Abstract
Microhomology-mediated end joining (MMEJ) joins DNA ends via short stretches [5–20 nucleotides (nt)] of direct repeat sequences, yielding deletions of intervening sequences. Non-homologous end joining (NHEJ) and single-strand annealing (SSA) are other error prone processes that anneal single-stranded DNA (ssDNA) via a few bases (<5 nt) or extensive direct repeat homologies (>20 nt). Although the genetic components involved in MMEJ are largely unknown, those in NHEJ and SSA are characterized in some detail. Here, we surveyed the role of NHEJ or SSA factors in joining of double-strand breaks (DSBs) with no complementary DNA ends that rely primarily on MMEJ repair. We found that MMEJ requires the nuclease activity of Mre11/Rad50/Xrs2, 3′ flap removal by Rad1/Rad10, Nej1, and DNA synthesis by multiple polymerases including Pol4, Rad30, Rev3, and Pol32. The mismatch repair proteins, Rad52 group genes, and Rad27 are dispensable for MMEJ. Sae2 and Tel1 promote MMEJ but inhibit NHEJ, likely by regulating Mre11-dependent ssDNA accumulation at DNA break. Our data support the role of Sae2 and Tel1 in MMEJ and genome integrity.
DNA double-strand breaks (DSBs) are extremely deleterious to a cell, and if left unrepaired or misrepaired, cell death will ensue (Khanna and Jackson 2001; Bassing and Alt 2004). All eukaryotes including the budding yeast Saccharomyces cerevisiae evolved multiple mechanisms to eliminate DSBs. Homologous recombination (HR) and non-homologous end joining (NHEJ) are the two DSB repair mechanisms that are characterized in some detail (Paques and Haber 1999). HR faithfully reconstitutes genetic information by copying from a template homologous to the sequence flanking the DSB (Sung et al. 2000). All three subclasses of HR, gene conversion, break-induced replication (BIR), and single-strand annealing (SSA), depend on functional RAD52 epistasis group genes (Paques and Haber 1999; Symington 2002). In contrast, NHEJ joins broken ends together with a few base pair deletions and/or insertions regardless of sequence homology (Critchlow and Jackson 1998; Daley et al. 2005b; Hefferin and Tomkinson 2005). NHEJ requires the evolutionary conserved KU70/KU80 and DNL4/LIF1 (Ligase 4 and Xrcc4 in mammals) proteins for end binding and ligation (Rathmell and Chu 1994; Taccioli et al. 1994; Nussenzweig et al. 1996; Bogue et al. 1997; Gu et al. 1997; Schar et al. 1997; Wilson et al. 1997; Boulton and Jackson 1998; Herrmann et al. 1998). Multiple factors including POL4, FEN1, and artemis process ends to configure them for ligation (Wilson and Lieber 1999; Wu et al. 1999; Tseng and Tomkinson 2004). In yeast, Mre11/Rad50/Xrs2 functions in end joining by aligning and tethering DNA ends for ligation (Chen et al. 2001), whereas in mammals such activities involve the scid gene product, a catalytic subunit of DNA-dependent protein kinase (DNA-PK) (Cary et al. 1997). Most recently, Cernunnos/XLF, a likely homolog of yeast NEJ1, has been identified in a screen for mutations that cause severe combined immunodeficiency in human patients (Ahnesorg et al. 2006; Buck et al. 2006).
In addition to these two mechanisms, a third DSB repair pathway that operates independently of Rad52 and Ku proteins has been described for some time. The presence of this mechanism is most evident when HR and/or NHEJ are not available for repair, e.g., owing to mutations in mouse Ku86 or deletion of yeast KU70 and RAD52 genes (Liang and Jasin 1996; Boulton and Jackson 1998; Yu and Gabriel 2003). Nevertheless, when a DNA break occurs with no complementary end sequence for alignment or homologous template sequence for recombination, Rad52- and KU-independent microhomology-mediated end joining (MMEJ) becomes the primary repair choice (Ma et al. 2003). In Drosophila melanogaster, P-element-excised DNA DSBs are primarily eliminated by a DmRad51 (spn-A)-mediated synthesis-dependent strand annealing (SDSA) mechanism. However, in spn-A mutants repair proceeds through a nonconservative pathway involving the annealing of microhomologies within the 17-nt overhangs produced by P-element excision (McVey et al. 2004). End-to-end fusion of short telomeres in an Arabidopsis ku tert mutant occurs by MMEJ (Heacock et al. 2004). In mammals, a backup end-joining pathway kinetically slower than the Ku-dependent pathway has been revealed by biochemical fractionation of extracts from cells lacking DNA-dependent protein kinase (Perrault et al. 2004). Similarly, slower end-joining activity independent of Ku proteins has been reported from studies in Xenopus extracts and chicken DT-40 cells (Gottlich et al. 1998; Takata et al. 1998; Labhart 1999). MMEJ is thus an evolutionarily well-conserved mechanism to eliminate DSBs even when NHEJ and HR can in principle carry out this function.
MMEJ is highly mutagenic and likely contributes to genome instability found in certain cancer cells. It involves base pairing between microhomologies that always leads to deletion of intervening sequence between the microhomologies (Ma et al. 2003; Daley and Wilson 2005). Moreover, MMEJ may be responsible for gross chromosomal rearrangements observed in yeast mutants with a compromised DNA damage response (Kolodner et al. 2002). Lack of NHEJ and/or HR also leads to a dramatic increase in chromosome abnormalities such as translocations and deletions in both yeast and mice (Davies et al. 1995; Pierce and Jasin 2001; Yu and Gabriel 2003). Remarkable similarity between junctional sequences of MMEJ repair and those from human disease-causing chromosomal translocations underscores the role of MMEJ in oncogenic chromosome rearrangements (Yu and Gabriel 2003). In accordance with this idea, repair of DNA breaks in human bladder cancer cells occurs by an alternative mutagenic end-joining pathway that is mediated by microhomology (Bentley et al. 2004). Most pathological DSBs occurring in vivo may not have complementary overhangs for base pairing or homologous sister chromatids in the G1 phase (Lieber 1999). MMEJ may thus constitute a critical means for cells to repair breaks in this phase of the cell cycle. Indeed, MMEJ contributes almost equally with Ku-dependent NHEJ to the repair of ionizing radiation-induced DNA damage (Ma et al. 2003).
Despite its impact on genome integrity, little is known about the genetic requirements of MMEJ. In budding yeast, MMEJ is independent of Rad52 and Ku proteins (Ma et al. 2003; Daley and Wilson 2005). In fact, Ku may be inhibitory to non-Ku-mediated end joining that includes MMEJ (Boulton and Jackson 1998). Biochemical fractionation of Ku-independent end-joining activity in Xenopus egg extracts uncovered roles for DNA ligase III, Polɛ, Fen1, and an unidentified bidirectional exonuclease activity in the error prone end-joining reaction (Gottlich et al. 1998). DNA ligase III was also found in HeLa cell extracts that catalyze Ku-independent backup end joining (Wang et al. 2003). Recently, we identified a role for the yeast Mre11 complex and Rad1, a component of the structure-specific Rad1/Rad10 endonuclease, in repair of DNA DSBs with no complementary end sequences. These repair events occurred primarily by MMEJ-mediated annealing of recessed microhomologies on either side of the broken DNA ends (Ma et al. 2003). Partial reduction of MMEJ in dnl4Δ mutants also suggests a role for Dnl4 and Cdc9 in the sealing of DNA ends during MMEJ (Ma et al. 2003).
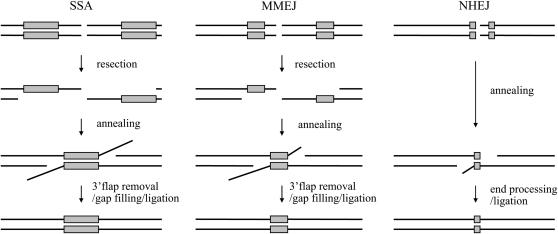
It is anticipated that SSA, NHEJ, and MMEJ all anneal DNA ends via recessed microhomology on opposite sides of two broken DNA ends and remove 3′ non-homologous tails prior to gap filling and ligation as illustrated in Figure 1. Therefore, MMEJ may be mechanistically similar to the Rad52-dependent SSA or Ku-dependent NHEJ mechanisms and likely shares some genetic components with these two processes. In support of this model, Rad1 and Mre11 participate in SSA, whereas Dnl4 functions in NHEJ (Ma et al. 2003). We predict that additional NHEJ or SSA factors may participate in MMEJ. To test this hypothesis, we surveyed the role of many factors known to function in either NHEJ or SSA in the repair by MMEJ of two breaks with no complementary end sequences. This study revealed a dozen genes involved in MMEJ and their catalytic functions during MMEJ reactions. The results also validate our model that MMEJ is mechanistically similar to SSA and Ku-dependent NHEJ.
Figure 1.—
A model of the biochemical steps that lead to MMEJ, NHEJ, and SSA. Following resection, MMEJ, NHEJ, and SSA all anneal microhomologies located at either side of the break, followed by end processing including 3′ flap removal, gap filling, and ligation of broken ends to complete repair.
MATERIALS AND METHODS
Strains and plasmids:
All strains are derivatives of SLY19, which has the genotype hoΔ MATα∷URA3∷HOcs hmlΔ∷ADE1 hmrΔ∷ADE1 ade1-100 leu2-3,112 lys5 trp1∷hisG ura3-52 ade3∷GAL∷HO (Ma et al. 2003) (supplemental Table 1 at http://www.genetics.org/supplemental). All single gene deletion mutants were constructed by using a PCR-derived KANMX module flanked by short terminal sequences homologous to the ends of each single gene open reading frame (Wach et al. 1994). Double mutant strains were constructed by first replacing the KANMX selectable marker from the single gene deletion using a DNA fragment containing the TRP1 marker and then deleting the second gene by one-step gene replacement using the now available KANMX module again. Strain SLY19-mre11Δ was transformed with centromeric plasmids carrying either MRE11, mre11-3, mre11-H125N, or mre11-P162S, or an empty vector to test for the effect of MRE11 mutations on MMEJ. Similarly, SLY19-pol4Δ was transformed with plasmids pTW300, pTW301, or pTW305 (Wilson and Lieber 1999) to investigate the role of Pol4 in MMEJ.
HO endonuclease induction:
HO endonuclease was induced according to the protocol described previously (Ma et al. 2003). Briefly, serial dilutions of cells grown to mid-log phase in YEP-glycerol at 30° were plated onto YEP-agar medium containing either galactose (YEP–GAL) or glucose (YEPD). The frequency of survival following an HO-induced DSB was calculated by dividing the number of colonies growing on YEP–GAL by the number of colonies growing on YEPD.
Analysis of repair events:
Colonies growing on YEP–GAL plates were replica plated onto synthetic complete (SC) medium lacking uracil, and mating type was determined by complementation using tester strains. Colonies displaying uracil auxotrophy and mating-type a were selected for analysis of repair events by amplifying the region spanning the repair junctions with PCR using a set of primers: pX (5′-GTAAACGGTGTCCTCTGTAAGG-3′) and p2 (5′-TCGAAAGATAAACAACCTCC-3′) and sequencing.
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (ChIP) was carried out as described previously with minor modifications (Sugawara et al. 2003). Briefly, cultures grown to 3 × 106 cells/ml in pre-induction medium (YEP–glycerol) were induced for HO endonuclease by addition of 2% (final concentration) galactose, crosslinked by 1% formaldehyde (final volume), and sonicated to an average size of 0.5 kb. The sonicated extracts were incubated with anti-RPA antibody (kindly provided by S. Brill) at 4° for 2 hr and then with protein G agarose bead for 1 hr. PCR condition and the primers used for ChIPs are described previously (Shim et al. 2005).
RESULTS
Identification of additional MMEJ genes that also function in NHEJ or SSA:
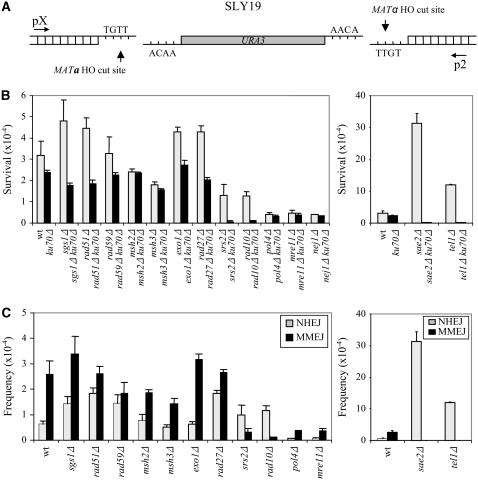
Predicted similarity between MMEJ, SSA, and Ku-dependent NHEJ (Figure 1) suggests that additional NHEJ or SSA factors besides the Mre11 complex, Rad1, and Dnl4 may contribute to MMEJ (Ma et al. 2003). Using an assay in S. cerevisiae that measures the repair of two simultaneous DSBs with no complementary end sequences, we surveyed the role of almost every NHEJ or SSA factor in MMEJ. Expression of a galactose-inducible HO gene inserted at the ADE3 locus simultaneously cleaves two inversely oriented HO recognition sites, a 117-bp sequence from MATa and the MATα sequence, separated by ∼2.0 kb of DNA containing the URA3 gene and Yα sequence (SLY19) (Figure 2A). Lack of homologous templates (HML and HMR) effectively eliminates repair of the DSBs by HR. Thus, cells survive these breaks primarily by MMEJ (∼80%) or less frequently by Ku-dependent NHEJ (∼20%), which renders the HO recognition sites resistant to additional enzyme cleavage (Ma et al. 2003).
Figure 2.—
MMEJ frequency in various mutants. (A) Cleavage at two HO sites in the MAT locus of SLY19 separated by ∼2 kb of URA3 sequence in opposite orientation generates noncomplementary breaks that are preferentially joined by Ku- and Rad52-independent MMEJ. The location of primers for PCR amplification and sequencing are shown by arrows. (B) Survival frequency after induction of HO breaks of each mutant strain (shaded bars) and the corresponding KU70 deletion derivatives (solid bars). The frequency of survival after an HO-induced DSB was calculated by dividing the number of colonies growing on YEP–GAL by the number of colonies growing on YEPD. Each value represents the average from at least three independent experiments ± standard deviation. (C) Junctional sequences observed in survivors of SLY19 and the mutant derivatives were used to calculate the frequency of repair events carried out by MMEJ or NHEJ. Typically, MMEJ involves >5 bp of imperfect microhomology, whereas NHEJ joins ends using <5 nt of complementary base pairing. The region spanning junctional sequences was PCR amplified with a set of primers [p2 and pX (A)] that anneal to the proximal and distal regions of the HO cleavage sites and align with the original sequence of SLY19. For each mutant, between 24 and 96 survivors were sequenced (see supplemental Figure 2).
SLY19 derivative strains deleted for EXO1, MSH2, MSH3, MSH6, NEJ1, POL4, RAD10, RAD27, RAD51, RAD59, SAE2, SGS1, or SRS2, all of which have roles in NHEJ, SSA, or both (Paques and Haber 1999; Symington 2002; Daley et al. 2005b), were induced for HO expression by plating onto galactose-containing medium (YEP–GAL). Survival rates were determined from the number of colonies growing on galactose-containing plates, normalized by that on glucose-containing medium (Figure 2B). To determine whether the repair of HO-induced breaks in SLY19 occurred by NHEJ or Ku-independent MMEJ, yeast strains deleted for both KU70 and each NHEJ or SSA gene were tested for their survival by MMEJ only. Types of repair events that occur in the survivors were determined by PCR amplification and sequencing of the region spanning repair junctions using a set of oligonucleotides that anneal to the regions 5′ to the 117-bp MATa cleavage site and 3′ to the MATα HO recognition site (Figure 2C and supplemental Table 2 at http://www.genetics.org/supplemental). Junctions using >5 bp of microhomology were attributed to the MMEJ product, while those using fewer than 5 bp of microhomology for base pairing were by Ku-dependent NHEJ (Ma et al. 2003).
The approach described above identified four new MMEJ genes, all of which have known roles in either NHEJ or SSA. Deletion of NEJ1, POL4, RAD10, or SRS2 from SLY19 or the KU70 deletion derivative of SLY19 (SLY19-ku70Δ) reduced cell survival from HO breaks, revealing their role in MMEJ (Figure 2B). Analysis of repair junctions from survivors confirmed their involvement in MMEJ, as the majority of junctions from strains deleted for these genes retain features indicative of their formation by Ku-dependent NHEJ using fewer than 5 bp of microhomology for base pairing, but junctions by MMEJ that use >5 bp of microhomology disappear from the mutants (Figure 2C). It is possible, however, that an assay analyzing repair events from unselected total cell population may yield a different outcome. Interestingly, deletion of SAE2 increased cell survival on YEP–GAL by ∼10-fold, although deletion of SAE2 and KU70 reduced cell survival from the HO breaks to a level similar to that of rad10Δ ku70Δ, suggesting that increased survival of the sae2Δ mutant arose from increased KU-dependent NHEJ, but not from enhanced MMEJ (Figure 2B). Survival rates and the sequence of repair junctions indeed indicated that the deletion of SAE2 severely reduced MMEJ, which was compensated for by elevated Ku-dependent NHEJ (Figure 2C). Deletion of mismatch repair genes (MSH2, MSH3, MSH6, and EXO1), RAD52 epistasis group genes (RAD51 and RAD59), FEN1, or SGS1 in either SLY19 or the SLY19-yku70Δ strains did not significantly (Student's t-test, P > 0.1) alter cell survival after DSB induction nor the type of repair events (mostly MMEJ) among survivors (Figure 2, B and C).
Provided that MMEJ and NHEJ occur primarily at different phases of the cell cycle, alteration of the cell cycle can account for the reduced MMEJ and a concomitant increase in NHEJ among the newly identified MMEJ mutants. This is unlikely, however, as the parallel experiments with the nocodazole-arrested MMEJ mutants at G2 prior to galactose addition revealed that the MMEJ deficiency was identical to that without synchronization (supplemental Figure 1 at http://www.genetics.org/supplemental). Flow cytometry also failed to detect substantial change in cell-cycle patterns among MMEJ mutants (supplemental Figure 2). The results support our hypothesis that MMEJ, NHEJ, and SSA often rely on the same gene products to catalyze similar reactions for DSB repair.
Mre11 resects DNA ends to reveal and anneal microhomology in MMEJ:
Having established a role for several genes and their gene products in MMEJ, we set out to determine how these proteins catalyze MMEJ reactions. Previously, we discovered that MMEJ depends on Mre11 and Rad50 and likely Xrs2 as well (Ma et al. 2003) (Figure 3). Biochemical studies have shown that Mre11 has double-strand exonuclease and single-strand endonuclease activities (Paull and Gellert 1998; Trujillo and Sung 2001) and a DNA hairpin endonuclease activity that can incise DNA at a single-strand/duplex junction (Paull and Gellert 1999; Trujillo and Sung 2001). We thus reasoned that Mre11 functions to resect DNA ends in the MMEJ reaction. Should the Mre11 complex provide the nuclease activity in MMEJ, we predicted that the mre11 mutations that inactivate the nuclease activity, such as the mre11-H125N (Moreau et al. 1999) and mre11-3 mutations (Bressan et al. 1998), will also impair MMEJ. The mre11-H125N mutant has been confirmed biochemically to be deficient for nuclease activity but maintains the ability to form a heterotrimeric complex with Rad50 and Xrs2 (Moreau et al. 1999).
Figure 3.—
The role of Mre11 nuclease activity in MMEJ. (A) SLY19-mre11Δ was transformed with yeast centromeric plasmids expressing MRE11, mre11-H125N, mre11-3, mre11-P162S, or an empty vector. Survival frequency and the relative contribution of MMEJ and NHEJ to survival after induction of HO breaks were determined by analysis of junctional sequences observed in survivors. (B) Survival frequency of SLY19-exo1Δ, SLY19-exo1Δ mre11Δ, or SLY19-mre11Δ expressing additional Exo1 from a GAL promoter. An empty vector containing the GAL promoter was used as a control. Each value represents the average from at least three independent experiments ± standard deviation.
The mre11 gene deletion derivative of SLY19 (SLY19-mre11Δ) was transformed with centromeric plasmids bearing mutant mre11 constructs to assess the effect of the mre11 mutations on cell survival after induction of persistent HO cleavage by plating cells onto galactose-containing medium (Figure 3A). The results were compared to SLY19-mre11Δ transformed with either the wild-type MRE11 expression construct or an empty vector. We also sequenced ∼100 PCR-amplified repair junctions from the survivors to ensure the repair events were mediated by 5 bp or more of microhomology, typical of MMEJ products. We found that expression of the nuclease defective mre11-H125N or mre11-3 mutations rescued lethality of the mre11 null mutant upon DSB induction (Figure 3A). However, analysis of repair junctions from survivors revealed that almost all were repaired by Ku-dependent NHEJ rather than MMEJ (Figure 3A and supplemental Table 2 at http://www.genetics.org/supplemental). Expression of an all but null allele of mre11-P162S (Chamankhah et al. 2000) markedly reduced cell survival from HO breaks both by MMEJ and NHEJ. Our interpretation of these results is that the nuclease activity of Mre11 is critical for MMEJ.
Overproduction of the 5′ to 3′ exonuclease Exo1 was previously shown to complement the resection defect of the mre11Δ mutant (Tsubouchi and Ogawa 2000; Moreau et al. 2001; Lee et al. 2002; Lewis et al. 2002). In addition, the 5′ to 3′ resection defect of the mre11Δ exo1Δ double mutant is more severe than that of the mre11Δ strain (Tsubouchi and Ogawa 2000). If Mre11 participates in MMEJ as a nuclease to create single-stranded DNA (ssDNA) at DSBs, Exo1p may rescue the MMEJ deficiency of the mre11Δ strain. We overexpressed EXO1 in SLY19-mre11Δ and assayed the MMEJ defect of this strain by measuring survival after HO cleavage and cataloging the repair events from sequencing the repair junctions. As shown in Figure 3B, overproduction of Exo1 but not the vector alone partially but consistently rescued the decreased survival of SLY19-mre11Δ. In addition, an mre11Δ exo1Δ double mutant showed a further reduction in survival after HO expression rather than the mre11Δ mutant alone (Figure 3B). We thus propose that one function of the Mre11 complex in MMEJ is degrading 5′ DNA ends to generate ssDNA to reveal microhomology for annealing. This suggests that in the absence of Mre11, other nuclease(s) such as Exo1 can provide less efficient end resection for the limited MMEJ reactions.
Rad1/Rad10 and Srs2 are involved in MMEJ:
Rad1 and Rad10 form a heterodimeric structure-specific endonuclease that removes 3′ flaps during HR (Sugawara et al. 1997). Rad1/Rad10 likely participates in MMEJ by catalyzing 3′ flap removal after annealing of microhomology between ssDNA (Ma et al. 2003). Consistent with their function as a complex, a yeast strain deleted for both RAD1 and RAD10 showed identical MMEJ deficiency as the rad1Δ and rad10Δ single mutant strains (Figure 2, B and C).
Srs2 suppresses recombination by displacing Rad51 from ssDNA (Krejci et al. 2003; Veaute et al. 2003). Srs2 also performs prorecombination functions during allelic recombination and SSA (Sugawara et al. 2000; Ira et al. 2003). In SSA, Srs2 may stabilize homologous joints and promote the removal of 3′ flap DNA with the Rad1/Rad10 complex by relieving superhelical stress from unwinding donor duplex DNA (Paques and Haber 1997). Alternatively, Srs2 can promote MMEJ by displacing Rad51 and thus inhibiting recombination of resected DNA ends. To distinguish between these two possibilities, we deleted RAD51 in an srs2Δ strain and investigated its ability to undergo MMEJ repair of an HO break. We found that deletion of RAD51 fully rescued the MMEJ repair defect in srs2Δ (Figure 4). This result suggests that Srs2 is involved in MMEJ as an antirecombinase by displacing Rad51 from ssDNA.
Figure 4.—
Lack of Rad51 rescues the MMEJ deficiency of srs2Δ. Survival frequency following induction of HO breaks of srs2Δ, rad51Δ, and srs2Δ rad51δ strains (shaded bars) and the corresponding KU70 deletion derivatives (solid bars) were determined as described in Figure 2. The average from at least three independent experiments ± standard deviation is shown.
Polymerase activity of Pol4 is essential for gap fill-in after annealing between microhomology:
MMEJ likely depends on DNA polymerase(s) to fill gaps of various sizes ranging from a few to several hundred nucleotides following 5′ nucleolytic degradation to expose microhomologies within the regions flanking a DSB (Figure 1). In addition to Pol4, we tested two other nonessential polymerases (Rev3 and Rad30), Pol32 (a subunit of Polδ), and poly (A) RNA polymerase Trf4 (Holbeck and Strathern 1997; McDonald et al. 1997; Gerik et al. 1998; Wang et al. 2000; Zhao et al. 2004, 2006), for their role in MMEJ by determining survival of single-deletion mutant strains following HO expression (Figure 5A). We found that deletion of POL4, POL32, RAD30, or REV3 all led to a 3- to 10-fold reduction in survival from the HO break because of a combined deficiency in NHEJ and MMEJ (pol4Δ) or the MMEJ defect only (pol32Δ, rad30Δ, and rev3Δ). Yeast strains lacking TRF4 did not confer any detectable MMEJ deficiency. Potential involvement of multiple polymerases in MMEJ prompted us to consider redundancy among polymerases in the gap-filling step of MMEJ. We tested this idea by constructing yeast strains with two or more polymerase gene deletions and assessed their MMEJ deficiency by analyzing repair events following HO cleavage. None of double mutant strains showed additional MMEJ deficiency beyond that of each single gene deletion mutant (Figure 5B). However, deletion of POL32, RAD30, and REV3 together almost completely wiped off any residual MMEJ among survivors (Figure 5B). The results thus reveal redundancy among these polymerases in MMEJ repair.
Figure 5.—
MMEJ requires multiple polymerases including two translesion bypass polymerases. Survival frequency (A) and the contribution of MMEJ and NHEJ to repair (B) among survivors after induction of HO breaks of strains lacking one or more nonessential DNA polymerases or KU70 were determined by analysis of junctional sequences observed in survivors. (C) Plasmids expressing POL4 or its mutant derivatives were introduced into SLY19-pol4Δ, and the frequency of MMEJ and NHEJ following induction of HO breaks was determined as in A. Each value represents the average from at least three independent experiments ± standard deviation.
Pol4 is not a processive polymerase (Bebenek et al. 2005). It mostly fills small gaps without synthesizing long stretches of nucleotides (Daley et al. 2005a). Lack of processivity led us to consider the possibility that Pol4 plays a role in MMEJ separate from its DNA polymerization activity. Previously, the BRCT domain of Pol4 was shown to interact with the Dnl4/Lif1 complex and to couple the gap filling and ligation reactions during Ku-dependent NHEJ (Tseng and Tomkinson 2002, 2004). Consistent with the partial dependency of MMEJ on Dnl4, we reasoned that Pol4 may facilitate the ligation step of MMEJ by recruiting Dnl4/Lif1 to the repair site. To test this hypothesis, a yeast centromeric plasmid expressing either the BRCT domain-truncated pol4 (pol4Δbr) mutation or the polymerase-dead pol4-D367E mutation (Wilson and Lieber 1999) was introduced into the POL4-deleted SLY19 derivative. Expression of pol4Δbr failed to rescue MMEJ repair in pol4Δ, suggesting that the BRCT domain is required for repair by MMEJ (Figure 5C). More importantly, expression of the polymerase-defective pol4-D367E mutation partially rescued the lethality of an HO break in the pol4Δ mutant by elevated Ku-dependent NHEJ but failed to recover the MMEJ deficiency. Analysis of repair events among survivors of HO breaks confirmed that most survivors repaired DSBs by NHEJ via 2 bp of microhomology, deleting 5 nt at the junctions without DNA synthesis (see supplemental Table 2 at http://www.genetics.org/supplemental). These results support the hypothesis that Pol4 is required for gap filling in MMEJ.
Sae2 and Tel1 promote MMEJ but inhibit NHEJ:
Sae2, along with the Mre11 complex, is required for cleavage of Spo11 bound to DNA ends to initiate meiotic recombination (McKee and Kleckner 1997; Prinz et al. 1997). Sae2 also contributes to SSA between direct repeat sequences by promoting the nuclease activity of the Mre11 complex and tethering broken DNA ends in mitotic cells (Clerici et al. 2005). Interestingly, deletion of SAE2 or expression of a nuclease defective mre11 allele (mre11-H125N) results in a very similar phenotype—an increase in Ku-dependent NHEJ but reduced MMEJ (Figures 2, B and C and 3A). We questioned whether Sae2 contributes to MMEJ by promoting Mre11-dependent nucleolytic degradation but inhibiting Ku-dependent NHEJ. If this is the case, we predict that overexpression of EXO1 may partially rescue the MMEJ deficiency of sae2Δ or sae2Δ mre11Δ and simultaneously reduce NHEJ. Increased expression of EXO1 had no impact on MMEJ or NHEJ in sae2Δ but significantly increased survival of sae2Δ mre11Δ mutants upon HO induction with a modest increase in MMEJ repair (Figure 6B). The results thus support the model that Sae2-dependent 5′ resection plays a regulatory role in modulating MMEJ and NHEJ.
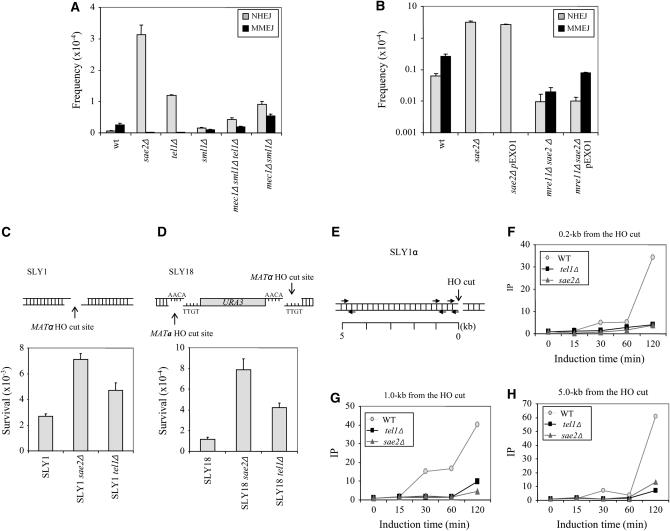
Figure 6.—
Roles of Sae2, Mec1, and Tel1 in MMEJ and NHEJ. Contribution of MMEJ and NHEJ to repair among survivors after induction of HO breaks of sae2Δ, tel1Δ, and mec1Δsml1Δ strains in A or sae2Δ and sae2Δ mre11Δ expressing additional Exo1 in B were determined as described in Figure 2. Survival frequency following induction of HO breaks in sae2Δ or tel1Δ derivatives of SLY1 in C and SLY18 in D, which carry one or two HO cleavage sites in direct repeat orientation in the MAT locus, were determined by plating cells on YEP–GAL normalized by the plating efficiency on YEPD. Each value represents the average from at least three independent experiments ± standard deviation. HO cleavage sites in the MAT locus in SLY1 or SLY18 are shown schematically above each graph. In E–H ChIP assays were used to assess the levels of RPA in sae2Δ or tel1Δ at the DSB using an anti-RPA antibody. Chromatin was isolated at the indicated time after galactose addition, crosslinked with formaldehyde, and fragmented by sonication. After immunoprecipitation and reverse crosslinking, purified DNA was analyzed by qPCR using three sets of primers that anneal 0.2 kb in F, 1 kb in G, and 5 kb in H to the DSB, as well as primers specific for the PRE1 gene situated on chromosome V as a control. PCR signals from each primer set at different durations of HO expression were quantified and plotted as a graph. IP represents the ratio of the RPA PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control. The positions of HO cut site and the location of primers are shown in E. Each point is the average of two separate experiments.
Sae2 is a phosphoprotein whose phosphorylation depends on the Mec1/Tel1 kinases (yeast ATR and ATM homologs, respectively) and DNA damage (Baroni et al. 2004). To test the effect of phosphorylation of Sae2 on MMEJ, we deleted TEL1 or MEC1/SML1 from SLY19 and measured their effect on MMEJ efficiency following two HO breaks. We discovered that there was a fivefold increase in NHEJ compared to wild type, but that rare MMEJ repair of two simultaneous HO breaks was reduced in the TEL1-deleted strain (Figure 6A). These data suggest that Sae2 and Tel1 constitute an important control for antagonistic modulation of MMEJ and NHEJ.
We also assessed the role of Sae2 and Tel1 in NHEJ by examining the effect of a sae2 or tel1 deletion in SLY1 and SLY18 strains (Figure 6, C and D). Repair of a single HO break in these two strains occurs primarily by Ku-dependent NHEJ. We found that deletion of SAE2 or TEL1 improved cell survival up to eightfold by Ku-dependent NHEJ (Figure 6, C and D). The results suggest that suppression of NHEJ by Sae2 is not unique to SLY19 and is independent of the MMEJ pathway.
Increase in NHEJ efficiency at the expense of decreased MMEJ from these mutants after persistent HO cleavage may be explained by the reduced 5′ resection (Clerici et al. 2005; Mantiero et al. 2007), as the extent of nucleolytic degradation may be an impediment for NHEJ (Ira et al. 2004). We thus determined the rate of resection at the DSB from sae2Δ or tel1Δ strain by measuring the level of replication protein A (RPA) binding, a trimeric single-strand binding protein, to ssDNA formed at or near the DSB with ChIP assay followed by qPCR using primer sets annealed to multiple regions at the MAT locus (Shim et al. 2005). We found that depletion of SAE2 or TEL1 reduced the replication protein A (RPA) binding to the DSB and thus decreased the formation of ssDNA (Figure 6, E–H).
DISCUSSION
To assess the mechanistic overlap between MMEJ, NHEJ, and SSA, we surveyed the majority of known NHEJ or SSA factors for their roles in MMEJ repair of two DSBs lacking complementary base overhangs. The analysis newly identified genes responsible for MMEJ. Those include NEJ1, POL4, POL32, RAD10, RAD30, REV3, SRS2, SAE2, and TEL1. In addition, this study provided some insight into their specific functions in MMEJ. We also found that nucleolytic degradation suppresses NHEJ, whereas it is essential for MMEJ. We propose that MMEJ is achieved by a series of distinct but mechanistically similar biochemical reactions that also occur in NHEJ and SSA.
Role of ssDNA formation at DNA DSBs in MMEJ and NHEJ:
MMEJ requires the Mre11 complex to repair two nearby DSBs without complementary end sequences (Ma et al. 2003). By examining the effect of mre11 mutations, which impair various enzymatic functions of the wild-type protein, we discovered that the nuclease activity of the Mre11 complex is critical for MMEJ. The importance of Mre11's nuclease activity in MMEJ is further supported by the observed partial complementation of MMEJ deficiency in mre11Δ mutant cells by overproduction of yet another nuclease Exo1p. Deletion of SAE2, a key component of meiotic DSB processing and mitotic SSA, or TEL1 also led to reduced MMEJ repair of two simultaneous DSBs. In addition, reduced MMEJ in the mre11Δ sae2Δ mutant is partially rescued by expressing EXO1 in excess. Together, these data provide compelling evidence that end processing by the Mre11 complex and Sae2 is a critical part of the MMEJ reaction. We therefore reasoned that 5′ resection activity should uncover microhomologies at either side of the DSB for annealing during the MMEJ reaction.
Although it is an important feature of the MMEJ repair reaction, 5′ resection actually suppresses NHEJ, as the nuclease defective mre11 mutation or a SAE2 deletion greatly stimulated Ku-dependent NHEJ. Previously, unprocessed DNA ends were identified as a preferred substrate for NHEJ (Frank-Vaillant and Marcand 2002). NHEJ is most active in the G1 phase of the cell cycle when the nucleolytic processing of DNA ends is less active (Ferguson et al. 2000; Aylon et al. 2004; Ira et al. 2004). Together, these results suggest that nucleolytic end processing affects both NHEJ and MMEJ but in opposite ways. It promotes MMEJ but limits NHEJ reactions. The inhibitory effect of 5′ resection on NHEJ suggests that yKu may be inhibitory to the MMEJ process. The absence of Ku accelerates nucleolytic degradation (Liang and Jasin 1996; Lee et al. 1998) and can stimulate the MMEJ reaction. The results also explain why two DSBs with no complementary end sequences are preferentially repaired by MMEJ rather than by NHEJ (Ma et al. 2003). The lack of complementary bases prevents DSB ends from annealing, making them more vulnerable to nuclease activity.
The Mre11 complex and Sae2 are modified by the Mec1 or Tel1 kinases upon DNA damage (Usui et al. 2001; Baroni et al. 2004). Loss of Tel1 (and to a lesser extent Mec1) increased NHEJ but reduced MMEJ. Interestingly, Sae2 phosphorylation was shown to be critical for processing and displacing Mre11 from DSB ends (Clerici et al. 2005, 2006). Considering the role of nucleolytic degradation in MMEJ and NHEJ, it seems logical to propose that phosphorylation of Sae2 modulates MMEJ by promoting DSB-induced DNA end resection.
Role of Srs2 and Rad1/Rad10 in MMEJ:
Following 5′ degradation of DSB ends to generate 3′ ssDNA, proper microhomologies anneal, and the 3′ flaps are removed for gap fill-in synthesis and ligation to complete MMEJ. We showed that both Rad1 and Rad10 are needed for MMEJ but dispensable for NHEJ. As Rad1 and Rad10 form an endonuclease complex that removes 3′ flaps during gene conversion or SSA (Fishman-Lobell and Haber 1992; Davies et al. 1995; Ferreira and Cooper 2004), we propose that this complex catalyzes 3′ flap removal in MMEJ.
We demonstrated that deletion of SRS2 displayed reduced MMEJ frequency, which can be reversed by simultaneous deletion of RAD51. We imagine that ssDNA formed by Mre11-dependent 5′ resection of DSBs will attract binding of single-strand binding proteins such as RPA or Rad51. Given its function as a strand-annealing protein between ssDNA and duplex DNA molecules (Sung 1994), Rad51 binding to ssDNA may interfere with annealing between single-stranded DSB ends during SSA or MMEJ. The inhibitory effect of Rad51 in SSA is supported by the finding that deletion of RAD51 significantly improves the efficiency of SSA (Ira and Haber 2002). Alternatively, Rad51 may interfere with binding of yet another single-strand binding protein that catalyzes annealing of ssDNA. Genetic and biochemical evidence strongly suggests that Srs2 is an antirecombinase, displacing Rad51 from ss- or double-stranded DNA (dsDNA) molecules (Krejci et al. 2003; Veaute et al. 2003). We propose that Rad51 displacement from ssDNA is necessary for efficient MMEJ, and Srs2 achieves this through its Rad51 displacement activity.
Multiple polymerases fill in gaps during MMEJ:
Analysis of MMEJ repair events suggests that cells frequently use microhomologies, with one only a few bp away but the other at 60 to 300 bp away from break. Provided that 5′ resection exposing microhomology proceeds symmetrically at either side of the break, MMEJ needs considerable DNA synthesis to fill these gaps for efficient MMEJ. Interestingly, we found that at least three DNA polymerases and one accessory factor of a major replicative polymerase are needed to accomplish efficient MMEJ. Why does MMEJ require multiple polymerases? One reason may be that each polymerase contributes to MMEJ at different cell-cycle phases, locations, or sequences. Indeed, Rad30 and Rev3 expression fluctuates during the cell cycle (Saccharomyces Genome Database, http://www.yeastgenome.org), whereas expression of Pol4 is constant throughout all phases. Alternatively, the requirement for multiple polymerases may reflect that MMEJ is achieved by the “two polymerase two step model” originally proposed to account for multiple polymerase involvement in translesion synthesis (Wang 2001). According to this model, one or more polymerases initiate gap synthesis, followed by extension synthesis catalyzed by yet another polymerase(s). Pol4 is well suited to initiate 3′-end synthesis from mismatches at annealed microhomologies because of its relaxed template dependency (Pardo et al. 2006). However, the low processivity of Pol4 prevents it from long gap fill-in synthesis. Indeed, recombinant Pol4 demonstrated very poor processivity in primer extension assays with <25% of a 5-nt gap filled in (Bebenek et al. 2005). Polδ, Rad30, Rev3, or yet another polymerase(s) may contribute to MMEJ by extending synthesis beyond the initial synthesis by Pol4. As deletion of Pol4 did not completely eliminate MMEJ, we also propose that another polymerase may initiate gap synthesis even in the absence of Pol4.
Among four polymerase mutants tested, pol32Δ is most defective in MMEJ. The results thus suggest that Pol32 plays the most critical role(s) among polymerases in MMEJ, and its function cannot be replaced by other polymerases. As one of the Polδ subunits, the major contribution of Pol32 in MMEJ may simply indicate the significance of Polδ-mediated DNA synthesis in MMEJ (Johansson et al. 2004). However, Pol32 is uniquely involved in UV-induced mutagenesis (Huang et al. 2000) and physically interacts with Srs2, Rad9, or alternate clamp loaders, Rad24, Ctf18, and Elg1 (Bellaoui et al. 2003; Tong et al. 2004). Pol32 has been proposed to load Polζ to the damaged sites (Gibbs et al. 2005). Additional studies are needed to reveal the precise function(s) of Pol32 in MMEJ.
Is MMEJ a variant of NHEJ or SSA?
Having identified additional common gene products that are required for MMEJ, NHEJ, and SSA, one may wonder if MMEJ is a variant of NHEJ or SSA or just a salvage pathway that cells rarely use rather than a separate repair mechanism. Lack of γ-irradiation sensitivity in XPF (mammalian homolog of yeast Rad1) mutants supports the view that MMEJ contributes little to survival from damage (Friedberg et al. 1995). The strong dependence of MMEJ on SSA or NHEJ components also suggests that there are many common biochemical steps between these processes, although they are not identical. However, our results provide an alternative explanation as to why the lack of MMEJ does not necessarily lead to an inability to repair DSBs. Reduced MMEJ in several mutants is compensated by a concomitant increase in NHEJ efficiency. Remarkable plasticity of MMEJ, which relies on multiple redundant factors for execution of biochemical steps, further complicates simple measurement of its contribution in damage tolerance. The consequences of MMEJ deficiency may be underestimated by the apparent lack of damage sensitivity. Additional studies are needed to accurately measure the consequences of MMEJ deficiency, which includes the cataloguing of repair events that occur in the absence of MMEJ repair. In fact, deletion of Brca1 or Chk2 alters the spectrum of repair events rather than dramatically changing the overall efficiency of damage repair (Zhong et al. 2002; Zhuang et al. 2006).
Acknowledgments
We thank Steve Brill, James Haber, Kyungjae Myung, Patrick Sung, and Tom Wilson for reagents and helpful comments. We also thank Debra Walther for editorial assistance. This work was supported by funds from the National Institutes of Health GM071011 and the pilot award from the San Antonio Cancer Institute.
References
- Ahnesorg, P., P. Smith and S. P. Jackson, 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313. [DOI] [PubMed] [Google Scholar]
- Aylon, Y., B. Liefshitz and M. KUPIEC, 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23: 4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni, E., V. Viscardi, H. Cartagena-Lirola, G. Lucchini and M. P. Longhese, 2004. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 24: 4151–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing, C. H., and F. W. Alt, 2004. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst.) 3: 781–796. [DOI] [PubMed] [Google Scholar]
- Bebenek, K., M. Garcia-Diaz, S. R. Patishall and T. A. Kunkel, 2005. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J. Biol.Chem. 280: 20051–20058. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., M. Chang, J. Ou, H. Xu, C. Boone et al., 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22: 4304–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, J., C. P. Diggle, P. Harnden, M. A. Knowles and A. E. Kiltie, 2004. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 32: 5249–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue, M. A., C. Wang, C. Zhu and D. B. Roth, 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity 7: 37–47. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan, D. A., H. A. Olivares, B. E. Nelms and J. H. Petrini, 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, D., L. Malivert, R. de Chasseval, A. Barraud, M. C. Fondaneche et al., 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299. [DOI] [PubMed] [Google Scholar]
- Cary, R. B., S. R. Peterson, J. Wang, D. G. Bear, E. M. Bradbury et al., 1997. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 94: 4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamankhah, M., T. Fontanie and W. Xiao, 2000. The Saccharomyces cerevisiae mre11(ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics 155: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., K. Trujillo, W. Ramos, P. Sung and A. E. Tomkinson, 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Clerici, M., D. Mantiero, G. Lucchini and M. P. Longhese, 2005. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 280: 38631–38638. [DOI] [PubMed] [Google Scholar]
- Clerici, M., D. Mantiero, G. Lucchini and M. P. Longhese, 2006. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 7: 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow, S. E., and S. P. Jackson, 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23: 394–398. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., and T. E. Wilson, 2005. Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell Biol. 25: 896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley, J. M., R. L. Laan, A. Suresh and T. E. Wilson, 2005. a DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 280: 29030–29037. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. b Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Davies, A. A., E. C. Friedberg, A. E. Tomkinson, R. D. Wood and S. C. West, 1995. Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J. Biol. Chem. 270: 24638–24641. [DOI] [PubMed] [Google Scholar]
- Ferguson, D. O., J. M. Sekiguchi, S. Chang, K. M. Frank, Y. Gao et al., 2000. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA 97: 6630–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M. G., and J. P. Cooper, 2004. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 18: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell, J., and J. E. Haber, 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2002. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell 10: 1189–1199. [DOI] [PubMed] [Google Scholar]
- Friedberg, E. C., G. C. Walker and W. Siede, 1995. DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Gerik, K. J., X. Li, A. Pautz and P. M. Burgers, 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 273: 19747–19755. [DOI] [PubMed] [Google Scholar]
- Gibbs, P. E., J. McDonald, R. Woodgate and C. W. Lawrence, 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlich, B., S. Reichenberger, E. Feldmann and P. Pfeiffer, 1998. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur. J. Biochem. 258: 387–395. [DOI] [PubMed] [Google Scholar]
- Gu, Y., S. Jin, Y. Gao, D. T. Weaver and F. W. Alt, 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. USA 94: 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock, M., E. Spangler, K. Riha, J. Puizina and D. E. Shippen, 2004. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefferin, M. L., and A. E. Tomkinson, 2005. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair. 4: 639–648. [DOI] [PubMed] [Google Scholar]
- Herrmann, G., T. Lindahl and P. Schar, 1998. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17: 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck, S. L., and J. N. Strathern, 1997. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M. E., A. de Calignon, A. Nicolas and F. Galibert, 2000. Pol32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38: 178–187. [DOI] [PubMed] [Google Scholar]
- Ira, G., and J. E. Haber, 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22: 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira, G., A. Malkova, G. Liberi, M. Foiani and J. E. Haber, 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani et al., 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require Cdk1. Nature 431: 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, E., P. Garg and P. M. Burgers, 2004. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 279: 1907–1915. [DOI] [PubMed] [Google Scholar]
- Khanna, K. K., and S. P. Jackson, 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27: 247–254. [DOI] [PubMed] [Google Scholar]
- Kolodner, R. D., C. D. Putnam and K. Myung, 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557. [DOI] [PubMed] [Google Scholar]
- Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy et al., 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Labhart, P., 1999. Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol. Cell. Biol 19: 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner et al., 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., D. A. Bressan, J. H. J. Petrini and J. E. Haber, 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair 1: 27–40. [DOI] [PubMed] [Google Scholar]
- Lewis, L. K., G. Karthikeyan, J. W. Westmoreland and M. A. Resnick, 2002. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160: 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F., and M. Jasin, 1996. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 271: 14405–14411. [DOI] [PubMed] [Google Scholar]
- Lieber, M. R., 1999. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells 4: 77–85. [DOI] [PubMed] [Google Scholar]
- Ma, J. L., E. M. Kim, J. E. Haber and S. E. Lee, 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell Biol. 23: 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero, D., M. Clerici, G. Lucchini and M. P. Longhese, 2007. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8:380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J. P., A. S. Levine and R. Woodgate, 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee, A. H., and N. Kleckner, 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey, M., D. Radut and J. J. Sekelsky, 2004. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168: 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, S., J. R. Ferguson and L. S. Symington, 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, S., E. A. Morgan and L. S. Symington, 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig, A., C. Chen, V. da Costa Soares, M. Sanchez, K. Sokol et al., 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382: 551–555. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, B., E. Ma and S. Marcand, 2006. Mismatch tolerance by DNA polymerase Pol4 in the course of nonhomologous end joining in Saccharomyces cerevisiae. Genetics 172: 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull, T. T., and M. Gellert, 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1: 969–979. [DOI] [PubMed] [Google Scholar]
- Paull, T. T., and M. Gellert, 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13: 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault, R., H. Wang, M. Wang, B. Rosidi and G. Iliakis, 2004. Backup pathways of NHEJ are suppressed by DNA-PK. J. Cell. Biochem. 92: 781–794. [DOI] [PubMed] [Google Scholar]
- Pierce, A. J., and M. Jasin, 2001. NHEJ deficiency and disease. Mol. Cell 8: 1160–1161. [DOI] [PubMed] [Google Scholar]
- Prinz, S., A. Amon and F. Klein, 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell, W. K., and G. Chu, 1994. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 91: 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schar, P., G. Herrmann, G. Daly and T. Lindahl, 1997. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 11: 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, E. Y., J. L. Ma, J. H. Oum, Y. Yanez and S. E. Lee, 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell Biol. 25: 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., F. Paques, M. Colaiacovo and J. E. Haber, 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., G. Ira and J. E. Haber, 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell Biol. 20: 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., X. Wang and J. E. Haber, 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12: 209–219. [DOI] [PubMed] [Google Scholar]
- Sung, P., 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science 265: 1241–1243. [DOI] [PubMed] [Google Scholar]
- Sung, P., K. M. Trujillo and S. Van Komen, 2000. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451: 257–275. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli, G. E., T. M. Gottlieb, T. Blunt, A. Priestley, J. Demengeot et al., 1994. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265: 1442–1445. [DOI] [PubMed] [Google Scholar]
- Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto et al., 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17: 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Trujillo, K. M., and P. Sung, 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276: 35458–35464. [DOI] [PubMed] [Google Scholar]
- Tseng, H. M., and A. E. Tomkinson, 2002. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 277: 45630–45637. [DOI] [PubMed] [Google Scholar]
- Tseng, H. M., and A. E. Tomkinson, 2004. Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J. Biol. Chem. 279: 47580–47588. [DOI] [PubMed] [Google Scholar]
- Tsubouchi, H., and H. Ogawa, 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 2221–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, T., H. Ogawa and J. H. Petrini, 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7: 1255–1266. [DOI] [PubMed] [Google Scholar]
- Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam et al., 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang, H., A. R. Perrault, Y. Takeda, W. Qin and G. Iliakis, 2003. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 31: 5377–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., 2001. DNA damage-induced mutagenesis: a novel target for cancer prevention. Mol. Interv. 1: 269–281. [PubMed] [Google Scholar]
- Wang, Z., I. B. Castano, A. De Las Penas, C. Adams and M. F. Christman, 2000. Pol kappa: A DNA polymerase required for sister chromatid cohesion. Science 289: 774–779. [DOI] [PubMed] [Google Scholar]
- Wilson, T. E., and M. R. Lieber, 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 274: 23599–23609. [DOI] [PubMed] [Google Scholar]
- Wilson, T. E., U. Grawunder and M. R. Lieber, 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498. [DOI] [PubMed] [Google Scholar]
- Wu, X., T. E. Wilson and M. R. Lieber, 1999. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96: 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., and A. Gabriel, 2003. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163: 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Z. Xie, H. Shen and Z. Wang, 2004. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 32: 3984–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., J. Wang, N. E. Geacintov and Z. Wang, 2006. Poleta, Polzeta and Rev1 together are required for G to T transversion mutations induced by the (+)- and (−)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells. Nucleic Acids Res. 34: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Q., C. F. Chen, P. L. Chen and W. H. Lee, 2002. BRCA1 facilitates micro-homology mediated end-joining of DNA double-strand breaks. J. Biol. Chem. 277:28641–28647. [DOI] [PubMed] [Google Scholar]
- Zhuang, J., J. Zhang, H. Willers, H. Wang, J. H. Chung et al., 2006. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 66: 1401–1408. [DOI] [PubMed] [Google Scholar]