Abstract
It is generally assumed that transposable elements, including endogenous retroviruses (ERVs), are silenced by DNA methylation/chromatin structure in mammalian cells. However, there have been very few experimental studies to examine the methylation status of human ERVs. In this study, we determined and compared the methylation status of the 5′ long terminal repeats (LTRs) of different copies of the human endogenous retrovirus (HERV) family HERV-E, which are inserted in various genomic contexts. We found that three HERV-E LTRs which function as alternative gene promoters in placenta are unmethylated in that tissue but heavily methylated in blood cells, where these LTRs are not active promoters. This difference is not solely due to global hypomethylation in placenta, since two general measures of methylation levels of HERV-E and HERV-K LTRs suggest only 10–15% lower overall HERV methylation in placenta compared to blood. Comparisons between methylation levels of the LTR-derived gene promoters and six random HERV-E LTRs in placenta showed that the former display significantly lower methylation levels than random LTRs. Moreover, the differences in methylation between LTRs cannot always be explained by their genomic environment, since methylation of flanking sequences can be very different from methylation of the LTR itself.
INTRODUCTION
Methylation of cytosines is one of the marks of transcriptionally inactive chromatin (1). Constitutively silenced chromatin, like pericentromeric heterochromatin, is heavily methylated in all cells throughout an organism's lifespan, whereas most promoters of genes are methylated only when permanent silencing is needed during development (2). Clearly, different loci in the genome are treated distinctly by the DNA methylation machinery and more generally by the heterochromatin assembly pathway. To what extent those differences are mediated by differences in composition of the DNA sequence, the genomic context or by DNA binding proteins remains generally unclear.
One class of sequences illustrating the complexities of methylation patterns is comprised of transposable elements (TEs). TEs are thought to be silenced by heavy CpG methylation in mammalian genomes (3). However, a number of reports suggest that the silencing of repeated sequences is not homogeneous throughout the genome or in all cells of an organism. For example, the human endogenous retrovirus (HERV) family HERV-K displays different methylation levels between copies and between cell lines (4). In Brassica napus, differences in methylation are also observed between random selected copies of S1, a SINE element (5). In humans, the promoter region of ERVWE1, a HERV-W copy encoding Syncytin-1 is specifically hypomethylated in placenta and methylated in other tissues (6). Syncytin-1 is a retroviral protein with fusogenic properties involved in the development of human placenta (7). However, it is unknown whether hypomethylation in this tissue is a characteristic specific to this co-opted copy or general to the HERV-W family. It is notable that DNA methylation, and the heterochromatic structure in general, have the property of spreading from the silenced target to the surrounding DNA (5,8,9). Such observations have led to the assumption that transposable elements should generally have the same methylation level as their flanking sequence (10). If this is true, different methylation levels between endogenous retroviral copies should reflect the methylation status of the surrounding DNA. This assumption, however, has not been extensively tested.
Placenta is of particular interest with respect to expression of endogenous retroviruses (ERVs) because many ERV families are transcribed in this tissue (11–16). One of the hypotheses proposed to explain this relatively high degree of ERV expression is that placenta has lower DNA methylation compared to other tissues (13,17). Indeed, in mouse blastocysts, the trophectoderm seems to be less methylated than the inner cell mass (18), whereas in human blastocysts the opposite situation has been reported (19). On the other hand, human term placentas show about 20% lower overall DNA methylation compared to brain, liver and peripheral blood lymphocytes (PBL) (20,21) and 10–27% lower methylation of Alu retroelements compared to that observed in spleen (22). Moreover, compared to other tissues, placenta has a higher frequency of HERV copies acting as alternative promoters to cellular genes (23,24). These gene promoters of retroviral origin initiate transcription in their long terminal repeat (LTR) and, in most cases; their activity is tissue-specific. Indeed, about 40% of the tissue-specific LTR-derived gene promoters which have been documented are active in placenta. This could be the result of the general hypomethylation of this tissue allowing the expression of many HERV copies and consequently of LTR-derived promoters as well. The main questions addressed in this study were (i) whether the methylation level of LTR-derived gene promoters is representative of the general methylation of HERVs in placenta and (ii) whether differences in methylation between copies can be explained by differing genomic environments. To answer to these questions, we analyzed the methylation status of three HERV-E LTRs active as alternative gene promoters and of six other highly related LTRs in different genomic contexts.
MATERIALS AND METHODS
Samples
Two term placenta samples were used for all bisulfite sequencing and combined bisulfite and restriction enzyme analysis (COBRA) experiments: P24 from a pre-eclamptic pregnancy, gestational age 32 weeks and P25, from a normal placenta, gestational age 40 weeks. For LTR-Mid1 only, two additional placental samples were used: P15 from an eclamptic pregnancy, gestational age 38 weeks and P33, from a pre-eclamptic pregnancy, gestational age 40 weeks. P21, a normal term placenta sample, gestational age 41 weeks, was used for the Southern blot. Different samples of normal PBLs were used for bisulfite methods (sequencing and COBRA) and Southern blotting.
COBRA and Bisulfite sequencing
Bisulfite conversion of DNA was performed using the EZ DNA methylation kit (Zymo Research), according to the manufacturer's protocol, with the following modification: genomic DNA was incubated with CT conversion reagent at 50°C for a total of nearly 4 h with 15 s pulses to 95°C every 15 min. Converted DNA was eluted in 20 μl elution buffer and 2 μl was used for PCR. All primers used are shown in Supplementary Table S1. To ensure that only the LTR of interest was amplified, the forward primer was located in the 5′ flanking sequence of this LTR. A first PCR (PCR1) was performed under the following conditions: 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, variable annealing temperatures depending on the primers for 1 min, and 72°C for 30 s. A final elongation step of 10 min was included in all reactions. In most cases, a second PCR (PCR2) was necessary to obtain enough PCR products for subsequent analysis. PCR2 was performed either by using the same primers as for PCR1 or with nested primers (Table S1). For PCR2, 2 μl of the PCR1 product was used as template. PCR2 conditions were the same as for PCR1 except that only 35 cycles were performed. PCR1 and PCR2 were performed on each sample in three independent reactions to control for PCR bias in the following COBRA. The products were analyzed by gel electrophoresis and purified with MiniElute (Qiagen), then digested individually with restriction enzymes which contain CpG dinucleotides in their recognition sites. If the CpG was methylated in the original sample, then the restriction enzyme site is maintained during bisulfite treatment and the PCR product will be digested. Enzymes used and the number of restriction sites in each LTR is given in Supplementary Table S2. In some cases, several CpG sites were tested by a single restriction enzyme (e.g. four CpG sites of LTR-EBR were tested by TaqI, see Table S2). In these situations, it is impossible to distinguish among methylation states of the different CpGs. Therefore, the same results of COBRA are indicated for all those CpGs in Figures 2 and 5 and they are marked by a connector line. Consequently, COBRA results when connected should be regarded as a sum of several CpG sites and not separately for each CpG site. All enzymes were purchased from New England Biolabs, and digests were performed following manufacturer's instructions. No PCR bias was observed between different PCRs performed on the same sample. Hence, the three PCR products from the same individual sample were pooled and cloned using the pGEM-T Easy kit (Promega). Sequencing was performed by McGill University and Genome Québec Innovation Centre Sequencing Platform. Only unique sequences (as determined by either unique CpG methylation pattern or unique non-conversion of non-CpG cytosines) are shown, and all sequences had a conversion rate >96%.
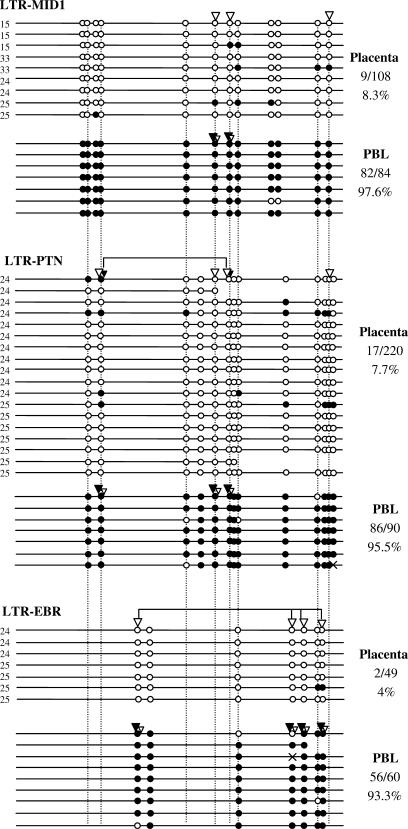
Figure 2.
Methylation of HERV-E LTR-derived gene promoters. Each line corresponds to a unique sequence of an LTR. Black circles correspond to methylated CpGs and white circles to unmethylated CpGs. Homologous CpGs between LTRs are connected by dashed lines. White and black triangles indicate unmethylated and methylated CpG sites, respectively, revealed by COBRA. Black and white triangles present at the same CpG site indicate that the population of amplified sequences contained both methylated and unmethylated CpGs at this site. In this case, the triangle's relative size corresponds to the relative band intensity of the digested and undigested forms. Connectors indicate CpG sites that were assayed by the same restriction enzyme in COBRA (see Materials and Methods section). Crosses correspond to mutated CpG sites. The number of methylated CpGs out of the total CpG sites is reported for each LTR. Numbers 24, 25, etc. on the left of each sequence indicate the corresponding placenta sample (P24, P25, etc).
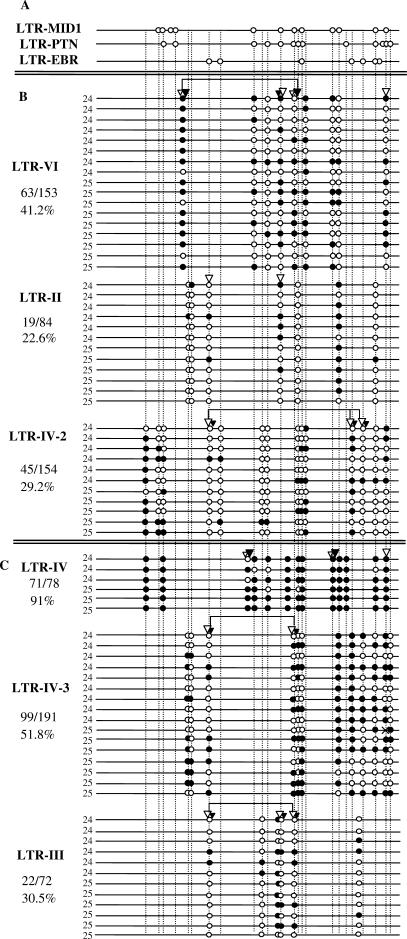
Figure 5.
Methylation of individual HERV-E LTRs in placenta. Symbols are as in Figure 2. Panel (A) shows only one sequence of each LTR-derived promoter of group A, in order to align the homologous CpG sites between all LTRs (see Figure 2 for complete data on methylation of group A LTRs). Panels (B) and (C) show the methylation status of the LTRs belonging to groups B and C as described in the text.
Genome-wide COBRA of HERVE-LTRs
To compare the methylation of HERV-E LTRs between placenta and PBL in a genome-wide way, the LTR2B-cons-fw: 5′-TGAGGGAAGAGAGAGATTTTT 3′ and the LTR2B-cons- rev: 5′- ATTATAAAAAAAAAAACTTTATTCAACTAA-3′ primers were chosen in a well-conserved region of the LTR2B consensus given by Repbase (25). (LTR2B is the Repbase nomenclature for a major subfamily of HERV-E LTRs to which LTR-MID1 and LTR-PTN belong). An in silico PCR was performed with the UCSC In-silico PCR program (UCSC Genome Browser) using as primer sequences the corresponding LTR2B-cons-fw and -rv sequences for non-bisulfite converted DNA. Seventy-eight HERV-E LTRs were predicted to be amplified. A multiple alignment of the 78 sequences showed presence of a SnaBI restriction site (TACGTA) in 42 sequences, a ClaI (ATCGAT) in 36 sequences and a DraI (TTTAAA) in all 78 sequences (two sites present in most of them and at least one was maintained). For band intensity quantification the ImageQuant 5.0 (Molecular Dynamics) program was used. To deduce the percent of methylated CpGs, the theoretical number of CpG-carrying sequences out of the total of amplified sequences as predicted by the In-silico PCR was used.
Methylation sensitive Southern blot
Genomic DNA from placenta (sample P21) and PBL was digested with the methylation sensitive enzyme (HpaII) and the methylation insensitive isoschisomer (MspI) overnight. Then, the ApoI restriction enzyme was added for 5 h. All digestions were performed according to the manufacturer's instructions (New England Biolabs). Digested DNA was separated in a 1% agarose gel and transferred to a Zeta-Probe GT Blotting Membrane (Bio-Rad). The probe corresponding to the gag region of HERV-K was amplified by PCR with probe-K-fw: AGCGTGGTCATTGAGGACA and probe-K-rev: AAAGCTGAGATAAGAGGCATATTT primers. The 150 bp long PCR product was cloned into the pGEM-T Easy vector, confirmed by sequencing and labeled with 32-P. Hybridization was performed in ExpressHyb Hybridization Solution (BD Biosciences) at 60°C. Washing conditions were: 2 × 1 min and 2 × 20 min at room temperature in 2 × SSC; 0.1% SDS and 3 × 20 min at 60°C in 0.5 × SSC; 0.1% SDS. Quantification of band intensity was performed as for the genome-wide COBRA. To calculate the percent of methylated CpGs, the percent of sequences not carrying CpGs was deduced from the non-digested band in the MspI digest.
Choice of individual LTRs
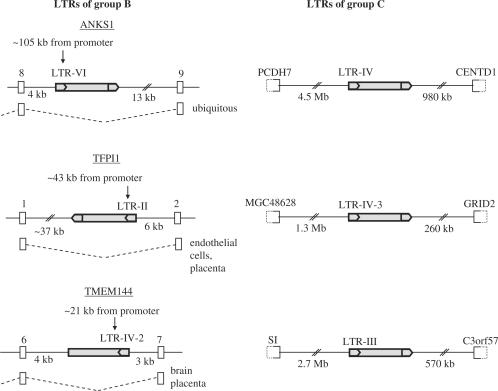
Individual LTRs were arbitrarily chosen as follows: the consensus sequence of the three LTR-derived promoters was derived by hand and placed upstream of the first 1260 bp of the HERV-E internal consensus sequence termed ‘Harlequin’ by Repbase (25). This file was used as a query to search the human genome for similar sequences using the BLAT alignment tool (26). Among the output sequences, LTRs were chosen based on the following criteria: their total length had to match the query and they had to be in the 5′extremity of a provirus. The genomic contexts also were taken into account: three LTRs located in introns of genes expressed in placenta were chosen (group B). Genes were considered to be expressed in placenta if corresponding placental expressed sequenced tag (ESTs) were reported in the UCSC Genome Browser (27). Three LTRs were chosen to be the furthest away from any known genes and EST-rich regions (group C). Group A comprises the three LTR-derived promoters. The percent identity between all LTRs analyzed here is shown in Supplementary Table S3.
Determination of transcribed HERV-E copies in placenta
Human genome annotation data including retroelements, ESTs and tissue information were obtained from the UCSC annotation database (Mar. 2006 GenBank freeze, hg18, Build 36.1). Because ESTs comprising only repetitive/retroelement sequences (denoted as ‘simple ESTs’) are not included in the UCSC annotation database, we also retrieved the original sequences of those ESTs from NCBI dbEST database (release version 154).
In the UCSC Genome Browser, some HERV-E copies are annotated only by the disrupted fragments. We computationally merged all the HERV-E fragments into complete copies based on the following criteria: a complete copy of HERV-E should have one of these structures: LTR-internal-LTR, LTR-internal, internal-LTR, LTR-only, internal-only. ‘LTR’ in the above structures were sequences annotated as LTR2, LTR2B or LTR2C; the ‘internal’ region annotated as either ‘HERVE’ or ‘Harlequin’; and all the merged fragments should be on the same chromosome, in the same orientation and with a distance of less than 10 kb between any two of them.
To determine which HERV-E copies are transcribed in placenta, we used two different strategies to deal with the EST data from the UCSC annotation database and the NCBI dbEST database, respectively. In the first case, the genomic coordinates of both HERV-Es and ESTs from placenta are known, and by comparing these coordinates, we identified all those HERV-Es which overlapped with ESTs in placenta and assigned them as specific HERV-E copies expressed in placenta. In the second case, there was no information on genomic coordinates available for those ‘simple ESTs’ from NCBI. However, since HERV-E copies are relatively old and diverged from other copies (e.g. Table S3), we successfully mapped most of these ESTs to the human genome with significant mapping results by using BLAT (standalone version: v. 33) (26). In order to reduce the computational workload, we did not try to map all ‘simple ESTs’ to the human genome, but only those derived from HERV-Es by repeat-masking them with the HERV-E consensus sequence and the CENSOR program (v 4.1) downloaded from RepBase (28) After the mapping, genomic coordinates of those ‘simple ESTs’ were obtained, and the same strategy described in the first case was applied to find specific HERV-E copies expressing these ESTs in placenta.
Statistics
Distribution free Mann–Whitney U-tests and Kruskal–Wallis H-tests were used for the statistical analysis.
RESULTS AND DISCUSSION
Description of the three LTR-derived promoters studied
The three LTR-derived gene promoters analyzed here are alternative promoters specifically active in placenta, keeping the coding sequences of the genes intact, and they are all derived from 5′ LTRs of HERV-E proviruses (29) (Figure 1).
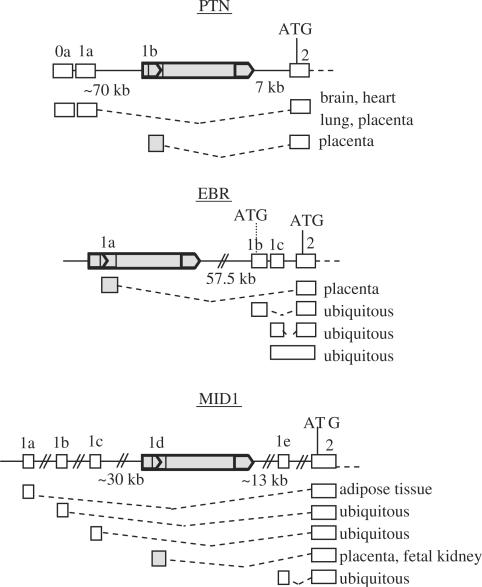
Figure 1.
Genomic context and organization of the HERV-E LTR-derived gene promoters. Grey boxes correspond to HERV-E sequences; grey arrows indicate HERV-E LTRs. White boxes represent gene exons. The different transcripts and their tissue specificity are shown. In each case, the use of different promoters does not affect the open reading frame.
The first is the LTR-derived promoter of Pleiotrophin (LTR-PTN) (30), a secreted heparin-binding cytokine with diverse functions involving mitogenic activity in fibroblasts, endothelial and epithelial cells, lineage-specific differentiation of glial progenitor cells, neurite outgrowth and angiogenesis (31). A promoter of genomic origin called hereinafter ‘native promoter’ drives expression in developing brain and in the adult testis, uterus, glia and neurons. The provirus providing the LTR-PTN promoter is located around 70 kb downstream of the native promoter and 7 kb upstream of the first coding exon. Although Schulte et al. (30) reported that the LTR-PTN was the only promoter responsible for placental expression, native-promoter driven transcripts in placenta are present in Genbank. Experiments with PTN-depleted choriocarcinoma cells suggest that placental expression of PTN might be important for trophoblast growth, invasion and angiogenesis in vivo.
The second LTR-derived gene promoter studied here is associated with the endothelin B receptor gene (LTR-EBR) (32). EBR is a G protein-coupled receptor of endothelin (33). In human placenta, endothelins are involved in fetoplacental circulation and can also act as growth factors (34,35). The HERV-E provirus providing the alternative promoter is located 57.5 kb upstream of the native promoter which has a ubiquitous expression pattern. The LTR-EBR is responsible for about 15% of the total amount of placental transcripts (36).
The last HERV-E LTR-derived promoter is linked to the Midline 1 gene (LTR-MID1) (37). MID1 encodes a microtubule-associated protein (38). Mutations in this gene are involved in the pathogenesis of Opitz syndrome, a genetic disorder affecting midline structures (39). This gene has five alternative promoters, three of them being widely expressed, one adipose tissue specific and the LTR-MID1 placental and fetal kidney specific (40). The provirus driving placental expression is located between two ubiquitous promoters: at 13 kb from the downstream promoter and 30 kb from the upstream promoter (37). The retroviral promoter drives 25% of the placental transcripts and 22% of the fetal kidney transcripts (40).
Methylation status of the HERV-E LTR-derived gene promoters is in accordance with their expression profile
As mentioned earlier, LTR-MID1, LTR-EBR and LTR-PTN are alternative promoters specifically expressed in placenta (30,32,40). To assess whether their expression correlates with their methylation levels, methylation of the three LTRs was determined by COBRA and bisulfite sequencing in placenta and PBL samples. Prior to this analysis, the expression profile of the three LTR-derived promoters was confirmed by RT-PCR in both tissues using the same samples as for the methylation analysis. All three promoters are active in placenta and silent in PBL (data not shown). The results of the COBRA and the bisulfite sequencing in placenta and PBL are shown in Figure 2. In placenta, these three LTRs are almost completely unmethylated. In contrast, they are heavily methylated in PBL. The results obtained by COBRA are in good agreement with those obtained by bisulfite sequencing demonstrating that the sequenced clones are representative of the pool of amplified sequences. The low methylation of all three LTR-derived promoters in placenta and their high degree of methylation in PBL indicates that methylation and expression profiles of the HERV-E LTR-derived promoters correlate. These results suggest that DNA methylation is involved in regulation of the three LTR-derived gene promoters.
HERVs in placenta are only slightly less methylated than in PBL
To clearly address the question of whether the strong hypomethylation of LTR-derived promoters in placenta reflects an overall hypomethylation of HERVs in this tissue, a Southern blot analysis with methylation-sensitive enzymes was attempted. However, use of a HERV-E specific probe resulted in smears and lack of any defined bands (data not shown). This is presumably due to the relatively high divergence (10–30%) between HERV-E copies. Since HERV-Es are too divergent for Southern blot analysis, a genome-wide COBRA analysis was performed. For this, primers corresponding to relatively conserved regions of the consensus LTR2B—a subfamily of HERV-E LTRs— were chosen allowing the amplification of 78 copies, as predicted by the UCSC In-silico PCR program (UCSC Genome Browser). Out of the 78 copies, 54% have a SnaBI restriction site and 46% a ClaI site, both sites including a CpG dinucleotide. A DraI site (TTTAAA) present on all 78 sequences was used to control that, when methylation is not a factor, the digestion profile was as expected according to the in silico predictions and was identical for both samples from placenta and PBL. COBRA of this heterogeneous amplicon (Figure 3A) yields a double band of lower size (bands number 2; Figure 3A) corresponding to the methylated sequences. The undigested band (band 1; Figure 3A) corresponds to LTR copies with either unmethylated CpG sites or those not containing the CpG site. Measurement of the relative intensity of bands 1 and 2 for each of the digestions of three independent experiments with SnaBI and one with ClaI shows 4–25% lower methylation in placenta compared to PBL. The average of the different measures suggests 14% lower methylation of HERV-E in placenta than in PBL. However, to correctly calculate these percentages, the precise proportion of sequences lacking the tested CpG sites is necessary. With this method, this proportion is theoretical, based on the prediction of the In-silico PCR program. This prediction could be somehow biased, since the primers used in silico correspond to non-bisulfite treated DNA and are different from the actual primers used in the COBRA. The experimental estimation of the proportion of the actual CpG lacking sequences in the pool of amplified sequences is impossible. Hence, another experimental procedure allowing a more accurate estimation of the percent of methylation was necessary, in order to confirm our results obtained by the genome-wide COBRA.
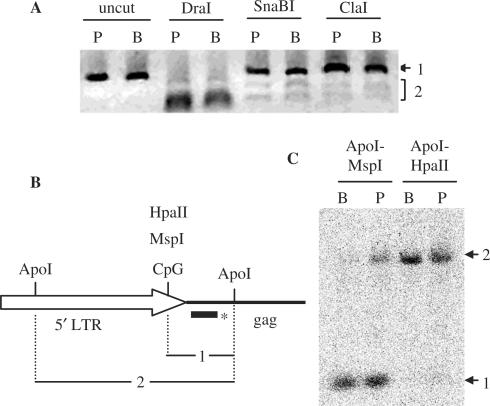
Figure 3.
Methylation of HERV-E and HERV-K sequences in placenta and PBL. (A) Genome-wide COBRA on HERV-E LTRs. Bisulfite treated DNA from placenta (P lanes) and PBL (B lanes) were used as template for PCR with primers amplifying HERV-E LTRs. PCR amplicons were treated with three restriction enzymes (SnaBI, ClaI and DraI). Bands 2 for SnaBI and ClaI digests correspond to sequences with methylated CpGs, whereas band 1 to a mix of sequences with unmethylated CpGs and sequences lacking the CpG site. The CpG free DraI restriction site is present in all sequences and was used as a control (see text). (B) and (C) methylation of HERV-K family determined by Southern blot. (B) Schematic representation of HERV-K 5′ LTR and gag region, with the restriction sites of the enzymes used. The bar with the asterisk corresponds to the probe overlapping the gag region. (C) Autoradiogram of the Southern blot. Lanes B are loaded with genomic DNA from PBL and lanes P with genomic DNA from placenta. Bands labeled 1 and 2 correspond to restriction fragments 1 and 2 shown in B.
For that purpose, the most recent family of HERVs, HERV-K [HML-2 subfamily, 1–2% divergence (41)], was chosen to carry out a Southern blot with methylation sensitive enzymes. MspI and its methylation-sensitive isoschizomer HpaII were used and the results are shown in Figure 3B and C. The majority of HERV-K copies seem to be methylated at the tested CpG site in the LTR both in placenta and PBL. Measurement of the relative intensity of bands 1 and 2 from two independent experiments and different exposure times, suggests 9–14% less methylation of HERV-K in placenta than in PBL with an average of 11%. This percentage is very similar to the one obtained by the genome-wide COBRA on HERV-E sequences.
Taken together, these results show a slightly lower methylation of two families of HERVs in placenta than in PBL. This observation could reflect a general phenomenon of HERV families. Moreover, our results are in accordance with the previous results mentioned earlier that found general placental hypomethylation of 10–27% compared to other tissues. However, the LTR-derived promoters are about 80% less methylated in placenta than in PBL indicating that these particular copies are not representative of the bulk of HERV-E copies regarding their methylation levels in the tissues tested.
Comparison of methylation levels between HERV-E LTRs in different genomic contexts
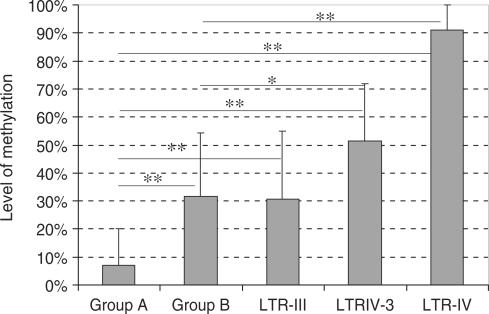
To compare the methylation levels of other HERV-E copies inserted in similar or different genomic contexts with those of the LTR-derived promoters, we chose six HERV-E proviruses without any known gene-promoting function (see Materials and Methods section) and the methylation of their 5′ LTRs in placenta was determined by both COBRA and bisulfite sequencing. We define ‘group A’ as the three LTR-derived gene promoters described previously and ‘group B’ as the three out of the six random LTRs located in introns of genes expressed in placenta (Figure 4). LTRs of groups A and B have similar genomic context: they are all located in introns of genes expressed in placenta with the exception of LTR-EBR which is located 57.7 kb upstream of the first exon of EBR gene (Figure 1). LTR-VI is located in an intron of the Ankyrin S1 (ANKS1) gene expressed in many tissues including placenta. LTR-II is located in an intron of the Tissue Factor Pathway Inhibitor 1 gene (TFPI1) expressed mainly in endothelial cells and also in placenta. LTR-IV-2 is located in an intron of the TMEM144 gene highly expressed in the central nervous system and providing a few ESTs in placenta. The other three LTRs are located far away from any annotated gene and are ‘group C’ (Figure 4). The results of methylation analysis of these LTRs are shown in Figure 5. LTRs of group B present 22.6–41.2% methylation and LTRs of group C 30.5–91% methylation. COBRA results are in good accordance with bisulfite sequencing results reflecting the global methylation of each LTR. Group A and B LTRs have no significant intra-group differences in their methylation levels in placenta whereas group C is very heterogeneous in terms of methylation. Hence, it cannot be considered as a homogeneous group regarding methylation. This could be due to the fact that the definition of this group as ‘far from genes’ is inexact: the presence of unknown genes or transcriptional activity in the vicinity of such LTRs cannot be excluded. For instance, LTR-III shows a methylation level (30.5%) similar to the levels displayed by LTRs of the group B. LTR-III could be located nearby an un-annotated transcriptionally active region. However, according to the UCSC Genome Browser, no obvious difference is observed between the insertion sites of group C LTRs with regard to EST occurrence in nearby regions or the conservation of the surrounding sequences between mammalian species. The other two LTRs of group C, LTR-IV and LTR-IV-3, have significantly higher methylation levels when compared to group B and group A (Figure 6). The comparison between groups A and B, both located in introns of genes expressed in placenta, shows that group A displays highly significant lower methylation than group B (P = 2e−06). This shows that groups A and B, although located in regions with similar transcriptional activity, differ in their methylation status. In conclusion, LTR-derived promoters have significantly lower methylation levels than any arbitrarily chosen LTR even when located in similar genomic contexts.
Figure 4.
Genomic context and organization of the HERV-E LTRs of groups B and C. Grey boxes correspond to HERV-E sequences; grey arrows indicate HERV-E LTRs. White boxes with solid lines represent gene exons and white boxes with dashed halves represent genes. LTRs of group B located in introns of genes expressed in placenta and other tissues. ANKS1: Ankyrin repeat and SAM domain—containing protein 1; TFP1: tissue factor pathway inhibitor; PCDH7: protocadherin 7 isoform c precursor; CENTD1: centaurin delta 1 isoform b; GRID2: glutamate receptor, ionotropic, delta 2; SI: sucrase-isomaltase (alpha-glucosidase); TMEM144: transmembrane protein 144.
Figure 6.
Methylation levels of different groups of HERV-E LTRs. Average methylation for group A, group B and for the individual LTRs of group C are shown. Bars correspond to the SD.* Corresponds to P < 0.01 and ** to P ≤ 0.001. Only significant differences are indicated.
Identification of HERV-E copies transcribed in placenta
Our study shows that HERV-E copies display widely variable methylation levels in placenta. It was unknown how many of the approximately 294 copies of the full-length HERV-E proviruses [out of ∼1200 total HERV-E copies (42)] are transcriptionally active in placenta and we chose the six random LTRs based on similarity and location criteria but with no knowledge of their expression status. To gain an estimate of the number of HERV-E copies transcribed in placenta and to assess whether the difference of methylation between copies correlates with expression, we extracted placental ESTs homologous to HERV-E derived sequence and then identified the specific copies of HERV-Es producing each of these ESTs (see Materials and Methods section). The results are shown in Table 1. The 162 placenta-specific ESTs were clearly identified as corresponding to HERV-E sequences but only 74 could be unambiguously mapped to 26 HERV-E loci. ESTs comprising only HERV-E sequences are described as ‘simple’ and those comprising non-HERV-E sequences as well are denoted as ‘chimeric’. ESTs starting inside the 5′LTR are likely to correspond to transcripts initiated by the LTR itself. Only 10 of the 26 copies are associated with this type of EST and are classified as ‘likely’ transcriptionally active copies. Among these, the proviruses corresponding to the three LTR-derived promoters were identified. ESTs corresponding to HERV-E's internal region could be the result of transcription initiated by the provirus itself or by upstream sequences. Therefore nine copies associated with these types of placental ESTs are classified as ‘possibly’ transcriptionally active. Among these copies, LTR-II and LTR-VI were identified, both members of group B. ESTs comprising the upstream flanking sequence of a 5′LTR or corresponding to copies deleted in their 5′ extremity are probably transcribed from a genomic promoter upstream of the provirus. Consequently, they are considered as ‘unlikely’ transcriptionally active copies. LTR-II provides five ESTs corresponding only to its internal sequence and four spliced ESTs composed of exons corresponding to the provirus and the flanking gene. Hence the LTR-II copy is considered as possibly active but also involved in read-through transcription. Interestingly, one of the ‘likely’ active copies (chromosome 1 : 166098952–166105765) seems to act as an alternative promoter for the IQWD1 gene of unknown function. Therefore, over one third of the ‘likely’ transcriptionally active copies correspond to LTR-derived alternative gene promoters.
Table 1.
HERV-E copies providing ESTs in placenta
| HERV-E | Chromosome: coordinates | Strand | Simple ESTs | Chimeric ESTs | Total ESTs | Comments | |
|---|---|---|---|---|---|---|---|
| Likely | Solo LTR | 1: 89420430–89421200 | − | 0 | 1 | 1 | |
| Full length | 1: 166098952–166105765 | + | 1 | 2 | 3 | Alternative promoter of IQWD1 | |
| 3′ deleted | 2: 34755763–34756702 | + | 0 | 1 | 1 | ||
| Solo LTR | 2: 96948142–96948605 | + | 0 | 1 | 1 | ||
| Full length | 7: 136597798–136604133 | − | 0 | 3 | 3 | LTR-PTN | |
| Full length | 13: 77442456–77448210 | − | 0 | 3 | 3 | LTR-EBR | |
| *Full length | 19: 58093589–58099917 | − | 2 | 0 | 2 | ||
| Full length | 21: 43225142–43230724 | + | 2 | 0 | 2 | ||
| Solo LTR | X: 3863806–3864286 | + | 1 | 0 | 1 | ||
| Full length | X: 10512832–10518380 | − | 3 | 2 | 5 | LTR-MID1 | |
| Possibly | Full length | 1: 204449081–204455474 | − | 2 | 0 | 2 | |
| **Full length | 2: 188083461–188090051 | + | 5 | 0 | 5 | LTR-II | |
| Full length | 6: 35074115–35080062 | + | 2 | 0 | 2 | LTR-VI | |
| Full length | 6: 111712066–111718525 | − | 1 | 0 | 1 | ||
| Full length | 10: 73369141–73376272 | + | 2 | 0 | 2 | ||
| Full length | 17: 23581671–23590492 | − | 11 | 0 | 11 | ||
| Full length | 17: 38565788–38573241 | − | 1 | 0 | 1 | ||
| 3′ deleted | 19: 63018482–63019644 | + | 1 | 0 | 1 | ||
| Full length | 20: 24847861–24861663 | + | 9 | 0 | 9 | ||
| Unlikely | Full length | 1: 1337494–1344095 | − | 0 | 2 | 2 | Genomic + 5′LTR |
| **Full length | 2: 188083461–188090051 | + | 0 | 4 | 4 | LTR-II internal region spliced into gene exons | |
| Internal sequence | 10: 15072826–15076892 | − | 0 | 1 | 1 | Flanking + internal sequence | |
| 5′ deleted | 11: 3451334–3459517 | − | 0 | 2 | 2 | Internal sequence + flanking | |
| Internal sequence | 13: 44848741–44856042 | − | 0 | 3 | 3 | HERVE duplications – gene exons | |
| Solo LTR | 16: 29617072–29617563 | + | 1 | 2 | 3 | Flanking + LTR | |
| *Full length | 19: 58093589–58099917 | − | 0 | 1 | 1 | Flanking + 5′LTR | |
| Solo LTR | 19: 60156353–60156881 | − | 0 | 1 | 1 | Flanking + 5′LTR, alternative prom NALP2 | |
| Internal sequence | 21: 43236635–43249084 | − | 1 | 0 | 1 | Internal sequence |
* and ** are proviruses that appear in two different categories because they provide two types of ESTs.
As reported by others (43), this study suggests that only a limited number of HERV-E loci are transcriptionally active. Notably, only two out of the 1200 HERV-E copies present in the genome, those at chromosome 17: 23581671–23590492 and chromosome 20: 24847861–24861663, provide 36% of the HERV-E placental ESTs (Table 1). Moreover, solitary LTRs represent 75% of all HERV-E copies but only 16% of the ‘likely’ or ‘possibly’ active copies correspond to this type of element. It seems that full length copies are more expressed than solitary LTRs as previously observed (43). It is also surprising that two LTRs (II and VI) out of the 294 full length copies identified in this screening as possibly active happened to be chosen at random for our methylation study of individual LTRs. This is very likely due to the criteria used for the choice of the individual LTRs: the sequence similarity to the LTR promoters (LTR-VI is the closest related to the LTRs MID-1 and PTN and LTR-II to LTR-EBR, see Table S3), the fact that they are part of full length proviruses and their localization in introns of genes expressed in placenta. All these criteria seem to increase the likelihood for an LTR to be active in placenta.
We attempted to determine whether a clear relation existed between level of methylation and level of expression as reflected by the number of ESTs found in placenta. Among the copies studied here, only ESTs corresponding to unmethylated copies (group A) or to copies with intermediary levels of methylation (group B) were found. Although these two groups differ by their methylation levels, no obvious difference was observed in the number of ESTs that they provide and the amount of data does not allow any statistical analysis. Therefore, experimental analysis of their expression is necessary to determine if differences in methylation levels as low as 20% affect expression. It has been reported that the same Intracisternal-A-Particle (IAP)-LTR promoter can be active or silenced in different individual mice when its methylation differs only by 20–30% (44). This could be due to the fact that, in some cases, LTR promoters seem to be active not because of their overall hypomethylation in a given tissue but because of their complete demethylation in a limited number of cells of this tissue. Indeed, the IAP-LTRs mentioned above, show not only a strong variability in methylation between mice but also a mosaic methylation between cells of the same mouse. A similar phenomenon could explain why our study identified ESTs corresponding to LTR-VI and LTR-II but not LTR-IV-2. The latter is the only one from group B not presenting any completely demethylated sequences. In addition, when different LTRs are compared, it is very likely that methylation or chromatin context is not the only factor responsible for the transcription levels; sequence divergence and heterogeneity for transcription factor binding sites will be involved as well (17). For example, when the promoter activity of two unmethylated HERV-K LTRs was compared in transient transfection assays, they displayed different levels of transcription (4). However, when their endogenous expression was tested in human teratocarcinoma cells, they displayed different levels of transcription but in the opposite sense, since the stronger LTR promoter is more methylated in the cell.
Comparison of methylation levels between LTRs and their flanking sequences
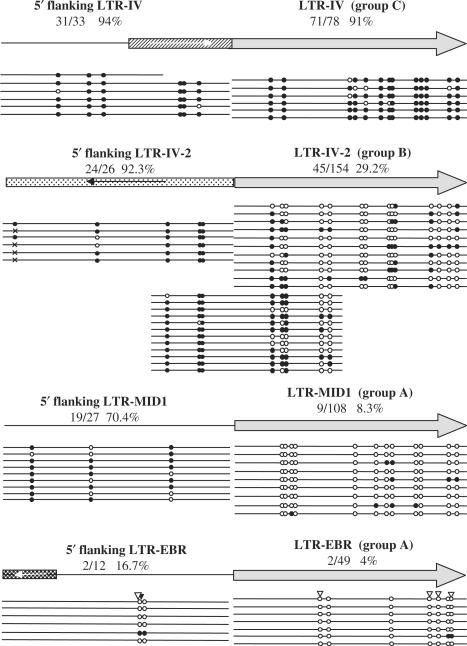
To determine if methylation of HERV-E LTRs is the same as their flanking sequence and if the difference in methylation between LTR-derived promoters (group A) and random LTRs located in similar genomic contexts (group B) is related to differences in methylation of their flanking regions, we determined the methylation status of flanking sequences of two LTRs from group A (LTR-MID1 and LTR-EBR), one LTR from group B (LTR-IV-2) and one LTR of group C (LTR-IV). Methylation of the CpG sites contained in 500 bp adjacent to the LTRs was determined by bisulfite sequencing and results are shown in Figure 7. LTR-IV (group C) has the same methylation level as its flanking sequence, whereas LTR-IV-2 (group B) and LTR-MID1 have much lower methylation levels compared to their flanking sequence (P < 0.001, Figure 7). For LTR-EBR and its flanking sequence, no significant difference in methylation level was revealed by bisulfite sequencing. However, a COBRA of LTR-EBR and its flanking region suggests a low methylation level of the flanking sequence and a complete absence of methylation for the LTR-EBR itself. The flanking sequences of LTR-IV-2 and LTR-MID1 show similar, high methylation levels (92.3% and 70.4%, respectively) even though they are located in introns of genes expressed in placenta. This discrepancy between levels of methylation of the LTRs and their flanking sequences is striking since it is believed that the main determinant of ERV methylation is the chromatin state of their insertion site (10). To confirm this pattern and eliminate the possibility that this observation results from eventual PCR biases, we amplified and sequenced 500 bp containing five CpGs of the LTR-IV-2 and three CpGs of its flanking sequence (Figure 7). Indeed, these clones confirm the pattern observed previously. For LTR-IV-2, methylation from the flanking sequence seems to spread over the first CpGs of the LTR but drops gradually towards the internal CpG sites. Methylation of LTR-MID1 seems to drop more abruptly at the border of its 5′ extremity resulting in a clear-cut difference of the methylation level between LTR-MID1 and its flanking sequence. This difference between LTR-IV-2 and LTR-MID1 could be explained by a less efficient spreading of methylation through LTR-MID1 because of a lower density of CpGs in the flanking sequence or/and to transcription factors binding to the LTR and protecting it from methylation (45,46). Indeed, LTR-MID1 is transcriptionally active, but no ESTs corresponding to LTR-IV-2 were identified by our screening of the NCBI database, suggesting this LTR is not active. Moreover, no difference in methylation between flanking sequences of different groups was observed: they all present high methylation levels except for one of the two members of group A (LTR-EBR). Considering CpG sites located in other repeats present in the flanking sequences, there is no evidence for more methylation of repeated DNA compared to unique sequences. In conclusion, very different levels of methylation are observed in flanking sequences, an observation that cannot be explained by the nature of the sequence (repeat or unique) or the transcriptional activity (intron or inter-genomic sequence). Our results suggest that there is no systematic correlation between methylation levels of HERV-E LTRs and their flanking sequences. Furthermore, the hypomethylation of the LTR-derived promoters compared to random LTRs cannot be explained by a difference in methylation of their flanking sequences.
Figure 7.
Methylation levels of HERV-E LTRs and their flanking sequences. Grey arrows correspond to HERV-E LTRs, with their flanking sequence represented on the left by a line for unique genomic sequences or by a box for repeated sequences. The dashed box represents an Alu element, the white box with black dots an L1 and the black box with the white dots an L2 element. The orientation of the repeats is shown by arrows. The methylation of each region is shown below by lines corresponding to the sequence of a single clone. Other symbols are as in Figure 2.
CONCLUDING REMARKS
In this study, we report methylation levels of nine HERV-E LTRs. We show that methylation levels of LTRs in placenta are widely variable, ranging from 4% to 91%. It seems that the genomic environment has an impact on the methylation level of the LTRs, since those located in well-defined genomic environments such as introns of genes expressed in placenta (groups A and B) have similar levels of methylation in intra-group comparisons. However, genomic environment alone cannot explain other differences observed; for example, the difference between LTR-derived promoters and LTRs located in introns of genes expressed in placenta. This statement is even clearer when flanking sequences of LTRs are compared. The methylation level of an LTR is not systematically similar to that of its flanking sequence, whether it is an LTR-derived promoter or a random LTR. Clearly, spreading of heterochromatin is not the only determinant of the degree of methylation of LTR promoters. Other factors such as transcription factors binding the LTRs are likely involved. Expression levels of individual LTRs will depend on both methylation/chromatin context as well as the retention of transcription factor binding sites. Indeed, our in silico screening for transcriptionally active HERV-Es in placenta show that very few of them are active, very likely because expression is dependent on the combination of factors mentioned earlier.
LTR-derived promoters occur more frequently in placenta than in any other tissue (23,24). This does not seem to be solely due to a general hypomethylation of the DNA in this tissue, since the LTR-derived promoters studied here are almost completely unmethylated and therefore do not reflect general methylation levels. Moreover, LTR-MID1 is located in a highly methylated flanking region showing that its lack of methylation does not result from a general ‘protection’ of the surrounding sequence against methylation. Consequently, the expression of LTR-derived promoters in placenta cannot be explained only as a secondary effect of the genomic environment. The HERV-E LTR-derived gene promoters studied here present tissue-specific methylation in accordance with their expression profile. Their epigenetic status and activity make them more analogous to tissue-specific promoters of genomic origin than to transposable elements.
Overwhelming evidence for essential roles of ERV-encoded proteins in placenta morphogenesis and evolution has been reported in recent years (47–49). However, there may be additional impact of ERVs in the placenta due to host gene regulation effects. It is clear that most genes essential for placental development are not specific for this tissue (50), suggesting that placenta specific regulatory networks may play an important role in morphogenesis and evolution of this organ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Louie van de Lagemaat for advice on the bioinformatics analysis and thank him and Sally Rogers for comments on the article. We thank Sylvain Charlat for advice on the statistical analysis. We also thank Patrik Medstrand (Lund University) for the placental samples and the Stem Cell Assay group (British Columbia Cancer Research Centre) for the PBL samples. Work in our laboratory was supported by a grant from the Canadian Institutes of Health Research with core support from the British Columbia Cancer Agency. DR was supported by a fellowship from the Multiple Sclerosis Society of Canada. Funding to pay the Open Access publication charges for this article was provided by Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 2.Craig JM. Heterochromatin–many flavours, common themes. Bioessays. 2005;27:17–28. doi: 10.1002/bies.20145. [DOI] [PubMed] [Google Scholar]
- 3.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 4.Lavie L, Kitova M, Maldener E, Meese E, Mayer J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2) J. Virol. 2005;79:876–883. doi: 10.1128/JVI.79.2.876-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnaud P, Goubely C, Pelissier T, Deragon JM. SINE retroposons can be used in vivo as nucleation centers for de novo methylation. Mol. Cell Biol. 2000;20:3434–3441. doi: 10.1128/mcb.20.10.3434-3441.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matouskova M, Blazkova J, Pajer P, Pavlicek A, Hejnar J. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell Res. 2006;312:1011–1020. doi: 10.1016/j.yexcr.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Frendo JL, Olivier D, Cheynet V, Blond JL, Bouton O, Vidaud M, Rabreau M, Evain-Brion D, Mallet F. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakimoto BT. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 9.Jahner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 10.Khodosevich K, Lebedev Y, Sverdlov ED. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 2004;32:e31. doi: 10.1093/nar/gnh035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, Pfeifer-Ohlsson S, Kato M, Larsson E, Rydnert J, Ohlsson R, Cohen M. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain human cellular sequences. J. Virol. 1987;61:2182–2191. doi: 10.1128/jvi.61.7.2182-2191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson DA, Goodchild NL, Saxton TM, Wood S, Mager DL. Evidence for a functional subclass of the RTVL-H family of human endogenous retrovirus-like sequences. J. Virol. 1993;67:2981–2989. doi: 10.1128/jvi.67.6.2981-2989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okahara G, Matsubara S, Oda T, Sugimoto J, Jinno Y, Kanaya F. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics. 2004;84:982–990. doi: 10.1016/j.ygeno.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Yi JM, Kim HM, Kim HS. Human endogenous retrovirus HERV-H family in human tissues and cancer cells: expression, identification, and phylogeny. Cancer Lett. 2006;231:228–239. doi: 10.1016/j.canlet.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Kim HS, Yi JM, Hirai H, Huh JW, Jeong MS, Jang SB, Kim CG, Saitou N, Hyun BH, et al. Human endogenous retrovirus (HERV)-R family in primates: chromosomal location, gene expression, and evolution. Gene. 2006;370:34–42. doi: 10.1016/j.gene.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Andersson AC, Yun Z, Sperber GO, Larsson E, Blomberg J. ERV3 and related sequences in humans: structure and RNA expression. J. Virol. 2005;79:9270–9284. doi: 10.1128/JVI.79.14.9270-9284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005;79:341–352. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 19.Fulka H, Mrazek M, Tepla O, Fulka J., Jr DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- 20.Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 22.Hellmann-Blumberg U, Hintz MF, Gatewood JM, Schmid CW. Developmental differences in methylation of human Alu repeats. Mol. Cell Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leib-Mosch C, Seifarth W, Schon U. Influence of Human Endogenous Retroviruses on Cellular Gene Expression. In: Sverdlov E, editor. Retroviruses and Primate Genome Evolution. 2005. pp. 123–143. Landes Bioscience. [Google Scholar]
- 24.Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, Mager DL. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet. Genome Res. 2005;110:342–352. doi: 10.1159/000084966. [DOI] [PubMed] [Google Scholar]
- 25.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 26.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taruscio D, Floridia G, Zoraqi GK, Mantovani A, Falbo V. Organization and integration sites in the human genome of endogenous retroviral sequences belonging to HERV-E family. Mamm. Genome. 2002;13:216–222. doi: 10.1007/s00335-001-2118-7. [DOI] [PubMed] [Google Scholar]
- 30.Schulte AM, Lai S, Kurtz A, Czubayko F, Riegel AT, Wellstein A. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl Acad. Sci. USA. 1996;93:14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deuel TF, Zhang N, Yeh HJ, Silos-Santiago I, Wang ZY. Pleiotrophin: a cytokine with diverse functions and a novel signaling pathway. Arch. Biochem. Biophys. 2002;397:162–171. doi: 10.1006/abbi.2001.2705. [DOI] [PubMed] [Google Scholar]
- 32.Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 2001;276:1896–1903. doi: 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- 33.Arai H, Nakao K, Takaya K, Hosoda K, Ogawa Y, Nakanishi S, Imura H. The human endothelin-B receptor gene. Structural organization and chromosomal assignment. J. Biol. Chem. 1993;268:3463–3470. [PubMed] [Google Scholar]
- 34.Handwerger S. Endothelins and the placenta. J. Lab. Clin. Med. 1995;125:679–681. [PubMed] [Google Scholar]
- 35.Fant ME, Nanu L, Word RA. A potential role for endothelin-1 in human placental growth: interactions with the insulin-like growth factor family of peptides. J. Clin. Endocrinol. Metab. 1992;74:1158–1163. doi: 10.1210/jcem.74.5.1373736. [DOI] [PubMed] [Google Scholar]
- 36.Landry JR, Mager DL. Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J. Virol. 2003;77:7459–7466. doi: 10.1128/JVI.77.13.7459-7466.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landry JR, Mager DL. Widely spaced alternative promoters, conserved between human and rodent, control expression of the Opitz syndrome gene MID1. Genomics. 2002;80:499–508. [PubMed] [Google Scholar]
- 38.Schweiger S, Foerster J, Lehmann T, Suckow V, Muller YA, Walter G, Davies T, Porter H, van Bokhoven H, et al. The Opitz syndrome gene product, MID1, associates with microtubules. Proc. Natl Acad. Sci. USA. 1999;96:2794–2799. doi: 10.1073/pnas.96.6.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opitz JM. G syndrome (hypertelorism with esophageal abnormality and hypospadias, or hypospadias-dysphagia, or “Opitz-Frias” or “Opitz-G” syndrome)–perspective in 1987 and bibliography. Am. J. Med. Genet. 1987;28:275–285. doi: 10.1002/ajmg.1320280203. [DOI] [PubMed] [Google Scholar]
- 40.Landry JR, Rouhi A, Medstrand P, Mager DL. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 2002;19:1934–1942. doi: 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- 41.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Lagemaat LN, Medstrand P, Mager DL. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 2006;7:R86. doi: 10.1186/gb-2006-7-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu L, Hornung D, Kurek R, Helen O, Blomberg J, Bergqvist A. Expression of human endogenous gammaretroviral sequences in endometriosis and ovarian cancer. AIDS Res. Hum. Retroviruses. 2006;22:551–557. doi: 10.1089/aid.2006.22.551. [DOI] [PubMed] [Google Scholar]
- 44.Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 46.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 47.Prudhomme S, Bonnaud B, Mallet F. Endogenous retroviruses and animal reproduction. Cytogenet. Genome Res. 2005;110:353–364. doi: 10.1159/000084967. [DOI] [PubMed] [Google Scholar]
- 48.Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- 49.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl Acad. Sci. USA. 2006;103:14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–130. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.