Abstract
Many DNA modification and repair enzymes require access to DNA bases and therefore flip nucleotides. Restriction endonucleases (REases) hydrolyze the phosphodiester backbone within or in the vicinity of the target recognition site and do not require base extrusion for the sequence readout and catalysis. Therefore, the observation of extrahelical nucleotides in a co-crystal of REase Ecl18kI with the cognate sequence, CCNGG, was unexpected. It turned out that Ecl18kI reads directly only the CCGG sequence and skips the unspecified N nucleotides, flipping them out from the helix. Sequence and structure conservation predict nucleotide flipping also for the complexes of PspGI and EcoRII with their target DNAs (/CCWGG), but data in solution are limited and indirect. Here, we demonstrate that Ecl18kI, the C-terminal domain of EcoRII (EcoRII-C) and PspGI enhance the fluorescence of 2-aminopurines (2-AP) placed at the centers of their recognition sequences. The fluorescence increase is largest for PspGI, intermediate for EcoRII-C and smallest for Ecl18kI, probably reflecting the differences in the hydrophobicity of the binding pockets within the protein. Omitting divalent metal cations and mutation of the binding pocket tryptophan to alanine strongly increase the 2-AP signal in the Ecl18kI–DNA complex. Together, our data provide the first direct evidence that Ecl18kI, EcoRII-C and PspGI flip nucleotides in solution.
INTRODUCTION
Base or nucleotide flipping is the displacement of a base in regular B-DNA from the helix into an extrahelical position. First observed by X-ray crystallography for the bacterial C5-cytosine methyltransferases M.HhaI (1) and M.HaeIII (2), nucleotide flipping (base extrusion) has been documented later for other methyltransferases (3–5), glycosylases (6–9), glycosyltransferases (10,11) and various DNA repair enzymes (12–17). Some enzymes, e.g. the methyltransferases, flip a nucleotide of only one DNA strand (1–5). Others, like endonuclease IV, alter the backbone conformations of both strands flipping the deoxyribose and nucleotide at an abasic site (13). Either way, nucleotide flips occur because enzymes need access to a DNA base to perform chemistry. For example, DNA methyltransferases transfer the methyl group to the extruded base, while glycosylases involved in DNA repair excise the extrahelical base (18). Typically, an amino acid side chain is intercalated into the DNA to fill in the ‘hole’ introduced after the base flipping event (6,12,19).
Nucleotide flipping in the co-crystals of restriction endonuclease Ecl18kI with cognate DNA came as a surprise (20). In a functional sense, Ecl18kI is a ‘standard’ Type II restriction endonuclease (REase): it recognizes pentanucleotide sequence CCNGG and cuts phosphodiester bonds on the 5′ sides of the outer cytosines to generate 5 nt 5′-overhangs (21). Although the endonuclease does not subject the central bases to any kind of modification, in the crystal structure these bases were clearly extrahelical and accommodated in pockets on Ecl18kI made by the side chain atoms of Arg57 on one face and the indole ring of Trp61 on the other face (Figure 1). Unlike in other complexes with flipped nucleotides, there was no ‘hole’ in the DNA and no amino acid intercalation. Instead, the DNA was compressed, so that the base pairs adjacent to the flipped nucleotides stacked directly against each other. The resulting DNA compression reduced the length of the interrupted 5 bp stretch CCNGG to the length of a 4 bp stretch CCGG and made the distance between the scissile phosphates in the Ecl18kI–DNA complex equal to the distance between the scissile phosphates in the NgoMIV complex with a continuous sequence GCCGGC (20,22). Therefore, we suggested that Ecl18kI uses base flipping to adapt the conserved sequence readout machinery for the interrupted target site and predicted that the evolutionary related REases EcoRII and PspGI that cut the related CCWGG sequence before the first C, might also flip nucleotides (23–25).
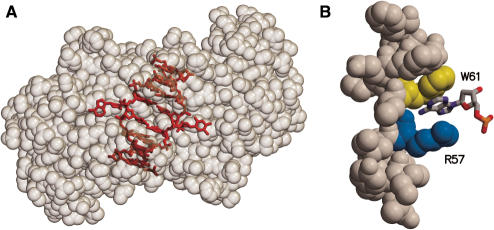
Figure 1.
Flipped nucleotides in the Ecl18kI–DNA complex structure (2FQZ). (A) General view of the Ecl18kI dimer–DNA complex. Protein is shown in spacefill. Residues 60–69 and 91–136 are removed for clarity. DNA is depicted in red. (B) Binding ‘pocket’ for the flipped out base. A flipped adenine base is accommodated in the ‘pocket’ made by the side chain atoms of Arg57 on one face and the indole ring of Trp61 on the other face.
Nucleotide flipping in solution by Ecl18kI, PspGI and EcoRII remains to be established. So far, it is only supported by the observation that PspGI accelerates deamination of the central cytosine in the incorrect CCCGG sequence, which differs from the canonical sequence at the center (26). 2-Aminopurine (2-AP) has often been used as a fluorescence probe to detect base flipping in solution (27–35). The 2-AP fluorescence is highly quenched in polynucleotides due to the stacking interactions with neighboring bases (36) and therefore increases strongly when the base is flipped out of the DNA helix (28,29). Here, we use 2-AP as a fluorescence probe for base flipping and provide the first direct evidence in solution that Ecl18kI, the C-terminal domain of EcoRII (EcoRII-C) and PspGI extrude the central base pair while interacting with their recognition sites.
MATERIALS AND METHODS
Oligonucleotides
2-AP containing oligodeoxynucleotides were obtained from Integrated DNA Technologies (HPLC grade, Coralville, USA), non-modified oligodeoxynucleotides were from Metabion (HPLC grade, Martinsried, Germany). In order to assemble oligoduplexes, appropriate oligodeoxynucleotides (Table 1) containing 2-AP or non-fluorescent control strands were mixed with a 1.05-fold molecular excess of complementary strands in the reaction buffer A (33 mM Tris-acetate, pH 7.9 at 25°C, 66 mM potassium acetate), heated to 85°C and allowed to cool slowly over several hours to room temperature. For the DNA binding and cleavage studies one strand of the 25 bp duplexes was 5′-end labeled with [γ-33P]ATP (Hartmann Analytic, Braunschweig, Germany) using a DNA labeling kit (Fermentas, Vilnius, Lithuania).
Table 1.
Oligoduplexes used in this study*
| Sequence | Oligoduplex |
|---|---|
| 5′ CGCACGCCTTCCTGGAAGCACACTA 3′ | Oligoduplex I |
| 3′ GCGTGCGGAAGG2CCTTCGTGTGAT 5′ | |
| 5′ CGCACGCCTTCCTGGAAGCACACTA 3′ | Oligoduplex II |
| 3′ GCGTGCGGA2GGACCTTCGTGTGAT 5′ | |
| 5′ CGCACGCCTTCCTGGAAGCACACTA 3′ | Oligoduplex III |
| 3′ GCGTGCGGAAGGACCTTCGTGTGAT 5′ |
*Ecl18kI/EcoRII-C/PspGI/MvaI recognition site is in boldface; 2, 2-aminopurine, is in boldface and underlined.
Proteins
The wt Ecl18kI, Ecl18kI mutant W61A, EcoRII-C, PspGI and MvaI proteins were purified and concentrations were determined by measuring absorbance at 280 nm as described in (24,25,37,38). All protein concentrations are indicated in terms of the dimer, except for MvaI, which is a monomer in solution (38).
Mutagenesis
The W61A mutant of Ecl18kI was obtained by the modified QuickChange Mutagenesis Protocol (39). Plasmid pET21b(+)_R.Ecl18kI [ApR] (20) was amplified by PCR using Pfu DNA polymerase (Fermentas, Vilnius, Lithuania) and two complementary (partially overlapping) primers obtained from Metabion (desalt grade, Martinsried, Germany) containing the desired mutation. After PCR the methylated parental (non-mutated) plasmid was digested with DpnI (Fermentas, Vilnius, Lithuania). Escherichia coli BL21(DE3) cells carrying the plasmids pVH1 [KnR] (with lacIq) and pHSG415ts [CmR] bearing the ecl18kIM gene (20) were transformed with the PCR product by the CaCl2 method. Plasmid DNA was isolated by the alkaline lysis procedure and purified using the GeneJET™ Plasmid Miniprep Kit (Fermentas, Vilnius, Lithuania). Sequencing of the entire gene of the mutant confirmed that only the designed mutation had been introduced.
Gel mobility shift assay
Gel shift analysis of DNA binding by wt proteins and Ecl18kI W61A mutant protein was performed by titrating 33P-labeled 25 bp oligoduplex (see Table 1) at 0.1 nM concentration with increasing amounts of protein. Kd values were evaluated as described elsewhere (40).
DNA cleavage activity
The DNA cleavage activities of wt Ecl18kI, Ecl18kI mutant W61A, EcoRII-C and PspGI were monitored using a 25 bp oligoduplex containing a 33P-label either in the top or the bottom DNA strand (Table 1). Cleavage rates of both strands were evaluated separately. Ecl18kI cleavage reactions were conducted at 20°C in the reaction buffer A, containing 10 mM MgCl2 and 0.1 mg/ml BSA using 200 nM of oligoduplex and 300 nM of protein. EcoRII-C and PspGI cleavage reactions were performed in the same buffer A at 25°C using 200 nM of oligoduplex and 1000 nM of protein. Aliquots were removed at timed intervals and quenched by mixing with loading dye [95% (v/v) formamide, 0.01% (w/v) bromphenol blue, 25 mM EDTA] before denaturing gel electrophoresis. The samples were analyzed and quantified as described in (41).
Fluorescence spectroscopy
All fluorescence measurements were acquired in photon counting mode on a Fluoromax-3 (Jobin Yvon, Stanmore, UK) spectrofluorometer equipped with Xe lamp. Sample temperatures were maintained at 25°C by a circulating water bath. Oligoduplexes I or II were used as 2-AP labeled DNA (Table 1). Emission spectra (340–420 nm) were recorded at an excitation wavelength λex = 320 nm with excitation and emission bandwidths of 2 and 8 nm, respectively. At least two scans were averaged for each spectrum. Sample emission spectra were collected in reaction buffer A in the presence and absence of calcium acetate on 250 nM DNA alone or 250 nM DNA mixed with a 5-fold excess of the protein to ensure saturation of the fluorescence signal. Control spectra used for the background subtraction corrections were collected under identical conditions except that oligoduplex III was used instead of the fluorescent DNA. The fluorescence emission value of the corrected spectrum was determined at the emission maximum (see Supplementary Table S1) for each sample. For the oligoduplex titration experiment, emission spectra of the 250 nM oligoduplex I with protein in a 0–2000 nM range were collected.
RESULTS
Probes for Ecl18kI triggered nucleotide flipping
We have used the 2-AP fluorescence assay to monitor base flipping in the Ecl18kI–DNA complex in solution. A number of 25 nt oligoduplexes that contain the fluorescent base analog 2-AP at different positions were designed (Table 1). In the oligoduplex I, 2-AP was incorporated within the CCNGG sequence instead of A in the central base position. In the oligoduplex II, 2-AP was introduced immediately adjacent to the target site. Like most restriction enzymes, Ecl18kI requires Mg2+ ions for DNA cleavage. In the absence of divalent cations, it forms a rather weak complex with cognate DNA (Supplementary Table S2). Addition of Ca2+ ions that do not support cleavage significantly increased Ecl18kI–DNA complex stability (24). Gel shift experiments revealed that 2-AP incorporation into the target sequence had no effect on the affinity of Ecl18kI for cognate DNA in the presence of Ca2+ ions (Figure 2). In the buffer supplemented with Mg2+ ions, Ecl18kI cleaved 2-AP containing and lacking oligoduplexes at identical rates (data not shown).
Figure 2.
Gel mobility shift analysis of the interactions between Ecl18kI and oligoduplexes. (A) Ecl18kI binding of the oligoduplex III containing the recognition sequence. (B) Ecl18kI binding of the cognate oligoduplex I containing 2-AP instead of the central A base. The binding reactions contained 33P-labeled 25 bp oligoduplex (0.1 nM) and the Ecl18kI at concentrations as indicated by each lane. Samples were analyzed by PAGE under non-denaturing conditions (see, Material and Methods Section). Gels were run in the presence of 5 mM of Ca2+ ions.
Ecl18kI in the presence of Ca2+ ions
We titrated the 2-AP containing oligonucleotides with Ecl18kI in the binding buffer supplemented with Ca2+ ions and monitored the change of the 2-AP fluorescence intensity at 367 nm (Figure 3A, Supplementary Table S1). The free oligoduplex containing 2-AP at the central position showed low signal because the fluorescence was quenched due to base stacking interactions (Figure 3B). When Ecl18kI bound at saturating concentrations to oligoduplex I, which contains 2-AP in the central position, the fluorescence intensity increased 6.5-fold. In contrast, only small changes were observed with oligoduplex II, which carries 2-AP outside of the target site (Figure 3B and C). The change of fluorescence intensity for the oligoduplex I suggests that the 2-AP stacking with DNA bases is disrupted. It is compatible with nucleotide flipping, which has been shown to enhance 2-AP fluorescence to varying extents in different systems (27–35).
Figure 3.
Fluorescence study of base flipping by Ecl18kI in solution. Titration of 250 nM 2-AP containing oligoduplex I with increasing amounts of Ecl18kI in the presence (A) and absence (D) of Ca2+ ions, respectively. Corrected 2-AP emission spectra of Ecl18kI–DNA complexes (1250 nM Ecl18kI and 250 nM oligoduplexes I or II) in the presence (B) and absence (E) of Ca2+ ions (see Materials and Methods Section for the details). Diagrams in (C) and (F) illustrate the maximum fluorescence intensity values of the corrected fluorescence emission spectra presented in (B) and (E), respectively.
Ecl18kI in the absence of Ca2+ ions
Gel shift analysis showed highly decreased binding of Ecl18kI to cognate DNA in the absence of Ca2+ ions (see, Supplementary Figure S1). However, we found that at much higher enzyme and DNA concentrations used in the fluorescence titration experiments, Ecl18kI formed a binary complex with cognate DNA in the absence of Ca2+ ions (Figure 3D). The Kd value obtained from the titration data was 52 ± 12 nM. In the Ca2+-free buffer, Ecl18kI binding to the oligoduplex I containing the 2-AP in the central position increased the fluorescence intensity ∼28.5-fold at saturating protein concentrations, while only small changes were observed with oligoduplex II (Figure 3E and F). The 2-AP signal in the buffer without Ca2+ ions was ∼4 times higher than the signal in the buffer supplemented with Ca2+ (Figure 3D). These results suggest that the structure of the complex formed in the presence of Ca2+ ions may differ from that formed without Ca2+.
Ecl18kI W61A mutation
2-AP fluorescence is often quenched in the hydrophobic environment of a protein (30,31,42,43). In the crystallographic complex of Ecl18kI with DNA, the flipped nucleotides are accommodated in pockets that are lined by tryptophan Trp61. In order to test whether Trp61 quenches 2-AP fluorescence, we replaced this residue with alanine. The Ecl18kI W61A variant did not cleave cognate oligoduplex III or the 2-AP containing oligoduplex I radioactively labeled at either strand, but it retained the ability to bind both oligoduplexes albeit at ∼10-fold decreased affinity according to the gel shift assay (see, Supplementary Table S2). Binding of Ecl18kI W61A to oligoduplex I in the presence of Ca2+ ions increased the 2-AP fluorescence intensity ∼125-fold (Figure 4) suggesting that the mutant was able to flip out the central nucleotide. 2-AP fluorescence in the ternary W61A–DNA–Ca2+ complex was ∼20 times higher than in the wt Ec18kI–DNA–Ca2+ complex (Figure 4). Thus, the W61A mutant data support the assumption that low 2-AP fluorescence in the ternary complex with the wt protein is due to the quenching of the extruded base by stacking interactions with the Trp61 residue. However, one cannot exclude that increased space in the binding pocket of the W61A mutant may allow a different orientation of the extrahelical 2-AP and affect the fluorescence intensity.
Figure 4.
2-AP fluorescence in the ternary complexes of the wt Ecl18kI and W61A mutant. Diagrams illustrate the fluorescence intensity values of the corrected fluorescence emission spectra at fluorescence maximum (367 nm for wt, 365 nm for W61A and 370 nm for oligoduplexes). Reactions contained 1250 nM protein and 250 nM oligoduplexes I or II and were measured in the presence of Ca2+ ions.
EcoRII-C and PspGI
The C-terminal domain of the EcoRII restriction enzyme and the PspGI restriction enzyme are specific for the CCWGG sequence (where W stands for A or T) and cleave it before the first C. It was suggested that Ecl18kI and EcoRII-C/PspGI may be evolutionarily related (23–25). This raises the intriguing question of whether EcoRII-C and PspGI also flip the central W nucleotides while interacting with their target sites.
The EcoRII structure, which was solved in the absence of DNA (44) shows a very similar fold to Ecl18kI (20) except that it has an extra N-terminal regulatory domain (Figure 5). Structural comparison between Ecl18kI and the C-terminal domain of EcoRII reveals that Arg57 and Trp61, which sandwich the flipped bases in the Ecl18kI–DNA cocrystal structure, spatially coincide with the Arg222 and Tyr226 of EcoRII suggesting that EcoRII may flip the central base similarly to Ecl18kI (20). Therefore, we analyzed base flipping by EcoRII-C in solution using the 2-AP fluorescence assay. EcoRII-C turned out to bind to the oligoduplex I containing the 2-AP in the central position (see, Supplementary Table S2) and binding was accompanied by ∼12-fold increase of fluorescence (Figure 6), suggesting that the base is extruded from the double helix.
Figure 5.
Ecl18kI, EcoRII and PspGI monomer structures. Central β-sheets are colored in red. N-terminal effector domain which is unique to EcoRII (44), is shown in gray. PspGI structural model (26) is shown.
Figure 6.
2-AP fluorescence in the ternary complexes of EcoRII-C, PspGI and MvaI. Diagrams show the fluorescence intensity values of the corrected fluorescence emission spectra at fluorescence maximum (360 nm for EcoRII-C and PspGI, 370 nm for MvaI and oligoduplexes). Reactions contained 1250 nM protein and 250 nM oligoduplexes I or II and were measured in the presence of Ca2+ ions.
The structure of the PspGI enzyme which recognizes the same CCWGG sequence as EcoRII but lacks the extra N-terminal domain is not yet known, but modeling studies (26) suggest significant similarities to Ecl18kI (Figure 5). Moreover, genetic studies support the PspGI model and provide indirect evidence that PspGI may flip central nucleotides within a sequence that matches its target site except for the presence of a G-C pair instead of the A-T pair at the center (26). We found that 2-AP at the central position of the recognition site does not change PspGI binding affinity (see, Supplementary Table S2). PspGI titration of 2-AP-containing oligonucleotide I showed an increase of 2-AP fluorescence and resulted in a 64-fold increase of the signal at saturation in comparison to the free oligoduplex (Figure 6). Thus, according to the 2-AP assay, PspGI should flip central nucleotides while interacting with its recognition site like Ecl18kI and EcoRII-C.
MvaI
Recently, we have solved the crystal structure of MvaI restriction enzyme that recognizes the CC/WGG sequence identical to that recognized by EcoRII and PspGI but cleaves it before the W nucleotide as indicated by the ‘/’ (38). In the MvaI–DNA complex structure, the DNA conformation does not deviate essentially from the canonical B-form and there is no evidence for base flipping. Binding studies in solution revealed that MvaI binds 2-AP-containing oligonucleotide I (see, Supplementary Table S2), however, this did not lead to an increase of 2-AP fluorescence (Figure 6).
DISCUSSION
Enzymes typically flip nucleotides to gain access to otherwise poorly accessible bases. Based on crystallographic information and sequence analysis, we have suggested that the restriction endonucleases Ecl18kI, EcoRII and PspGI employ base flipping in a novel way to achieve specificity for their targets and to adjust their cleavage patterns.
2-AP as a probe
Here, we used 2-AP fluorescence as a probe to monitor base flipping by Ecl18kI, EcoRII-C and PspGI in solution. Fluorescence probes to monitor nucleotide flipping in solution have to balance the conflicting requirements for mimicry of natural nucleotides and environment-sensitive fluorescence in a wavelength range not obscured by background signal from the other nucleotides and protein. 2-AP represents a good compromise in this respect. At neutral pH it makes a Watson–Crick base pair with T, which is only slightly weaker than the natural A-T pair (45). We have found that Ecl18kI, EcoRII-C and PspGI binding is not sensitive to the modification, hence 2-AP is a good surrogate for A in experiments with these enzymes.
Evidence for Ecl18kI-triggered nucleotide flipping
2-AP fluorescence is strongly quenched if the base is stacked in DNA and increases when the stacking is perturbed. Therefore, a 2-AP fluorescence does not necessarily indicate nucleotide flipping, since it could also be attributed to a less drastic DNA unstacking deformation. However, the much higher 2-AP fluorescence increase in the W61A–DNA–Ca2+ ternary complex compared to the wt–DNA–Ca2+ ternary complex (Figure 4) strongly suggests that in the latter complex the fluorophore comes close to the indole ring of Trp61 for efficient quenching. Moreover, the lack of activity of the Ecl18kI W61A mutant in the presence of Mg2+ ions and the nearly 10-fold reduced affinity to DNA are also consistent with a loss of interactions between the flipped nucleotide and the indole ring of Trp61.
In contrast to the effect of the W61A mutation, which can be readily attributed to the different hydrophobicities of tryptophan and alanine, the effect of Ca2+ ions on the 2-AP fluorescence is more difficult to interpret. According to the gel shift assay, the binary Ecl18kI–DNA complex is much weaker than the ternary Ecl18kI–DNA–Ca2+ complex (Supplementary Table S2). Nevertheless, 2-AP fluorescence in the binary complex is much higher than in the ternary complex in presence of Ca2+ ions (Figure 3). Moreover, the fluorescence increase in the binary complexes of wt Ecl18kI and W61A mutant is comparable (data not shown). Perhaps the flipped bases are firmly trapped in the quenching pockets of the enzyme in the presence of Ca2+ ions, but retain mobility and therefore higher fluorescence in the absence of Ca2+ ions?
Evidence for EcoRII and PspGI-triggered nucleotide flipping
Ecl18kI and EcoRII/PspGI are evolutionarily related and recognize target sequences that differ only in the central base pair. The strong 2-AP fluorescence increase upon addition of EcoRII-C and PspGI supports the idea that these enzymes also flip the central nucleotides of their target sequences. The 2-AP fluorescence intensity differences (Figure 6) likely reflect the nature of the enzyme pockets that accommodate the flipped bases. A structure-based alignment indicates that these pockets are lined by Trp61 in Ecl18kI, Tyr226 in EcoRII and Phe64 in PspGI. In the absence of crystal structures of DNA complexes of EcoRII and PspGI, it remains unclear whether the differences in 2-AP fluorescence in the enzyme–DNA complexes are purely due to different hydrophobicities, or whether changes in the orientation of the aromatic side chains or other alterations around the flipped nucleotides contribute to the observed effects.
Unlike Ecl18kI, which accepts any base pair at the center of its recognition sequence, EcoRII and PspGI cleave only target sequences with a central A-T pair. Modeling argues against the possibility that discrimination against a G-C pair could be due to the base-specific hydrogen bonding interactions in the EcoRII/PspGI DNA complexes. Instead, the strength of the hydrogen bonding interaction of the central base pair may determine specificity. Cytosine deamination experiments, however, provide indirect evidence that PspGI flips the central nucleotides in the sequence CCCGG, which is not efficiently cleaved by PspGI (26). As rates for flipping and back-flipping are not yet known, it is conceivable that the detailed balance between these two processes decides which substrates are cleaved by Ecl18kI, EcoRII and PspGI.
SUMMARY
The results of the 2-AP fluorescence assay provide direct evidence that Ecl18kI, EcoRII and PspGI unstack bases at the center of their recognition sequences and flip them into pockets that are formed by the enzymes (20). The new results show that prior findings from the co-crystal structure of Ecl18kI with DNA are relevant in solution. Moreover, the new results complement genetic experiments on PspGI, which had provided indirect evidence that this restriction enzyme flips the central cytosine in the sequence CCCGG, which is related to the PspGI target sequence, but not cleaved by PspGI. Finally, the change of the 2-AP fluorescence signal upon Ecl18kI/EcoRII-C/PspGI binding paves the way for stopped flow experiments to measure base flipping in real time (27,30,31,33–35,46–50) and time resolved fluorescence studies (43,51) to identify possible intermediates on the base flipping pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Giedre Tamulaitiene and Elena Manakova for the help with protein purification, Giedrius Sasnauskas for the critical reading of manuscript and Saulius Klimasauskas, Dalia Daujotyte and Aneta Kaczmarczyk for the comments and discussion. Research in the V. S. laboratory was supported by the grant from the Lithuania Science and Studies Foundation (T-07264). M.B. thanks the European Molecular Biology Organization (EMBO) and HHMI for a Young Investigator Award and the UNESCO/Polish Academy of Sciences Cellular and Molecular Biology Network for financial support. V.S. and M.B. acknowledge support by the European Commission 5th Framework Programme project “Center of Excellence in Molecular Bio-Medicine Contract no: QLK6-CT-2002-90363” (Warsaw) and ‘Center of Excellence - Biocell’ Contract no: QLK2-CT-2002-30575 (Vilnius), respectively. Authors also thank New England Biolabs for the PspGI clone, Fermentas for the MvaI clone and Monika Reuter (Institute of Virology, Berlin, Germany) for the EcoRII clone. Funding to pay the Open Access publication charge was provided by Lithuania Science and Studies Foundation (T-07264).
Conflict of interest statement. None declared.
REFERENCES
- 1.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 2.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995;82:143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 3.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 4.Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J. Mol. Biol. 2006;358:559–570. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 7.Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 8.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 9.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lariviere L, Morera S. A base-flipping mechanism for the T4 phage beta-glucosyltransferase and identification of a transition-state analog. J. Mol. Biol. 2002;324:483–490. doi: 10.1016/s0022-2836(02)01091-4. [DOI] [PubMed] [Google Scholar]
- 11.Lariviere L, Sommer N, Morera S. Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase. J. Mol. Biol. 2005;352:139–150. doi: 10.1016/j.jmb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 13.Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 14.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 15.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 16.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 18.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 20.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Den’mukhametov MM, Zakharova MV, Kravets AN, Pertsev AV, Sineva EV, Repik AV, Beletskaia IV, Gromova ES, Solonin AS. Characteristics of a plasmid bearing a gene of a restriction modification type II system – the SsoII isoschizomer. Mol. Biol. (Mosk) 1997;31:831–838. [PubMed] [Google Scholar]
- 22.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 23.Pingoud V, Kubareva E, Stengel G, Friedhoff P, Bujnicki JM, Urbanke C, Sudina A, Pingoud A. Evolutionary relationship between different subgroups of restriction endonucleases. J. Biol. Chem. 2002;277:14306–14314. doi: 10.1074/jbc.M111625200. [DOI] [PubMed] [Google Scholar]
- 24.Tamulaitis G, Solonin AS, Siksnys V. Alternative arrangements of catalytic residues at the active sites of restriction enzymes. FEBS Lett. 2002;518:17–22. doi: 10.1016/s0014-5793(02)02621-2. [DOI] [PubMed] [Google Scholar]
- 25.Pingoud V, Conzelmann C, Kinzebach S, Sudina A, Metelev V, Kubareva E, Bujnicki JM, Lurz R, Luder G, et al. PspGI, a type II restriction endonuclease from the extreme thermophile Pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003;329:913–929. doi: 10.1016/s0022-2836(03)00523-0. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter M, Divvela P, Pingoud V, Bujnicki J, Bhagwat AS. Sequence-dependent enhancement of hydrolytic deamination of cytosines in DNA by the restriction enzyme PspGI. Nucleic Acids Res. 2006;34:3762–3770. doi: 10.1093/nar/gkl545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 28.Allan BW, Reich NO. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35:14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 29.Holz B, Klimasauskas S, Serva S, Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gowher H, Jeltsch A. Molecular enzymology of the EcoRV DNA-(Adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol. 2000;303:93–110. doi: 10.1006/jmbi.2000.4127. [DOI] [PubMed] [Google Scholar]
- 31.Szegedi SS, Reich NO, Gumport RI. Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase. Nucleic Acids Res. 2000;28:3962–3971. doi: 10.1093/nar/28.20.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy YV, Rao DN. Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol. 2000;298:597–610. doi: 10.1006/jmbi.2000.3673. [DOI] [PubMed] [Google Scholar]
- 33.Malygin EG, Evdokimov AA, Zinoviev VV, Ovechkina LG, Lindstrom WM, Reich NO, Schlagman SL, Hattman S. A dual role for substrate S-adenosyl-L-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase. Nucleic Acids Res. 2001;29:2361–2369. doi: 10.1093/nar/29.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebert K, Hermann A, Schlickenrieder M, Jeltsch A. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 2004;341:443–454. doi: 10.1016/j.jmb.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 35.Walker RK, McCullough AK, Lloyd RS. Uncoupling of nucleotide flipping and DNA bending by the t4 pyrimidine dimer DNA glycosylase. Biochemistry. 2006;45:14192–14200. doi: 10.1021/bi060802s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 37.Shlyakhtenko LS, Gilmore J, Portillo A, Tamulaitis G, Siksnys V, Lyubchenko YL. 2007. Direct Visualization of EcoRII-DNA Triple Synaptic Complex by Atomic Force Microscopy. in press. [DOI] [PubMed] [Google Scholar]
- 38.Kaus-Drobek M, Czapinska H, Sokolowska M, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically. Nucleic Acids Res. 2007;35:2035–2046. doi: 10.1093/nar/gkm064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamulaitis G, Mucke M, Siksnys V. Biochemical and mutational analysis of EcoRII functional domains reveals evolutionary links between restriction enzymes. FEBS Lett. 2006;580:1665–1671. doi: 10.1016/j.febslet.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Allosteric communication network in the tetrameric restriction endonuclease Bse634I. J. Mol. Biol. 2006;363:800–812. doi: 10.1016/j.jmb.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 42.Scavetta RD, Thomas CB, Walsh MA, Szegedi S, Joachimiak A, Gumport RI, Churchill ME. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res. 2000;28:3950–3961. doi: 10.1093/nar/28.20.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenz T, Bonnist EY, Pljevaljcic G, Neely RK, Dryden DT, Scheidig AJ, Jones AC, Weinhold E. 2-Aminopurine flipped into the active site of the adenine-specific DNA methyltransferase M.TaqI: crystal structures and time-resolved fluorescence. J. Am. Chem. Soc. 2007;129:6240–6248. doi: 10.1021/ja069366n. [DOI] [PubMed] [Google Scholar]
- 44.Zhou XE, Wang Y, Reuter M, Mucke M, Kruger DH, Meehan EJ, Chen L. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 2004;335:307–319. doi: 10.1016/j.jmb.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 45.Goodman MF, Ratliff RL. Evidence of 2-aminopurine-cytosine base mispairs involving two hydrogen bonds. J. Biol. Chem. 1983;258:12842–12846. [PubMed] [Google Scholar]
- 46.Allan BW, Beechem JM, Lindstrom WM, Reich NO. Direct real time observation of base flipping by the EcoRI DNA methyltransferase. J. Biol. Chem. 1998;273:2368–2373. doi: 10.1074/jbc.273.4.2368. [DOI] [PubMed] [Google Scholar]
- 47.Vilkaitis G, Merkiene E, Serva S, Weinhold E, Klimasauskas S. The mechanism of DNA cytosine-5 methylation. Kinetic and mutational dissection of Hhai methyltransferase. J. Biol. Chem. 2001;276:20924–20934. doi: 10.1074/jbc.M101429200. [DOI] [PubMed] [Google Scholar]
- 48.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. Presteady-state analysis of a single catalytic turnover by Escherichia coli uracil-DNA glycosylase reveals a "pinch-pull-push" mechanism. J. Biol. Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 49.Bellamy SR, Krusong K, Baldwin GS. A rapid reaction analysis of uracil DNA glycosylase indicates an active mechanism of base flipping. Nucleic Acids Res. 2007;35:1478–1487. doi: 10.1093/nar/gkm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuznetsov NA, Koval VV, Nevinsky GA, Douglas KT, Zharkov DO, Fedorova OS. Kinetic conformational analysis of human 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2007;282:1029–1038. doi: 10.1074/jbc.M605788200. [DOI] [PubMed] [Google Scholar]
- 51.Neely RK, Daujotyte D, Grazulis S, Magennis SW, Dryden DT, Klimasauskas S, Jones AC. Time-resolved fluorescence of 2-aminopurine as a probe of base flipping in M.HhaI-DNA complexes. Nucleic Acids Res. 2005;33:6953–6960. doi: 10.1093/nar/gki995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.