and terminated by heating for 5 min at 95°C. After cooling, reactions were mixed with 10 μL of 20 mg/mL oyster glycogen, and 140 μL of 7% KCl and polyglucans were precipitated by adding 750 μL of 100% methanol. The solvent was incubated for 5 min at 0°C and centrifuged at 20,000g for 10 min at 0°C. The pellet was mixed with 230 μL of 4.3% of KCl and precipitated by adding 750 μL of 100% methanol. These steps of mixing with the KCl solution and precipitation with methanol were performed three times more under the same conditions. The pellet was suspended with 500 μL of distilled water and the amount of the radioactive label incorporated was determined by liquid scintillation counting. All assays were performed in triplicate. Preliminary experiments demonstrated that the amount of 14C incorporated into methanol-precipitable glucans was linear with the amount of protein and the reaction time (approximately 20 min) in the assays.
The protein concentration of the crude enzyme extract was estimated using protein assay kit (no. 500-0006, Bio-Rad) according to the instructions provided.
Extraction of Proteins from the Developing and Mature Endosperm and Estimation of the Amount of Protein
One developing endosperm each taken at DAF 7, 16, and 25 of the SSIIIa mutant lines (ss3a-1 and ss3a-2) and the wild type (‘Nip’ and ‘T65’) was individually homogenized using plastic pestle in 5 volumes of a cold grinding solution containing 50 mm imidazol-HCl (pH 7.4), 8 mm MgCl2, 50 mm 2-mercaptoethanol, and 12.5% (v/v) glycerol. Mature endosperm samples were crashed with pliers then hand powdered using a mortar and pestle prior to homogenization. The homogenate was centrifuged at 20,000g at 4°C for 10 min, and the supernatant was set aside. The pellet was washed twice with 2 volumes of cold grinding solution and the pooled supernatants (80–150 μL) were used as the SP fraction. The residual pellet was homogenized in 3 volumes of a cold SDS solution containing 55 mm Tris-HCl (pH 6.8), 2.3% SDS, 5% 2-mercaptoethanol, and 10% glycerol. The homogenate was centrifuged at 20,000g at 4°C for 10 min and the supernatant was set aside. The pellet was washed twice more with 2 volumes of a cold SDS solution, and the pooled supernatants (80–170 μL) were used as the LBP fraction. The residual pellet (starch granules) was washed with 1 mL of DW and twice with 1 mL of acetone and dried under pressure. The starch granules (about 3 mg) were suspended with 10 volumes of an SDS solution and boiled for 7 min. After cooling, 10 volumes of the SDS solution were added while stirring. The slurry was centrifuged at 20,000g for 10 min at 4°C. The supernatant was set aside, and the pellet was resuspended in 10 volumes of an SDS solution and recentrifuged. The pooled supernatants (30–55 μL) were used as the TBP fraction.
The amount of SSI and GBSSI protein was estimated according to the methods of Fujita et al. (2006).
RT-PCR
RT-PCR of SSI, SSIIIa, and GBSSI gene of developing rice grains (DAF 10) in ss3a-1 and ‘Nip’ were performed according to the methods and conditions of Ohdan et al. (2005).
Analysis of Starch Granules of Endosperm
Estimation of α-polyglucan of rice seeds, pasting properties of endosperm starch measured by RVA, x-ray diffraction measurement, the measurement of thermal properties of endosperm starch by DSC, and observation of starch granules by SEM (JEOL-5600) were performed as described previously (Fujita et al., 2003, 2006).
For observation of endosperm cross section, rice seeds were dried completely under low pressure and cut across the short axis with razor blade. The surface was sputter coated with gold and observed by SEM.
Determination of the Mr of amylopectin was done by HPSEC-MALLS-RI according to the method of Fujita et al. (2003).
Preparation of Starch Granules or Amylopectin for Gel Filtration
Starch granules were prepared from polished rice following the cold-alkali method (Yamamoto et al., 1973, 1981). Rice amylopectin of SSI mutant was isolated and purified from rice starch granules using n-butanol according to Schoch (1954) as modified by Takeda et al. (1986).
Molecular Size Separation of Starch and Amylopectin by Sephacryl S-1000SF Chromatography
Molecular size separation of starch and amylopectin was performed by the method of Kubo et al. (1999). After chromatography, an aliquot of each fraction was used for the measurement of carbohydrate content by the phenolic sulfuric method and for the measurement of λmax value of glucan-iodine complex.
Debranching of Starch and Amylopectin with Isoamylase and Fractionation by Gel Filtration Chromatography
Starches and amylopectin were debranched with crystalline Pseudomonas isoamylase (no. 100780, Seikagaku-kogyo) by the method of Ikawa et al. (1981). Debranched materials were fractionated by gel filtration on a column (300 × 20 mm) of Toyopearl HW55S connected in series to three columns (300 × 20 mm) of Toyopearl HW50S according to the method of Inouchi et al. (1999). Carbohydrate content and λmax value of glucan-iodine complex of each fraction was measured by the method described above.
Chain-Length Distribution of Endosperm Amylopectin
Extraction of starch from mature and developing rice endosperm for amylopectin chain-length distribution was performed according to the methods of Fujita et al. (2001).
The chain-length distributions of α-polyglucans from endosperm were analyzed using the capillary electrophoresis methods of 0'Shea and Morell (1996) and Fujita et al. (2001) in a P/ACE MDQ carbohydrate system (Beckman Coulters).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AP004660 and AY100469.
Acknowledgments
The authors are grateful to Dr. Sayuri Akuzawa (Tokyo University of Agriculture, Tokyo) for helpful discussions, to Dr. Perigio B. Francisco Jr. (University of West Australia) for reading the manuscript, and to Mr. Takashi Ohdan (Core Research for Evolutional Science and Technology, Japan Science and Technology) and Mr. Tomokazu Ushijima (Kyushu University) for technical supports.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Naoko Fujita (naokof@akita-pu.ac.jp).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Abel GJW, Springer F, Willmitzer L, Kossman J (1996) Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J 10 981–991 [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54 207–233 [DOI] [PubMed] [Google Scholar]
- Beckles DM, Smith AM, ap Rees T (2001) A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol 125 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer CD, Liu KC (1985) The interaction of endosperm genotype and genetic background. Starch/Stärke 37 73–79 [Google Scholar]
- Boyer CD, Preiss J (1981) Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol 67 1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, James MG, Myers AM (2000) Purification and characterization of soluble starch synthases from maize endosperm. Arch Biochem Biophys 373 135–146 [DOI] [PubMed] [Google Scholar]
- Davis JH, Kramer HH, Whistler RL (1955) Expression of the gene du in the endosperm of maize. Agron J 47 232–235 [Google Scholar]
- Denyer K, Dunlap F, Thorbjornsen T, Keeling P, Smith AM (1996) The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol 112 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56 623–632 [DOI] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, van der Koornhuyse N, Maddelein ML, Fournet B, Ball S (1992) Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol 174 3612–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rossner U, Martin C, Smith AM (1999) A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J 17 251–261 [Google Scholar]
- Flipse E, Keetels CJAM, Jacobsen E, Visser RGF (1996) The dosage effect of the wild type GBSS allele is linear for GBSS activity but not for amylose content: absence of amylose has a distinct influence on the physico-chemical properties of starch. Theor Appl Genet 92 121–127 [DOI] [PubMed] [Google Scholar]
- Fujita N, Hasegawa H, Taira T (2001) The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.). Plant Sci 160 595–602 [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB Jr, Nakakita M, Harada K, Minaka N, Nakamura Y (1999) Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta 208 283–293 [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Suh SD, Wong KS, Jane JL, Ozawa K, Takaiwa F, Inaba Y, Nakamura Y (2003) Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol 44 607–618 [DOI] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Edwards A, Pilling E, Robinson HL, Fahy B, Seale R, Kato L, Donald AM, Geigenberger P, Martin C, et al (2002) Role of granule-bound starch synthase in determination of amylopectin structure and starch granule morphology in potato. J Biol Chem 277 10834–10841 [DOI] [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM (1998) Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4 118–122 [DOI] [PubMed] [Google Scholar]
- Hirose T, Terao T (2004) A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220 9–16 [DOI] [PubMed] [Google Scholar]
- Hizukuri S (1986) Polymodal distribution of the chain-lengths of amylopectins, and its significance. Carbohydr Res 147 342–347 [Google Scholar]
- Hizukuri S (1995) Starch, analytical aspects. In AE Eliasson, ed, Carbohydrates in Food. Marcel Dekker, Lund, Sweden, pp 347–429
- Horibata T, Nakamoto M, Fuwa H, Inouchi N (2004) Structural and physicochemical characteristics of endosperm starches of rice cultivars recently bred in Japan. J Appl Glycosci (1999) 51 303–313 [Google Scholar]
- Ikawa Y, Glover DV, Sugimoto Y, Fuwa H (1981) Some structural characteristics of starches of maize having a specific genetic background. Starch/Stärke 33: 9–13
- Inouchi N, Glover DV, Sugimoto Y, Fuwa H (1991) DSC characteristics of gelatinization of starches of single-, double-, and triple-mutants and their normal counterpart in the inbred Oh43 maize (Zea mays L.) background. Starch/Stärke 43 468–472 [Google Scholar]
- Inouchi N, Glover DV, Takaya T, Fuwa H (1983) Development changes in fine structure of starches of several endosperm mutants of maize. Starch/Stärke 35 371–376 [Google Scholar]
- Inouchi N, Nishi K, Tanaka S, Asai M, Kawase Y, Hata Y, Konishi Y, Yue S, Fuwa H (1999) Characterization of amaranth and quinoa starches. J Appl Glycosci (1999) 46 233–240 [Google Scholar]
- Jane JL, Chen YY, Lee LF, McPherson AE, Wong KS, Radosavljevic M, Kasemsuwan T (1999) Effects of amylopectin branch chain-length and amylose content on the gelatinization and pasting properties of starch. Cereal Chem 76 629–637 [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y (1999) The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol 121 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Hirochika H (2001) Applications of retrotoransposons as genetic tools in plant biology. Trends Plant Sci 6 127–134 [DOI] [PubMed] [Google Scholar]
- Li Z, Mouille G, Kosar-Hashemi B, Rahman S, Clarke B, Gale KR, Appels R, Morell MK (2000) The structure and expression of the wheat starch synthase III gene: motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Landschutze V, Kossmann J (1999) Simultaneous antisense inhibition of two starch-synthase isoforms in potato tubers leads to accumulation of grossly modified amylopectin. Biochem J 338 515–521 [PMC free article] [PubMed] [Google Scholar]
- Maddelein ML, Libessart N, Bellanger F, Delrue B, D'Hulst C, van der Koornhuyse N, Fontaine T, Wieruszeski JM, Decq A, Ball S (1994) Toward an understanding of the biogenesis of the starch granule. J Biol Chem 269 25150–25157 [PubMed] [Google Scholar]
- Mangelsdorf PC (1947) The inheritance of amylaceous sugary endosperm and its derivatives in maize. Genetics 32 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J, Sidebottom C, Debet M, Martin C, Smith AM (1996) Identification of the major starch synthase in the soluble fraction of potato tubers. Plant Cell 8 1121–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol 43 718–725 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Francisco PB Jr, Hosaka Y, Satoh A, Sawada T, Kubo A, Fujita N (2005) Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol 58 213–227 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yuki K, Park SY, Ohya T (1989) Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol 30 833–839 [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H (2001) Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol 127 459–472 [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56 3229–3244 [DOI] [PubMed] [Google Scholar]
- Okuda M, Aramaki I, Koseki T, Satoh H, Hashizume K (2005) Structural characteristics, properties, and in vitro digestibility of rice. Cereal Chem 82 361–368 [Google Scholar]
- O'Shea MG, Morell MK (1996) High resolution slab gel electrophoresis of 8-amino-1,3, 6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis 17 681–688 [DOI] [PubMed] [Google Scholar]
- Ral JP, Colleoni C, Wattebled F, Dauvillee D, Nempont C, Deschamps P, Li Z, Morell MK, Chibbar R, Purton S, et al (2006) Circadian clock regulation of starch metabolism establishes GBSSI as a major contributor to amylopectin synthesis in Chlamydomonas reinhardtii. Plant Physiol 142 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo N, Yu C, Park C-S, Baik Y, Park IM, Cho M-H, Bhoo SH, An G, Hahn FR, Jeon J-S (2007) Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep 26 1083–1095 [DOI] [PubMed] [Google Scholar]
- Sano Y (1984) Differential regulation of waxy gene expression in rice endosperm. Theor Appl Genet 68 467–473 [DOI] [PubMed] [Google Scholar]
- Sato K (1984) Starch granules in tissues of rice plants and their changes in relation to plant growth. JARQ 18 78–86 [Google Scholar]
- Satoh H, Omura T (1979) Induction of mutation by treatment of fertilized egg cell with N-methyl-N-nitrosourea in rice. J Fac Agric Kyushu Univ 24 165–174 [Google Scholar]
- Schoch TJ (1954) Purification of starch and the starch fractions. Methods Enzymol 3 5–6 [Google Scholar]
- Singletary GW, Banisadr R, Keeling PL (1997) Influence of gene dosage on carbohydrate synthesis and enzymatic activities in endosperm of starch-deficient mutants of maize. Plant Physiol 113 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Denyer K, Martin C (1997) The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol 48 67–87 [DOI] [PubMed] [Google Scholar]
- Takeda Y, Hizukuri S, Juliano BO (1986) Purification and structure of amylose from rice starch. Carbohydr Res 148 299–308 [Google Scholar]
- Takeda Y, Hizukuri S, Juliano BO (1987) Structures of rice amylopectins with low and high affinities for iodine. Carbohydr Res 168 79–88 [Google Scholar]
- Tsai CY (1974) The function of waxy locus in starch synthesis in maize endosperm. Biochem Genet 11 83–96 [DOI] [PubMed] [Google Scholar]
- Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y (2002) Mapping of a gene responsible for the difference in amylopectin structure between japonioca-type and indica-type rice varieties. Theor Appl Genet 104 1–8 [DOI] [PubMed] [Google Scholar]
- Wang YJ, White P, Pollak L, Jane J (1993. a) Characterization of starch structures of 17 maize endosperm mutant genotype with Oh43 inbred line background. Cereal Chem 70 171–179 [Google Scholar]
- Wang YJ, White P, Pollak L, Jane J (1993. b) Amylopectin and intermediate materials in starches from mutant genotypes of the Oh43 inbred line. Cereal Chem 70 521–525 [Google Scholar]
- Yamamoto K, Sawada S, Onogaki I (1973) Properties of rice starch prepared by alkali method with various condition. J Jap Soc Starch Sci 20 99–104 [Google Scholar]
- Yamamoto K, Sawada S, Onogaki I (1981) Effects of quality and quantity of alkali solution on the properties of rice starch. J Jap Soc Starch Sci 28 241–244 [Google Scholar]
- Yamanouchi H, Nakamura Y (1992) Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol 33 985–991 [Google Scholar]
- Yeh JY, Garwood DL, Shannon JC (1981) Characterization of starch from maize endosperm mutants. Starch/Stärke 33 222–230 [Google Scholar]
- Zhang X, Myers AM, James MG (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]