Abstract
The RPW8 locus of Arabidopsis thaliana confers broad-spectrum resistance to powdery mildew pathogens. In many A. thaliana accessions, this locus contains two homologous genes, RPW8.1 and RPW8.2. In some susceptible accessions, however, these two genes are replaced by HR4, a homolog of RPW8.1. Here, we show that RPW8.2 from A. lyrata conferred powdery mildew resistance in A. thaliana, suggesting that RPW8.2 might have gained the resistance function before the speciation of A. thaliana and A. lyrata. To investigate how RPW8 has been maintained in A. thaliana, we examined the nucleotide sequence polymorphisms in RPW8 from 51 A. thaliana accessions, related disease reaction phenotypes to the evolutionary history of RPW8.1 and RPW8.2, and identified mutations that confer phenotypic variations. The average nucleotide diversities were high at RPW8.1 and RPW8.2, showing no sign of selective sweep. Moreover, we found that expression of RPW8 incurs fitness benefits and costs on A. thaliana in the presence and absence of the pathogens, respectively. Our results suggest that polymorphisms at the RPW8 locus in A. thaliana may have been maintained by complex selective forces, including those from the fitness benefits and costs both associated with RPW8.
DURING the long-time conflict between plants and potential pathogens, plants have evolved disease resistance (R) genes to detect the invasion of infectious pathogens and trigger effective defenses (Chisholm et al. 2006). In the past 15 years, >60 plant R genes have been isolated (Xiao 2006), of which the majority encode nucleotide-binding site (NBS) and leucine-rich-repeat (LRR) domains. The NBS-LRR genes constitute the largest R gene class and are abundant and ubiquitously expressed in all higher plants (Dangl and Jones 2001; McHale et al. 2006). A less frequent class of R genes comprises members of extracellular (e) LRR-containing receptor-like proteins (eLRR-RLPs) (Jones et al. 1994; Hammond-Kosack and Jones 1997) and receptor-like kinases (eLRR-RLKs) (Song et al. 1995; Sun et al. 2004). These two classes of LRR-containing R proteins are thought to be intracellular or cell-surface receptors that detect pathogen-derived virulence proteins (referred to as Avr effectors if recognized by R proteins) through direct or indirect interaction (Dangl and Jones 2001). The remaining characterized R genes encode proteins that either resemble the overall structure or a domain of the above two classes with some degree of structural variations, or have a novel protein structure that does not show significant homology to any other R proteins (Xiao 2006). Therefore, in terms of protein structures, they are atypical R genes in contrast to the typical LRR-encoding R genes.
The evolution and maintenance of plant R genes has become a research focus in recent years. Different mechanisms for sequence evolution have been documented for R genes (Michelmore and Meyers 1998; Meyers et al. 2005). However, the type and strength of selection acting on specific R genes is not well characterized. In conjunction with the recent advances in understanding of the molecular mechanisms of R-Avr interaction, several recent evolutionary analyses suggest that the mode of R-Avr recognition may profoundly influence the patterns of R-Avr coevolution (Dangl and McDowell 2006; Dodds et al. 2006).
While the simplest “arms-race” model used for describing the coevolution between plants and pathogens predicts directional selection or selective sweeps, a recent genomewide survey of R gene polymorphisms in Arabidopsis did not detect convincing evidence for a recent selective sweep for any of the R genes analyzed (Bakker et al. 2006). For some R genes in Arabidopsis, balancing selection appears to play a central role in molecular evolution (Stahl et al. 1999; Tian et al. 2002; Shen et al. 2006). For example, Arabidopsis RPM1 and RPS5, whose protein products detect their Avr proteins indirectly by association with the host target proteins of the Avr effectors (Mackey et al. 2002; Shao et al. 2003), are subject to balancing selection on resistance and susceptible alleles. At these loci, there are simple presence/absence polymorphisms (for the entire R-protein coding regions) that respectively correspond to the resistant and susceptible phenotypes. The R-Avr recognition in both cases appears to be of ancient origin and has been maintained for millions of years by balancing selection, presumably in a frequency-dependent fashion (Stahl et al. 1999; Tian et al. 2002). Relatively low genetic diversity with simple resistance/susceptibility allelism has been found at the Arabidopsis RPS2 locus, which was also interpreted as being consistent with balancing selection (Caicedo et al. 1999; Mauricio et al. 2003). Similar to RPM1 and RPS5, RPS2 recognizes its cognate Avr (avrRpt2) through indirect interaction (Axtell and Staskawicz 2003; Mackey et al. 2003).
On the other hand, some R proteins may recognize cognate Avr proteins by direct physical interaction (Jia et al. 2000; Deslandes et al. 2003; Dodds et al. 2006). These R genes seem to have been under diversifying selection for amino acid differentiation to generate new R proteins, which could recognize modified Avr effectors. This R-Avr coevolution would result in high genetic diversity at the R and the corresponding Avr loci (Dangl and McDowell 2006; Dodds et al. 2006). Compelling evidence for diversifying selection comes from recent studies by Ellis et al. (1999) on the flax L locus and the cognate AvrL567 locus in the flax rust pathogen, Melampsora lini. The L locus encodes at least 11 R alleles (including L5, L6, and L7) capable of recognizing distinct Avr genes belonging to different loci, including AvrL567, in the pathogen (Ellis et al. 1999). The AvrL567 locus also contains multiple Avr genes that are recognized by the R alleles at the L locus (Dodds et al. 2004). More significantly, they demonstrated that the R proteins L5, L6, and L7 physically interact with the corresponding Avr proteins in the yeast two-hybrid system in a specific manner that matches the specificity of the genetic interaction observed (Flor 1956; Dodds et al. 2006). These results strongly suggest diversifying selection at the R and the corresponding Avr loci for high levels of amino acid sequence polymorphism. Such high amino acid sequence diversity has also been observed at the Arabidopsis R genes RPP13 and RPP1 (Botella et al. 1998; Rose et al. 2004) and at the corresponding Avr genes ATR13 and ATR1 in Hyaloperonospora parasitica (Allen et al. 2004; Rehmany et al. 2005), implying that these two R-Avr pairs may be engaged in direct interaction.
The Arabidopsis thaliana RPW8 locus from accession Ms-0 confers broad-spectrum resistance to powdery mildew (Xiao et al. 2001). This locus contains two homologous genes, RPW8.1 and RPW8.2, both of which contribute to resistance. All tested Arabidopsis accessions contain three homologs of RPW8, i.e., HR1, HR2, and HR3, that are closely linked to the RPW8 locus (Xiao et al. 2001, 2004). Based on the presence/absence of RPW8.1 and RPW8.2, there are two basic Arabidopsis haplotypes at the RPW8 locus: one contains both RPW8.1 and RPW8.2 and the other contains HR4 in replacing RPW8.1 and RPW8.2 (Xiao et al. 2001, 2004). HR4 shares the most recent common ancestor with RPW8.1, and they might be orthologous (Xiao et al. 2004). RPW8.1 and RPW8.2 (hereafter referred to as RPW8 unless otherwise indicated) are unique because they confer broad-spectrum resistance to polyphagous Erysiphe pathogens that cause powdery mildew disease on many different plant species and they encode novel proteins showing no significant homology to other proteins (Xiao et al. 2001). How powdery mildew pathogens cause disease and how RPW8 detects the pathogens and induces resistance in A. thaliana are not clear. However, RPW8 appears to activate defense through a conserved signaling pathway that is also utilized by a subset of NBS-LRR R genes (Xiao et al. 2003, 2005). Our previous evolutionary analysis indicated that the origin of the RPW8 locus is relatively young, probably after the separation of Arabidopsis from the Brassica lineages and that RPW8.1 and RPW8.2 evolved from an HR3-like progenitor gene by duplication and functional diversification (Xiao et al. 2004). However, it is not known how divergent RPW8 alleles have evolved and been maintained in the A. thaliana populations. In the present study, we analyze the intraspecific sequence polymorphism at RPW8.1 and RPW8.2 to examine the evolutionary mechanism of RPW8 in A. thaliana. We relate the disease reaction phenotypes to the evolutionary history of the RPW8.1 and RPW8.2 alleles and identify allelic mutations that likely contribute to phenotypic variations. More significantly, we provide evidence that gene expression of RPW8 is associated with both fitness benefits and costs and that activation of defense-related cell death in the absence of the pathogen may account for the fitness cost of RPW8 expression.
MATERIALS AND METHODS
Plant materials:
Fifty-one accessions of A. thaliana from different geographical locations (Figure 1) were selected for sequence determination for RPW8.1 and RPW8.2, or HR4, if present, and HR3 (for 26 accessions). Nucleotide sequences of RPW8.1 and RPW8.2 or HR4 in 32 of the 51 accessions were previously determined in Xiao et al. (2004), but have not been analyzed at the nucleotide level. Seeds of A. thaliana accessions were obtained from the Arabidopsis Biological Resource Center or the Nottingham Arabidopsis Stock Centre. The six A. lyrata accessions were provided by Charles Langley, University of California, Davis.
Figure 1.—
Geographic distribution of the surveyed A. thaliana accessions. (A) The accessions containing either RPW8.1 and/or RPW8.2 encoding identical protein sequences as those of Ms-0 alleles are indicated by solid arrows; accessions containing RPW8.1 and RPW8.2 alleles encoding proteins different from those of Ms-0 are indicated by shaded arrows; accessions lacking RPW8.1 and RPW8.2 but having HR4 are indicated by dashed arrows. Numbers in A match numbers in B. (B) Names, disease reaction phenotypes (R, resistant; I, intermediate; S, susceptible), genotypes (R1, RPW8.1 identical to that of Ms-0 at the protein level; r1, RPW8.1 divergent from that of Ms-0; R2, RPW8.2 identical to that of Ms-0 at the protein level; r2, RPW8.2 divergent from that of Ms-0), and locations of 51 accessions. Accessions with an asterisk may contain powdery mildew R genes different from RPW8.1 and RPW8.2.
Assessment of disease phenotypes:
Most of these accessions were analyzed for their disease reaction phenotypes in response to powdery mildew isolates Erysiphe cruciferarum UEA1 and E. cichoracearum UCSC1 (Adam et al. 1999; Xiao et al. 2004). The newly obtained accessions were tested with E. cichoracearum UCSC1 three times using the method previously described (Xiao et al. 2003, 2005). Among the 51 accessions surveyed, there was a range of disease reaction (DR) phenotypes from very resistant to very susceptible. We used three categories to simplify the data analysis: resistant (R) (no visible fungus, HR, DR score 0–1), intermediate (I) (some fungus with <30% leaf coverage, with or without a slower HR, DR score 1 or 1–2), and susceptible (S) (profuse fungus with >30% leaf coverage, no HR, DR score 2 or 2–3 or higher).
DNA sequence determination and analysis:
Gene-specific primers (sequences available upon request) were used for PCR amplification of the target genes. PCR products were purified and sequenced from both strands. DNA sequences were aligned using AlignX function of Vector NTI Suite (Invitrogen) and corrected manually. Amino acid sequences were deduced from the nucleotide sequences by Vector NTI and aligned by AlignX. DnaSP version 4.0 was used for calculation of nucleotide polymorphism and divergence (Jukes-Cantor corrected) (Rozas and Rozas 1999). The molecular evolutionary genetic analysis program version 3.1 (Kumar et al. 2004) was used to generate phylogenetic trees based on nucleotide sequences. Trees generated by neighbor-joining (using Jukes-Cantor distance or p distance), maximum parsimony, or minimum evolution (using Jukes-Cantor distance or p distance) were very similar, and the trees constructed by neighbor-joining were presented.
Estimation of divergence time:
To estimate the divergence time for the resistant and divergent/susceptible RPW8.1 and RPW8.2 alleles, we inferred the synonymous mutation rate for RPW8.2 to be 2.12 × 10−8 per synonymous substitution per site per year, based on the divergence time (T) of 5.3 million years ago (MYA) for the separation of A. lyrata and A. thaliana (Koch and Kiefer 2005) and the synonymous substitution (ds) of 0.2247 between AlRPW8.2 and AtRPW8.2/Ms-0 (Xiao et al. 2004). We then applied this mutation rate to estimate the divergence time for both RPW8.1 and RPW8.2 using the formula ds/2T = synonymous mutation rate.
Neutrality tests:
Neutrality tests were performed using DnaSP version 4.0 (Rozas and Rozas 1999). P-values to obtain the observed Tajima's D and Fu and Li's D and F (Tajima 1989; Fu and Li 1993) were calculated based on 10,000 replicates of coalescent simulations assuming no recombination. Observed test statistics were further tested using empirical distribution in A. thaliana populations (Nordborg et al. 2005). In McDonald and Kreitman's test (McDonald and Kreitman 1991), Col-0 HR4, AlRPW8.2, and AlHR3 were used as outgroups for RPW8.1, RPW8.2, and HR3, respectively. HKA test (Hudson et al. 1987) was performed on 23 accessions from which both RPW8.2 and HR3 were sequenced, using sequences from A. lyrata as an outgroup. We were unable to do the HKA test for RPW8.1 because it is absent from A. lyrata.
Transgene analysis:
The genomic DNA fragments containing the coding sequence of AtRPW8.1 plus 1000 bp upstream of the ATG start codon from Ms-0, Sy-0, Ler, or Ws-0, and AtRPW8.2 plus 1000 bp upstream of the ATG start codon from Ms-0, Can-0, Ler, or Ws-0, and AlRPW8.2 (99m9) were amplified with Pfu-turbo with gene-specific primers and cloned into the binary vector pSMB (Mylne and Botella 1998) under control of the 35S promoter. A genomic fragment containing the AlRPW8.2 coding sequence plus 753 bp upstream of the ATG start codon (which is the whole intergenic region between AlHR3 and AlRPW8.2) from Al99m9 was amplified and cloned into binary vector pBIN19-plus. All these constructs were introduced to Col-gl (Col-0 harboring the glabrous mutation 1). Homozygous transgenic lines were generated and tested for their DR phenotypes in response to E. cichoracearum UCSC1.
Tests of fitness costs:
More than 20 Col-gl lines transgenic for a 6.2-kb genomic fragment from Ms-0 containing both AtRPW8.1 and AtRPW8.2 under control of their native promoters were generated, of which 7 contained a single copy of the transgene. Six of the 7 lines showed no defects in growth and development and no sign of spontaneous HR-like cell death (at least not visible to the naked eye) under normal growth conditions, but had powdery mildew-induced HR and resistance similar to that in Ms-0. The relative mRNA levels of AtRPW8.1 from 3 homozygous lines (i.e., S5, T5, and T7) were measured in comparison with Ms-0 by real-time quantitative RT-PCR using the procedures previously described (Xiao et al. 2003). The locations of the T-DNA transgenes in these 3 lines were determined by thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) (Sessions et al. 2002). The experiments were carried out in two different environments: a growth room and a greenhouse. For the test in a growth room in 2003, only line S5 was used to compare with Col-gl for vegetative growth (dry mass of the rosette leaves of the entire plant) under three conditions: no infection, early (heavy) infection, and late (light) infection by powdery mildew E. cichoracearum UCSC1, and for measuring seed yield in the absence of the pathogen. Plants were cultivated in an autoclaved soil mixture consisting of 2 vol of John Innes compost 3 (Gem Gardening, Lancashire, UK), 2 vol horticultural grit (Gem Gardening), 2 vol peat (Shamrock, Newbridge, Ireland), and 1 vol vermiculate (Vermiperl, Lincolin, UK). The growth conditions were 22°, 65–75% relative humidity (RH), and ∼125 μmol · m−2 · sec−1 light (fluorescent lamps) intensity. Unless otherwise indicated, 2-week-old, short-day grown seedlings were transplanted into round pots (5 cm in diameter) and first kept in short day (8 hr light, 16 hr dark) for 2 weeks and then shifted to long day (16 hr light, 8 hr dark) until sample collection or seed maturation. Plants were irrigated regularly and supplied with fertilizer (1/2 teaspoon of Miracle-Gro in 1 liter of water for 32 pots) twice (at 4 and 6 weeks old) during the entire growth period. These experiments were repeated twice with similar results.
For the experiments in a greenhouse in 2005, three transgenic lines, S5, T5, T7, and Col-gl were used for measuring seed yield in the absence of any powdery mildew pathogens. To assess the effect of the same AtRPW8 transgene in a different genetic background, the AtRPW8 transgene from S5 was introduced to Ler background by backcrossing for five generations. This line (denoted as S5/Ler) was then used for comparison with Ler wild type for seed yield. Seeds were sown in Sunshine Mix 1 soil (Maryland Plant & Suppliers, Baltimore) and cold treated (4° for 2 days) before moving out to 22°, 75% RH, short day (8 hr light at ∼125 μmol · m−2 · sec−1, 16 hr dark). Two weeks later, seedlings were transplanted into 1/2-in. square pots filled with Sunshine Mix 1 soil and kept in the greenhouse under ∼22°, 65–75% RH and natural light conditions. Individual plants from different genotypes were placed in the same trays as randomly as possible. Plants were irrigated once with GNATROL (Greenfire, Sacramento, CA) to control fungus gnats, and were supplied with Miracle-Gro once at the same concentration as used in the growth room. Seeds from individual plants were collected and weighed after maturation.
Other analyses:
Trypan blue staining for cell death (Xiao et al. 2003), mRNA extraction, and RT-PCR (Xiao et al. 2004) were performed as previously described. TAIL-PCR (Sessions et al. 2002) was used to determine the location of the AtRPW8 transgenes in S5, T5-3, and T7-10 lines.
RESULTS
A. lyrata RPW8.2 confers powdery mildew resistance in A. thaliana:
The syntenic RPW8 locus of A. lyrata contains an orthologous gene (AlRPW8.2) of the A. thaliana RPW8.2 (AtRPW8.2) (Xiao et al. 2004). AlRPW8.2 and AtRPW8.2 share 81.2% and 61% sequence identity at the nucleotide and the amino acid level, respectively. We asked whether AlRPW8.2 is a functional powdery mildew R gene in A. lyrata. To address this question, we first challenged six A. lyrata accessions (99m6, 99m7, 99m8, 99m9, 99m11, and 99m23) with four powdery mildew isolates reported in Xiao et al. (2004). All of the six accessions were moderately to highly resistant to these pathogens (see supplemental Table 1 at http://www.genetics.org/supplemental/). We then sequenced the AlRPW8.2 alleles from the six accessions and found they were nearly identical, with only eight silent substitutions (four in the exons and four in the singular intron). Because A. thaliana accession Col-0 lacks AtRPW8.1 and AtRPW8.2, it is ideal for testing if AlRPW8.2 is a functional R gene. We expressed AlRPW8.2 in Col-0 by its native promoter. More than 60% (14 of 22) transgenic lines obtained were resistant to E. cichoracearum UCSC1 (Figure 2A), indicating that AlRPW8.2 is indeed a functional R gene. We also overexpressed AlRPW8.2 by the strong viral 35S promoter in Col-0 and found that 3 of 20 transgenic lines developed severe spontaneous HR-like cell death (SHL) and plants of these 3 lines were tiny in size (Figure 2B). This is reminiscent of the results from overexpression of AtRPW8.1 and AtRPW8.2 by their native promoters (Xiao et al. 2003). Thus, AlRPW8.2 is capable of inducing cell death and powdery mildew resistance in A. thaliana. These results suggest that AlRPW8.2 can contribute to powdery mildew resistance in A. lyrata and that the resistance function of RPW8.2 might have evolved before the speciation of A. lyrata and A. thaliana.
Figure 2.—
AlRPW8.2 confers powdery mildew resistance in Col-0. (A) A 2.3-kb genomic fragment containing the AlRPW8.2 gene and its native promoter was introduced to Col-gl background and the transgenic plants were inoculated with E. cichoracearum USCS1. Disease phenotypes were scored and typical infected leaves were photographed at 10 days post-inoculation (dpi). (B) The AlRPW8.2 gene was expressed under control of the 35S promoter in Col-gl background. Plants of ∼10% (3 of 29) transgenic lines showed SHL and 4-week-old T3 plants of one line with the most severe SHL were shown together with the wild type. Arrows indicate dead or dying leaves. Bar, 1 cm.
Intraspecific genetic variation at RPW8:
Previously we determined the sequences of the RPW8.1 and RPW8.2 alleles from 32 A. thaliana accessions mostly collected from Europe and found that there were two basic haplotypes: Ms-0-like and Col-0-like based on the presence and absence of RPW8.1 and RPW8.2 (Xiao et al. 2004). To systematically investigate the levels of genetic variation at this locus, we further conducted nucleotide sequence analyses on RPW8.1 and RPW8.2 from 51 worldwide samples (the 32 accessions described in Xiao et al. 2004 plus 19 accessions from wider geographic areas, including the USA and Japan; Figure 1). Among the 51 A. thaliana accessions analyzed, 43 contain RPW8.1 and RPW8.2, and the remaining 8 accessions lack RPW8.1 and RPW8.2 but contain HR4.
Genetic variation at RPW8.1:
The 43 RPW8.1 alleles have the same overall gene structure (two exons split by a single intron) and they all encode full-length proteins. The length of the complete alignment of the 43 alleles was 718 nucleotides, including 281 bp for the first exon, 208 bp for the intron, and 229 bp for the second exon. As shown in Table 1 and Figure 3, there were 31 nucleotide segregating sites, of which 9 were singletons. Among the segregating sites, the number of sites causing nonsynonymous substitutions was 15. The average nucleotide diversity (π = 0.012) and the number of segregating sites per base pair (S = 0.047) of RPW8.1 were close to the average values derived from a set of 27 NBS-LRR R genes recently surveyed (Bakker et al. 2006), which were both significantly higher than the empirical distribution of polymorphism in 876 randomly distributed genomic regions (Nordborg et al. 2005). The 43 RPW8.1 alleles distinguished 17 haplotypes and encoded 12 distinct proteins (Figure 3).
TABLE 1.
Nucleotide polymorphism and divergence at the RPW8 locus of Arabidopsis thaliana
| Gene (no. of alleles) | Parameter | Entire gene | Coding region | Synonymous | Nonsynonymous |
|---|---|---|---|---|---|
| RPW8.1 (43) | No. of sitesa | 644 | 444 | 103.29 | 340.71 |
| Segregating sites | 31 | 24 | 9 | 15 | |
| π | 0.0124 | 0.0127 | 0.0141 | 0.0124 | |
| E[π]b | 0.0147 | 0.0151 | 0.0167 | 0.0147 | |
| Tajima's D | 0.37 | 0.04 | |||
| RPW8.2 (43) | No. of sitesa | 652 | 519 | 114.64 | 404.36 |
| Segregating sites | 33 | 32 | 7 | 25 | |
| π | 0.0099 | 0.0118 | 0.0125 | 0.0116 | |
| E[π]b | 0.0118 | 0.0140 | 0.0148 | 0.0138 | |
| Tajima's D | −0.63 | −0.28 | |||
| HR4 (8) | No. of sites | 848 | 555 | 116.42 | 438.58 |
| Segregating sites | 4 | 3 | 0 | 2 | |
| π | 0.0020 | 0.0023 | 0.0000 | 0.0030 | |
| E[π]b | 0.0129 | 0.0147 | 0.0000 | 0.0188 | |
| Tajima's D | 0.48 | 0.46 | |||
| HR3 (26) | No. of sitesa | 1004 | 639 | 136.13 | 502.87 |
| Segregating sites | 92 | 9 | 4 | 5 | |
| π | 0.01 88 | 0.0021 | 0.0060 | 0.0010 | |
| Tajima's D | −0.94 | −1.41 |
Excluding all gaps; π, the average nucleotide diversity.
Expected average nucleotide diversity was calculated on the basis of allele frequencies (Innan and Tajima 1997).
Figure 3.—
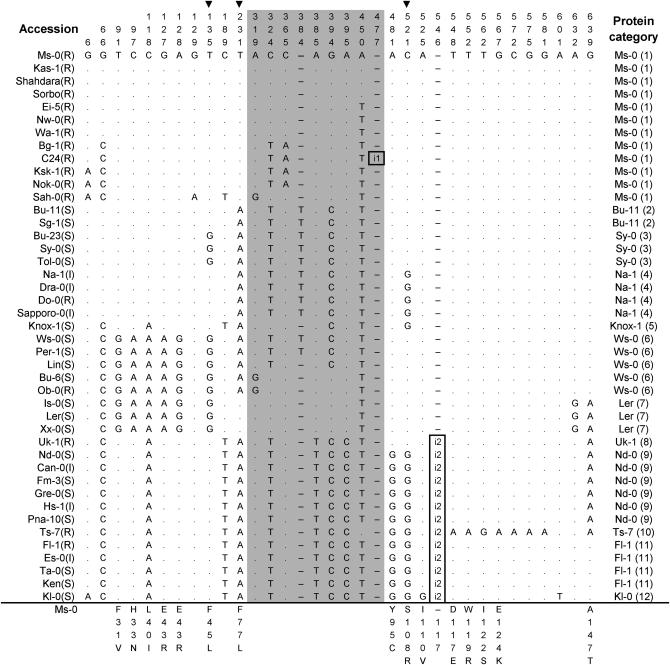
Polymorphic sites of RPW8.1 aligned against the Ms-0 RPW8.1 allele (GenBank accession no. AF273059). A dot indicates an identical nucleotide and a dash indicates a gap. Shaded are the substitutions in the intron. The numbers at the top indicate the nucleotide position relative to the start codon of the Ms-0 alleles. Amino acid replacements caused by nucleotide substitutions are indicated at the bottom. The arrowhead at the top indicates the nonsynonymous substitution that may affect functionality of the proteins (site that showed statistically significant association with the DR phenotype at the 5% level after Bonferroni correction). The disease phenotypes (R, resistant; I, intermediate; S, susceptible) are indicated in the parentheses after the names of the accessions. Identical deduced proteins are represented by one allele only and the numbers in the parentheses indicate the group numbers. Boxed are two insertions: i1, an insertion of 10 bp (GTTTATCTTT); and i2, an insertion of 63 bp (GATCAATGGGACGATATCAAAGAAATCAAGGCCAAGATATCTGAAACGGACACTAAACTTGCT), which was an intragenic duplication (from nucleotide 546–608) and resulted in an insertion of 21 amino acids (DQWDDIKEIKAKISETDTKLA).
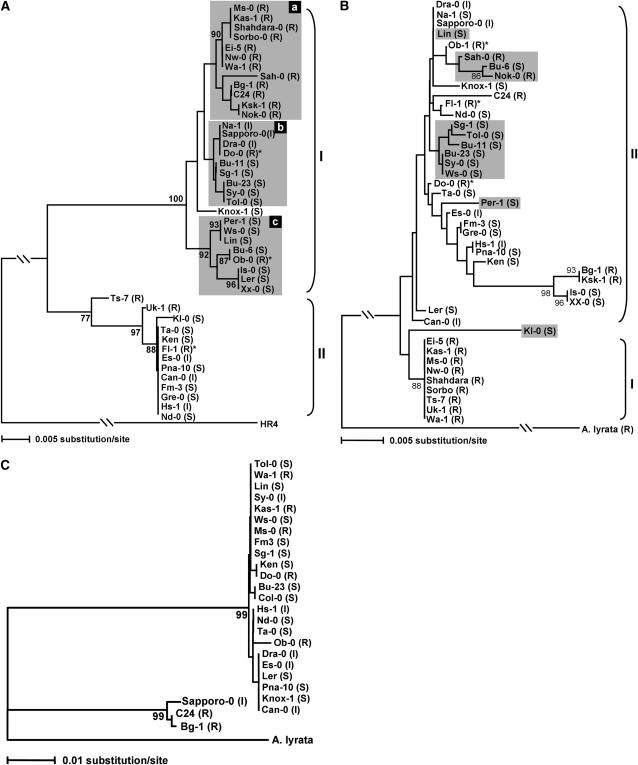
As shown in the phylogenetic tree based on the nucleotide polymorphism in the entire genes (Figure 4A), these alleles can be divided into two major clades. Clade I comprises 30 alleles that can be further divided into three groups (a–c). Group a contains 12 resistant alleles that encode proteins identical to RPW8.1 of Ms-0 from which the RPW8 locus was isolated (Xiao et al. 2001). Group b contains 9 alleles that are similar to the Ms-0-(like) alleles, with one to three amino acid replacements. Among the nine accessions from which the 9 alleles are derived, one was resistant to powdery mildew, three had an intermediate phenotype, and five were susceptible (Table 2, Figure 4A). Group c contains 8 alleles carrying five unique nonsynonymous substitutions in the first exon, and all the accessions except Ob-0 were fully susceptible, suggesting that these are nonfunctional alleles and that resistance of Ob-0 may be conferred by RPW8.2/Ob-0 or other R loci. Clade II contains 13 alleles all characterized by a 63-bp intragenic duplication at the nucleotide position 546, which resulted in an insertion of 21 amino acids in the C-terminal tails of the proteins (Figure 3). Seven of the accessions were susceptible, and resistance of Uk-1 and Ts-7 can be attributed to the presence of the functional RPW8.2 alleles (see later text). The associations between these clades and their DR phenotypes were highly significant (χ2 = 26.6, P < 0.0001), confirming the functional significance of RPW8.1 to the powdery mildew resistance in A. thaliana. The minimum number of recombination events (Hudson and Kaplan 1985) among the 43 alleles detected was eight. The Knox-1 allele might have resulted from a recombination between an Ms-0-like allele (such as Na-1) in Clade Ib, and any allele in Clade II (Figure 3).
Figure 4.—
Neighbor-joining tree of 43 RPW8.1 alleles (A), RPW8.2 alleles (B), and 26 HR3 alleles (C) constructed on the basis of nucleotide sequences of the entire genes by MEGA3.1 using Jukes-Cantor or p distance. Bootstrap proportions of 500 bootstrap replicates >70 are indicated under the branches. Alleles with an asterisk may not be responsible for the resistance (R) phenotype. (A) The RPW8.1 tree was rooted using HR4, which shows the highest similarity with RPW8.1. Shaded areas highlight the subgroups in Clade I. (B) The RPW8.2 tree was rooted using AlRPW8.2. Shaded alleles contain insertions or deletions that resulted in frameshifts. (C) The HR3 tree was rooted using AlHR3.
TABLE 2.
Genotype-phenotype relationship concerning the RPW8 locus and powdery mildew resistance
| Genotype at RPW8.1
|
Genotype at RPW8.2
|
||||||
|---|---|---|---|---|---|---|---|
| Phenotypic class | Accession no. | Clade-Ia | Clade-Ib | Clade-Ica | Clade-II | Clade-I | Clade-II |
| Resistant | 17 | 12 | 1 | 1 | 3 | 9 | 8 |
| Intermediate | 6 | 0 | 3 | 0 | 3 | 0 | 6 |
| Susceptible | 20 | 0 | 5 | 8 | 7 | 0 | 20 |
RPW8.1/Knox-1 is not subgrouped (Figure 4A) but is added to Clade-Ic for simplicity.
Genetic variation at RPW8.2:
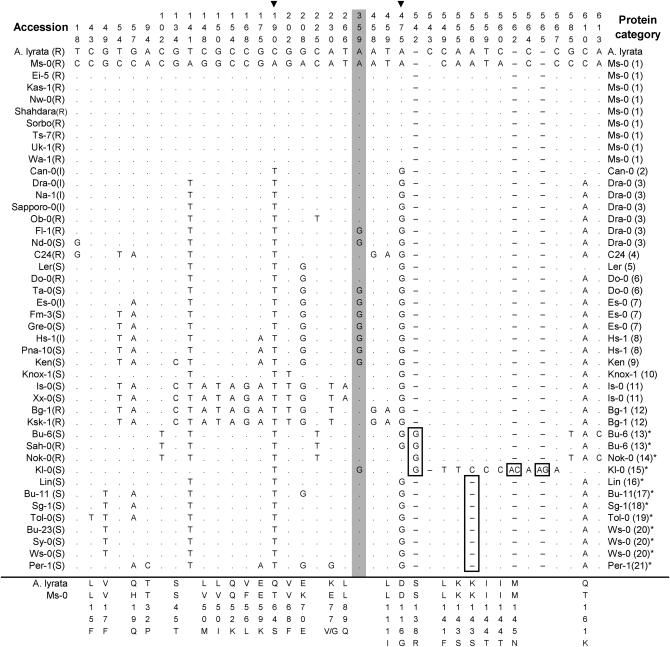
The level of genetic variation at RPW8.2 was similar to that at RPW8.1. All the 43 RPW8.2 alleles had a similar overall gene structure. The length of the complete alignment of the 43 alleles was 653 nucleotides, including 296 bp for the first exon, 128 bp for the intron and 229 bp for the second exon. As shown in Table 1 and Figure 5, there were 33 nucleotide polymorphic sites (excluding four indels), of which 9 were singletons. Among the 25 segregating sites in the coding region, 18 were nonsynonymous substitutions, whereas only 7 were synonymous substitutions. The average nucleotide diversity (π = 0.010) and the number of segregating sites per base pair (S = 0.048) of RPW8.2 is also close to that of RPW8.1. The 43 RPW8.2 alleles distinguish 26 haplotypes, which encode 21 different proteins. Except for the RPW8.2/Kl-0 allele, the remaining 42 alleles can be divided into two clades in the phylogenetic tree (Figure 4B), even though the division of the two clades is not as reliable as in the RPW8.1 tree. Clade I comprises 9 alleles identical to RPW8.2/Ms-0, and all of the nine accessions carrying the Clade I allele were resistant. Clade II comprises 33 alleles that differ from RPW8.2/Ms-0 by 2 (e.g., RPW8.2/Can-0) to 14 (e.g., RPW8.2/Is-0) nonsynonymous substitutions (Figure 5). The majority (20 out of 34) of the accessions that carry the Clade II RPW8.2 alleles were susceptible, indicating that these alleles are less functional or nonfunctional (Table 2). The associations between the clades in RPW8.2 and the DR phenotypes were highly significant (χ2 = 17.4, P = 0.0002). The RPW8.2/Kl-0 allele differs from all other alleles by two small indels and 4 singleton nonsynonymous substitutions in the second exon. Twelve alleles (shaded in Figure 4B) including RPW8.2/Kl-0 contain a single base pair indel (at the nucleotide position 542 or 556) that resulted in frameshift and a truncation of 28–34 amino acids at the C-termini of the deduced proteins. The minimum number of recombination events (Hudson and Kaplan 1985) among the 43 alleles was eight.
Figure 5.—
Polymorphic sites of RPW8.2 aligned against the Ms-0 RPW8.2 allele (GenBank accession no. AF273059). This figure was made in the same way as Figure 3 except that the AlRPW8.2 sequence was used as an ancestral reference. Proteins with * have 29–34 aa deletion at the C-termini due to early stop codons introduced by the single nucleotide insertion (boxed G) or deletion (boxed —).
Genetic variation at HR4 (At3g50480):
Eight of the 51 A. thaliana accessions (15.7%, accessions Fr-3, Is-1, Nie-0, Sg-2, Sh-0, Wc-1, Wt-4, and Col-0) contain HR4 in the place of RPW8.1 and RPW8.2. They were generally more susceptible to the powdery mildew isolates (Xiao et al. 2004). Sequence analysis revealed that there were only two alleles among the 8 accessions, and the average genetic diversity was low (π = 0.002). The low level of genetic variation in HR4 was expected a priori, because of the small fraction of allelic class in the populations (Innan and Tajima 1997). The expected nucleotide diversity at the HR4 (0.013 = 0.002 × 51/8), based on the allele frequency (Innan and Tajima 1997), was close to that at RPW8.1 (0.015) and RPW8.2 (0.012, Table 1), suggesting that there is no recent selective sweep among these loci. Nie-0 is identical to Col-0, while the remaining six alleles are identical to each other, differing from the Col-0 allele by two nonsynonymous substitutions (GCol-0 145A and T161A, resulting in E49K and V54E, respectively), one silent (C567T) substitution, and a deletion of 42 bp (GATACAAGTCGACCAATGGACCGATATCAAAGAAATGAAGGC), which is exactly the third copy of the five tandem duplicated segments in the 3′ end of HR4/Col-0 (Xiao et al. 2004). This suggests that an unequal intragenic recombination occurred between the Col-0-like alleles. Interestingly, unlike RPW8.1- and RPW8.2-containing accessions that are distributed throughout the surveyed geographical areas, all the HR4-containing accessions are from Germany except for Col-0 (annotated as from USA; however, see discussion) (Figure 1).
Genetic variation at HR3 (At3g50470):
Based on PCR amplification, all of the 51 accessions surveyed contain HR3 (data not shown). For comparisons, we sequenced the HR3 alleles from 26 out of 51 accessions. The HR3 allelic sequences showed a very low level of nucleotide diversity in the coding region (π = 0.002). Twenty two of the 26 alleles encode identical proteins, and the remaining 4 alleles encode proteins that differ from the others by only one or two amino acid replacements (Figure 4C and supplemental Figure 1 at http://www.genetics.org/supplemental/), indicating that HR3 is highly conserved. This is consistent with purifying selection inferred from a low ratio (0.17) of nonsynonymous substitution rate (Ka) and synonymous substitution rate (Ks) between AlHR3 and AtHR3 (Xiao et al. 2004). There are two types of introns among the 26 alleles (type I and type II, see supplemental Figure 1). These introns shared only ∼67% homology between each other and ∼70% to the intron of AlHR3. The highly divergent nucleotide sequences between two types of intron suggest relaxed selective constraint in this region and an old common ancestor between alleles carrying different types of intron in HR3. There was no significant association between these haplotypes and their DR phenotypes (χ2 = 4.8, P = 0.09).
Correlation between sequences and phenotypes:
Alleles of RPW8.1 and RPW8.2 in the resistant accessions are not scattered throughout the genealogies, but rather are grouped together (see above), suggesting a correlation between functionality of the two RPW8 genes and powdery mildew resistance phenotype. However, assessing the functionality of the individual RPW8 alleles is difficult because both RPW8.1 and RPW8.2 could function independently and probably additively to confer non-race-specific resistance to powdery mildew (Xiao et al. 2001; S. Xiao, unpublished data). Based on our classification (see materials and methods), 17 of 51 accessions surveyed were resistant to E. cichoracearum (Ec)-UCSC1, 6 were intermediate, and 28 were susceptible. As shown in Figures 3 and 5, 7 of the 17 resistant accessions (Ms-0, Shahdara, Sorbo, Kas-1, Ei-5, Nw-0, and Wa-1) contained nearly identical RPW8.1 (only one silent substitution A450T in the intron in Ei-5, Nw-0, and Wa-1) and identical RPW8.2 alleles. Thus, they are identical to Ms-0 in terms of the RPW8.1 and RPW8.2 protein sequences. Five accessions (i.e., C24, Sah-0, Ksk-1, Nok-0, and Bg-1) contained RPW8.1 alleles that differ from the Ms-0 alleles in four to six silent substitutions in the exon and the intron but encode identical proteins, while their RPW8.2 alleles were divergent from the Ms-0 allele at both the nucleotide and amino acid levels. Two resistant accessions (Uk-1 and Ts-7) contained RPW8.2 alleles identical to those of Ms-0, but their RPW8.1 alleles were divergent. Therefore, the DR phenotypes of these 14 accessions could be at least in part attributable to the presence of the functional RPW8.1 and/or RPW8.2 alleles. The remaining 3 resistant accessions (Do-0, Ob-0, and Fl-1) contain divergent RPW8.1 and RPW8.2 alleles, suggesting that either these alleles may also be functional or resistance in these 3 accessions is controlled by genes that differ from the RPW8 genes.
Figure 7.—
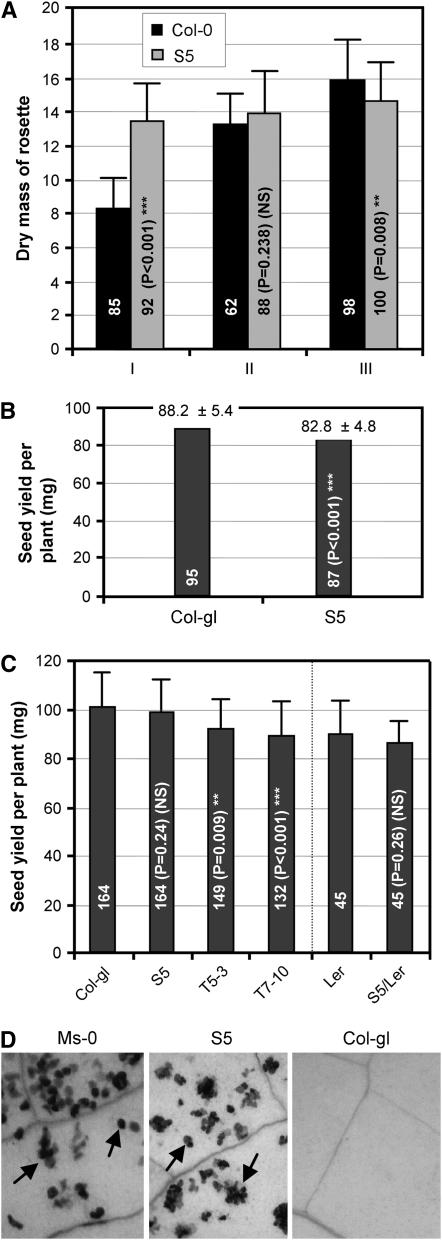
Fitness cost associated with RPW8. The experiments were carried out under two environmental conditions: a growth room (A and B) and a greenhouse (C). The number of plants of an indicated genotype used in these experiments was shown inside the shaded column and the P-value from the paired (wild type vs. a transgenic line) Student's t-test is also shown inside the column. *Significant at P < 0.05; **significant at P < 0.01; ***significant at P < 0.001; NS, not significant. For detailed experimental conditions, see materials and methods. (A) Arabidopsis seedlings were inoculated with E. cichoracearum UCSC1 when they were 2 weeks old (I) or 4 weeks old (II) or remained uninoculated (III). Plants were shifted from short day (8 hr light) to long day (16 hr light) when they were 3 weeks old and all rosette leaves were sampled for measuring dry mass at 6 weeks old. (B) Seedlings of Col-gl and S5 were shifted from short day to long day at 3 weeks old and were maintained in long day until seed maturation in the absence of the pathogen. (C) Seedlings from indicated genotypes were grown in a growth chamber under short day for 2 weeks and then transferred to a greenhouse for further growth until seed maturation. (D) Leaves of 6-week-old, long-day grown plants were stained with trypan blue for cell death. Arrows indicate single and clustered mesophyll cells.
Also as shown in Figures 3 and 5, among the 6 accessions with an intermediate DR phenotype, 3 (Dra-0, Na-1, and Sapporo-0) contained identical RPW8.1 and RPW8.2 alleles that differ from those of Ms-0 alleles by only two amino acids, FMs-077L (nucleotide/amino acid of Ms-0 is placed first here in all polymorphic sites) and S108R, for RPW8.1 and three amino acids, T64S, D116G, and T161K, for RPW8.2 at the protein level. Accession Can-0 contained an RPW8.2 allele that differs from the Ms-0 allele by only two amino acids (T64S and D116G) and a more divergent RPW8.1 allele. The sequence and phenotypic data together suggest that these slightly different RPW8 alleles may be (partially) functional. Accessions Hs-1 and Es-0 contained divergent RPW8.1 and RPW8.2 alleles carrying four and five nonsynonymous substitutions that were also present in 7 and 10 susceptible accessions, respectively, implying that the RPW8 locus may not be responsible for the intermediate phenotypes of the 2 accessions.
Among the 28 susceptible accessions, 5 (Bu-11, Bu-23, Sg-1, Sy-0, and Tol-0) contain RPW8.1 alleles with only one (F77L) or two amino acid replacements (F45L and F77L), implying that either these two residues are critical to the resistance function, or possibly these (partially) functional alleles are not sufficiently expressed. Fifteen accessions either contained more divergent RPW8.1 and RPW8.2 alleles with three or more nonsynonymous substitutions and/or indels that resulted in an insertion of 21 amino acids in RPW8.1 or a truncation of ∼30 amino acids at the C-terminus of RPW8.2. The extreme susceptibility of the remaining 8 accessions was associated with the absence of RPW8.1 and RPW8.2 and the presence of HR4 (Figure 1) (Xiao et al. 2004). Since HR4 showed the highest homology to RPW8.1 (72.5%; the homology between HR4-RPW8.2 is only 50%) and shared two unique indels with RPW8.1 that are not present in other members of the RPW8 gene family (Figure 2 in Xiao et al. 2004), it is reasonable to consider HR4 and RPW8.1 to be orthologous, albeit not typical. Thus, there are two basic haplotypes at the RPW8 locus, RPW8.1- (and RPW8.2-) containing and HR4-containing haplotypes, and the sequence diversity at the RPW8 locus can account for a major (∼80%) proportion but not all of the phenotypic variations among the 51 A. thaliana accessions (Table 2).
Neutrality tests:
Based on the nucleotide sequence polymorphisms at the RPW8 locus, we applied several statistical tests to examine the selective neutrality (see materials and methods for details). They include tests based on allele frequencies (Tajima 1989; Fu and Li 1993), polymorphism level differences between loci [HKA test; (Hudson et al. 1987)], the distribution of synonymous and nonsynonymous polymorphism, and divergence (McDonald and Kreitman 1991). Although none of the tests detected statistical significance for natural selection at the 5% level in RPW8.1, RPW8.2, and HR3 (data not shown), Tajima's D value (Tajima 1989) was negative and the lowest in the coding sequences of HR3 (D = −1.41, Table 1), agreeing with a previous study that suggested purifying selection at HR3 (Xiao et al. 2004). In contrast, coding sequences of RPW8.1 and RPW8.2 showed no sign of purifying selection or selective sweep (D ∼−0.28–0.04, Table 1).
To further test the selective neutrality on the maintenance of RPW8, we applied the Tajima's and Fu and Li's tests to the nucleotide sequence alignment including both the 43 RPW8.1 alleles and 8 HR4 alleles, assuming that these two genes are orthologous (Xiao et al. 2004). The average genetic divergence [Jukes-Cantor corrected DXY, (Nei 1987)] between RPW8.1 and HR4 alleles was 0.29. Although Tajima's D did not detect statistical significance (D = 0.85), Fu & Li's D* and F* were both positive (D* = 2.08, F* = 1.93) and showed statistical significance by both the coalescent simulation and the empirical distribution (D*: P < 0.001; F*: P < 0.05, see materials and methods). These results suggest that if HR4 and RPW8.1 have been maintained as orthologous alleles in A. thaliana populations, their genetic differences cannot be explained by the simplest neutral mutation model, and that balancing selection may be acting on these two haplotypes.
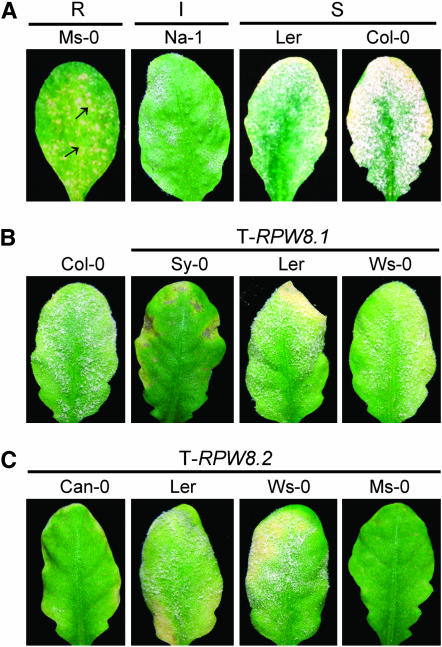
RPW8.1/Sy-0 and RPW8.2/Can-0 alleles confer enhanced resistance when overexpressed:
To obtain genetic evidence to support the idea that some alleles slightly divergent from the Ms-0 alleles at the protein level may be (partially) functional, we cloned three Clade I RPW8.1 alleles [Sy-0 (Clade Ib), Ler (Clade Ic), and Ws-0 (Clade Ic)] and three Clade II RPW8.2 alleles (Can-0, Ler, Ws-0 = Sy-0) by placing the genomic fragments containing the genomic sequence of the genes and 1000-bp native promoter (Np) sequence upstream of their start codons downstream of the 35S promoter to enhance the expression of the transgenes. We found that ∼25% (14/57) of the transgenic lines containing 35S∷Np-RPW8.1/Sy-0 and ∼16% (5/32) of the transgenic lines containing 35S∷Np-RPW8.2/Can-0 exhibited enhanced resistance to Ec-UCSC1 with a DR score of 0–1 to 1–2 and a slower HR (Figure 6, B and C), slightly less resistant compared to plants expressing the corresponding Ms-0 alleles. These results indicate that RPW8.1/Sy-0 and RPW8.2/Can-0 are at least partially functional. However, none of transgenic lines (>30 for each construct) containing 35S∷Np-RPW8.1/Ler, 35S∷Np-RPW8.1/Ws-0, 35∷Np-RPW8.2/Ler, or 35S∷Np-RPW8.2/Ws-0 examined showed obviously enhanced resistance (Figure 6, B and C), suggesting that these alleles are not functional. It is worth noting that RPW8.1/Sy-0 differed from RPW8.1/Ms-0 by two amino acids (F45L and F77L), both of which showed statistically significant associations with the DR phenotype (Figure 3). RPW8.2/Can-0 differed from RPW8.2/Ms-0 by two amino acids (T64S and D116G), and these sites were also significantly associated with the DR phenotype (Figure 5). These results suggest that these four sites may be important for full function of RPW8.1 and RPW8.2 but additional, functionally relevant amino acid replacements are required for nullification of the functions of these two proteins.
Figure 6.—
Some RPW8 alleles are (partially) functional. Six-week-old plants were inoculated with E. cichoracearum USCS1 and a typical infected leaf from each indicated genotype was photographed at 12 dpi and presented. (A) Disease reaction phenotypes of four representative A. thaliana accessions. R, resistant; I, intermediate; S, susceptible. Arrows indicate HR lesions. (B and C) Genomic sequences of RPW8.1 (B) or RPW8.2 (C) including its native promoter (1 kb upstream of the ATG start codon) from indicated accessions were placed downstream of the 35S promoter and the constructs were introduced into Col-gl. T3 homozygous lines were tested with the pathogen together with Col-gl wild type.
Expression of RPW8.1 and RPW8.2 incurs fitness costs:
In the above analyses, we detected a general association between the genotypes at RPW8.1 and/or RPW8.2 and the DR phenotypes of the accessions surveyed. This was based on the assumption that RPW8.1 and RPW8.2 are expressed in all the accessions. To provide evidence for this assumption, we examined the expression of RPW8.1 and RPW8.2 in 14 accessions along with Ms-0 and Col-gl transgenic for RPW8 (line S5; see below) by RT-PCR and found that all had expression of both RPW8.1 and RPW8.2 (see supplemental Figure 2 at http://www.genetics.org/supplemental/).
The high levels of genetic and phenotypic variations in RPW8 invite an important question: why have so many divergent alleles been maintained in the A. thaliana populations? We hypothesized that expression of the RPW8 functional genes renders fitness benefits to plants when infected by the pathogens; however, it may incur fitness costs. Hence, in the absence of the pathogens, natural selection may favor accumulation of deleterious mutations in or complete elimination of the two genes. We took a transgene approach to test this idea of fitness costs associated with RPW8 expression. We generated >30 Col-gl lines transgenic for a 6.2-kb genomic fragment containing both RPW8.1 and RPW8.2 and their promoters from Ms-0 and selected three lines (S5, T7-10, and T5-3) that were homozygous for a single copy transgene and did not develop SHL in soil. We then determined the locations of the T-DNA by TAIL-PCR (Sessions et al. 2002). We found that the transgenes in S5 and in T7-10 were inserted in intergenic regions and the transgene in T5-3 was inserted in the intron of At1g43970, an expressed gene with unknown function (see supplemental Figure 3 at http://www.genetics.org/supplemental/). Compared with Col-gl, these three transgenic lines showed no noticeable developmental defects and more importantly 5-week-old plants grown on MS-agar medium in which RPW8 expression is suppressed (Xiao et al. 2003) had no difference in the amount of vegetative growth (data not shown). We then measured their mRNA levels of RPW8.1 relative to that in Ms-0 by quantitative RT-PCR. The mRNA levels of RPW8.1 in the three transgenic lines were about the same as in Ms-0 in 3-week-old seedlings, and were ∼2.5–4 times of that in Ms-0 in the 6-week-old plants (see supplemental Figure 3B), presumably due to the self-transcriptional amplification nature of the RPW8 genes in mature plants (Xiao et al. 2003).
Experiments for measuring likely fitness costs of RPW8-expression were done under two environmental conditions. We first compared Col-gl and Col-gl transgenic line S5 for plant vegetative growth and seed yield in Ec-UCSC1-inoculated and -uninoculated plants in a growth room. For plants inoculated at an early stage (2 weeks old), the transgenic plants were performing much better at the time (6 weeks old) when the rosette leaves were sampled for measuring dry mass (see supplemental Figure 4A at http://www.genetics.org/supplemental/). On average, S5 plants had ∼40% more dry mass than Col-gl plants (P < 0.001 by Student's t-test, Figure 7A), indicating fitness benefits of RPW8-conferred resistance. For those inoculated at 4 weeks old, there was no significant difference in plant size and dry mass (P = 0.238, Figure 7A; supplemental Figure 4B), implying a balance between benefits and costs conferred by RPW8 upon pathogen infection at this timing point. However, for uninoculated plants, S5 plants were slightly smaller in size, had 7.5% less dry mass (P = 0.008, Figure 7A, supplemental Figure 4C) and 6.1% lower seed yield compared with Col-gl plants (P < 0.001, Figure 7B). To consolidate these results, we carried out a larger-scale experiment for measuring seed yield in a greenhouse. Besides using S5, T5-3, and T10-7 transgenic lines, we also introduced the RPW8 transgene from S5 to Ler background, which probably contains nonfunctional RPW8.1 and RPW8.2 alleles (Figures 3, 5, and 6, B and C), by backcrossing for five generations and then the homozygous line S5/Ler was compared to Ler wild-type plants. As shown in Figure 7C, S5 and S5/Ler had slightly lower seed yield compared to Col-gl and Ler wild types, respectively, but the difference was not statistically significant (P = 0.24). However, the average seed yield of T5-3 and T7-10 was 92.2% and 85.3% of that of Col-gl, respectively, and these differences were statistically significant (P < 0.01, Figure 7C), suggesting that T5-3 and T7-10 plants had reduced fitness that may be attributable to the expression of the RPW8 genes. The diminished difference in seed yield between S5 and Col-gl in the latter experiment could be due to differences in overall growth conditions (see discussion).
We previously observed that enhanced expression of RPW8 leads to spontaneous cell death lesions and constitutive activation of defense gene expression (Xiao et al. 2003). One probable cause of fitness costs in RPW8-expressing plants is the constitutive activation of cell death and defense under certain environmental conditions. To test this, we examined the leaves of >20 6-week-old plants of Ms-0, S5, and Col-gl grown in long-day (16 hr light, 8 hr dark) conditions for sign of cell death by trypan blue staining and for expression of PR-1, a reporter gene for defense activation. We detected death of individual as well as clustered mesophyll cells in mature leaves of Ms-0 and S5 plants, but not in leaves of Col-gl plants (Figure 7D). We also found that the mRNA levels of PR-1 in Ms-0 and S5 were 3.5 and 4.0 times higher, respectively, than that of Col-gl (data not shown). We did not, however, observe cell death and difference in PR-1 expression in plants of these three genotypes grown in short day (8 hr light and 16 hr dark) or on MS-agar medium during the same time frame (Xiao et al. 2003; data not shown). This, together with our earlier observations (Xiao et al. 2003, 2005) indicates that fitness costs of RPW8 expression may be attributable to the energy consumption caused by constitutive activation of a defense-related cell death pathway by RPW8 in the absence of pathogens under permissive conditions, such as long day in soil.
DISCUSSION
In this study, we made two major observations regarding the maintenance of a unique R gene locus in A. thaliana. First, we found relatively high levels of genetic variation at the RPW8 locus, suggesting no recent selective sweep for RPW8. Second, we found that gene expression of RPW8 has both benefits and costs on the fitness of individuals, depending on the presence and absence of the pathogens. These results suggest that polymorphisms at the RPW8 locus in A. thaliana may have been shaped and maintained by complex selective forces, including those from the fitness benefits and costs associated with RPW8.
Genetic variation at the RPW8 locus:
RPW8 represents a complex R gene locus identified in A. thaliana. The relatively high genetic diversity in the RPW8.1 and RPW8.2 coding regions (π = ∼0.01; Table 1) is in sharp contrast to that in the coding regions of HR3 (π = 0.002), the presumable progenitor of the RPW8 gene family located at the same genomic region (Xiao et al. 2004). This result is consistent with our previous study, in which a strong selective constraint on HR3 was suggested between A. lyrata and A. thaliana (Xiao et al. 2004). Within A. thaliana, the HR3 alleles are highly conserved in amino acid sequence (see supplemental Figure 1 at http://www.genetics.org/supplemental/). For the coding region of HR3, the ratio of πnon (average nucleotide diversity at nonsynonymous sites) to πsyn (average nucleotide diversity at synonymous sites) is only 0.174 (πnon = 0.001 and πsyn = 0.006, Table 1), confirming the strong selective constraint on the amino acid changes in HR3. In RPW8.1 and RPW8.2, on the other hand, πsyn is about two times larger than πsyn in HR3, whereas πnon is >10 times larger in both RPW8.1 and RPW8.2 than in πnon HR3 (Table 1). The ratios of πnon:πsyn are 0.880 in RPW8.1 and 0.929 in RPW8.2, suggesting that the amino acid changes in RPW8 are relatively free from selective constraints in A. thaliana. It is unclear why the HR3 nonfunctional gene with respect to powdery mildew resistance is under the strong selective constraint but the functional RPW8 genes are not in A. thaliana. Our preliminary data indicate that HR3 might be involved in the basal defense mechanism of A. thaliana, which may explain the selective constraint on HR3 (S. Xiao, unpublished data). At the RPW8 locus, the functional redundancy due to gene duplication might, in part, be responsible for the high genetic variation in A. thaliana.
Evolution of the RPW8.2 resistance function:
Inferring the time when RPW8.1 gained resistance function is difficult, because RPW8.1 is absent from A. lyrata, presumably due to a deletion event (Xiao et al. 2004). The presence of an ortholog of AtRPW8.2 in A. lyrata provides an opportunity to address such a question regarding RPW8.2. Our demonstration that AlRPW8.2 confers powdery mildew resistance when expressed by the native promoter (Figure 2) suggests that AlRPW8.2 may be functionally equivalent to AtRPW8.2. There are two likely scenarios for the evolutionary history of the RPW8.2 function: (i) RPW8.2 gained resistance function before the divergence of A. lyrata and A. thaliana, or (ii) AlRPW8.2 and AtRPW8.2 evolved resistance function independently after the speciation by convergent evolution. If the first scenario is true, functional AlRPW8.2 and AtRPW8.2 alleles would more likely be conserved at those sites that encode amino acids that are critical for resistance. Sequence alignment (Figure 5) showed that AlRPW8.2 possesses the same nucleotides as the nine functional AtRPW8.2 alleles at 17 (including 12 nonsynonymous) sites, whereas AlRPW8.2 possesses the same nucleotide at only 3 sites (including 2 nonsynonymous) with the remaining (likely) susceptible AtRPW8.2 alleles in Clade II (Figure 4B). At the remaining 5 sites, AlRPW8.2 differs from both. Thus, AlRPW8.2 is in general more similar to the resistant AtRPW8.2 alleles at the sites important for function. Based on this inference, we favor the first scenario, that RPW8.2 evolved the resistance function before the speciation. The eight amino acid changes (V17F, S45T, Q52K, E59K, V68F, L89Q, L111I, and D116G) between the functional and likely susceptible AtRPW8.2 alleles but conserved between AlRPW8.2 and AtRPW8.2/Ms-0 may be important for the resistance function of RPW8.2 (Figure 5). Among those, the D116G mutation is especially interesting, since the change from aspartic acid (D) to glycine (G) happened in all but two divergent alleles that encode truncated, thus most likely nonfunctional, proteins. Indeed, association test suggested statistically significant correlation between the DR phenotypes and alleles that encode different amino acids at this site (Figure 5). Therefore, the aspartic acid residue is probably important for full function of RPW8.2. However, since overexpression of RPW8.2/Can-0, which harbors D116G and an additional mutation (T64S), resulted in enhanced resistance (Figure 6C), the D116G mutation may not completely abolish RPW8.2 resistance function; rather it may affect RPW8.2 function in an incremental manner. A similar situation was found for the F77L mutation in RPW8.1 (Figure 3). Future site-directed in vitro mutagenesis should help assess functional importance of this aspartic acid and other residues encoded by the segregating sites.
Functional and genetic divergence of RPW8 in A. thaliana:
Classifying a particular allele into resistant (functional) or susceptible (nonfunctional) is difficult for RPW8.1 and RPW8.2, because they could function independently (Xiao et al. 2001) and there may exist other powdery mildew R genes unlinked to the RPW8 locus (Adam and Somerville 1996; Xiao et al. 1997). While we could infer that alleles encoding identical proteins as the Ms-0 alleles are resistant alleles and alleles from susceptible accession are most likely susceptible, we were not sure whether alleles that are derived from three resistance accessions (Do-0, Fl-1, and Ob-0) and six moderately resistant accessions (Can-0, Dra-0, Es-0, Hs-1, Na-1, and Sapporo) are resistant or susceptible alleles as they are divergent from those of Ms-0 at the protein level. Despite this ambiguity, we noticed several features of the genetic variations at RPW8. First, we found that resistant alleles of both RPW8.1 and RPW8.2 tend to cluster together in the RPW8.1-Clade Ia and the RPW8.2-Clade I (Table 2, Figure 4, A and B), respectively, and that the (likely) susceptible alleles are more diversified, implying that (i) there are more sequence constraints on the functional RPW8 genes, and (ii) resistant and susceptible alleles have been separated for a relatively long time. Similar observation was made for RPS2 and interpreted as an indication of long-time maintenance of the resistant and susceptible alleles in the populations (Caicedo et al. 1999; Mauricio et al. 2003). We estimated the average divergence time of the resistant vs. susceptible/divergent alleles to be on average 0.5 MYA for RPW8.1 and 0.4 MYA for RPW8.2 (see materials and methods), indeed reflecting the relatively long evolutionary time for the maintenance of the resistant/susceptible alleles at the RPW8 locus.
The relatively long-time maintenance of the RPW8.1 and RPW8.2 alleles may also be reflected by the worldwide distribution of the accessions carrying RPW8 (Figure 1). The HR4-containing accessions, on the other hand, are exclusively from German populations except for Col-0 from USA. Among the 22 accessions from Germany (Figure 1), 7 (32%) are the HR4-containing, and only 6 (27%) are resistant to powdery mildew pathogens. These proportions are contrasting to 3% (1/29) and 62% (18/29), respectively, among the other populations when intermediate phenotype is considered as resistant phenotype. The deviations of German populations from the rest were statistically significant (P = 0.014 and 0.0058, respectively, by the Chi-square test of independence), indicating that RPW8 might be selected against especially strongly in the German populations, possibly due to a shift of the fitness cost–benefit balance related to activities of powdery mildew pathogens on this plant.
Furthermore, the only HR4-containing accession from outside of Germany, Col-0, might actually have been collected from Germany too. Col-0 is annotated in both the Nottingham Arabidopsis Stock Centre and the Arabidopsis Biological Resource Center to have an origin from Columbia, Missouri. However, George P. Rédei at the University of Missouri, who is among the earliest scientists using Arabidopsis as the model genetic material, recalled that he had received seeds from F. Laibach (the first Arabidopsis collector) in Germany in 1955 and named one line 5-13 as Col-0 (Rédei 1992; George P. Rédei, personal communication), which agrees with our speculation based on the information from the HR4 gene.
The second feature regarding the nucleotide polymorphism at RPW8.1 and RPW8.2 is that compared with the resistant alleles, all nonsynonymous polymorphisms at RPW8.1 and RPW8.2 are exclusively biallelic between resistant and susceptible alleles (Figures 3 and 5). This biallelic polymorphism is in contrast to that at RPP13, in which 55% of the polymorphic codons in the LRR domain of RPP13 (which is considered to be under diversifying selection) encode three or more amino acids and more than one-quarter encode for four or more amino acids (Rose et al. 2004). This pattern of polymorphism at RPW8 might reflect functional attenuation of the RPW8 alleles, rather than diversification as shown at RPP13. This is in agreement with our sequence-phenotypic data, in which RPW8.1 and RPW8.2 alleles that are less divergent from the resistant alleles (with one or two amino acid replacements) may be partially functional (Figure 6, B and C), whereas more divergent alleles tend to be nonfunctional. Similar allelic polymorphism was reported for RPS2. Apart from resistant and susceptible alleles (Caicedo et al. 1999; Mauricio et al. 2003), the Rps2/Po-1 allele appeared to be partially functional against averRpt2-expressing bacterial strain when ectopically expressed in the Col-0 background (Banerjee et al. 2001).
Given that resistant alleles of RPW8.1 or RPW8.2 are highly similar and clustered together, one would reason that the resistant alleles are relatively young compared with the more diversified susceptible alleles. This is contradictory to the inference that RPW8.2 may have gained resistance function before the speciation of the two Arabidopsis species (above). A plausible explanation is that resistant alleles have been conserved and maintained in populations exposed to powdery mildew pathogens due to the benefits they confer to plants, and that the formation and maintenance of susceptible alleles reflect natural selection against the resistant alleles in the absence of powdery mildew pathogens due to the fitness costs associated with them. In this scenario, both resistant and susceptible alleles may have been maintained in a spatiotemporal manner, and the derived alleles (i.e., susceptible alleles) can have multiple origins. In addition, susceptible alleles must have been created by loss-of-function mutations, and they might evolve more or less in the neutral manner after loss of function. The maintenance of the susceptible HR4 alleles in replacing RPW8.1 (and RPW8.2) in eight accessions is also supportive of the evolutionary disadvantage of RPW8, at least in some circumstances. Thus, this scenario well explains the high levels of genetic variation and the lack of evidence for natural selection in the sequence variation, although further study is required to test this evolutionary hypothesis.
Fitness costs of RPW8 expression:
It has been assumed that long-time maintenance of resistant and susceptible polymorphisms at several R loci in A. thaliana populations is determined by two opposing forces, fitness benefits and fitness costs, both of which are associated with expression of the R genes (Stahl et al. 1999; Tian et al. 2002; Mauricio et al. 2003). Indeed, fitness costs have been demonstrated for RPM1 in the absence of the cognate pathogen (Tian et al. 2003), suggesting that at least some NBS-LRR R genes may incur fitness costs in the absence of pathogens. The features of sequence polymorphisms at the RPW8 locus prompted us to ask the question: does RPW8 expression really incur fitness costs in the absence of the pathogens?
Our experimental data (Figure 7) strongly support the conclusion that expression of the functional RPW8.1 and RPW8.2 genes indeed incurs fitness costs. Although we cannot formally exclude the possibility that reduced yield in the three RPW8 transgenic lines may be caused by T-DNA insertion-introduced genomic perturbance, we have strong reasons to believe that the reduced yield is associated with RPW8 expression, because the reduction of seed yield was not only detectable but it also seemed to be correlating with the levels of RPW8 expression (Figure 7). In addition, we showed that S5 plants had increased fitness when challenged by the pathogen at an early developmental stage (Figure 7A), further linking the plant performance with expression of the functional RPW8 genes. Therefore, although there is no statistical evidence of natural selection from the RPW8.1 and RPW8.2 sequences, our results suggest that fitness costs associated with resistant RPW8.1 and RPW8.2 alleles may explain selective advantage of susceptible RPW8.1 and RPW8.2 alleles and the HR4 gene in the absence of the pathogen that led to formation and maintenance of those alleles in A. thaliana populations. However, it should be pointed out that the fitness costs we detected with RPW8 as a transgene in this analysis do not necessarily reflect the exact levels of fitness costs in natural A. thaliana accessions carrying the RPW8 gene. Also, we have not directly tested if the maintenance of susceptible or partially resistant alleles is indeed due to reduced fitness costs in plants expressing those alleles. Comparative analysis of the performance of plants expressing the resistant or susceptible RPW8 alleles in the same background is needed to consolidate our conclusion.
Previously, we showed that enhanced expression of the RPW8 genes led to SHL and greatly reduced plant stature, and suppression of RPW8-expression by growing plants on MS-agar medium led to suppression of SHL and restoration of normal plant size (Xiao et al. 2003, 2005). These observations provided indirect evidence for RPW8-dosage-dependent fitness costs. We also found that high-light or long-day conditions enhanced RPW8 expression, whereas high humidity and high temperature attenuated RPW8 expression (Xiao et al. 2003). This environmental regulation of RPW8 expression may explain why S5 showed clear fitness costs when plants were grown in a growth room (in the earlier experiments) but was not significantly different from Col-gl when plants were grown in a greenhouse (in the later experiment): RPW8 expression was attenuated under the environmental conditions in the greenhouse, and so were the associated fitness costs. Recognition of this environmental regulation of RPW8 expression and its associated fitness costs has an important implication. That is: fitness costs and, as a corollary, fitness benefits (in the presence of the pathogens), are not only determined by the strength of the functionality of the RPW8.1 and RPW8.2 alleles but also influenced by physical environment conditions besides the pathogens that influence their expression levels.
Source of fitness costs:
Our recent work suggested that RPW8 may activate a conserved defense-related cell death pathway through a salicylic acid (SA)-dependent feedback amplification circuit (Xiao et al. 2003, 2005). Detection of hypersensitive response-like cell death (Figure 7D) and PR1 expression in leaves of 6-week-old plants of S5 and the naturally RPW8-expressing accession Ms-0 grown in soil in the absence of any pathogens, along with the observation that enhanced RPW8 expression leads to spontaneous cell death and reduced plant size (Xiao et al. 2003) strongly supports our speculation that reduced fitness in the RPW8 transgenic lines was due to RPW8-expression-triggered constitutive activation of an SA-dependent defense pathway under certain environmental conditions. This is consistent with the recent observations that constitutive activation of the SA-dependent defenses in several A. thaliana mutants incurred fitness costs in the absence of pathogens (Heidel et al. 2004) and that normal pathogen-inducible SA-dependent defenses may be beneficial to plants under pathogen pressure (Heidel and Dong 2006). Our results in this work, together with similar results from others, suggest that activation of the SA-dependent defenses in plants is costly and thus R genes as triggers at the top of the signaling pathway are under stringent selection to balance the fitness benefits and costs associated with the expression of the R genes according to the temporally and spatially variable physical and pathogen environments.
Acknowledgments
We thank Magnus Nordborg and two anonymous reviewers for useful comments, Charles Langley for seeds of A. lyrata, and the Arabidopsis Biological Resource Center at Ohio State University and the Nottingham Arabidopsis Stock Center for seeds of A. thaliana. We thank John Turner for his support at the early stage of this work, the greenhouse staff of University of Maryland (College Park) for assistance in growing Arabidopsis plants, and Jerome Regier and Xiao lab members for their valuable comments on the manuscript. This work was supported by the National Research Initiative of the United States Department of Agriculture (grant no. 2005-35319-15656 to S.X.).
References
- Adam, L., and S. C. Somerville, 1996. Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9: 341–356. [DOI] [PubMed] [Google Scholar]
- Adam, L., S. Ellwood, I. Wilson, G. Saenz, S. Xiao et al., 1999. Comparison of Erysiphe cichoracearum and E. cruciferarum and a survey of 360 Arabidopsis thaliana accessions for resistance to these two powdery mildew pathogens. Mol. Plant Microbe Interact. 12: 1031–1043. [DOI] [PubMed] [Google Scholar]
- Allen, R. L., P. D. Bittner-Eddy, L. J. Grenville-Briggs, J. C. Meitz, A. P. Rehmany et al., 2004. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306: 1957–1960. [DOI] [PubMed] [Google Scholar]
- Axtell, M. J., and B. J. Staskawicz, 2003. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377. [DOI] [PubMed] [Google Scholar]
- Bakker, E. G., C. Toomajian, M. Kreitman and J. Bergelson, 2006. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18: 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, D., X. Zhang and A. F. Bent, 2001. The leucine-rich repeat domain can determine effective interaction between RPS2 and other host factors in Arabidopsis RPS2-mediated disease resistance. Genetics 158: 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella, M. A., J. E. Parker, L. N. Frost, P. D. Bittner-Eddy, J. L. Beynon et al., 1998. Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10: 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., B. A. Schaal and B. N. Kunkel, 1999. Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S. T., G. Coaker, B. Day and B. J. Staskawicz, 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Dangl, J. L., and J. D. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Dangl, J. L., and J. M. McDowell, 2006. Two modes of pathogen recognition by plants. Proc. Natl. Acad. Sci. USA 103: 8575–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L., J. Olivier, N. Peeters, D. X. Feng, M. Khounlotham et al., 2003. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100: 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P. N., G. J. Lawrence, A. M. Catanzariti, M. A. Ayliffe and J. G. Ellis, 2004. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16: 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P. N., G. J. Lawrence, A. M. Catanzariti, T. Teh, C. I. Wang et al., 2006. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J. G., G. J. Lawrence, J. E. Luck and P. N. Dodds, 1999. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H. H., 1956. The complementary genic systems in flax and flax rust. Adv. Genet. 8: 29–54. [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K. E., and J. D. Jones, 1997. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 575–607. [DOI] [PubMed] [Google Scholar]
- Heidel, A. J., and X. Dong, 2006. Fitness benefits of systemic acquired resistance during Hyaloperonospora parasitica infection in Arabidopsis thaliana. Genetics 173: 1621–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel, A. J., J. D. Clarke, J. Antonovics and X. Dong, 2004. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168: 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguade, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., and F. Tajima, 1997. The amounts of nucleotide variation within and between allelic classes and the reconstruction of the common ancestral sequence in a population. Genetics 147: 1431–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., S. A. McAdams, G. T. Bryan, H. P. Hershey and B. Valent, 2000. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D. A., C. M. Thomas, K. E. Hammond-Kosack, P. J. Balint-Kurti and J. D. Jones, 1994. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793. [DOI] [PubMed] [Google Scholar]
- Koch, M. A., and M. Kiefer, 2005. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species—Capsella rubella, Arabidopsis lyrata subsp Petraea, and A. thaliana. Am. J. Bot. 92: 761–767. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Mackey, D., B. F. Holt, III, A. Wiig and J. L. Dangl, 2002. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Y. Belkhadir, J. M. Alonso, J. R. Ecker and J. L. Dangl, 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Mauricio, R., E. A. Stahl, T. Korves, D. Tian, M. Kreitman et al., 2003. Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana. Genetics 163: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- McHale, L., X. Tan, P. Koehl and R. W. Michelmore, 2006. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., S. Kaushik and R. S. Nandety, 2005. Evolving disease resistance genes. Curr. Opin. Plant Biol. 8: 129–134. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., and B. C. Meyers, 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8: 1113–1130. [DOI] [PubMed] [Google Scholar]
- Mylne, J., and J. R. Botella, 1998. Binary vectors for sense and antisense expression of arabidopsis ESTs. Plant Mol. Biol. Rep. 16: 257–262. [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei, G. P., 1992. A heuristic glance at the past of Arabidopsis genetics, pp. 1–15 in Methods in Arabidopsis Research, edited by C. Koncz, N.-H. Chua and J. Schell. World Scientific, Singapore.
- Rehmany, A. P., A. Gordon, L. E. Rose, R. L. Allen, M. R. Armstrong et al., 2005. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17: 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, L. E., P. D. Bittner-Eddy, C. H. Langley, E. B. Holub, R. W. Michelmore et al., 2004. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics 166: 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Sessions, A., E. Burke, G. Presting, G. Aux, J. McElver et al., 2002. A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F., C. Golstein, J. Ade, M. Stoutemyer, J. E. Dixon et al., 2003. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shen, J., H. Araki, L. Chen, J. Q. Chen and D. Tian, 2006. Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172: 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W. Y., G. L. Wang, L. L. Chen, H. S. Kim, L. Y. Pi et al., 1995. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- Stahl, E. A., G. Dwyer, R. Mauricio, M. Kreitman and J. Bergelson, 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671. [DOI] [PubMed] [Google Scholar]
- Sun, X., Y. Cao, Z. Yang, C. Xu, X. Li et al., 2004. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37: 517–527. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., M. B. Traw, J. Q. Chen, M. Kreitman and J. Bergelson, 2003. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77. [DOI] [PubMed] [Google Scholar]
- Xiao, S., 2006. Current perspectives on molecular mechanisms of plant disease resistance, pp. 317–333 in Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues, Vol. III, edited by J. Teixeira da Silva. Global Science Books, London.
- Xiao, S., S. Ellwood, K. Findlay, R. P. Oliver and J. G. Turner, 1997. Characterization of three loci controlling resistance of Arabidopsis thaliana accession Ms-0 to two powdery mildew diseases. Plant J 12: 757–768. [DOI] [PubMed] [Google Scholar]
- Xiao, S., S. Ellwood, O. Calis, E. Patrick, T. Li et al., 2001. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120. [DOI] [PubMed] [Google Scholar]
- Xiao, S., S. Brown, E. Patrick, C. Brearley and J. G. Turner, 2003. Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S., B. Emerson, K. Ratanasut, E. Patrick, C. O'Neill et al., 2004. Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol. Biol. Evol. 21: 1661–1672. [DOI] [PubMed] [Google Scholar]
- Xiao, S., O. Calis, E. Patrick, G. Zhang, P. Charoenwattana et al., 2005. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 42: 95–110. [DOI] [PubMed] [Google Scholar]