Abstract
To identify novel regulators of nervous system development, we used the GAL4-UAS misexpression system in Drosophila to screen for genes that influence axon guidance in developing embryos. We mobilized the Gene Search (GS) P element and identified 42 lines with insertions in unique loci, including leak/roundabout2, which encodes an axon guidance receptor and confirms the utility of our screen. The genes we identified encode proteins of diverse classes, some acting near the cell surface and others in the cytoplasm or nucleus. We found that one GS line drove misexpression of the NF-κB transcription factor Dorsal, causing motor axons to bypass their correct termination sites. In the developing visual system, Dorsal misexpression also caused photoreceptor axons to reach incorrect positions within the optic lobe. This mistargeting occurred without observable changes of cell fate and correlated with localization of ectopic Dorsal in distal axons. We found that Dorsal and its inhibitor Cactus are expressed in photoreceptors, though neither was required for axon targeting. However, mutation analyses of genes known to act upstream of Dorsal revealed a requirement for the interleukin receptor-associated kinase family kinase Pelle for layer-specific targeting of photoreceptor axons, validating our screen as a means to identify new molecular determinants of nervous system development in vivo.
NERVOUS system function relies upon patterned development of neurons and controlled establishment of synaptic connections. Axons of developing neurons are guided by instructive cues to their appropriate target areas, where they then select their correct synaptic partners. Understanding how axons are guided by these cues and how they distinguish appropriate targets from inappropriate ones is a central issue in developmental neurobiology. Drosophila melanogaster has proven a successful model with which to apply genetics to this issue and study fundamental and evolutionarily conserved molecular mechanisms that underlie axon guidance. Both forward and reverse genetics approaches have been used to identify mutants with disrupted patterning of axon tracts in either the embryonic ventral nerve cord (VNC) or the developing visual system. Despite considerable successes in identifying novel genes required for axon guidance and targeting, forward genetic screening approaches can be limited by technical challenges and by genetic redundancies which obscure the detection of mutant phenotypes, undoubtedly leaving many genes and molecular pathways remaining to be described. We have exploited P-element transposition in Drosophila, combined with the GAL4-UAS gene misexpression system, to identify genes that can influence axon guidance and reveal genes that may not emerge readily from forward genetic screens. We have mobilized the Gene Search (GS) P element, which supports GAL4-directed expression of genes that flank the site of insertion (Toba et al. 1999), and screened for genes whose misexpression can influence the stereotypic patterning of embryonic axon tracts. Here we report the results of this screen, and describe a collection of GS lines highly enriched for genes whose misexpression is likely to disrupt neuronal morphology or function. We have experimentally pursued one of these lines (GSd447) in detail because we found that it selectively caused the axons of specific subsets of motor neurons and photoreceptors to bypass their correct termination sites. We focused primarily on its effects in the developing visual system.
The adult compound eye of Drosophila is composed of nearly 800 units called ommatidia. Each ommatidium contains eight photoreceptors, also known as R cells (R1–R8). Development of the adult visual system begins in larval stages: in late third instar, the axons of the R cells project from the eye-antennal imaginal disc, through the optic stalk, and into the optic lobe of the brain where they exhibit layer-specific neuronal targeting. R1–R6 axons terminate in the lamina plexus layer, while R7 and R8 cells project past the lamina and terminate in the medulla (Tayler and Garrity 2003). The R1–R6 axons follow the pioneering R8 axons, but terminate between two layers of glia, the lamina epithelial glia and the marginal glia (Perez and Steller 1996; Poeck et al. 2001) in response to an unknown molecular stop signal.
The response of R cells to this stop signal has recently begun to be appreciated. Where it has been tested directly, R1–R6 targeting defects have been identified in flies carrying mutations in the genes encoding the receptor tyrosine kinase Off-track (Otk) (Cafferty et al. 2004), the SH2–SH3 adaptor molecule Dreadlocks (Dock) (Garrity et al. 1996), the Ste-like kinase Misshapen (Msn) (Ruan et al. 1999), and the cytoskeletal regulator Bifocal (Bif) (Ruan et al. 2002). The mutant phenotypes, combined with biochemical data, suggest that a cascade of Otk, Dock, Msn, and Bif activity occurs in response to a stop signal received by the growth cones of R1–R6 photoreceptor cells, leading to reduced growth cone motility and termination in the lamina.
Transcriptional regulation of this pathway controlling layer-specific neuronal targeting of R1–R6 axons remains unclear, although the genes brakeless (bks) and runt have been implicated (Rao et al. 2000; Kaminker et al. 2002). Bks is a putative zinc-finger transcription factor and bks mutations cause R1–R6 cells to bypass the lamina and project to the medulla (Rao et al. 2000). Bks represses the transcription factor Runt in R2 and R5 cells; Runt is normally expressed in R7 and R8 cells only. However, Runt misexpression in R2 and R5, or its derepression in bks mutants, causes all R1–R6 to behave like R7 and R8 cells and project to the medulla (Kaminker et al. 2002). Exactly how these transcription factors regulate effectors of R-cell targeting remains to be determined, and it is not unreasonable to predict that there are additional transcriptional regulatory mechanisms that contribute to R-cell targeting.
We show here that GSd447 causes misexpression of the dorsal (dl) gene, which encodes an NF-κB transcription factor involved in embryonic patterning (Morisato and Anderson 1995), humoral immunity (Engstrom et al. 1993), hematopoiesis (Qiu et al. 1998), and muscle development (Halfon et al. 1995). Signaling to Dorsal has perhaps been best characterized in patterning the dorsal–ventral axis of Drosophila embryos, where activation of the Toll receptor by Spätzle (Morisato and Anderson 1994) signals through a multiprotein complex including Toll, Weckle, MyD88, and Tube (Sun et al. 2002; Sun et al. 2004; Chen et al. 2006) to the serine-threonine protein kinase Pelle, a homolog of the mammalian interleukin receptor-associated kinase (IRAK) (Hecht and Anderson 1993; Shelton and Wasserman 1993; Janssens and Beyaert 2003). Activation of Pelle triggers the dissociation of Dorsal from its inhibitor Cactus, causing Cactus to be degraded, and leading to the nuclear import of Dorsal and the ordered expression of Dorsal-responsive genes to polarize and pattern the embryo along the dorsal–ventral axis (Govind et al. 1996; Drier et al. 2000). Our analysis of Dorsal and other components of the NF-κB signaling pathway led to the discovery that Pelle is required for R-cell targeting in the larval visual system.
MATERIALS AND METHODS
A prescreen for lethality caused by GS-directed misexpression in the nervous system:
To generate novel insertions to promote gene misexpression in Drosophila, the P{GS1} P element (Toba et al. 1999) was mobilized from the second chromosome in animals also carrying a source of transposase (Δ2-3,Ki). Males of the genotype CyO,P(w+)GS1/+;Δ2-3,Ki/+ were mated to w1118 virgin females in vials, and male progeny without Cy or Ki whose eye color differed from the source GS line (suggesting mobilization to a different autosomal locus) were singly mated to w;T(2;3)apXa females. Male progeny (<5) from each of these crosses were then mated to scrt11-6-Gal4/TM3,Sb virgin females, and for each line we determined whether adult lethality resulted from GAL4-UAS-driven misexpression. TM3,Sb male progeny with an eye color were then used to recover the chromosomes bearing the newly-mobilized P(w+)GS. The new insertions were mapped and stocks were established using lacZ-marked balancer chromosomes.
Immunohistochemical screen for axon guidance defects:
The new GS stocks were re-mated to scrt11-6-Gal4/TM3,Sb, Ac-lacZ to confirm lethality, and the progeny were screened for defects in the pattern of expression of Fasciclin 2 (Fas2), which is expressed weakly in the cell bodies and strongly along the axons of a subset of CNS interneurons and all motor neurons. Embryos at stages 13–17 were collected at 25°, dissected to reveal the VNC, fixed in 4% paraformaldehyde, and stained according to standard procedures with an anti-Fas2 monoclonal antibody (1D4, dilution 1:50) from the Developmental Studies Hybridoma Bank (DSHB). If necessary, they were co-labeled with rabbit anti-β-Gal (1:1000, MP Biomedicals) to identify animals carrying the lacZ balancer.
Inverse polymerase chain reaction (PCR):
We used inverse PCR to identify sequences of genomic DNA immediately flanking each new GS insertion site, essentially as described by the Berkeley Drosophila Genome Project (http://www.fruitfly.org/about/methods/inverse.pcr.html) and the Gene Disruption Project (http://flypush.imgen.bcm.tmc.edu/pscreen/). Briefly, genomic DNA from flies carrying the GS P element was digested with one of several restriction enzymes (HinPI, HpaII, EcoRI, PstI) and ligated to form a circular DNA. PCR primers targeting GS P-element sequences were then used to amplify the flanking genomic DNA from the circularized template. The primer combinations used were EY.3.F/EY.3.R, Pry1/Pry4, and Pry1/Pry2. The primer sequences used were: EY.3.F, CCTTTCACTCGCACT TATTG; EY.3.R, GTGAGACAGCGATATGATTGT; Pry1, CCTTAGCAT GTCCGTGGGGTTTGAAT; Pry4, CAATCATATCGCTGTCTCACTCA; and Pry2, CTTGCCGACGGGACCACCTTATGTTATT. The DNA sequences of the amplified flanking fragments were then determined, and their location in the Drosophila genome was precisely mapped using BLAST searches at the FlyBase website against Version 5.1 of the Drosophila genome annotation. For simplicity of reporting, the sequence tags provided in supplemental Table 1 (http://www.genetics.org/supplemental/) have been trimmed to 60 bp or less. For each GS line, the site of insertion is the first nucleotide of the sequence provided.
Drosophila stocks and genetics:
Fly stocks obtained from the Bloomington Stock Center: GAL4-ninaE.GMR (GMR-GAL4), w1118, dl1, dl4, cact4, cact1, Df(2L)TW119, Df(2L)H20, Df(2L)cact-255rv64, cactchi64, Df(2L)r10, tub2, Df(3R)XM3, Tl4, TlrK344. Fly stocks from alternative published sources: scrt11-6-Gal4 (Boyle et al. 2006), GAL4109-68, Mt14-GAL4, and lz-GAL4 (Kaminker et al. 2002), UAS-nGFP (Thomas and van Meyel 2007), UAS-dlA (also known as UASp-dorsal) (Matova and Anderson 2006), ro-tau-lacZ (Garrity et al. 1999), Dif1 and Dif2 (Rutschmann et al. 2000), pll7, pll2, and pll25 (Towb et al. 2001), Df(2L)J4 (Meng et al. 1999).
To analyze the projections of the R2–R5 axons, a ro-tau-lacZ transgene on the third chromosome was crossed in and the progeny larvae stained with α-β-galactosidase. For mutations of pll, which resides on the third chromosome, a recombinant carrying pll7 and ro-tau-lacZ was generated. Developing eye discs were rendered mosaic for Df(2L)J4 using the FLP-FRT-mediated mitotic recombination and ey-FLP transgene. A recombinant chromosome carrying FRT40A and Df(2L)J4 was generated, and large patches of mutant tissue were generated in the presence of a recessive cell-lethal mutation that eliminated twin-spots, and were confirmed with anti-Dorsal immunochemistry.
To construct UAS-dlB-HA, a 3.56-kb fragment containing the entire dl-B cDNA and a hemagglutinin (HA) epitope tag was excised with XbaI from pJM699 (Gross et al. 1999), then cloned into Xba1-cut pUAST. Germ-line transformation of Drosophila embryos was then carried out using standard microinjection methods (Spradling and Rubin 1982). Expression was confirmed by anti-HA immunochemistry.
Immunohistochemistry for visual system:
Eye–brain complexes of wandering third instar larvae were dissected and fixed in 4% paraformaldehyde for 50 min and then processed for routine immunochemistry according to established protocols. The following mouse monoclonal primary antibodies were used: 24B10 (α-Chaoptin) 1:200 (DSHB), 9E10 (α-myc) 1:50 (DSHB), α-Prospero 1:200 (DSHB), α-Boss 1:2000 (Van Vactor et al. 1991), α-Bks 1:100 (Rao et al. 2000), α-Repo 1:100 (DSHB), α-β-galactosidase 1:100 (DSHB), α-Dorsal 7A4 1:200 for misexpression, and 1:50 for endogenous (DSHB), α-Cactus 3H12 1:50 (DSHB). The following polyclonal primary antibodies were used: rabbit α-Otk 1:50 (Pulido et al. 1992) and guinea pig anti-Runt (1:200) (Kosman et al. 1998). Anti-mouse or anti-rabbit HRP-conjugated secondary antibodies were used, followed by development in DAB as above, except in the studies of endogenous Dorsal, Cactus, and Runt immunolocalization, in which Alexa 488-labeled secondary antibodies were used. HRP immunochemistry was visualized using Nomarski Optics on a Zeiss Axioskop 2 Microscope. Confocal microscopy was performed using a Yokogawa spinning disk confocal system (Perkin-Elmer) and an Eclipse TE2000-U microscope (Nikon). Z-series images were collected using Metamorph software (Molecular Devices).
Chemical mutagenesis for suppressors of GSd447 misexpression:
To confirm whether dl or another gene located ∼2.5 kb away, CG5050, was responsible for the GSd447 misexpression phenotypes, we conducted chemical mutagenesis to suppress the misexpression effects. Starved GSd447 male flies were exposed to 25 mm ethylmethanesulfonate (EMS) overnight, then crossed to GMR-GAL4 virgin females, transferring to new food twice per day for 10 days. After the progeny of the above cross had eclosed, adult males were then screened for suppression of the glazed eye of GSd447 misexpressors. After screening >45,000 flies, 2 males were isolated in which the exterior eye phenotype had been suppressed, from which balanced stocks were established. Fly genomic DNA was prepared from GSd447*C/Df(2L)H20 and GSd447*D/Df(2L)H20 mutant flies. PCR was then performed using primer pairs to generate predicted fragments from the Dorsal coding sequence. The fragments were gel extracted and sequenced on both strands to reveal mutations.
Cuticle preparation:
The new mutations dlC447 and dlD447 were crossed to Df(2L)H20, and hemizygous female progeny were then crossed to w1118 males. Embryos from this cross were left to develop for 48 hr at 20°, then dechorionated in 50% bleach and mounted in Hoyer's/lactic acid medium. Slides were set to dry overnight at 60° (Stern and Sucena 2000), then visualized using dark-field microscopy and scored for degree of dorsalization (Roth et al. 1991).
RESULTS
A misexpression screen to identify genes capable of influencing axon guidance:
In our initial screen, 1127 single male flies carrying independent insertions of the GS P element were crossed to scrt11-6-Gal4, a PGawB insertion in the scratch locus that expresses GAL4 throughout the central (Figure 1A) and peripheral nervous systems of Drosophila embryos. We reasoned that GS insertions that resulted in significant defects of axon or dendrite development would cause lethality when crossed to scrt11-6-Gal4. Therefore, we prescreened for lethality prior to eclosion, and identified and confirmed 142 new independent insertion lines. We performed inverse PCR to identify the site of insertion for each of these lines, and the results are presented in supplemental Table 1 (http://www.genetics.org/supplemental/). Seven of these 142 lines (GSd246, GSd319, GSd406, GSd439, GSd462, GSd464, and GSd481) appear to harbor two insertions, although we have not confirmed which is responsible for lethality. For each insertion, we have identified in supplemental Table 1 the gene closest to the site of insertion. For insertions situated equally between two genes (GSd079, GSd318, GSd402, GSd417, GSd434, GSd464), both genes are listed in supplemental Tables 1 and 2. There were four lines (GSd329, GSd406, GSd464, and GSd489) in which the insertion occurred in the 3′ end of the gene, suggesting it could drive the expression of an antisense RNA for that gene.
Figure 1.—
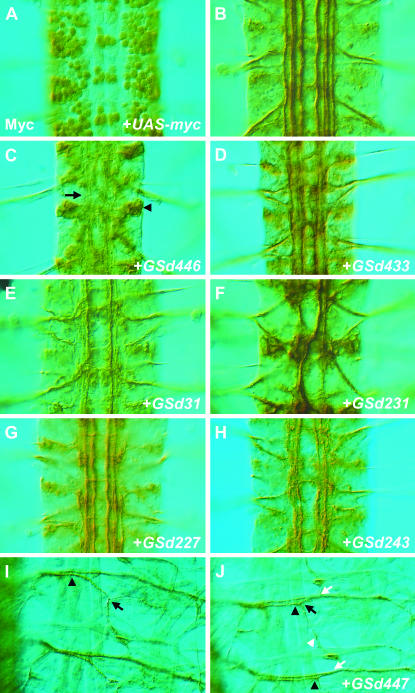
Selected examples of Fas2 phenotypes caused by misexpression of GS lines using scrt11-6-Gal4. (A) The nuclear targeted reporter UAS-nGFP, which also encodes five copies of the myc epitope, was used to demonstrate the expression driven by scrt11-6-Gal4 throughout the central and peripheral nervous systems of Drosophila embryos. Shown here is a dorsal view of three abdominal segments of an embryonic ventral nerve cord at stage 16, immunostained for myc. (B–J) Anti-Fas2 immunochemistry to reveal Fas2-positive axon bundles in the CNS (B–H) and motor neuron projections (I–J). (B) Three segments of a scrt11-6-Gal4/+ animal immunostained for Fas2, showing the characteristic Fas2-positive axon bundles traveling the length of the longitudinal connectives. (C) Misexpression of GSd446 causes many of the Fas2 bundles to become missing/disrupted or otherwise diffuse and less distinct (arrow). Fas2 staining was also mislocalized to the cell body (arrowhead). Additional phenotypes included inappropriate midline crossing, as in D–F and H, selective loss, or disruption of specific Fas2 axon bundles (E, G), hyperfasciculation (F), and misplaced axon bundles within the longitudinal connectives (F, H). (I) In the muscle field innervated by Fas2-positive motor neurons, SNa normally separates from the ISN at a point directly over the ventral muscle group (arrowhead), then bifurcates into two branches that innervate distinct muscle targets (black arrow). (J) Misexpression of GSd447 causes SNa to take an alternate trajectory that aligns with ISN for longer than usual (black arrowheads). Both the initial separation of SNa from ISN and the bifurcation of SNa (black arrow) appear intact, with a posterior-directed branch (white arrowhead) extending to muscle eight. However, after bifurcation the dorsal-directed branch of SNa often inappropriately rejoins ISN (white arrows), forming a fascicle that projects axons to more dorsal muscle targets.
To assess the neuronal defects caused by these genetic manipulations, we dissected embryos in which scrt11-6-Gal4 was used to drive expression of each GS line and immunostained for the marker Fas2. By comparison with control animals (Figure 1, B and I), we assessed (1) the overall integrity of the nervous system, (2) the patterns of the Fas2-positive axon bundles within the CNS, and (3) the Fas2-positive motor axon projections in the periphery. We found that 56 of the 142 lines (39%) produced axonal defects when misexpressed in the nervous system (Table 1). There were 10 genes for which multiple GS insertion lines were generated, showing that our screen was consistent and reproducible, since independent insertions gave rise to similar phenotypes. Taking these into account, 42 of 142 (30%) GS lines had axon defects and had unique insertion sites. Some of these insertion sites lie between genes, and some lines have two inserts; in these cases it is difficult to predict which gene is most likely to be responsible for the phenotypes. Therefore, in the 42 lines, we have annotated 51 genes that lie close to the GS insertion sites. Of these 51 genes, 38 (75%) have been studied previously. We note that 17 (33%) have been previously shown to have a role in nervous system development (Table 2), demonstrating the screen's ability to identify such genes. Interestingly, 13 (25%) of the genes we found are novel (Table 2). Further characterization of these genes may reveal new molecular determinants of nervous system development and axon guidance. We also found that the screen was not limited in its ability to identify genes encoding proteins of diverse classes: some are predicted to act at or near the cell surface, others in the cytoplasm or nucleus (Table 2).
TABLE 1.
Phenotypes observed with anti-Fas2 immunohistochemistry
| Fas2 bundles in the VNC were:
|
Motor nerves that were:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Line | Closest gene | Missing or disrupted | Fused | Diffuse or less distinct | Misplaced within connective | Crossing the midline | Missing or thinned | Thickened | Stalled or misrouted | Other observations |
| GSd031 | IM1 | X | ||||||||
| GSd033 | mir-313 | X | ISNd | ISNb | ||||||
| GSd034 | ttk | X | X | ISNb, ISNd, SNa, SNc | Fas2 staining mislocalized to cell body | |||||
| GSd057 | lig | X | ISNb, ISNd | |||||||
| GSd079 | EST: EN1075/EC37335 | X | ||||||||
| GSd201 | pnt | X | ||||||||
| GSd204 | Kr-h1 | Cells of VNC appear loosely held together | ||||||||
| GSd209 | apt | X | ||||||||
| GSd210 | Akap200 | X | X | VNC thickened | ||||||
| GSd211 | CG7518 | X | ISNb, ISNd, SNa, SNc | Fas2 staining mislocalized to cell body | ||||||
| GSd212 | CG17836 | X | ||||||||
| GSd214 | for | X | ||||||||
| GSd217 | CG32223 | X | X | |||||||
| GSd219 | bl | X | X | |||||||
| GSd220 | ken | X | ||||||||
| GSd226 | cbt | X | ||||||||
| GSd227 | Btk29A | X | ||||||||
| GSd229 | pnt | X | ISNb, SNc | |||||||
| GSd231 | Rho1 | X | X | |||||||
| GSd233 | foxo | X | X | X | ||||||
| GSd236 | pnt | X | X | X | ||||||
| GSd243 | Hsromega | X | X | X | SNa, SNc | |||||
| GSd246 | (1)Kr-h1, (2)dimm | X | ISNb, ISNd, SNa, SNc | |||||||
| GSd247 | mir-313 | X | X | |||||||
| GSd248 | spen | X | X | |||||||
| GSd302 | nos | X | SNa, SNc | ISNb | ||||||
| GSd312 | hdc | X | SNa | ISNb | ||||||
| GSd315 | faf | X | Synaptic boutons enlarged | |||||||
| GSd317 | H | X | ISNb | ISNb, SNa | ||||||
| GSd318 | CG2469/CG2277 | X | ISNb | ISNb | ||||||
| GSd319 | (1)CG30389, (2)GstE1 | X | X | |||||||
| GSd325 | bnl | ISNb, ISNd, SNa, SNc | VNC severely disrupted, muscle defects | |||||||
| GSd328 | for | X | ||||||||
| GSd402 | EST:EN05557/EN06658 | X | SNa, SNc | ISNb, ISNd | ||||||
| GSd404 | hdc | X | ISNb, ISNd | |||||||
| GSd405 | mirr | X | X | ISNb | ||||||
| GSd406 | (1)foxo, (2)slam (antisense) | X | X | ISNb | ||||||
| GSd411 | foxo | X | ||||||||
| GSd420 | pnt | X | ISNb, ISNd | ISNb, ISNd | ||||||
| GSd427 | mam | X | X | ISNb | Fas2 staining mislocalized to cell body | |||||
| GSd428 | PFE | X | ISNb, ISNd, SNa, SNc | |||||||
| GSd431 | Rho1 | X | X | X | ISNb, ISNd, SNa, SNc | |||||
| GSd433 | lea (Robo2) | X | X | X | SNa | ISNb, ISNd, SNc | ||||
| GSd439 | (1)Trl, (2)tara | X | ISNb, ISNd, SNa, SNc | |||||||
| GSd446 | ttk | X | X | ISNb, ISNd, SNa, SNc | Fas2 staining mislocalized to cell body | |||||
| GSd447 | dl | ISNb | SNa | |||||||
| GSd450 | CG2617 | X | X | ISNb, ISNd, SNa, SNc | VNC thinned, Fas2 staining mislocalized to cell body | |||||
| GSd458 | psq | X | ||||||||
| GSd462 | (1)ttk, (2)Stat92E | X | ISNb, ISNd, SNa, SNc | Fas2 staining mislocalized to cell body | ||||||
| GSd468 | ttk | X | X | ISNb, ISNd, SNa, SNc | ||||||
| GSd469 | woc | X | X | |||||||
| GSd481 | (1)esg, (2)imd/Dp1 | X | ||||||||
| GSd482 | ken | X | X | X | SNa | ISNb, ISNd, SNc | ||||
| GSd486 | CG3624 | ISNb, ISNd, SNa, SNc | Cells of VNC appear loosely held together | |||||||
| GSd496 | CG9582 | X | X | ISNb, ISNd, SNa, SNc | Fas2 staining mislocalized to cell body | |||||
| GSd497 | mirr | X | ISNb | |||||||
ISN, intersegmental nerve; SN, segmental nerve.
TABLE 2.
Genes closest to the GS insertions that cause observable phenotypes with anti-Fas2
| Proposed site of protein activity | Molecular function | Demonstrated role in Drosophila neural development |
|---|---|---|
| Nucleus | ||
| apt | bZIP transcription factor, RNA binding | Takasu-Ishikawa et al. (2001) |
| bl | KH domain protein, RNA binding, | |
| cbt | C2H2 zinc finger transcription factor | |
| dimm | bHLH transcription factor | Hewes et al. (2003) |
| dl | NF-κB-like transcription factor | |
| Dp1 | Multi-KH-domain DNA binding protein | |
| esg | Zinc-finger transcriptional repressor | |
| foxo | forkhead transcription factor | |
| H | Transcription repressor | Bang and Posakony (1992) |
| Hsromega | Noncoding RNA | |
| IM1 | Immune induced molecule | |
| ken | BTB/POZ domain transcription factor | Allen et al. (2007) |
| Kr-h1 | Zinc-finger protein | |
| mam | Transcriptional coactivator | Kania et al. (1995) |
| mir-313 | microRNA | |
| mirr | Homeodomain transcription factor | McNeill et al. (1997) |
| pnt | ETS domain transcription factor | O'Neill et al. (1994) |
| psq | BTB/POZ domain transcription factor | Weber et al. (1995) |
| Stat92E | Transcription factor | Li et al. (2003) |
| tara | Nuclear protein of trithorax group | |
| Trl | GAGA transcription factor | Bejarano and Busturia (2004) |
| ttk | BTB/POZ domain transcription factor | Xiong and Montell (1993) |
| woc | Zinc-finger transcription factor | |
| Cytoplasm | ||
| Akap200 | A-kinase anchor protein | |
| Btk29A | Non-receptor tyrosine kinase | |
| faf | Ubiquitin-specific protease | Fischer-Vize et al. (1992) |
| GstE1 | Glutathione transferase | |
| hdc | Cysteine-rich cytoplasmic protein | |
| imd | Death domain protein | |
| lig | Novel protein | |
| nos | Translation factor | Ye et al. (2004) |
| Rho1 | GTPase | Lee et al. (2000) |
| spen | RRM-motif protein | Kuang et al. (2000) |
| Membrane-associated | ||
| for | Cyclic nucleotide-dependent kinase | Renger et al. (1999) |
| slam | Novel protein | |
| Transmembrane | ||
| lea | Transmembrane receptor | Simpson et al. (2000) |
| Rajagopalan et al. (2000) | ||
| PFE | Transmembrane receptor kinase | |
| Secreted | ||
| bnl | Growth factor | |
| Unknown | ||
| CG17836 | ||
| CG2469/CG2277 | ||
| CG2617 | ||
| CG30389 | ||
| CG32223 | ||
| CG3624 | ||
| CG7518 | ||
| CG9582 | ||
| EST: EN1075/EC37335 | ||
| EST:EN05557/EN06658 | ||
The CNS phenotypes, ranging from misprojections of axon subsets to severe CNS disorganization, could be categorized as follows: the central axon bundles were (1) missing or diffuse (Figure 1, C, E, and G), (2) crossing the midline inappropriately (Figure 1, D, F, and H), (3) fused (Figure 1F), or (4) misplaced within the longitudinal connectives (Figure 1H). Often more than one phenotypic category was observed per misexpression line. For motor axons in the periphery, we assessed the morphology and projection pattern of the “a” and “c” branches of the segmental nerve (SN), and the “b” and “d” branches of the intersegmental nerve (ISN). We cataloged whether these motor axons were: (1) missing/thinned, (2) thickened, or (3) stalled/misrouted (Table 1). In many cases the defects were widespread, while in some instances the defects were quite specific for particular motor nerve branches (Figure 1J).
Misexpression of GSd447 causes the axons of motor neurons and R cells to bypass correct termination sites:
We chose to focus on one line in particular, GSd447, which caused highly specific axon guidance defects of embryonic SNa motor axons when expressed with scrt11-6-Gal4. Rather than projecting normally to their muscle targets (Figure 1I), SNa axons formed a discrete bundle alongside the ISN, often bypassing their normal targets completely (Figure 1J) and following a path normally taken by the ISN to muscles located more dorsally. We wondered whether GSd447 could similarly affect another neuronal cell type, and therefore we tested its effects in photoreceptors (R cells). The developing visual system is another important and well-studied model system to study axon guidance and target selection. Expressed via the eye-specific driver GMR-Gal4, GSd447 resulted in robust axon fasciculation and targeting defects in which R-cell axons appeared to bypass their correct targets in the optic lobes of third-instar larvae (see below). We determined that GSd447 is inserted immediately upstream of the dorsal (dl) gene, encoding a nuclear factor kappa B (NF-κB) transcription factor. We examined dl mutant embryos and found no motor axon pathfinding defects. Since dl has a significant maternal contribution that could obscure defects in zygotic mutants, we studied the developing visual system instead.
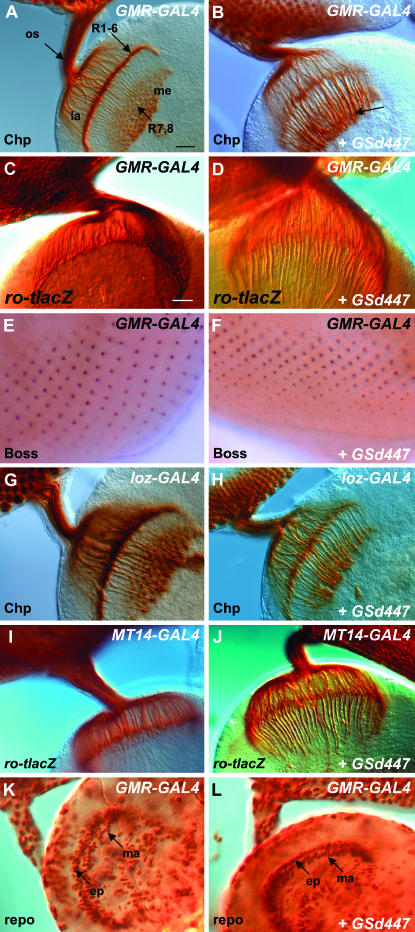
To characterize the axon guidance defects associated with GSd447 in the visual system, this line was crossed with GMR-GAL4, a driver active in all cells of the developing larval eye imaginal disc posterior to the morphogenetic furrow. In GMR-GAL4/+ controls (Figure 2A), a normal pattern of R-cell axon projections is observed using the monoclonal antibody 24B10, which detects the membrane protein Chaoptin on the surface of all R cells. In contrast, severe defects of this pattern are caused by misexpression of GSd447 (Figure 2B). The lamina layer of the brain is severely depleted and the bundles that project through the lamina to the medulla are thickened. In addition, the axon terminals often fail to expand. Therefore GSd447-mediated misexpression in the eye results in severely disrupted patterning of R-cell innervation.
Figure 2.—
The effects of GSd447 misexpression on the developing visual system. (A, B) R-cell axons can be specifically labeled with an antibody to Chaoptin (Chp). They project from the eye imaginal disc, through the optic stalk (os) and into the optic lobe where R1–R6 axons terminate in the lamina plexus (la), while R7 and R8 axons terminate in the medulla (me). GMR-GAL4 drives expression in all cells of the eye posterior to the morphogenetic furrow. Expression of GMR-GAL4 alone (genotype: GMR-GAL4/+) has no effect on the normal pattern of projections. Bars in A and C, 20 μm. (B) Misexpression of GSd447 results in severe axon guidance defects (GMR-GAL4/GSd447). The lamina is depleted and the axon bundles that project into the medulla are thickened (arrow), suggesting R1–R6 axons are bypassing normal targets in the lamina layer. (C, D) Axon targeting to the lamina can be assessed with the ro-tlacZ reporter, which labels the R2–R5 axons using α−β-galactosidase immunochemistry. No axon targeting defects of the R2–R5 axons are seen in GMR-GAL4/+ controls, but large bundles of R2–R5 axons project past the lamina in GMR-GAL4/GSd447 individuals. (E, F) Immunostaining for Boss, and R8-specific marker, is unaltered in GMR-GAL4/GSd447 individuals. (G, H) Lz-GAL4 expresses specifically in R1, R6, and R7. Control animals (Lz-GAL4/+) have no axon targeting defects, but in Lz-GAL4/+; GSd447/+ larvae the lamina is thin, and axon bundles in the medulla are thickened. (I, J) MT14-GAL4 is specific for R2, R5, and R8. MT14-GAL4/+ animals have normal targeting of the R2–R5 axons as revealed by ro-tlacZ (I), but misexpression of GSd447 results in severe mistargeting of the R2–R5 axons past the lamina (GSd447/+; Mt14-GAL4/+) (J). (K) An antibody to the glial specific protein, Reversed Polarity (Repo), reveals the normal positioning of the two layers of laminar glia, the epithelial glia (ep) and marginal (ma) glia, in GMR-GAL4/+ controls. These glia present incoming R-cell axons with a stop signal to which R1–R6 cells respond. (L) Misexpression of GSd447 does not affect the positioning of the epithelial (ep) or marginal (ma) glia, which remain in ordered rows within the optic lobe.
To more clearly visualize the defects caused by GSd447, the ro-tau-lacZ reporter transgene was used to reveal only the axon projections of R2, R3, R4, and R5. Immunostaining with an antibody to β-galactosidase allows the visualization of R2–R5 axons, which normally terminate within the lamina. In control animals for GMR-GAL4 (Figure 2C) or GSd447 (not shown) alone, normal targeting of R2–R5 axons to the lamina was observed. However, misexpression of GSd447 with GMR-GAL4 caused bundles of axons that normally terminate in the lamina to project through it to deeper regions in the developing optic lobe (Figure 2D). One question that arises is whether mistargeting is caused by R2–R5 cells having adopted an R7 or R8 identity, and consequently an R7 or R8 pattern of axon projection to the medulla. To address this issue, we studied R7 and R8 cell-specific identity markers in the developing eye imaginal disc of third instar larvae. Using immunochemistry for bride of sevenless (Figure 2, E and F), an R8-specific cell-surface protein (Hart et al. 1993; Cafferty et al. 2004) and Prospero (data not shown), a nuclear protein found in R7 cells (Kauffmann et al. 1996), we found no evidence to indicate that GSd447-induced misprojections of R2–R5 neurons are due to adoption of R7 or R8 cell fate.
Characterization of targeting defects caused by misexpression of GSd447:
Since GMR-GAL4 is expressed in all cells of the eye posterior to the morphogenetic furrow, we used R-cell-specific GAL4 lines to test whether the axon mistargeting was due to misexpression specifically in R cells or to expression in other cells of the developing eye disc. We found a depleted lamina layer and axon targeting defects upon misexpression of GSd447 using Loz-GAL4 (Figure 2H), which is specific for R1, R6, and R7, or using Mt14-GAL4 (Figure 2J), which is specific for R2, R5, and R8. Therefore, the mistargeting is caused by expression within R cells and likely affects all R1–R6 neurons. In contrast, GSd447 misexpression using GAL4109-68, which drives expression in R8 only, did not produce any axon targeting defects (not shown), suggesting that R cells that normally target to the medulla are refractory to the misexpression of GSd447.
To begin to explore the mechanism through which misexpression of GSd447 in R cells causes an axon targeting defect, two antibodies were used to test the involvement of molecules known to influence R-cell axon guidance and whose mutant phenotypes are reminiscent of that caused by misexpression of GSd447. It has been hypothesized that the receptor tyrosine kinase Otk is part of the receptor complex controlling R-cell termination in the lamina, and that the transcription factor Bks regulates R1–R6 termination. Neither Otk nor Bks expression was affected by misexpression of GSd447 using GMR-GAL4 (not shown). In addition, we also tested the possibility that GSd447 misexpression might upregulate the expression of Runt. Runt is normally limited to R7 and R8 but Runt misexpression in R1–R6 phenocopies the misprojection defects we observed with GSd447 (Kaminker et al. 2002). However, Runt expression was not altered upon misexpression of GSd447 (not shown).
Mutations in the genes nonstop (Poeck et al. 2001) and jab1/csn5 (Suh et al. 2002), which encode a ubiquitin-specific protease and a subunit of the COP9 signalosome complex, respectively, result in the disorganization of glial cells and have a drastic effect on R-cell axon guidance. To test whether GSd447 misexpression led to incorrect positioning of the glial cells in the lamina we studied the glial-cell-specific marker Reversed Polarity (Repo) (Xiong and Montell 1995; Rangarajan et al. 1999). In contrast to nonstop and jab1/csn5 mutants, the four layers of glial cells—satellite, epithelial, marginal, and medulla glia—were positioned normally in GMR-GAL4 controls (Figure 2K) and animals misexpressing GSd447 (Figure 2L). In particular, the epithelial and marginal glial cells, which prefigure the lamina plexus and present a stop signal to incoming R1–R6 axons, formed neat rows that were aligned as in controls.
In summary, although the effects of GSd447 misexpression on axon targeting resemble those of bks, otk, nonstop, and jab1/csn5 mutants, bks and otk expression are unaltered, and the glial cells were found to be positioned correctly. Runt expression is also unaffected. Thus, these known causes of axon mistargeting in the visual system cannot obviously explain the mechanism causing R-cell axons to be mistargeted by GSd447.
Misexpression of Dorsal-A and Dorsal-B results in axon targeting defects:
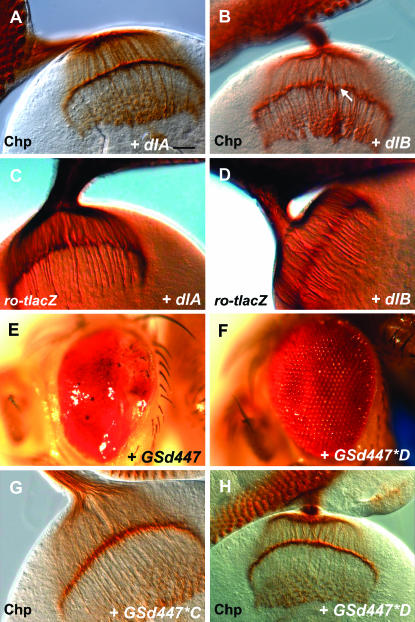
The GSd447 P element is located 22 nucleotides upstream of the most 5′ exon of dorsal. Dorsal is characterized by an N-terminal Rel homology domain which possesses domains for dimerization, DNA binding, and interactions with its inhibitor Cactus. In addition, it has a C-terminal transactivation domain. Dimers of NF-κB proteins can function as either transcriptional activators or repressors. Two differentially spliced forms of Dorsal, known as Dorsal-A and Dorsal-B, can heterodimerize and act synergistically to enhance transcription from reporter genes in vitro (Gross et al. 1999). These isoforms are identical in their N-terminal Rel homology domains (amino acids 1–329), but have divergent C-termini that encode unique transactivation domains (Isoda et al. 1992; Gross et al. 1999). In addition, Dorsal-A possesses a nuclear localization sequence (NLS) from amino acids 329–340, while Dorsal-B does not. Dorsal-B may heterodimerize with Dorsal-A and thereby achieve nuclear entry (Gross et al. 1999). It is possible that both Dorsal-A and Dorsal-B could be driven by GSd447. Therefore, either one may be responsible for the R-cell axon mistargeting. To test this, transgenic flies carrying UAS-dlA or UAS-dlB (dlB) constructs were obtained to determine whether either could recapitulate the defects observed with GSd447 driven by GMR-GAL4. Visualizing R-cell axons with α-Chaoptin, we found that neither UAS-dlA (Figure 3A), which had no effect, nor UAS-dlB (Figure 3B), which had mild effect, were as severe as GSd447 (Figure 2B). To examine axon targeting in the lamina more specifically, ro-tau-lacz was used as a reporter to visualize only the R2–R5 axons. Using this more sensitive marker, we found that misexpression of UAS-dlA (Figure 3C) or UAS-dlB (Figure 3D) each caused R2–R5 axons to be mistargeted to the medulla, although UAS-dlB was more severe.
Figure 3.—
The effects of GSd447 are caused by misexpression of the dorsal gene. (A) Misexpression of Dorsal-A (GMR-GAL4/UAS-dlA) does not appear to result in an axon targeting defect when photoreceptors are stained using α-Chaoptin (Chp). Bar, 20 μm. (B) Misexpression of Dorsal-B (GMR-GAL4/+; UAS-dlB/+) causes gaps in the lamina (arrow) and thickened bundles that project into the medulla. (C) Using ro-tau-lacZ, some R2–R5 axons project past the lamina when Dorsal-A is misexpressed (GMR-GAL4/UAS-dlA). (D) Many R2–R5 axons mistarget past the lamina when Dorsal-B is misexpressed (GMR-GAL4; UAS-dlB/+). (E) GMR-GAL4/GSd447 flies have eyes that are small, misshapen, and glazed in appearance. Anterior is left and posterior is right. (F) Two mutagenized lines were identified in which these defects were suppressed and the eye returned to normal, as shown here in the case of GMR-GAL4/GSd447*D. (G, H) R-cell axon mistargeting is suppressed by mutations generated in genetic screen, shown here using α-Chaoptin (G,H).
Our findings suggested an important role for the dl gene in explaining the GSd447 misexpression phenotype, and raised several possibilities: (1) that dl-B is largely responsible for the axon targeting defects of the GSd447 misexpression phenotype; (2) that both Dorsal-A and Dorsal-B can mistarget axons, but that the UAS-dlB transgene is expressed at higher levels than the UAS-dlA transgene; or (3) that both dl-A and dl-B synergize to give rise to the full effect observed with GSd447. To distinguish between these possibilities, we performed a chemical mutagenesis screen for suppressors of GSd447 misexpression effects. We exploited the fact that misexpression of GSd447 with GMR-GAL4 results in a smaller eye with a glazed appearance (Figure 3E). In this screen, GSd447 adult males were exposed to the mutagen EMS and then crossed to GMR-GAL4 females. A total of 45,000 adult progeny of this cross were screened to identify two lines that showed suppression of this phenotype to a relatively normal eye structure (Figure 3F). Both lines, referred to as GSd447*C (Figure 3G) and GSd447*D (Figure 3H), suppressed the axon targeting defects nearly completely when subsequently tested with α-Chaoptin. We used PCR to confirm that the GS447 P element remained at the dl locus in each of these lines.
Genetic suppressors of GSd447 misexpression harbor mutations of dl:
In tests of both maternal effect lethality and dorsalization in cuticle preparations, we found that both GSd447*C and GSd447*D failed to complement the dl1 allele, or a deficiency that encompassed the dl gene (Df(2L) H20), and led to strongly dorsalized (D0) progeny (data not shown). Surprisingly, GSd447*C was found to be a dominant mutation since heterozygous mothers were often found to give rise to embryos that were moderately dorsalized (D2).
To confirm that GSd447*C and GSd447*D bear mutations in dl, the dl gene was sequenced from genomic DNA isolated from hemizygous (GSd447*C/Df(2L)H20) and (GSd447*D/Df(2L)H20) flies. PCR primers were designed to span all coding sequences of the dl gene, both dl-A and dl-B isoforms. The GSd447*C mutation was found to be a missense mutation (Arg-63→Cys). This mutation is identical to one previously reported which also displayed dominant female sterility: Arg-63 lies in the DNA-binding domain of dorsal (Isoda et al. 1992). The GSd447*D mutation was found to be a nonsense mutation in the dl-A isoform only (Gln-453→ STOP). It results in a truncation of the C-terminus of dl-A, in a region known to encode the transactivation domain. No other mutations in the dl gene were found in these lines.
Misexpression of GSd447 drives increased expression of dorsal:
Since the lines GSd447*C and GSd447*D harbor mutations of dl, we have named these alleles dlC447 and dlD447, respectively. We used an anti-Dorsal monoclonal antibody to confirm that Dorsal could be misexpressed in GSd447 animals with GMR-GAL4 and to determine whether misexpression was compromised in dlC447 and dlD447. This antibody recognizes an epitope in the C-terminal half of Dorsal (Whalen and Steward 1993), and is therefore specific for the Dorsal-A isoform and cannot recognize the Dorsal-B isoform. For clarity, we continue to distinguish the isoforms of Dorsal as detected by this antibody.
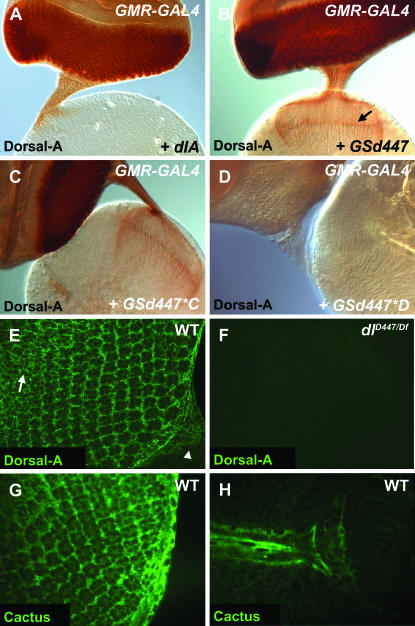
Using UAS-dlA as a positive control (Figure 4A), we found that GSd447 drove high levels of Dorsal-A expression in a GAL4-dependent manner (Figure 4B). The GSd447*C mutation does not appear to affect Dorsal-A immunoreactivity (Figure 4C), but GSd447*D abolishes it nearly completely (Figure 4D). In R cells, the ectopic Dorsal immunoreactivity was found in the cell body and axons, through the optic stalk and into the brain. It is noteworthy that while UAS-dlA misexpression caused strong Dorsal expression in cell bodies and proximal axons, expression was negligible in distal axons in the lamina plexus and medulla (Figure 4A). In contrast, Dorsal expression driven with GS447 and dlC447 was readily detectable along the entire length of axons (Figure 4, B and C). Finally, dlD447 lost all ectopic immunoreactivity, a finding that is consistent with our molecular data showing that dlD447 encodes a truncated protein and that the α-Dorsal-A antibody binds an epitope in the C-terminus (Whalen and Steward 1993).
Figure 4.—
Dorsal and Cactus are expressed in the developing eye imaginal disc. α-Dorsal-A was used to stain the eye–brain complex (A–D). (A) Misexpression of UAS-dlA results in staining of the eye posterior to the morphogenetic furrow; however, little staining progresses past the optic stalk and into axon terminals (GMR-GAL4/UAS-dlA). (B) Misexpression of GSd447 resulted in intense staining for Dorsal-A in cell bodies and along entire axons to their terminals (GMR-GAL4/GSd447) (arrow). (C, D) The GSd447*C mutation remains immunoreactive for Dorsal (GMR-GAL4/GSd447*C) and retains localization to distal axons, while the GSd447*D mutation does not. (E) Confocal image of eye disc showing that Dorsal-A is expressed in the cell bodies of R cells in addition to cone cells and peripodial cells in wild-type (WT, genotype: w1118) eye discs. Anterior is upper left and posterior is lower right, with the beginning of the optic stalk marked by an arrowhead. The optical slice shown here largely cuts through a plane at the level of the R-cell nuclei. In more anterior positions, Dorsal-A is also expressed in undifferentiated cells prior to their recruitment into ommatidia (arrow). (F) In dl mutants (dlD447/Df(2L)TW119), expression is no longer observed, proving the specificity of the antibody staining pattern for Dorsal (F). (G, H) In WT (genotype: w1118) eye discs at third instar stages, Cactus was expressed in the cell bodies of photoreceptor neurons (G) in addition to cone cells, glial cells of the optic stalk (H), and peripodial cells.
Dorsal and its inhibitor Cactus are expressed in R cells:
We next analyzed whether Dorsal was endogenously expressed in the developing larval visual system. We used the anti-Dorsal antibody and fluorescence immunochemistry to stain wild-type animals. Confocal microscopy was used to analyze the fluorescently labeled tissues. Moderately intense staining was present in what appears to be many, if not all, cells of the developing eye disc and optic stalk of wild-type animals (Figure 4E). Notably, Dorsal is localized to the cell bodies of R cells, cone cells, and peripodial cells. There were traces of Dorsal immunoreactivity found in R-cell axons in the optic stalk or in the axon terminals in the optic lobe (not shown). A large number of undifferentiated cells anterior to the morphogenetic furrow were also immunopositive (Figure 4E). We examined animals hemizygous for dl (dlD447/Df(2L)TW119) and found no labeling, demonstrating the anti-Dorsal staining pattern to be specific (Figure 4F). For R cells and cone cells, which have little cytoplasm, it is difficult to determine whether the staining was throughout the cytoplasm or in close proximity to the plasma membrane. In peripodial cells, however, the staining was evident throughout the cytoplasm. None of the staining was found to be nuclear, suggesting the majority of Dorsal is held in the cytoplasm, perhaps bound in inactive complexes with Cactus, a known inhibitor of Dorsal and other Rel proteins. Previous studies have shown that Cactus is distributed at low levels throughout the brain and nerve cord of Drosophila larvae, and is notably enriched in the mushroom bodies (Cantera et al. 1999b). We confirmed the reported expression of Cactus in the brain (data not shown) and found that Cactus is also expressed in the R-cell bodies (Figure 4G) and in cone cells (not shown). We also found strong Cactus labeling of the peripodial cells surrounding the eye disc and of the glial cells in the optic stalk (Figure 4H). Traces of Cactus were also evident in the optic lobe, though the diminished labeling there was difficult to characterize. Nevertheless, both Dorsal and Cactus are expressed in the developing visual system, and both are expressed in R cells.
dl mutants do not have axon targeting defects:
We have provided evidence that misexpression of Dorsal resulted in R-cell axon targeting defects, that mutations of dl suppress those defects, and that Dorsal and its inhibitor Cactus are expressed in R cells. To test the hypothesis that Dorsal could function in R-cell targeting, we examined dl mutants. Hemizygous (dl1/Df(2L)TW119) third-instar larvae were analyzed for axon-targeting defects with α-Chaoptin immunochemistry. No defects were observed relative to controls (not shown). The same was true for dlC447/Df(2L)H20 and dlD447/Df(2L)H20 hemizygotes (Figure 5, A and B). To use a more sensitive assay for R2–R5 axons, ro-tau-lacZ was tested in animals carrying heteroallelic combinations of five different alleles or deficiencies of dl, including dlC447, dlD447, dl1, dl4, and Df(2L)TW119. We examined a minimum number of 10 hemispheres (n) for each genotype, and results for dlC447/Df(2L)TW119 (n = 23)and dlD447/Df(2L)TW119 (n = 12) are presented (Figure 5, C and D). However, none of the allele combinations showed R2–R5 axon targeting defects. These findings suggested to us that dl alone is not required for correct axon targeting in the Drosophila visual system.
Figure 5.—
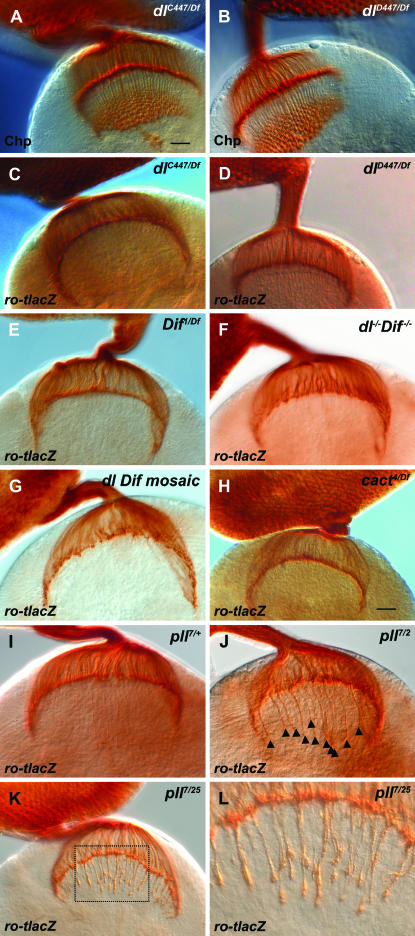
Mutations of pll, though not other members of the dorsal signaling pathway, cause photoreceptor mistargeting. (A, B) Neither dlC447 nor dlD447 hemizygotes have obvious disruptions of R-cell patterning when stained with α-Chaoptin (Chp). The genotypes shown are dlC447/Df(2L)H20) (A) and dlD447/Df(2L)H20) (B). Bar, 20 μm. (C–L) Ro-tau-lacZ was used to visualize the projections of the R2–R5 axons. No R2–R5 axon targeting defects are observed in dl or Dif hemizygotes. The following genotypes are shown: (C) dlC447/Df(2L)TW119; (D) dlD447/Df(2L)TW119; (E) Dif1/Df(2L)J4. Nor are there targeting defects in those rare dl Dif double mutants that reach wandering third instar (F, genotype Dif1/Df(2L)J4) or mosaic animals with large patches of the developing eye disc rendered doubly mutant for both Dif and dl (G, genotype: eyFLP;FRT40A,l(2)/FRT40A, Df(2L)J4). (H) Cactus mutants (cact4/Df(2L)r10255) also do not have R2–R5 axon targeting defects. In contrast with the above results, and compared here with pll7/+ heterozygote controls (I), >50% of pll mutant hemispheres show R2–R5 axon mistargeting (arrowheads in J). The genotypes shown are pll7/pll2 (J) and pll7/pll25 (K, L). (L) At higher magnification, many thickened axon bundles penetrate the lamina along its entire length.
Analyses of mutants of the NFκB signaling pathway in flies:
We explored the possibility that the Dorsal misexpression phenotype might underscore the importance of another NF-κB/Rel transcription factor in visual system development. In Drosophila there are three NF-κB transcription factors, Dorsal, Dif (Dorsal-related immunity factor), and Relish. Each functions in innate immunity, although only Dif and Dorsal function through the Toll receptor, while Relish responds to activation through the PGRP-LC/Imd receptor complex. Since Dorsal is most closely related to Dif, and because Dif has been shown previously to be expressed in the larval nervous system (Cantera et al. 1999b), we tested Dif1/Df(2L)J4 hemizygotes (n = 14, Figure 5E), Dif2/Df(2L)J4 hemizygotes (n = 10), and Dif1/Dif2 (n = 10) animals and found they all appeared similar to wild type. Several studies have reported that Dorsal and Dif exhibit partial functional redundancy in innate immunity and hematopoiesis, where double mutants of Dif and dl can be rescued by expressing either Dif or Dorsal (Lemaitre et al. 1995; Manfruelli et al. 1999; Meng et al. 1999; Matova and Anderson 2006). To test this possibility in the visual system, we examined the small deficiency Df(2L)J4, which deletes both genes (the dl and Dif genes lie next to one another at the cytological location 36C7-9). Animals carrying Df(2L)J4 in combination with a larger deficiency (Df(2L)J4/Df(2L)TW119) are null homozygotes for both dl and Dif. Those that survive to third instar are exceedingly rare, presumably due to compromised innate immunity. However, in the few that we did observe (n = 6), there were no defects in R2–R5 axon projections (Figure 5F). In addition, we examined the projection pattern of Df(2L)J4-deficient mutant R-cell axons in an otherwise heterozygous or wild-type target region by using genetic mosaic analysis. When large patches (>90% eye tissues) of Df(2L)J4 mutant R cells were generated by eye-specific mitotic recombination(Newsome et al. 2000) there were no defects of R2–R5 axon targeting observed (n = 16) (Figure 5G). Together, the data indicate that neither Dif nor dl is required for axon targeting at these stages. We also examined mutants of Relish, the third known NF-κB/Rel transcription factor, but again no obvious defects were observed (not shown).
Cactus is expressed in R cells (Figure 4, G and H), and Cactus binds and inhibits Dorsal. Since misexpression of dl using GMR-GAL4 leads to axon guidance defects, we hypothesized mutations of cactus might mimic this effect. To test the requirement for Cactus in correct R-cell axon targeting, heteroallelic combinations of cactus mutations were analyzed with ro-tau-lacZ. Several different combinations of cactus alleles and deficiencies were analyzed. Compared with controls (not shown), cactus4/Df(2L)r10 hemizygotes (n = 22) (Figure 5H), cactus4/Df(2L)cact-255 hemizygotes (n = 8), and cactus4/cactus1 (n = 8) animals showed no defects, with R2–R5 axons targeting properly to the lamina. It is therefore interesting to observe that, with respect to axon targeting, loss of cactus is not equivalent to the overexpression of Dorsal. We also found no targeting defects of R2–R5 axons in mutants of the receptor Toll or the adaptor protein Tube.
In embryonic dorsal–ventral patterning the kinase Pelle plays a pivotal role in transmitting signaling from the Toll receptor to Dorsal. We tested pll mutants and, in contrast to heterozygous controls (pll7/+, Figure 5I), pll mutants showed mistargeting of R2–R5 axons into the medulla (Figure 5, J and K). In control animals labeled for R2–R5 (ro-taulacZ/+), one occasionally sees thin, single axons (up to 10 per hemisphere) that penetrate the lamina and protrude into the medulla, regardless of genotype. When they occur, these ectopic projections in control animals mostly penetrate through the center of an otherwise smooth lamina (Figure 5I). In contrast, over half of the hemispheres from pll mutants showed a mistargeting defect that was both qualitatively and quantitatively distinct from controls. Thickened bundles of axons along the entire length of the lamina were observed to penetrate into the medulla (Figure 5, J and K), while the lamina itself was not smooth and somewhat irregular. Based on these criteria, and blind to genotype, we examined a larger number of animals and scored the number of phenotypically abnormal hemispheres in pll mutants and controls. In pll7/pll25 mutants (Figure 5, J and K), 55% (12/22) hemispheres were abnormal. The same penetrance was observed in pll7/pll2 mutants [22/41 hemispheres (54%)]. This represents a threefold increase over heterozygous controls (pll7/+), in which only 14/79 (17%) of hemispheres were scored abnormal. Though the penetrance of the phenotype was identical in the two different heteroallelic combinations we examined, the pll7/pll25 appeared to be qualitatively more severely affected. These results indicate that the IRAK family kinase Pelle is required for correct targeting of R-cell axons during larval development.
DISCUSSION
We conducted a screen using the GAL4-UAS misexpression system to identify novel molecular mechanisms underlying axon guidance and targeting. We generated a collection of 142 lines that carry independent GS P-element insertions. GS is a particularly potent UAS-based vector, seven times more effective than related elements such as EP (Rorth 1996), and each line in our collection was preselected for having lethal effects when misexpression was directed to the entire embryonic nervous system. During our prescreen we scored lethality at eclosion. Therefore, it remains unclear whether embryonic neural misexpression is sufficient to explain the lethality in every GS line. Nevertheless, our screen for neuronal morphology using Fas2 would indicate that our prescreened collection of 142 lines represents a significant enrichment for genes likely to disrupt neuronal morphology or function. In 30% of the 142 lines, the screen identified Fas2 phenotypes due to unique loci. Compared with other misexpression screens whose hit rate was considerably lower (Abdelilah-Seyfried et al. 2000; Kraut et al. 2001; McGovern et al. 2003; Nicolai et al. 2003; Kanuka et al. 2005; Zhang et al. 2006), it would appear that the lethal prescreen did indeed enrich for genes causing visible nervous system defects.
While providing added potency, the bidirectional nature of the GS element stipulates that genes located on either side of the insertion may be responsible for the misexpression effect, and that the culprit cannot be accurately predicted based on insertion site alone. In many cases it will take both molecular and functional analyses, such as those described here for GSd447, to convincingly demonstrate the causal gene. This is because many of the insertions sit between loci, and because GS can often initiate transcripts at considerable distances from annotated gene promoters. In addition, the Drosophila genome annotations are in some instances incomplete, as gene predictions often miss 5′ exons that are not represented in cDNA sequences and ESTs. In supplemental Table 1 (http://www.genetics.org/supplemental/) we have provided 60 bp of flanking sequence for each insertion to assist BLAST searches of future updates to the annotations of the Drosophila genome.
In our screen we identified 42 genes that can influence patterning of the Fas2-positive axons. Many of these genes encode proteins thought to be secreted, membrane-associated, and cytoplasmic, though most encode nuclear proteins. Perhaps this is not surprising since nuclear factors may influence the expression of multiple downstream effectors, thereby increasing the probability they may elicit defects of nervous system development. The identification of leak (lea), otherwise known as roundabout2 (robo2), which encodes a transmembrane receptor involved in guidance of axons in response to the secreted guidance cue Slit, confirms that we can isolate genes involved in axon guidance with this technique. In addition, the screen also found genes that influence neurogenesis (mam, H) and neuronal differentiation (ttk, pnt, mirr). Determining whether any of the uncharacterized genes identified in our screen are also required for axon guidance or neural differentiation will require further experimentation.
Here we have focused on the characterization of line GSd447. Misexpression of GSd447 caused the axons of motor neurons and R cells to bypass their correct termination sites. In R cells, we have found that these targeting defects are a consequence of misexpression of Dorsal, and do not appear to result from changes of R-cell fate determination. Since UAS-dlA was less potent than UAS-dlB in recapitulating the effects of GSd447, we proposed three possibilities: (1) that Dorsal-B alone is largely responsible for the GSd447 misexpression effect; (2) that either Dorsal-A or Dorsal-B can mistarget axons, and that the effect is dose-dependent; or (3) that dl-A and dl-B synergize. Our EMS-induced mutation dlD447, predicted to truncate only the Dorsal-A isoform while leaving the Dorsal-B isoform intact, indicates that Dorsal B alone is not responsible for mistargeting axons in GSd447.
The EMS-induced mutation dlC447 provides evidence that the activity of Dorsal in mistargeting axons is likely to occur through its well-established role as a nuclear transcription factor, since dlC447 encodes a DNA-binding domain mutant. Interestingly, the number of mistargeted axons resulting from GSd447 was far higher than caused by misexpression of UAS-dl-A, and this correlated with elevated expression of Dorsal in distal R-cell axons and their terminals. Perhaps the targeting of Dorsal to distal axons is simply a consequence of high levels of expression, but we do not favor this idea since Dorsal protein accumulation in cell bodies caused by misexpression of UAS-dl-A was comparable to that caused by GSd447 and should therefore have been readily detectable in axon terminals. Alternatively, it is possible that the endogenous dl transcript driven by the GS insertion contains noncoding regulatory elements that promote expression or targeting in distal axons. In support of the idea that mistargeting correlates with the localization of Dorsal to distal axons, we found substantial targeting defects caused by misexpression of an epitope-tagged version of Dorsal-B (UAS-dlB-HA, Figure 3, B and D), which has no nuclear localization sequence and can be readily observed along the entire length of R-cell axons (E. Mindorff, J. Yang and D. van Meyel, unpublished observations). A parsimonious explanation for the requirement for Dorsal-A despite its relative weakness, and the potency of Dorsal-B, is that perhaps Dorsal-A and Dorsal-B form heterodimers, with Dorsal-B promoting subcellular localization of Dorsal-A/Dorsal-B heterodimers to distal axons, and Dorsal-A promoting mistargeting following pathway activation and nuclear entry.
Misexpression of Dorsal has been shown to influence neuron number and axon growth and guidance in the mushroom bodies of the brain (Nicolai et al. 2003). In addition, misexpression of Cactus reduces the number of olfactory neurons in the fly antennal lobe (Zhang et al. 2006). Despite our findings that Dorsal and Cactus are expressed in R cells, and that Dorsal misexpression has dramatic consequences on axon targeting, our studies of dl, Dif, and Relish mutations found no requirement for any of these genes in this process. We also found that cactus mutants did not phenocopy the effect of Dorsal misexpression, suggesting that mutation of this cytoplasmic inhibitor is not sufficient to activate endogenous Dorsal and elicit mistargeting. However, we did find that the effects of Dorsal misexpression can be enhanced in animals heterozygous for a cactus mutation (E. Mindorff and D. van Meyel, unpublished observations), suggesting that Cactus is functionally capable of inhibiting Dorsal activity in R cells and supporting our evidence for a role for Dorsal in generating the misexpression phenotypes.
As yet, no direct role for the Dorsal pathway in regulating axon guidance has been shown in Drosophila. In Toll and dl mutants, motor neurons innervate incorrect targets, although this effect is thought to result from cell non-autonomous functions of these genes in muscles or glia (Halfon et al. 1995; Cantera et al. 1999a). A role for NF-κB signaling in murine nervous system development has been suggested by experiments in which inhibition of NF-κB, or its transcriptional activity, substantially reduced the size and complexity of neurite arbors of cultured sensory neurons and layer two pyramidal neurons in organotypic slices (Gutierrez et al. 2005). It is interesting to note that both Dorsal and Cactus specifically localize to synapses of the larval neuromuscular junction, where immunoreactivity diminishes in response to either electrical stimulation or glutamate administration (Bolatto et al. 2003). Mammalian NF-κB can also be found in synapses and glutamate stimulates redistribution of the NF-κB p65 subunit from synapses to nuclei in mouse hippocampal neurons (Kaltschmidt et al. 1993; Guerrini et al. 1995; Meberg et al. 1996; Wellmann et al. 2001). Therefore, NF-κB may be poised in nerve terminals and synapses to relay activity-dependent signals to the nucleus to effect changes in gene transcription. Accordingly, it is interesting to note that inhibition of NF-κB prevents induction of long-term depression and significantly reduces long-term potentiation (Albensi and Mattson 2000), and that mice deficient for the synaptic NF-κB subunit p65 have a deficit in spatial learning (Meffert et al. 2003). It is also notable that, in Drosophila, Dif and Cactus are expressed throughout the nervous system and at high levels in mushroom bodies, the learning and memory center of Drosophila (Cantera et al. 1999b). It is possible that misexpression of Dorsal in R cells, coupled with its targeting to axon terminals, activates inappropriate or precocious NF-κB signaling that is normally reserved for later steps of synaptic maturation or function.
Given the lack of loss-of-function evidence for a role of NF-κB signaling in R-cell axon targeting, our identification of the requirement for the serine-threonine kinase Pelle remains enigmatic. At present, we can only speculate on the mechanism by which it influences R cells. It has been proposed that, during embryonic patterning, Pelle not only activates signaling to Dorsal but also provides negative feedback to the Toll receptor to dampen the signaling response (Towb et al. 2001). In R cells, perhaps Pelle provides negative feedback to prevent inappropriate activation through Dorsal signaling; this may explain why mutations of pll mimic the effects of Dorsal misexpression. Alternatively, although its central role in Nf-κB signaling has been well established, Pelle is an IRAK family kinase which may have activity on other substrates, perhaps those already implicated as components of the molecular stop signal for R cells. It is interesting to note that Pelle and Dorsal have been shown to physically interact in yeast two-hybrid assays (Edwards et al. 1997; Yang and Steward 1997). It is possible that misexpression of Dorsal sequesters Pelle from functional, physiological interactions with other proteins. The mechanism by which Pelle regulates layer-specific targeting of photoreceptor axons remains to be determined, and further analyses of candidate genes found in our screen will be required to identify additional novel regulators of nervous system development in vivo.
Acknowledgments
The authors thank John Thomas, in whose lab the misexpression screen was initially conceived and conducted while D.D.O. and D.J.V. were trainees. We also thank Yong Rao, Par Towb, Keith Murai, Stefan Thor, Matthias Landgraf, and the members of the van Meyel, Thomas, and Rao labs for helpful advice and discussions. Thanks also go to the Bloomington Stock Center, the Berkeley Drosophila Genome Project, the Developmental Studies Hybridoma Bank, the Drosophila Genomics Resource Center, and to our colleagues in the fly community for stocks and reagents, in particular Kathryn Anderson, Nina Matova, Dominique Ferrandon, Shubha Govind, Par Towb, Steve Wasserman, Tony Ip, Ruth Steward, Utpal Banerjee, Larry Zipursky, Jean-Marc Reichhart, Yasushi Hiromi, Andrea Brand, and Laura Nilson. D.J.V. is a New Investigator of the Canadian Institutes for Health Research (CIHR). This research was supported by grants from the CIHR and Canadian Foundation for Innovation.
References
- Abdelilah-Seyfried, S., Y. M. Chan, C. Zeng, N. J. Justice, S. Younger-Shepherd et al., 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albensi, B. C., and M. P. Mattson, 2000. Evidence for the involvement of TNF and NF-kappaB in hippocampal synaptic plasticity. Synapse 35: 151–159. [DOI] [PubMed] [Google Scholar]
- Allen, M. J., J. A. Drummond, D. J. Sweetman and K. G. Moffat, 2007. Analysis of two P-element enhancer-trap insertion lines that show expression in the giant fibre neuron of Drosophila melanogaster. Genes Brain Behav. 6: 347–358. [DOI] [PubMed] [Google Scholar]
- Bang, A. G., and J. W. Posakony, 1992. The Drosophila gene Hairless encodes a novel basic protein that controls alternative cell fates in adult sensory organ development. Genes Dev. 6: 1752–1769. [DOI] [PubMed] [Google Scholar]
- Bejarano, F., and A. Busturia, 2004. Function of the Trithorax-like gene during Drosophila development. Dev. Biol. 268: 327–341. [DOI] [PubMed] [Google Scholar]
- Bolatto, C., S. Chifflet, A. Megighian and R. Cantera, 2003. Synaptic activity modifies the levels of Dorsal and Cactus at the neuromuscular junction of Drosophila. J. Neurobiol. 54: 525–536. [DOI] [PubMed] [Google Scholar]
- Boyle, M., A. Nighorn and J. B. Thomas, 2006. Drosophila Eph receptor guides specific axon branches of mushroom body neurons. Development 133: 1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferty, P., L. Yu and Y. Rao, 2004. The receptor tyrosine kinase Off-track is required for layer-specific neuronal connectivity in Drosophila. Development 131: 5287–5295. [DOI] [PubMed] [Google Scholar]
- Cantera, R., T. Kozlova, C. Barillas-Mury and F. C. Kafatos, 1999. a Muscle structure and innervation are affected by loss of Dorsal in the fruit fly, Drosophila melanogaster. Mol. Cell. Neurosci. 13: 131–141. [DOI] [PubMed] [Google Scholar]
- Cantera, R., E. Roos and Y. Engstrom, 1999. b Dif and cactus are colocalized in the larval nervous system of Drosophila melanogaster. J. Neurobiol. 38: 16–26. [PubMed] [Google Scholar]
- Chen, L. Y., J. C. Wang, Y. Hyvert, H. P. Lin, N. Perrimon et al., 2006. Weckle is a zinc finger adaptor of the toll pathway in dorsoventral patterning of the Drosophila embryo. Curr. Biol. 16: 1183–1193. [DOI] [PubMed] [Google Scholar]
- Drier, E. A., S. Govind and R. Steward, 2000. Cactus-independent regulation of Dorsal nuclear import by the ventral signal. Curr. Biol. 10: 23–26. [DOI] [PubMed] [Google Scholar]
- Edwards, D. N., P. Towb and S. A. Wasserman, 1997. An activity-dependent network of interactions links the Rel protein Dorsal with its cytoplasmic regulators. Development 124: 3855–3864. [DOI] [PubMed] [Google Scholar]
- Engstrom, Y., L. Kadalayil, S. C. Sun, C. Samakovlis, D. Hultmark et al., 1993. kappa B-like motifs regulate the induction of immune genes in Drosophila. J Mol. Biol. 232: 327–333. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize, J. A., G. M. Rubin and R. Lehmann, 1992. The fat facets gene is required for Drosophila eye and embryo development. Development 116: 985–1000. [DOI] [PubMed] [Google Scholar]
- Garrity, P. A., Y. Rao, I. Salecker, J. McGlade, T. Pawson et al., 1996. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell 85: 639–650. [DOI] [PubMed] [Google Scholar]
- Garrity, P. A., C. H. Lee, I. Salecker, H. C. Robertson, C. J. Desai et al., 1999. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron 22: 707–717. [DOI] [PubMed] [Google Scholar]
- Govind, S., E. Drier, L. H. Huang and R. Steward, 1996. Regulated nuclear import of the Drosophila rel protein dorsal: structure-function analysis. Mol. Cell. Biol. 16: 1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, I., P. Georgel, P. Oertel-Buchheit, M. Schnarr and J. M. Reichhart, 1999. Dorsal-B, a splice variant of the Drosophila factor Dorsal, is a novel Rel/NF-kappaB transcriptional activator. Gene 228: 233–242. [DOI] [PubMed] [Google Scholar]
- Guerrini, L., F. Blasi and S. Denis-Donini, 1995. Synaptic activation of NF-kappa B by glutamate in cerebellar granule neurons in vitro. Proc. Natl. Acad. Sci. USA 92: 9077–9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, H., V. A. Hale, X. Dolcet and A. Davies, 2005. NF-kappaB signalling regulates the growth of neural processes in the developing PNS and CNS. Development 132: 1713–1726. [DOI] [PubMed] [Google Scholar]
- Halfon, M. S., C. Hashimoto and H. Keshishian, 1995. The Drosophila Toll gene functions zygotically and is necessary for proper motoneuron and muscle development. Dev. Biol. 169: 151–167. [DOI] [PubMed] [Google Scholar]
- Hart, A. C., H. Kramer and S. L. Zipursky, 1993. Extracellular domain of the boss transmembrane ligand acts as an antagonist of the sev receptor. Nature 361: 732–736. [DOI] [PubMed] [Google Scholar]
- Hecht, P. M., and K. V. Anderson, 1993. Genetic characterization of tube and pelle, genes required for signaling between Toll and dorsal in the specification of the dorsal-ventral pattern of the Drosophila embryo. Genetics 135: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewes, R. S., D. Park, S. A. Gauthier, A. M. Schaefer and P. H. Taghert, 2003. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development 130: 1771–1781. [DOI] [PubMed] [Google Scholar]
- Isoda, K., S. Roth and C. Nusslein-Volhard, 1992. The functional domains of the Drosophila morphogen dorsal: evidence from the analysis of mutants. Genes Dev. 6: 619–630. [DOI] [PubMed] [Google Scholar]
- Janssens, S., and R. Beyaert, 2003. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol. Cell 11: 293–302. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt, C., B. Kaltschmidt and P. A. Baeuerle, 1993. Brain synapses contain inducible forms of the transcription factor NF-kappa B. Mech. Dev. 43: 135–147. [DOI] [PubMed] [Google Scholar]
- Kaminker, J. S., J. Canon, I. Salecker and U. Banerjee, 2002. Control of photoreceptor axon target choice by transcriptional repression of Runt. Nat. Neurosci. 5: 746–750. [DOI] [PubMed] [Google Scholar]
- Kania, A., A. Salzberg, M. Bhat, D. D'Evelyn, Y. He et al., 1995. P-element mutations affecting embryonic peripheral nervous system development in Drosophila melanogaster. Genetics 139: 1663–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanuka, H., T. Hiratou, T. Igaki, H. Kanda, E. Kuranaga et al., 2005. Gain-of-function screen identifies a role of the Sec61alpha translocon in Drosophila postmitotic neurotoxicity. Biochim. Biophys. Acta 1726: 225–237. [DOI] [PubMed] [Google Scholar]
- Kauffmann, R. C., S. Li, P. A. Gallagher, J. Zhang and R. W. Carthew, 1996. Ras1 signaling and transcriptional competence in the R7 cell of Drosophila. Genes Dev. 10: 2167–2178. [DOI] [PubMed] [Google Scholar]
- Kosman, D., S. Small and J. Reinitz, 1998. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 208: 290–294. [DOI] [PubMed] [Google Scholar]
- Kraut, R., K. Menon and K. Zinn, 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11: 417–430. [DOI] [PubMed] [Google Scholar]
- Kuang, B., S. C. Wu, Y. Shin, L. Luo and P. Kolodziej, 2000. split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development 127: 1517–1529. [DOI] [PubMed] [Google Scholar]
- Lee, T., C. Winter, S. S. Marticke, A. Lee and L. Luo, 2000. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron 25: 307–316. [DOI] [PubMed] [Google Scholar]
- Lemaitre, B., M. Meister, S. Govind, P. Georgel, R. Steward et al., 1995. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J. 14: 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., W. Li, H. C. Calhoun, F. Xia, F. B. Gao et al., 2003. Patterns and functions of STAT activation during Drosophila embryogenesis. Mech. Dev. 120: 1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfruelli, P., J. M. Reichhart, R. Steward, J. A. Hoffmann and B. Lemaitre, 1999. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 18: 3380–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matova, N., and K. V. Anderson, 2006. Rel/NF-kappaB double mutants reveal that cellular immunity is central to Drosophila host defense. Proc. Natl. Acad. Sci. USA 103: 16424–16429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern, V. L., C. A. Pacak, S. T. Sewell, M. L. Turski and M. A. Seeger, 2003. A targeted gain of function screen in the embryonic CNS of Drosophila. Mech. Dev. 120: 1193–1207. [DOI] [PubMed] [Google Scholar]
- McNeill, H., C. H. Yang, M. Brodsky, J. Ungos and M. A. Simon, 1997. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes Dev. 11: 1073–1082. [DOI] [PubMed] [Google Scholar]
- Meberg, P. J., W. R. Kinney, E. G. Valcourt and A. Routtenberg, 1996. Gene expression of the transcription factor NF-kappa B in hippocampus: regulation by synaptic activity. Brain Res. Mol. Brain Res. 38: 179–190. [DOI] [PubMed] [Google Scholar]
- Meffert, M. K., J. M. Chang, B. J. Wiltgen, M. S. Fanselow and D. Baltimore, 2003. NF-kappa B functions in synaptic signaling and behavior. Nat. Neurosci. 6: 1072–1078. [DOI] [PubMed] [Google Scholar]
- Meng, X., B. S. Khanuja and Y. T. Ip, 1999. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 13: 792–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisato, D., and K. V. Anderson, 1994. The spatzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell 76: 677–688. [DOI] [PubMed] [Google Scholar]
- Morisato, D., and K. V. Anderson, 1995. Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu. Rev. Genet. 29: 371–399. [DOI] [PubMed] [Google Scholar]
- Newsome, T. P., B. Asling and B. J. Dickson, 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127: 851–860. [DOI] [PubMed] [Google Scholar]
- Nicolai, M., C. Lasbleiz and J. M. Dura, 2003. Gain-of-function screen identifies a role of the Src64 oncogene in Drosophila mushroom body development. J. Neurobiol. 57: 291–302. [DOI] [PubMed] [Google Scholar]
- O'Neill, E. M., I. Rebay, R. Tjian and G. M. Rubin, 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78: 137–147. [DOI] [PubMed] [Google Scholar]
- Perez, S. E., and H. Steller, 1996. Migration of glial cells into retinal axon target field in Drosophila melanogaster. J. Neurobiol. 30: 359–373. [DOI] [PubMed] [Google Scholar]
- Poeck, B., S. Fischer, D. Gunning, S. L. Zipursky and I. Salecker, 2001. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron 29: 99–113. [DOI] [PubMed] [Google Scholar]
- Pulido, D., S. Campuzano, T. Koda, J. Modolell and M. Barbacid, 1992. Dtrk, a Drosophila gene related to the trk family of neurotrophin receptors, encodes a novel class of neural cell adhesion molecule. EMBO J. 11: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, P., P. C. Pan and S. Govind, 1998. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development 125: 1909–1920. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, S., E. Nicolas, V. Vivancos, J. Berger and B. J. Dickson, 2000. Crossing the midline: roles and regulation of Robo receptors. Neuron 28: 767–777. [DOI] [PubMed] [Google Scholar]
- Rangarajan, R., Q. Gong and U. Gaul, 1999. Migration and function of glia in the developing Drosophila eye. Development 126: 3285–3292. [DOI] [PubMed] [Google Scholar]
- Rao, Y., P. Pang, W. Ruan, D. Gunning and S. L. Zipursky, 2000. brakeless is required for photoreceptor growth-cone targeting in Drosophila. Proc. Natl. Acad. Sci. USA 97: 5966–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renger, J. J., W. D. Yao, M. B. Sokolowski and C. F. Wu, 1999. Neuronal polymorphism among natural alleles of a cGMP-dependent kinase gene, foraging, in Drosophila. J. Neurosci. 19: RC28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, S., Y. Hiromi, D. Godt and C. Nusslein-Volhard, 1991. cactus, a maternal gene required for proper formation of the dorsoventral morphogen gradient in Drosophila embryos. Development 112: 371–388. [DOI] [PubMed] [Google Scholar]
- Ruan, W., P. Pang and Y. Rao, 1999. The SH2/SH3 adaptor protein dock interacts with the Ste20-like kinase misshapen in controlling growth cone motility. Neuron 24: 595–605. [DOI] [PubMed] [Google Scholar]
- Ruan, W., H. Long, D. H. Vuong and Y. Rao, 2002. Bifocal is a downstream target of the Ste20-like serine/threonine kinase misshapen in regulating photoreceptor growth cone targeting in Drosophila. Neuron 36: 831–842. [DOI] [PubMed] [Google Scholar]
- Rutschmann, S., A. C. Jung, C. Hetru, J. M. Reichhart, J. A. Hoffmann et al., 2000. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12: 569–580. [DOI] [PubMed] [Google Scholar]
- Shelton, C. A., and S. A. Wasserman, 1993. pelle encodes a protein kinase required to establish dorsoventral polarity in the Drosophila embryo. Cell 72: 515–525. [DOI] [PubMed] [Google Scholar]
- Simpson, J. H., T. Kidd, K. S. Bland and C. S. Goodman, 2000. Short-range and long-range guidance by slit and its Robo receptors. Robo and Robo2 play distinct roles in midline guidance. Neuron 28: 753–766. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P-elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Stern, D. L., and E. Sucena, 2000. Preparation of larval and adult cuticles for light microscopy, pp. 601–615 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Suh, G. S., B. Poeck, T. Chouard, E. Oron, D. Segal et al., 2002. Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron 33: 35–46. [DOI] [PubMed] [Google Scholar]
- Sun, H., B. N. Bristow, G. Qu and S. A. Wasserman, 2002. A heterotrimeric death domain complex in Toll signaling. Proc. Natl. Acad. Sci. USA 99: 12871–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., P. Towb, D. N. Chiem, B. A. Foster and S. A. Wasserman, 2004. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. EMBO J. 23: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu-Ishikawa, E., M. Yoshihara, A. Ueda, M. B. Rheuben, Y. Hotta et al., 2001. Screening for synaptic defects revealed a locus involved in presynaptic and postsynaptic functions in Drosophila embryos. J. Neurobiol. 48: 101–119. [PubMed] [Google Scholar]
- Tayler, T. D., and P. A. Garrity, 2003. Axon targeting in the Drosophila visual system. Curr. Opin. Neurobiol. 13: 90–95. [DOI] [PubMed] [Google Scholar]
- Thomas, G. B., and D. J. van Meyel, 2007. The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression. Development 134: 591–600. [DOI] [PubMed] [Google Scholar]
- Toba, G., T. Ohsako, N. Miyata, T. Ohtsuka, K. H. Seong et al., 1999. The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towb, P., A. Bergmann and S. A. Wasserman, 2001. The protein kinase Pelle mediates feedback regulation in the Drosophila Toll signaling pathway. Development 128: 4729–4736. [DOI] [PubMed] [Google Scholar]
- Van Vactor, D. L., Jr., R. L. Cagan, H. Kramer and S. L. Zipursky, 1991. Induction in the developing compound eye of Drosophila: multiple mechanisms restrict R7 induction to a single retinal precursor cell. Cell 67: 1145–1155. [DOI] [PubMed] [Google Scholar]
- Weber, U., V. Siegel and M. Mlodzik, 1995. pipsqueak encodes a novel nuclear protein required downstream of seven-up for the development of photoreceptors R3 and R4. Embo J 14: 6247–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann, H., B. Kaltschmidt and C. Kaltschmidt, 2001. Retrograde transport of transcription factor NF-kappa B in living neurons. J. Biol. Chem. 276: 11821–11829. [DOI] [PubMed] [Google Scholar]
- Whalen, A. M., and R. Steward, 1993. Dissociation of the dorsal-cactus complex and phosphorylation of the dorsal protein correlate with the nuclear localization of dorsal. J. Cell Biol. 123: 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, W. C., and C. Montell, 1993. tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 7: 1085–1096. [DOI] [PubMed] [Google Scholar]
- Xiong, W. C., and C. Montell, 1995. Defective glia induce neuronal apoptosis in the repo visual system of Drosophila. Neuron 14: 581–590. [DOI] [PubMed] [Google Scholar]
- Yang, J., and R. Steward, 1997. A multimeric complex and the nuclear targeting of the Drosophila Rel protein Dorsal. Proc. Natl. Acad. Sci. USA 94: 14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, B., C. Petritsch, I. E. Clark, E. R. Gavis, L. Y. Jan et al., 2004. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 14: 314–321. [DOI] [PubMed] [Google Scholar]
- Zhang, D., W. Zhou, C. Yin, W. Chen, R. Ozawa et al., 2006. Misexpression screen for genes altering the olfactory map in Drosophila. Genesis 44: 189–201. [DOI] [PubMed] [Google Scholar]