Abstract
We applied a computational pipeline based on comparative genomics to bacteria, and identified 22 novel candidate RNA motifs. We predicted six to be riboswitches, which are mRNA elements that regulate gene expression on binding a specific metabolite. In separate studies, we confirmed that two of these are novel riboswitches. Three other riboswitch candidates are upstream of either a putative transporter gene in the order Lactobacillales, citric acid cycle genes in Burkholderiales or molybdenum cofactor biosynthesis genes in several phyla. The remaining riboswitch candidate, the widespread Genes for the Environment, for Membranes and for Motility (GEMM) motif, is associated with genes important for natural competence in Vibrio cholerae and the use of metal ions as electron acceptors in Geobacter sulfurreducens. Among the other motifs, one has a genetic distribution similar to a previously published candidate riboswitch, ykkC/yxkD, but has a different structure. We identified possible non-coding RNAs in five phyla, and several additional cis-regulatory RNAs, including one in ε-proteobacteria (upstream of purD, involved in purine biosynthesis), and one in Cyanobacteria (within an ATP synthase operon). These candidate RNAs add to the growing list of RNA motifs involved in multiple cellular processes, and suggest that many additional RNAs remain to be discovered.
INTRODUCTION
Recent discoveries of novel structured RNAs (1–4) indicate that such RNAs are common in cells. To assist in discovering additional structured RNAs, we have developed an automated pipeline that can identify conserved RNAs within bacteria (5). This pipeline assembles the potential 5′ untranslated regions (UTRs) of mRNAs of homologous genes, and uses the CMfinder (6) program to predict conserved RNA structures, or ‘motifs’, within each set of UTRs. Automated homology searches are then employed to find additional examples of these motifs, which CMfinder uses to improve the secondary structure model and sequence alignment of each motif. The output is a set of alignments with a predicted RNA secondary structure, and these alignments are subsequently analyzed manually to make improvements to the model and to assess which motifs merit further study.
Although other automated searches for RNAs have been performed (7–15), our pipeline (5) is distinguished from these by three features. First, our pipeline uses CMfinder, which can discover a motif even when some input sequences either do not contain the motif or include motif representatives carrying unrelated sequence domains. CMfinder also can produce a useful alignment even with low sequence conservation, however, the algorithm will exploit whatever sequence similarity is present. Second, our pipeline integrates homology searches to automatically refine the alignment and structural model for each motif. Third, since our pipeline aligns UTRs of homologous genes, it is well suited to find cis-regulatory RNAs, and its dependence on sequence conservation is further reduced.
Indeed, using the bacterial phylum Firmicutes as a test case, we previously demonstrated that this pipeline makes useful predictions for virtually all known cis-regulatory RNAs (5). The pipeline also finds motifs that are likely to be trans-encoded, or ‘non-coding RNAs’ (ncRNAs), when these happen to be upstream of homologous genes.
In the present report, we describe the use of this pipeline to find structured RNAs amongst all bacteria whose genomes have been sequenced. We describe 22 novel motifs that are likely to be conserved, structured RNAs. We were particularly interested in discovering riboswitches, a type of structured RNA usually found in mRNAs that directly senses a specific small molecule and regulates gene expression (16,17). Subsequent experiments have confirmed that two of these motifs are novel riboswitches. The first binds SAH (S-adenosylhomocysteine) (J.X.W., D. Rivera, E.R.L., R.R.B., in preparation; Figure 1) and the second is a SAM (S-adenosylmethionine)-binding riboswitch in Streptomyces coelicolor and related species (Z.W., E.E. Regulski, R.R.B., unpublished data). Experimental evidence with another riboswitch candidate indicates that it senses molybdenum cofactor or ‘Moco’ (E.E. Regulski, R. Moy, R.R.B., unpublished data).
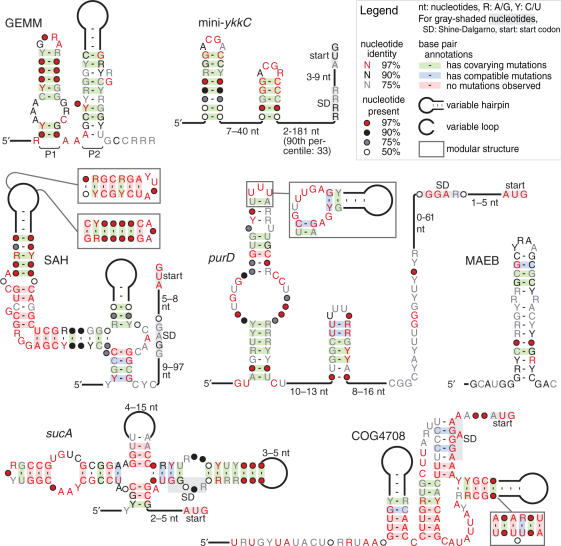
Figure 1.
Consensus sequences and structures are depicted for seven of the 22 motifs identified. Other motifs are presented as Supplementary Data, as are the alignments on which these diagrams are based. Calculations for conservation of nucleotide identity/presence and evidence of covariation are described in the ‘Materials and methods’ section. Proposed base pairs with more than 5% non-canonical or missing nucleotides are not classified as covarying. Note that the levels of nucleotide conservation are affected both by biochemical constraints on the motif and by phylogenetic diversity; motifs with limited range (e.g. the COG4708 motif) will appear more conserved. Some covarying positions in variable-length stems are not shown.
A candidate of particular interest is the Genes for the Environment, for Membranes, and for Motility (GEMM) motif, which has properties that are typical of riboswitches. For example, GEMM is a widespread and highly conserved genetic element that, in 297 out of 309 cases, is positioned such that it is likely to be present in the 5′ UTR of the adjacent open reading frame (ORF). The genes putatively regulated by GEMM are typically related to sensing and reacting to extracellular conditions, which suggests that GEMM might sense a metabolite produced for signal transduction or for cell–cell communication.
Some characteristics of all 22 predicted RNA motifs are summarized in Table 1, and we discuss the features and possible biological roles of some candidates in the ‘Results’ section. Additional information on all candidates, including annotated multiple sequence alignments, and collections of taxonomy and nearby genes, are presented in Supplementary Data. Raw pipeline predictions are accessible at http://bliss.biology.yale.edu/cmfinder_pipeline_output.
Table 1.
Summary of putative structured RNA motifs
| Motif | RNA? | Cis? | Switch? | Phylum/class | M,V | Cov. | # | Non-cis |
|---|---|---|---|---|---|---|---|---|
| GEMM | Y | Y | y | Widespread | V | 21 | 322 | 12/309 |
| Moco | Y | Y | Y | Widespread | M,V | 15 | 105 | 3/81 |
| SAH | Y | Y | Y | Proteobacteria | M,V | 22 | 42 | 0/41 |
| SAM-IV | Y | Y | Y | Actinobacteria | V | 28 | 54 | 2/54 |
| COG4708 | Y | Y | y | Firmicutes | M,V | 8 | 23 | 0/23 |
| sucA | Y | Y | y | β-proteobacteria | 9 | 40 | 0/40 | |
| 23S-methyl | Y | y | n | Firmicutes | 12 | 38 | 1/37 | |
| hemB | Y | ? | ? | β-proteobacteria | V | 12 | 50 | 2/50 |
| (anti-hemB) | (n) | (n) | (37) | (31/37) | ||||
| MAEB | ? | Y | n | β-proteobacteria | 3 | 662 | 15/646 | |
| mini-ykkC | Y | Y | ? | Widespread | V | 17 | 208 | 1/205 |
| purD | y | y | ? | ε-proteobacteria | M | 16 | 21 | 0/20 |
| 6C | y | ? | n | Actinobacteria | 21 | 27 | 1/27 | |
| alpha-transposases | ? | N | N | α-proteobacteria | 16 | 102 | 39/99 | |
| excisionase | ? | ? | n | Actinobacteria | 7 | 27 | 0/27 | |
| ATPC | y | ? | ? | Cyanobacteria | 11 | 29 | 0/23 | |
| Cyano-30S | Y | Y | n | Cyanobacteria | 7 | 26 | 0/23 | |
| lacto-1 | ? | ? | n | Firmicutes | 10 | 97 | 18/95 | |
| lacto-2 | y | N | n | Firmicutes | 14 | 357 | 67/355 | |
| TD-1 | y | ? | n | Spirochaetes | M,V | 25 | 29 | 2/29 |
| TD-2 | y | N | n | Spirochaetes | V | 11 | 36 | 17/36 |
| coccus-1 | ? | N | N | Firmicutes | 6 | 246 | 112/189 | |
| gamma-150 | ? | N | N | γ-proteobacteria | 9 | 27 | 6/27 |
‘RNA’ = functions as RNA (as opposed to dsDNA), ‘Cis’ = cis-regulatory, ‘Switch’ = riboswitch. Evaluation: ‘Y’ = certainly true, ‘y’ = probably true, ‘?’ = possible, ‘n’ = probably not, ‘N’ = certainly not. These evaluations were conducted prior to experimental examinations. Criteria for classification as an RNA include evidence of covariation and variable-length or modular stems. Evidence of covariation is strongest with covarying nucleotide positions for which surrounding sequence conservation permits high confidence that the covarying positions are correctly aligned, and was assessed manually based on alignments. Probable cis-regulatory motifs were consistently located upstream of homologous genes, or a set of genes with related functions, and often had features typical of known gene-control mechanisms (a transcription terminator or stem sequestering the Shine–Dalgarno sequence). Likely riboswitches were motifs that were classified as an RNA and a cis-regulatory element, showed evidence of high conservation of nucleotides at some positions, exhibited a complex secondary structure (not just a hairpin) and were associated with genes that were judged likely to be controlled by a small molecule. Motifs are characterized in detail according to these criteria in Supplementary Data. Remaining columns are ‘Phylum/class’ (phylum containing the motif, or class for Proteobacteria), ‘M,V’ (‘M’ = has modular stems, which are stems that are only sometimes present, ‘V’ = variable-length stems), ‘Cov.’ = number of covarying paired positions (see ‘Methods’ section; note that it is not advisable to rank motifs solely by this number, but rather the alignment as a whole should be evaluated), ‘#’ = number of representatives, ‘Non-cis’ = X/Y where X is number of representatives that are not in a 5′ regulatory configuration to a gene and Y is the number of representatives within sequences that have annotated genes (some RefSeq sequences lack annotations). Moco and SAM-IV riboswitch data will be presented in future reports. Gamma-150 and coccus-1 are only in the supplement.
MATERIALS AND METHODS
Identification of candidate RNA motifs
The potential 5′ UTRs of genes classified in the Conserved Domain Database (18) version 2.08 were used as input for our computational pipeline (5). Completed genome sequences and gene positions were taken from RefSeq (19) version 14, but we eliminated genomes whose gene content was highly similar to other genomes. Our UTR extraction algorithm (20) accounted for the fact that a UTR might not always be immediately upstream of the gene due to operon structure.
For each conserved domain, the collected sequences of potential UTRs were given as input to CMfinder (6) version 0.2, which produced local multiple sequence alignments of structurally conserved motifs within the UTR sets. These alignments were then used to search for additional homologs within annotated intergenic regions, except that intergenic regions were extended by 50 nt on both ends to account for misannotated ORFs. This homology search was performed with the RaveNnA program version 0.2f (21–23) with ML-heuristic filters (23), Covariance Model (24) in global mode implemented by Infernal (25) version 0.7 and an E-value (26) cutoff of 10. CMfinder then refined its initial alignment using these new homologs. Known RNAs were detected based on the Rfam Database (27) version 7.0. Predictions were scored based on phylogenetic conservation of short sequences (20), diversity of species and structural criteria. The pipeline algorithm is described in more detail elsewhere (5). The Supplementary Data includes a list of software and databases used.
We split bacterial genomic sequence data into groups, and performed UTR extraction, motif prediction and homology search on each group separately. This was motivated by the fact that UTRs in different phyla often do not contain the same motif. However, phyla with few sequenced members were coalesced based on taxonomy to try to assemble sufficient sequence data to predict a motif. Phyla with only one or two sequenced members (e.g. Chloroflexi) were ignored. The following eight groups were used: (1) Firmicutes, (2) Actinobacteria, (3) α-proteobacteria, (4) β-proteobacteria, (5) γ-proteobacteria, (6) δ- and ε-proteobacteria, (7) Cyanobacteria and (8) Bacteroidetes, Chlorobi, Chlamydiae, Verrucomicrobia and Spirochaetes.
Analysis of candidate motifs and homology search strategies
We then selected promising motifs, further analyzed them by performing additional homology searches and edited their alignments with RALEE (28). We used NCBI BLAST (29), Mfold (30), Rnall (31) (to identify rho-independent transcription terminators), CMfinder and RaveNnA to assist in these analyses. Information on metabolic pathways associated with motifs was retrieved from KEGG (32). To expand alignments by identifying additional structured elements, we extended alignments on their 5′ and 3′ ends by 50–100 nt, and realigned as necessary using either CMfinder or by manual inspection.
The additional searches for homologs used RaveNnA in several ways. We found both global- and local-mode (25) Covariance Model searches gave complementary results. Sequence databases used in manually directed searches were the ‘microbial’ subset of RefSeq version 19 (19), and environmental shotgun sequences from acid mine drainage (33) (GenBank accession AADL01000000) and Sargasso Sea (34) (AACY01000000). Additional marine shotgun sequences were used for the sucA and ATPC motifs (35).
Because the full set of sequences is roughly 3.2 billion nucleotides, searches can report many false positives, especially for shorter motifs. When appropriate, we searched four kinds of subsets of these sequences. First, we did not always use the environmental sequence data. Second, we sometimes searched only genomes in the bacterial group (e.g. β-proteobacteria) from which the motif was originally derived. Third, we sometimes searched only intergenic regions (extended by 50 nt as before). Fourth, we used the BLAST program tblastn to search for genes homologous to those associated with the motif. RaveNnA searches were then conducted on the 2 kb upstream of these matches (which is expected to contain the 5′ UTR), and 200 nt downstream (since apparent coding homology might extend upstream of the true ORF, causing BLAST to misidentify the start codon). Using the motif's bacterial group as the BLAST database facilitated the discovery of highly diverged homologs. For example, a search upstream of purD genes in ε-proteobacteria revealed homologs of the purD motif (see later) with a truncated stem. Additionally, with such small databases, we can forego the ML-heuristic filters in RaveNnA. When the full sequence set is used as the BLAST database, it can help to find homologs in other phyla.
Rejection of motifs
Motifs were rejected from further study when they failed to show features that are characteristic of structured RNAs. To help reject motifs with spurious predictions of structure, we performed homology searches using sequence information only, by removing all base pairs in the predicted structure. Sequence-based matches that do not conserve the structure indicate that the predicted structure is incorrect. However, such homologs can be missed by Covariance Models, which assume the structure is conserved. Several motifs were rejected using this strategy when sequence homologs revealed that the proposed structure was, in fact, poorly conserved.
We also generally rejected repetitive elements, which we defined as elements appearing many times per genome and showing extremely high sequence conservation, but little structure conservation. Although some of these repetitive elements could correspond to structured RNAs, there is a little support for such a hypothesis without good evidence of covariation.
Establishing the extent of conservation and covariation for consensus diagrams
To establish the extent of conservation reflected in consensus diagrams (e.g. in Figure 1), sequences were weighted to de-emphasize highly similar homologs. Weighting used the GSC algorithm (36), as implemented by Infernal (25), and weighted nucleotide frequencies were then calculated at each position in the multiple sequence alignment. To classify base pairs as covarying, the weighted frequency of Watson–Crick or G–U pairs was calculated. However, aligned sequences in which both nucleotides were missing or where the identity of either nucleotide was uncertain (e.g. was ‘N’, signifying any of the four bases) were discarded. Classification as a covarying position was made if two sequences had Watson–Crick or G–U pairs that differ at both positions amongst sequences that carry the motif. If only one position differed, the occurrence was classified as a compatible mutation. However, if the frequency of non-Watson–Crick or G–U pairs was more than 5%, we did not annotate these positions as covarying or as compatible mutations.
RESULTS
Evaluation and analysis of novel RNA motifs
Promising structured RNA motifs predicted by the CMfinder pipeline were examined manually to refine the consensus sequence and structural models (see ‘Materials and methods’ section) and to provide information on possible function. Key findings for each candidate are summarized in Table 1. Of the 22 motifs identified, seven are depicted in Figure 1 and the remainder are depicted in Supplementary Data.
Our decision to select a candidate RNA motif for further study was based on a qualitative evaluation of conservation of both sequence and structure, covariation and gene context (e.g. whether or not the motif is consistently upstream of a specific gene family). Conserved sequence is important because structured RNAs usually have many conserved nucleotides in regions that form complex tertiary structures or are under other constraints. Structured RNAs sometimes have variable-length stems or ‘modular stems’ (stems that are present in some but not all representatives). The fact that both sides of a stem either appear together or neither appear is analogous to covariation, and is evidence that the structure is conserved.
Cis-regulatory RNAs such as riboswitches are regions of mRNAs that regulate gene expression. In bacteria, most cis-regulatory RNAs occur in the 5′ UTR of the mRNA under regulation. Although it is not possible to reliably predict the transcription start site, we declare representatives of a motif as positioned in a ‘5′ regulatory configuration’ to a gene when the element could be in the 5′ UTR of an mRNA (if the transcription start site is 5′ to the element). When most or all representatives of a motif are in a 5′ regulatory configuration to a gene, this is evidence that the motif might have a cis-regulatory function.
Cis-regulatory RNAs often have one of two noteworthy structural features: rho-independent transcription terminators, or stems that overlap the Shine–Dalgarno sequence (bacterial ribosome-binding site) (16). Rho-independent transcription terminators usually consist of a strong hairpin followed by four or more U residues (37). Regulatory RNA domains can control gene expression by conditionally forming the terminator stem. Similarly, conditionally formed stems can overlap the Shine–Dalgarno sequence, thereby regulating genes at the translational level (16).
Some motifs identified in this study consist of a single or tandem hairpins. It is possible that some of these are protein-binding motifs in which a homodimeric protein binds to a given DNA-based element in opposite strands. For convenience, we also describe such motifs as having a 5′ regulatory configuration, even though they might not form structured RNAs or function at the mRNA level.
The GEMM motif
GEMM is widespread in bacteria and appears to have a highly conserved sequence and structure suggestive of a function that imposes substantial biochemical constraints on the putative RNA. We found 322 GEMM sequences in both Gram-positive and Gram-negative bacteria. It is common in δ-proteobacteria, particularly in Geobacter and related genera. Within γ-proteobacteria, it is ubiquitous in Alteromonadales and Vibrionales. It is also common in certain orders of the phyla Firmicutes and Plantomycetes. Prominent pathogens with GEMM include the causative agents of cholera and anthrax.
Out of 309 GEMM instances where sequence data includes gene annotations, GEMM is in a 5′ regulatory configuration to a gene in 297 cases, implying a cis-regulatory role. Genes presumably regulated by GEMM display a wide range of functions, but most genes relate to the extracellular environment or to the membrane, and many are related to motility.
GEMM consists of two adjacent hairpins (paired regions) designated P1 and P2 (Figure 1). P1 is highly conserved in sequence and structure, and consists of 2- and 6-bp stems separated by a 3-nt internal loop and capped by a terminal loop. The internal loop is highly conserved, and the terminal loop is almost always a GNRA tetraloop (38). The P1 stem exhibits considerable evidence of covariation at several positions, and is highly conserved in structure over a wide range of bacteria. This fact, and the more modest covariation and variable-length stems of P2, provide strong evidence that GEMM functions as a structured RNA. The sequence linking P1 and P2 is virtually always AAA, with only two exceptions in 322 examples.
The P2 hairpin shows more modest conservation than P1. When the P1 tetraloop is GAAA, a GNRA tetraloop receptor usually appears in P2. This receptor is often the well-known 11-nt motif, which might be favored by GAAA loops (39), but some sequences could be novel tetraloop receptors. When P1 has a GYRA tetraloop, the receptor-like sequence is almost never present, although a bulge nearer the P2 base is sometimes found (Figure 2).
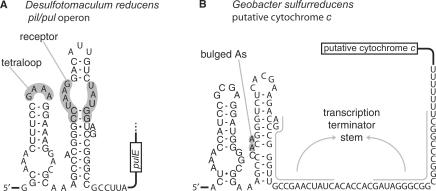
Figure 2.
Common features of GEMM motifs. Two GEMM instances were selected to illustrate common features, although these two examples do not represent the full 322 GEMMs (see Supplementary Data). (A) This putative RNA contains a canonical GNRA tetraloop and receptor (gray regions). Almost 50% of GEMM instances contain a likely tetraloop receptor. Only the first gene in the downstream operon is shown. (B) Some GEMM RNAs lack the tetraloop receptor, but there are two extra bulged A residues (gray shading) that are found in roughly half of the sequences lacking a receptor. Gray overlined nucleotides can fold to form a stem of a rho-independent transcription terminator (followed by 3′-trailing Us). This terminator appears to compete with the 3′ part of the P2 stem (right-most hairpin). 78 of 322 GEMM instances have predicted transcription terminators overlapping P2.
Many instances of GEMM include a rho-independent transcription terminator hairpin. The 5′ side of the terminator stem often overlaps (and presumably competes with) the 3′ side of the P2 stem (Figure 2B). If GEMM is a riboswitch, ligand binding could stabilize the proposed P1 and P2 structure, thus preventing the competing transcription terminator from forming. In this model, higher ligand concentrations will increase gene expression. One third of GEMM representatives in δ-proteobacteria, and some in other taxa, are in a ‘tandem’ arrangement, wherein one instance appears 3′ and nearby to another in the same UTR. Such arrangements of regulatory RNAs are implicated in more sophisticated control of gene expression than is permitted by a single regulatory RNA configuration (40–42).
An understanding of the biological role of GEMM will likely shed light on the broad variety of microbial processes that it appears to regulate. In fact, GEMM is implicated in two systems that are already the object of several studies in the species Vibrio cholerae and in Geobacter sulfurreducens. Vibrio cholerae causes cholera in humans, but spends much of its lifecycle in water, where it can adhere to chitin-containing exoskeletons of many crustaceans. Chitin, a polymer of GlcNAc (N-acetylglucosamine), has been shown to affect expression of many V. cholerae genes (43). GEMM appears to regulate two of these chitin-induced genes. The first, gbpA, is important for adhering to chitin beads (43) and human epithelial cells (44), as well as infection of mice (44).
The second chitin-induced gene is tfoXVC. Remarkably, chitin induces natural competence in V. cholerae (45), and tfoXVC expression is essential for this competence. V. cholerae has two genes that match the CDD models COG3070 and pfam04994 that correspond to separate tfoX domains. Both domains yield RPSBLAST (46) E-values better than 10−25. One of these is tfoXVC (locus VC1153). GEMM appears to regulate the other, which we call tfoXGEMM (VC1722). Thus, in V. cholerae and related bacteria, GEMM might participate in chitin-induced competence, or even regulate competence in environments not containing elevated chitin concentrations.
Geobacteria sulfurreducens and related δ-proteobacteria can generate ATP by oxidizing organic compounds, using metal ions such as Fe(III) as electron acceptors (47). GEMM is associated with pili assembly genes in Geobacter species. Pili in G. sulfurreducens have been shown to conduct electricity (48), and are thus a part of the process of reducing metal ions.
Moreover, GEMM appears to regulate seven cytochrome c genes in G. sulfurreducens. Although this bacteria has 111 putative cytochrome c genes, five of the seven GEMM-associated genes have been identified in previous studies, and might have special roles. OmcS (Outer-Membrane Cytochrome S) is one of two proteins that are highly abundant on the outer membrane of G. sulfurreducens, and is required for reducing insoluble Fe(III) oxide, but not for soluble Fe(III) citrate (49). OmcG and OmcH are necessary for production of OmcB, an essential cytochrome c in many conditions (50). OmcA and OmcT are associated with OmcG, OmcH or OmcS. Only four other Omc annotations remain in G. sulfurreducens that have no direct GEMM association: OmcB, OmcC, OmcE and OmcF.
Unlike known riboswitches, GEMM is associated with a great diversity of gene functions (Table 2). This observation indicates that, if GEMM is a riboswitch, it is not serving as a typical feedback sensor for control of a metabolic pathway. Rather, GEMM more likely senses a second-messenger molecular involved in signal transduction or possibly cell–cell communication (51). In this model, different bacteria use GEMM and its signaling molecule to control different processes. The fact that many GEMM-associated genes encode signal transduction domains could suggest a mechanism by which many of the signal transduction proteins are regulated. Preliminary biochemical results indicate that GEMM RNAs indeed serve as aptamer components of a new-found riboswitch class (N.S., E.R.L, R.R.B., unpublished data).
Table 2.
Gene families that appear to be regulated by GEMM in more than one instance
| Functional role | Gene families |
|---|---|
| Pili and flagella | cpaA, flgB, flgC, flgG, fliE, fliG, fliM, motA, motB, papC, papD, pilM, pilO, pilQ, pulF |
| Secretion (related to pili/flagella) | fhaC, fliF, fliI, hofQ |
| Chemotaxis regulator | cheW, cheY, methyl-accepting chemotaxis protein, Cache domain (classically associated with chemotaxis receptors in bacteria) |
| Signal transduction | PAS domain, histidine kinase, HAMP (Histidine kinases, Adenylyl cyclases, Methyl binding proteins, Phosphatases), HD-GYP domain, GGDEF domain |
| Chitin | chitin/cellulose binding domain, chitinase, carbohydrate-binding protein |
| Membranes | lysin domain (involved in cell wall remodeling, but might have general peptidoglycan binding function), uncharacterized outer membrane proteins and lipoproteins, putative collagen binding protein |
| Peptides | non-ribosomal peptide synthase, condensation domain (synthesis of peptide antibiotics), transglutaminase-like cysteine protease, subtilase (superfamily of extracellular peptidases) |
| Other | tfoX (regulator of competence), cytochrome c |
The SAH motif
The SAH motif is highly conserved in sequence and structure (Figure 1), showing covariation within predicted stem regions, including modular and variable-length stems. The SAH motif is found in a 5′ regulatory configuration to genes related to SAH (S-adenosylhomocysteine) metabolism, primarily in β- and some γ-proteobacteria, and especially the genus Pseudomonas. SAH is a part of the S-adenosylmethionine (SAM) metabolic cycle, whose main components include the amino acid methionine. SAH is a byproduct of enzymes that use SAM as a cofactor for methylation reactions. Typically, SAH is hydrolyzed into homocysteine and adenosine. Homocysteine is then used to synthesize methionine, and ultimately SAM.
High levels of SAH are toxic to cells because SAH inhibits many SAM-dependent methyltransferases (52). Therefore cells likely need to sense rising SAH concentrations and dispose of this compound before it reaches toxic levels. The genes that the SAH motif associates with are S-adenosylhomocysteine hydrolase (ahcY), cobalamin-dependent methionine synthase (metH) and methylenetetrahydrofolate reductase (metF), which synthesizes a methyl donor used in methionine synthesis. This genetic arrangement of the SAH motif and its high degree of conservation are consistent with a role in sensing SAH and activating the expression of genes whose products are required for SAH destruction. Indeed, biochemical and genetic evidence supports the hypothesis that this motif is an SAH-sensing riboswitch (J.X.W., D. Rivera, E.R.L. and R.R.B., in preparation).
The COG4708 motif
This motif is found upstream of COG4708 genes in some species of Streptococcus and in Lactococcus lactis, although some instances of the COG4708 gene family in Streptococcus lack the putative RNA motif. COG4708 genes are predicted to encode membrane proteins.
Although the COG4708 motif is highly constrained phylogenetically and has only six unique sequences, it shows covariation, modular stems and variable-length stems (Figure 1). The motif has a pseudoknot that overlaps the putative Shine–Dalgarno sequences of COG4708 genes, which suggests that the motif encodes a cis-regulator of these genes.
We recently characterized a riboswitch that senses the modified nucleobase preQ1 (53). Since this riboswitch is associated with COG4708, we proposed that COG4708 is a transporter of a metabolite related to preQ1. Therefore, we hypothesize that the COG4708 motif is also a preQ1-sensing riboswitch. Preliminary experiments support this hypothesis (M. Meyer, A.R. and R.R.B., unpublished data). The COG4708 motif shares no similarity in sequence or structure with the previously characterized preQ1-sensing riboswitch (53).
The sucA motif
The sucA motif is only found in a 5′ regulatory configuration to sucA genes, which are likely co-transcribed with the related downstream genes sucB/aceF and lpd. The products of these three genes synthesize succinyl-CoA from 2-oxoglutarate in the citric acid cycle. All detected instances of the sucA motifs are in β-proteobacteria in the order Burkholderiales. Although many nucleotides in the sucA motif are strictly conserved, those that are not show covariation and contain very few non-canonical base pairs (Figure 1). The motif has stems that overlap the putative Shine–Dalgarno sequence, so the sucA motif probably corresponds to a cis-regulatory RNA. Note that the exact position of the putative Shine–Dalgarno sequence is inconsistent among sucA motif instances, so is not well reflected in Figure 1 (see alignment in Supplementary Data). The relatively complex structure of the sucA motif suggests that it might be a riboswitch. However, it is difficult to evaluate its degree of sequence and structure conservation since the motif is not broadly distributed.
The 23S-methyl motif
This motif is consistently upstream of genes annotated as rRNA methyltransferases that probably act on 23S rRNA. The one exception occurs when 23S-methyl RNA is roughly 3 kb from the 23S rRNA methyltransferase ORF, with other genes on the opposite strand in the intervening sequence. The 23S-methyl motif is confined to Lactobacillales, which is an order of Firmicutes.
The 23S-methyl motif consists of two large hairpins. The second hairpin ends in a run of Us and appears to be a rho-independent transcription terminator. Both stems have considerable covariation, providing strong evidence that they are part of a functional RNA. Although the structural model shows that many paired positions sometimes have non-canonical base pairs, each instance of the motif consists predominantly of energetically favorable pairs, as shown in Supplementary Data. The presence of a putative transcription terminator suggests that this is a cis-regulatory RNA. Since 23S rRNA methyltransferase interacts with an RNA substrate, it might autoregulate its expression using the 23S-methyl motif, in a manner similar to autoregulation of ribosomal protein genes (54).
The hemB/anti-hemB motif
This motif is found in a variety of β-proteobacteria, especially Burkholderia. There is some ambiguity as to the DNA strand from which it might be transcribed, because its structure exhibits comparable covariation and conservation in both directions. In one direction, it is often upstream of hemB genes. It could be a cis-regulatory RNA in this direction, but there are two genes, not homologous to each other, that are immediately downstream of hemB motif representatives and these are positioned on the wrong strand to be controlled in the usual manner of cis-regulatory RNAs. In the other direction (anti-hemB), the motif is not typically in the 5′ UTRs of genes. The anti-hemB motif ends in a transcription terminator hairpin. Many genes downstream of anti-hemB instances are on the opposite strand, therefore we propose that anti-hemB could encode a non-coding RNA.
MAEB (metabolism-associated element in Burkholderia)
This motif consists of a single hairpin with several conserved positions (Figure 1). It is widespread in Burkholderia, a genus of β-proteobacteria. It typically occurs multiple times in succession (2–6 copies) with conserved linker sequences, but ranges to as many as 12 copies in two instances. In 141 occurrences of single or repetitive MAEB motifs, 132 are in a 5′ regulatory configuration to a gene. In fact, many of these genes are directly involved in primary metabolism (e.g. genes involved in biosynthesis, catabolism or transport of small molecules), and not genes such as DNA repair, replication, signaling or motility. Out of the 46 conserved domains (excluding hypothetical genes) downstream of MAEB in more than one instance, at least 42 are annotated as participating in primary metabolism (see Supplementary Data for list). There are many bacterial genes not involved in primary metabolism, so these data suggest a functional association with metabolic gene control.
There is a possible relationship between MAEB and cellular response to abundant glycine. MAEB is frequently associated with gcvP and gcvT, which are part of the glycine cleavage system, wherein excess glycine feeds into the citric acid cycle. MAEB is also associated with several citric acid cycle genes (see Supplementary Data). However, MAEB is associated with some other genes with a more tenuous relationship to glycine or the citric acid cycle. It is tempting to infer a relationship to the glycine cleavage system because the highest number of MAEB repeats are associated with the gcv genes in this system. Moreover, there exists at least one riboswitch class that binds glycine (42), but this class is present in only one copy per genome in organisms with MAEB, possibly leaving a role in glycine regulation for MAEB.
Representatives of the MAEB motif exhibit covariation that preserves base pairing, but others carry mutations that disrupt pairing. This fact suggests that it could, in fact, be a DNA-sequence that binds a protein dimer, such that each protein unit binds to opposite strands. However, one characteristic is inconsistent with this hypothesis. Nucleotides at one pair of symmetric positions are conserved as purines (A or G) in both sides of the stem. Since purines are never Watson–Crick pairs, they could not have the same identity on opposite strands. Although it is expected that instances of a DNA-binding motif will differ, the symmetric purines imply that the motif itself (and not merely the instances) has a distinct pattern on opposite strands.
Although we cannot rule out the possibility that MAEB could be a repetitive element, its association with metabolic genes argues against this hypothesis. It is also possible that MAEB is part of a protein-binding RNA like CsrB. CsrB is an RNA with roughly 18 hairpins, each of which can bind one CsrA protein subunit (55).
The mini-ykkC motif
The mini-ykkC motif consists of two tandem hairpins whose stems show considerable covariation and whose loops have characteristic ACGR motifs (Figure 1). Mini-ykkC is widespread in α-, β- and γ-proteobacteria, with additional examples in other taxa. We named this motif mini-ykkC because it appears to be a cis-regulator of a set of genes similar to that of the previously described ykkC/yxkD motif (7) (hereafter termed ‘ykkC’). The Supplementary Data lists all eight conserved domains common to ykkC and mini-ykkC. However, the structures of ykkC and mini-ykkC appear to be unrelated.
The ykkC motif is a highly structured and broadly conserved motif that was proposed previously to be a promising riboswitch candidate. The simple structure of mini-ykkC, however, is uncharacteristic of most other riboswitches, though its broad phylogeny suggests a function that dictates broad conservation. Mini-ykkC appears to be a cis-regulatory element because it is associated with a relatively narrow set of gene functions and it is near to their coding sequences (90% are within 33 nt of the Shine–Dalgarno sequence). We propose that mini-ykkC serves the same (but currently unknown) role as the ykkC motif, although the mechanisms used to control gene expression could be different.
We note that there might be instances of mini-ykkC with only one hairpin. However, we did not explore this possibility because the simplicity of the single hairpin would lead to a prohibitively high false positive rate in genome-scale searches. This issue is not a problem for the full, two-hairpin motif (see Supplementary Data).
The purD motif
The purD motif is found upstream of all purD genes in fully sequenced ε-proteobacteria (e.g. Campylobacter and Helicobacter). The purD gene encodes GAR (phosphoribosylglycinamide) synthetase, which is involved in purine biosynthesis. The purD motif shows covariation and modular stems, although it also exhibits some mutations that disrupt base pairing (Figure 1).
To test the hypothesis that the purD motif represents a riboswitch aptamer, we used in-line probing assays (56) to test for binding of the RNA against a panel of available purine compounds, including GAR (see Supplementary Data for list). Our assays showed no evidence of structural modulation induced by any of these compounds (data not shown). Although these data fail to support the hypothesis that the purD motif is a riboswitch, its consistent association with the purD gene at least implies a cis-regulatory role.
The 6C motif
This motif is widespread among Actinobacteria, and consists of two hairpins, where the loop of each contains a run of at least 6 Cs. The 6C motif exhibits significant covariation in its stems. 6C motif instances are usually moderately close (200–300 nt) to genes predicted to be related to chromosome partitioning and pilus assembly. However, given its distance, it is not clear whether 6C is functionally related to these genes.
Transposon- and excisionase-associated motifs
We also found one transposase-associated motif in α-proteobacteria and another associated with Xis excisionases in Actinobacteria, although the excisionase genes could be misclassified (see Supplementary Data). Both motifs consist primarily of a single hairpin with a 10-to-15-bp stem. Both motifs also exhibit much covariation, which suggests they form functional, structured RNAs. The excisionase motif is in a 5′ regulatory configuration to the excisionase gene.
However, both motifs could also be dsDNA sites recognized by protein dimers, where each subunit binds to sites on opposite strands. Alternately, either motif could conceivably function as a structural dsDNA element. A hairpin element, combined with other factors, could favor a structure with two intra-strand hairpins embedded in dsDNA, a ‘cruciform’-like structure that is the preferred target for proteins in distinct, though related contexts (57,58). DNA-binding motifs in Xis were also described (59), although no motifs containing hairpins were reported.
The ATPC motif
The ATPC motif occurs in some Cyanobacteria in an ATP synthase operon, between genes encoding the A and C subunits. ATPC motif instances are found in all sequenced strains of Prochlorococcus marinus, and certain species of Synechococcus. The motif consists primarily of a three-stem junction. Previous studies have proposed hairpin-like structures in Cyanobacterial ATP synthase operons, but not more complicated shapes, and in different locations from the ATPC motif (60).
The cyano-30S motif
This motif occurs in some Cyanobacteria and is in a 5′ regulatory configuration to genes encoding 30S ribosomal protein S1. It consists of two hairpins that are dissimilar to each other, and a pseudoknot wherein five nucleotides in the P1 loop base-pair with nucleotides just beyond the 3′ end of P2. Although there are several mutations that disrupt pairing in P1 and P2, there are also many compensatory mutations in these stems. Moreover, the pairing in the pseudoknot covaries and is present in all representatives.
Given the gene context, we expect that this motif mimics the ligand of the downstream ribosomal protein gene (54), and that the product of this gene thereby controls its own expression. Although we commented on ribosomal protein gene autoregulation previously in Firmicutes (5), we generally ignored ribosomal-gene-associated RNA motifs in the present study because many have already been characterized. However, the identification of the cyano-30S motif supports the view that such RNAs are found in a wide variety of phyla.
Lactobacillales motifs
The lacto-1 and lacto-2 motifs are confined to the order Lactobacillales. The lacto-1 motif has some covariation, but some mutations disrupt base pairing, so its assignment as a structured RNA is uncertain. Some instances intersect a variable region of the S(MK) (or SAM-III) (61) riboswitch between the main hairpin and the Shine–Dalgarno sequence. The lacto-2 motif consists of a large hairpin with many internal loops, some of which have highly conserved sequences. Although some mutations disrupt pairing, there is a considerable amount of covariation, which suggests that the lacto-2 motif instances are probably structured RNAs.
TD (Treponema denticola) motifs
Two predicted motifs have several instances in Treponema denticola, but are not found in any other sequenced bacteria. The motifs, TD-1 and TD-2 have 28 and 36 representatives, respectively. Seven TD-1 motif representatives overlap reverse complements of instances of TD-2, and share the two 5′-most hairpins. Although it is possible that the two motifs could be merged, it is not obvious how, because there is significant variation in the non-overlapping instances.
Both motifs show covariation and either variable-length or modular stems. However, the modest but noticeable number of mutations that disrupt pairing reduces confidence that they are functional RNAs. The TD-1 motif is usually in a 5′ regulatory configuration to genes, although the wide array of poorly characterized genes makes it difficult to suggest a coherent cis-regulatory function. The TD-2 motif does not share the 5′ regulatory configuration, so it could correspond to a non-coding RNA.
DISCUSSION
Using the CMfinder-based comparative genomics pipeline, we found 22 novel putative RNA motifs. Two have already been experimentally confirmed as riboswitches. For several others, covariation and other characteristics suggest that they are functional structured RNAs, and we have proposed possible functions for many of the motifs. Thus, our pipeline appears to be useful for discovering novel RNAs, which in turn will contribute to our understanding of RNA biochemistry and bacterial gene regulation.
Our findings here and previously (5) demonstrate that the CMfinder-based pipeline is usually able to recover RNAs that are widespread, possess a highly conserved and extensive secondary structure, are roughly 60 nt or more in length, and are associated with homologous genes. Three candidate riboswitches have these characteristics (GEMM, Moco and SAH). The remaining three candidates, SAM-IV, the COG4708 motif and the sucA motif are more narrowly distributed than most known riboswitches in that none of these motifs is found outside a single order in taxonomy level. This observation suggests that many of the undiscovered riboswitch classes have more narrow phylogenetic distributions than those discovered previously.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Nicholas Carriero and Robert Bjornson for technical assistance and members of the Breaker laboratory for helpful discussions. This work was funded by grants from the NIH (GM 068819, R01 HG02602) and NSF (EIA-0323510, DBI-0218798). J.E.B. was a Howard Hughes Medical Institute predoctoral fellow. RNA science in the Breaker laboratory is also supported by the HHMI. This work extensively used computers from the Yale Life Sciences High Performance Computing Center (NIH grant RR19895-02). Funding to pay the Open Access publication charges for this article was provided by HHMI.
Conflict of interest statement. None declared.
REFERENCES
- 1.He L, Hannon GJ. microRNAs: small RNAs with a big role in gene regulation. Nat Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Kima VN, Nam J-W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 4.Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 5.Yao Z, Barrick JE, Weinberg Z, Neph S, Breaker RR, Tompa M, Ruzzo WL. A computational pipeline for high throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput. Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Z, Weinberg Z, Ruzzo WL. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 7.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seliverstov AV, Putzer H, Gelfand MS, Lyubetsky VA. Comparative analysis of RNA regulatory elements of amino acid metabolism genes in Actinobacteria. BMC Microbiol. 2005;5:54. doi: 10.1186/1471-2180-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCutcheon JP, Eddy SR. Computational identification of non-coding RNAs in Saccharomyces cerevisiae by comparative genomics. Nucleic Acids Res. 2003;31:4119–4128. doi: 10.1093/nar/gkg438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coventry A, Kleitman DJ, Berger B. MSARI: multiple sequence alignments for statistical detection of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:12102–12107. doi: 10.1073/pnas.0404193101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washietl S, Hofacker IL, Lukasser M, Hüttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 17.Batey RT. Structures of regulatory elements in mRNAs. Curr. Opin. Struct. Biol. 2006;16:299–306. doi: 10.1016/j.sbi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruitt K, Tatusova T, Maglott D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neph S, Tompa M. MicroFootPrinter: a tool for phylogenetic footprinting in Prokaryotic Genomes. Nucleic Acids Res. 2006;34:W366–W368. doi: 10.1093/nar/gkl069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinberg Z, Ruzzo WL. 2004. pp. 243–251. RECOMB04: Proceedings of the Eighth Annual International Conference on Computational Molecular Biology, Faster genome annotation of non-coding RNA families without loss of accuracy San Diego, CA.
- 22.Weinberg Z, Ruzzo WL. Exploiting conserved structure for faster annotation of non-coding RNAs without loss of accuracy. Bioinformatics. 2004;20:i334–i341. doi: 10.1093/bioinformatics/bth925. [DOI] [PubMed] [Google Scholar]
- 23.Weinberg Z, Ruzzo WL. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics. 2006;22:35–39. doi: 10.1093/bioinformatics/bti743. [DOI] [PubMed] [Google Scholar]
- 24.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddy SR. Infernal User's Guide. 2005. ftp://ftp.genetics.wustl.edu/pub/eddy/software/infernal/Userguide.pdf.
- 26.Klein RJ, Eddy SR. RSEARCH: finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4:44. doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S. RALEE-RNA ALignment Editor in Emacs. Bioinformatics. 2005;21:257–259. doi: 10.1093/bioinformatics/bth489. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan X-F, Xu D. Intrinsic terminator prediction and its application in Synechococcus sp. WH8102. J. Comp. Sci. Tech. 2004;20:465–482. [Google Scholar]
- 32.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 34.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 35.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, et al. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerstein M, Sonnhammer ELL, Chothia C. Volume changes in protein evolution. J. Mol. Biol. 1994;236:1067–1078. doi: 10.1016/0022-2836(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 37.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 38.Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: building blocks of a modular biomolecule. Q. Rev. Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- 39.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 41.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 43.Meibom KL, Li XB, Nielsen AT, Wu C-Y, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. PNAS. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirn TJ, Jude BA, Taylor RK. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature. 2005;438:863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 45.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 46.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, et al. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 48.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 49.Mehta T, Coppi MV, Childers SE, Lovley DR. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 2005;71:8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim B-C, Qian X, Leang C, Coppi MV, Lovley DR. Two putative c-type multiheme cytochromes required for the expression of OmcB, an outer membrane protein essential for optimal Fe(III) reduction in Geobacter sulfurreducens. J. Bact. 2006;188:3138–3142. doi: 10.1128/JB.188.8.3138-3142.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Ueland PM. Pharmacological and biochemical aspects of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase. Pharmacol. Rev. 1982;34:223–253. [PubMed] [Google Scholar]
- 53.Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, et al. A riboswitch selective for the queuosine precursor preQ(1) contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 54.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 55.Liu MY, Gui G, Wei B, Preston JF, III, Oakford L, Yüksel Ü, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 56.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potamana VN, Shlyakhtenkob LS, Oussatchevaa EA, Lyubchenkob YL, Soldatenkov VA. Specific binding of Poly(ADP-ribose) polymerase-1 to cruciform hairpins. J. Mol. Biol. 2005;348:609–615. doi: 10.1016/j.jmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 58.Posey JE, Pytlos MJ, Sinden RR, Roth DB. Target DNA structure plays a critical role in RAG transposition. PLoS Biol. 2006;4:350. doi: 10.1371/journal.pbio.0040350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gottfried P, Kolot M, Yagil E. The effect of mutations in the Xis-binding sites on site-specific recombination in coliphage HK022. Mol. Genet. Genomics. 2001;266:584–590. doi: 10.1007/s004380100573. [DOI] [PubMed] [Google Scholar]
- 60.Curtis SE. Structure, organization and expression of cyanobacterial ATP synthase genes. Photosynth. Res. 1988;18:223–244. doi: 10.1007/BF00042986. [DOI] [PubMed] [Google Scholar]
- 61.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.