Abstract
Genetic analyses of the domestication syndrome have revealed that domestication-related traits typically have a very similar genetic architecture across most crops, being conditioned by a small number of quantitative trait loci (QTL), each with a relatively large effect on the phenotype. To date, the domestication of sunflower (Helianthus annuus L.) stands as the only counterexample to this pattern. In previous work involving a cross between wild sunflower (also H. annuus) and a highly improved oilseed cultivar, we found that domestication-related traits in sunflower are controlled by numerous QTL, typically of small effect. To provide insight into the minimum genetic changes required to transform the weedy common sunflower into a useful crop plant, we mapped QTL underlying domestication-related traits in a cross between a wild sunflower and a primitive Native American landrace that has not been the target of modern breeding programs. Consistent with the results of the previous study, our data indicate that the domestication of sunflower was driven by selection on a large number of loci, most of which had small to moderate phenotypic effects. Unlike the results of the previous study, however, nearly all of the QTL identified herein had phenotypic effects in the expected direction, with the domesticated allele producing a more crop-like phenotype and the wild allele producing a more wild-like phenotype. Taken together, these results are consistent with the hypothesis that selection during the post-domestication era has resulted in the introduction of apparently maladaptive alleles into the modern sunflower gene pool.
PLANT domestication typically involves intense directional selection, which produces large changes in quantitative traits, often accompanied by some degree of reproductive isolation between wild and domesticated taxa. Crop evolution thus allows for the investigation of basic evolutionary phenomena such as the phenotypic response of populations to long-term directional selection, the genetic consequences of recent selective sweeps, and the limitations imposed on selection response by genetic architecture (e.g., Stuber et al. 1980; Wang et al. 1999; Bost et al. 2001). Unlike researchers studying most wild systems, students of domestication often enjoy historical insights into the likely timing of selection, as well as the types of traits that have been subjected to selection. Common domestication traits include: increased seed or fruit size, more determinate growth and flowering, suppression of natural seed dispersal, and loss of self-incompatibility. Termed the “domestication syndrome” (Harlan 1992), these traits make crop plants easier to cultivate and result in more valuable products for human use.
Genetic analyses of the domestication syndrome have revealed that these traits have a similar genetic architecture across most crops (e.g., Doebley et al. 1990; Doebley and Stec 1991, 1993; Paterson et al. 1991; Koinange et al. 1996; Xiong et al. 1999). More specifically, crop-related traits are typically conditioned by a small number of quantitative trait loci (QTL), each with a relatively large effect on the phenotype (reviewed in Ross-Ibarra 2005). Perhaps the most well-known example of this is maize, wherein just five genomic regions account for the majority of the phenotypic differentiation between teosinte and maize (Doebley and Stec 1991, 1993). Doebley and Stec (1991, p. 294) argued that if “evolution is opportunistic, one would predict that major shifts in the morphological traits of plants could be controlled by the full range of genetic mechanisms from few genes with large effects to many genes with small effects.” They further argued that “The relative importance in plant evolution of these contrasting modes of inheritance remains to be determined.”
Although wild populations have been shown to respond to selection in a variety of ways (e.g., Bradshaw et al. 1998; Fishman et al. 2002), the pattern of few QTL of large effect is nearly universal in crop plants. In fact, there is just one counterexample—the evolution of domesticated sunflower, Helianthus annuus L. (Burke et al. 2002). In terms of the phenotypic response to cultivation, sunflower is a very typical crop. Human-mediated selection has resulted in a dramatic increase in apical dominance relative to its wild progenitor (common sunflower, also H. annuus), an increase in seed size, and the loss of natural seed dispersal, seed dormancy, and self-incompatibility. However, when these traits were investigated at a genetic level in a cross between common sunflower and an elite oilseed cultivar, they were found to be under the control of a large number of QTL of predominantly minor effect, with only 5% of all QTL detected accounting for ≥25% of the segregating phenotypic variation. Traits of obvious importance for domestication, such as seed weight, branching, and shattering, all lacked QTL of major effect, and seed dormancy was later shown to be under similarly complex genetic control in a different crop × wild mapping population (Gandhi et al. 2005).
The foregoing results suggest that sunflower may indeed be an exception to the rule, thereby supporting the notion that evolution under domestication is an opportunistic process, making use of whatever genetic variation happens to be available (Doebley and Stec 1991). However, a subsequent study of seed oil content and composition revealed that these original findings may have been influenced by the complex postdomestication breeding history of sunflower (Burke et al. 2005). In fact, it now seems clear that certain portions of the cultivated sunflower genome experienced post-domestication selective sweeps, meaning that the use of a modern inbred line as the cultivar parent in the original study likely confounded the effects of selection during domestication with the effects of selection during the subsequent period of breeding and improvement.
Here, we report the results of an investigation of the genetic architecture of sunflower domestication utilizing a cross between common sunflower and a primitive Native American domesticate. This work is thus designed to provide insight into the genetic changes that were necessary for the initial transformation of the weedy common sunflower into a useful crop plant. We have focused primarily on a suite of domestication-related traits that have previously been analyzed and are thus able to make a direct comparison to the results of earlier research.
MATERIALS AND METHODS
Mapping population:
The mapping population described in this study was derived from a common × domesticated sunflower cross. The wild parent used in this cross was drawn from the same population (Ann1238) in Keith County, Nebraska, that served as the source of the wild parent in the previous QTL analysis of sunflower domestication (Burke et al. 2002). This population is located within the same general range of the common sunflower that is thought to have given rise to domesticated sunflower (Harter et al. 2004). The domesticated parent was the Hopi sunflower (USDA PI 432504), which was selected for analysis because it represents one of the two most primitive extant cultivated sunflower lineages (Tang and Knapp 2003; Harter et al. 2004; Wills and Burke 2006). A single, self-compatible F1 individual from the initial wild × domesticated cross was self-pollinated to produce the F2 generation. F2 seeds were nicked with a razor blade and allowed to germinate on moist filter paper prior to being sown in flats. Seedlings were then transplanted into pots and grown under 16-hr days in the greenhouse. The final mapping population consisted of 378 F2 individuals. Fifteen individuals each from the Hopi landrace and the Ann1238 population were grown along with the mapping population to estimate the phenotypic means of the parental lines when grown under these conditions.
Phenotypic trait measurements:
Thirteen domestication-related traits that have been shown to differ between wild and domesticated sunflower were measured in all 378 F2 plants as well as in the 15 individuals from each parental line (or their selfed progeny in the case of seed traits; Table 1). The number of days to flowering was recorded for each individual. At flowering, the number of rays, disc diameter of the primary head, stem length, length and width of largest leaf, and stem diameter 3 cm above the soil were recorded for each individual. Leaf size was calculated as length × width. The primary head on each individual was bagged to prevent pollination from neighboring plants and rubbed to ensure self-pollination until florets ceased to emerge (∼9 days). Plants were maintained in the greenhouse until their seeds were mature, at which time the primary head was harvested, and the number of heads and branches were recorded for each individual. Primary heads were then dried for 3 days at 40°. To quantify shattering of the capitulum, the dried heads were dropped three times from a height of 12 cm. The total number of seeds released from the capitulum was then recorded, the heads were threshed, and the total seed output was recorded. Shattering was scored as the percentage of seeds released and 100-seed weight was estimated for each line. Seeds were then stored at 4° for 3 months. To quantify seed dormancy, 20 seeds from each selfed F2 individual with sufficient seed output were sown in pots at a soil depth of 2 cm and allowed to germinate in a growth chamber under 16-hr days with constant bottom watering. The number of germinated seeds was recorded each day, and pots were monitored for 100 days. Seeds that failed to germinate during the course of this trial were scored as having germinated on the 100th day, and the mean number of days until germination was calculated for each F2 line.
TABLE 1.
Comparison of 14 traits between a primitive sunflower domesticate (the Hopi landrace) and its wild progenitor (Helianthus annuus var. annuus)
| Trait | Hopi landrace | Common sunflower |
|---|---|---|
| Days to flower | 100.0 ± 4.4 | 84.8 ± 4.4 |
| Stem diameter (mm) | 21.7 ± 0.7 | 10.8 ± 0.7 |
| Height (cm) | 358 ± 14.0 | 171 ± 16.9 |
| No. main stem leaves | 47.5 ± 1.0 | 21.7 ± 1.9 |
| Leaf size (cm2) | 687 ± 23.8 | 335 ± 27.7 |
| No. branches | 0.4 ± 0.2 | 9.4 ± 1.6 |
| No. heads | 1 ± 0.0 | 4.2 ± 0.6 |
| Disk diameter (mm) | 75.3 ± 4.3 | 23.3 ± 1.9 |
| No. ray flowers | 42.5 ± 3.8 | 23.7 ± 1.6 |
| Self-compatibility | Yes | No |
| Achene weight (g/100) | 2.9 ± 1.1 | 0.6 ± 0.1 |
| Shattering | No | Yes |
| Seed dormancy | No | Yes |
All values are expressed as mean ± standard error
Genotyping:
Total genomic DNA was extracted from a sample of leaf tissue from each F2 individual using the Qiagen DNeasy plant mini kit (Qiagen, Valencia, CA). Genotyping for the genetic map was then carried out for 111 variable codominant loci, including 108 simple-sequence repeats (SSRs) that were previously mapped in sunflower (Tang et al. 2002, 2003; Yu et al. 2003; Lai et al. 2005). The SSRs were fluorescently labeled with 6FAM, TET, HEX, or VIC either by direct labeling of the 5′ end of the forward primer or using a modification of the three-primer PCR methodology presented by Schuelke (2000), previously adapted for sunflower by Wills et al. (2005). This technique involves incorporation of an arbitrarily selected sequence (the M13 Forward [−29] sequencing primer, 5′-CAC GAC GTT GTA AAA CGA C-3′) to the 5′ end of the forward primer. PCR products are then labeled by including a fluorescently tagged (6FAM, TET, or HEX) M13 forward (−29) primer in the reaction mixture. All reactions were performed in 10 μl total volume containing 10 ng of template DNA, 30 mm Tricine pH 8.4-KOH, 50 mm KCl, 2mm MgCl2, 100 μm of each dNTP, 0.02 μm forward primer, 0.1 μm of both the reverse primer and the fluorescently labeled M13F primer, and 2 units of Taq polymerase. When PCR was carried out with directly labeled primer pairs, the M13F primer was left out, and the forward primer was increased to 0.1 μm. Cycling conditions were as follows: initial denaturation at 95° for 3 min, followed by 10 cycles of 30 sec at 94°, 30 sec at 58° (annealing temperature was reduced by one degree per cycle), 45 sec at 72°, followed by 30 cycles of 30 sec at 94°, 30 sec at 48°, 45 sec at 72°, and a final extension time of 20 min at 72°.
Amplification products were visualized on either an MJ Research BaseStation automated DNA sequencer (South San Francisco, CA) or an Applied Biosystems 3730xl DNA analyzer (Foster City, CA). MapMarker 1000 ROX size standard (BioVentures, Murfreesboro, TN) was included in each lane to allow for accurate determination of fragment size. Alleles were called using the software package Cartographer (MJ Research) for the BaseStation runs or GeneMarker (SoftGenetics, State College, PA) for the 3730 data. The final map included three additional, previously unpublished markers: HT39, HT135, and HT1490 (S. Tang and S. J. Knapp, unpublished data). HT39 amplicons were visualized via SSCP gel electrophoresis using 0.5× MDE gels that were run for 14 hr at 4 W (Slabaugh et al. 1997) followed by silver staining (Sanguinetti et al. 1994). HT135 exhibited a length polymorphism in this cross and was scored in the same manner as the SSRs described above. HT1490 was not length polymorphic and could not be reliably scored via SSCP analysis. Thus, this locus was sequenced, and a restriction polymorphism corresponding to an Fnu4HI restriction site was found to be segregating within the mapping population. The forward primer was therefore 5′ end labeled with 6FAM, and each individual was amplified as described above. All PCR amplicons were then digested at 37° overnight with 1 unit of Fnu4HI (New England Biolabs, Ipswich, MA). The PCR–RFLP products were then run on an Applied Biosystems 3730xl and scored using GeneMarker.
Map construction:
The linkage map was constructed using MAPMAKER 3.0/EXP (Lander et al. 1987; Lincoln and Lander 1992). Initial linkage groups were identified using the “group” command with LOD > 5.0 and θ < 0.2. Preliminary map orders within groups were then set based on the results from previous sunflower mapping studies (Burke et al. 2002; Tang et al. 2002, 2003; Yu et al. 2003; Lai et al. 2005). Final map orders were then confirmed using the “ripple” and “compare” commands, such that the map orders presented herein reflect the statistically most likely order on the basis of the data at hand.
QTL analysis:
The initial QTL analysis followed the same general approach as outlined by Burke et al. (2002). Because shattering was scored as a proportion, the data for this trait were arcsine-square root transformed prior to analysis using JMP 4 (SAS Institute, Cary, NC). Composite interval mapping (CIM) (Zeng 1993, 1994) was then performed as implemented by the program Zmapqtl (model 6) of the software package QTL Cartographer version 1.17 (Basten et al. 1994, 2004). CIM was run with a 10-cM window and five background cofactors. Tests were performed at 2-cM intervals, and cofactors were selected via forward–backward stepwise regression using the program SRmapqtl. Genomewide threshold values (α = 0.05) for declaring the presence of QTL were estimated from 1000 permutations for each trait (Churchill and Doerge 1994; Doerge and Churchill 1996). A likelihood-ratio decline of 9.21 (equivalent to a LOD decline of 2.0) between adjacent peaks on a linkage group was taken as evidence of multiple linked QTL and one-LOD support limits for the position of each QTL were calculated from the CIM results. The degree of dominance of the Hopi allele at each locus was calculated as the dominance effect divided by the additive effect (d/a), and the following arbitrary thresholds were used to classify the mode of gene action for each QTL: underdominant ≤ −1.25 < recessive ≤ −0.75 < partially recessive ≤ −0.25 < additive ≤ 0.25 < partially dominant ≤ 0.75 < dominant < 1.25 ≤ overdominant. Finally, to allow a direct comparison to the results of Burke et al. (2002), we used arbitrary percentage of variance explained (PVE) thresholds of 10 and 25% to classify QTL as having “minor,” “intermediate,” or “major” effects.
Multiple interval mapping (MIM) (Kao and Zeng 1997; Kao et al. 1999) was then used to search for epistatic interactions amongst the QTL identified via CIM. The CIM results were used as the initial model for the MImapqtl module in QTL Cartographer (Basten et al. 1994, 2004), and the maximum number of allowable pairwise interactions was set to 19. Only those interactions that significantly improved the fit of the model were retained. As recommended by the authors, significance was determined on the basis of the information criterion IC(k) = −2(log(L) − k c(n)/2), where c(n) = log(n) as the penalty function and a threshold of 0.0.
RESULTS
Linkage analysis:
The map coalesced into the expected 17 linkage groups and covered a total of 906.4 cM with an average intermarker distance of 8.2 cM. As has been previously observed for common × cultivated sunflower crosses (e.g., Burke et al. 2002), this map showed evidence of suppressed recombination, with common marker intervals exhibiting nearly 20% compression when compared against the sunflower reference map, which is based on a cross between two elite inbred lines (RHA280 × RHA801) (Tang et al. 2002; S. Tang and S. J. Knapp, unpublished data). On the basis of a comparison of shared markers, coverage of the map described herein is equal to or exceeds that of the map constructed by Burke et al. (2002) for 16 of the 17 linkage groups (LGs). The one exception was a portion of the top of LG17, which we were unable to cover due to a lack of polymorphism. However, no QTL have been detected previously in this region, so this small gap in coverage is unlikely to influence our overall findings.
QTL analysis:
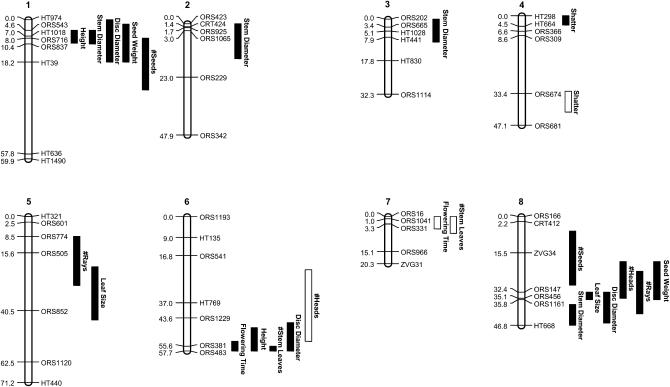
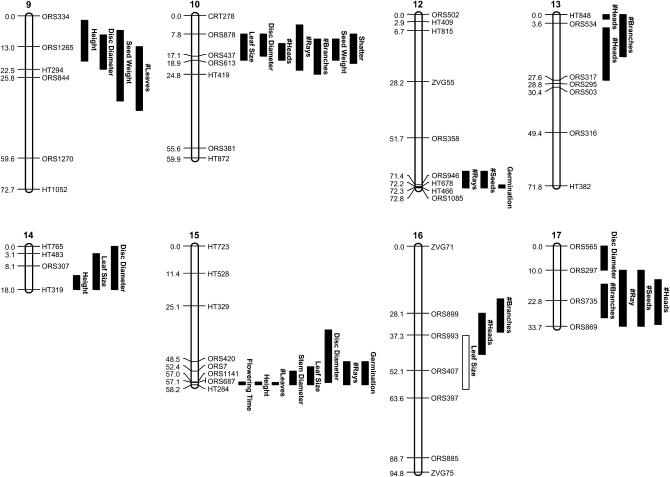
For the 13 domestication-related traits that we analyzed, CIM detected 61 QTL (Table 2; Figure 1). The number of QTL per trait ranged from 2 to 8 (mean = 4.7) (Figure 2A) with shattering, and the number of heads produced being the only traits with multiple QTL on a single linkage group. QTL were found on all linkage groups with the exception of LG11, and the one-LOD support intervals, which provide an approximate confidence interval for the true location of a given QTL, ranged from 1.1 to 30.8 cM (mean = 13.0 cM). Multiple overlapping QTL were observed on most linkage groups; the exceptions were LG2, LG3, and LG4, as well as LG11, which (as noted above) was devoid of QTL.
TABLE 2.
Putative QTL positions, effect magnitudes, and modes of gene action for 13 traits using composite interval mapping in an F2 population derived from a cross between a primitive sunflower domesticate (the Hopi landrace) and its wild progenitor (H. annuus var. annuus)
| Trait | Linkage groupa | Positionb | Nearest marker | One-LOD intervalc | Additive effectd | Dominance ratioe | PVEf | Previously identified?g |
|---|---|---|---|---|---|---|---|---|
| Days to flower | 6 | 57.6 | ORS483 | 53.6–57.7 | 4.6 | −0.23 | 7.6 | Yes |
| 7 | 1.0 | ORS1041 | 0–5.3 | −2.6 | −0.11 | 2.5 | Yes | |
| 15 | 57.1 | ORS687 | 57–58.2 | 10.4 | −0.49 | 46.9 | No | |
| Stem diameter | 1 | 7.0 | HT1018 | 4.6–10.4 | 1.2 | 0.72 | 10.0 | Yes |
| 2 | 1.7 | ORS925 | 0–15.0 | 0.7 | −0.03 | 3.0 | No | |
| 3 | 3.4 | ORS665 | 0–9.9 | 1.1 | −0.33 | 6.5 | Yes | |
| 8 | 43.8 | HT668 | 37.8–46.8 | 1.3 | 0.84 | 8.0 | No | |
| 15 | 56.4 | ORS1141 | 52.4–58.2 | 1.7 | 0.16 | 15.7 | No | |
| Height | 1 | 8.0 | ORS716 | 4.6–10.0 | 29.6 | 0.45 | 11.9 | No |
| 6 | 57.6 | ORS483 | 47.6–57.7 | 23.1 | 0.01 | 6.4 | Yes | |
| 9 | 10.0 | ORS1265 | 2.0–19.0 | 15.7 | 0.56 | 3.0 | No | |
| 14 | 16.1 | HT319 | 10.1–18.0 | 11.5 | 1.19 | 3.0 | No | |
| 15 | 57.1 | ORS687 | 57.0–58.2 | 53.2 | −0.31 | 39.4 | No | |
| No. main stem leaves | 6 | 57.6 | ORS483 | 55.6–57.7 | 2.7 | 0.09 | 4.9 | Yes |
| 7 | 1.0 | ORS1041 | 0–7.3 | −1.8 | 0.31 | 2.7 | Yes | |
| 9 | 19.0 | HT294 | 13.0–39.8 | 2.6 | −0.23 | 5.3 | Yes | |
| 15 | 57.1 | ORS687 | 57.1–58.2 | 8.0 | −0.49 | 57.0 | No | |
| Leaf size | 5 | 31.6 | ORS852 | 21.6–44.5 | 64.8 | −0.71 | 9.1 | Unknown |
| 8 | 35.6 | ORS1161 | 32.4–35.8 | 56.4 | −0.14 | 5.6 | No | |
| 10 | 15.8 | ORS437 | 7.8–18.9 | 46.7 | 0.64 | 4.4 | No | |
| 14 | 10.1 | ORS307 | 3.1–18.0 | 36.4 | 1.34 | 4.9 | No | |
| 15 | 57.0 | ORS1141 | 50.5–58.2 | 42.2 | 0.97 | 3.7 | No | |
| 16 | 45.4 | ORS407 | 37.4–60.1 | −31.6 | −1.96 | 5.1 | No | |
| No. branches | 10 | 17.1 | ORS437 | 9.8–24.8 | −1.3 | 0.26 | 4.6 | No |
| 13 | 0 | HT848 | 0–17.6 | −1.4 | −0.1 | 5.2 | No | |
| 16 | 30.1 | ORS899 | 22.0–36.1 | −1.3 | −0.87 | 7.0 | No | |
| 17 | 22.0 | ORS735 | 16.0–30.1 | −0.2 | 11.47 | 8.4 | No | |
| No. heads | 6 | 41.1 | ORS1229 | 22.8–53.6 | 0.8 | −0.43 | 3.1 | No |
| 8 | 29.5 | ORS147 | 19.5–35.2 | −1.3 | −0.06 | 5.9 | No | |
| 10 | 15.8 | ORS437 | 11.8–18.9 | −2.4 | 0.59 | 28.1 | No | |
| 13a | 0 | HT848 | 0–2.0 | −1.2 | −0.2 | 5.3 | No | |
| 13b | 15.6 | ORS317 | 5.6–27.6 | −1.3 | 0.03 | 6.5 | No | |
| 16 | 34.1 | ORS993 | 28.1–45.4 | −0.5 | −2.19 | 3.4 | No | |
| 17 | 24.8 | ORS735 | 14.0–32.8 | −0.4 | 2.31 | 3.4 | Yes | |
| Disc diameter | 1 | 14.4 | HT39 | 0–18.3 | 2.7 | 0.67 | 4.4 | No |
| 6 | 53.6 | ORS381 | 45.6–57.7 | 2.3 | −1.09 | 4.9 | No | |
| 8 | 35.2 | ORS456 | 32.4–45.8 | 4.1 | 0.23 | 9.0 | No | |
| 9 | 17.0 | ORS1265 | 8.0–22.5 | 3.8 | 0.16 | 7.7 | No | |
| 10 | 13.8 | ORS437 | 7.8–17.1 | 4.4 | 0.74 | 13 | No | |
| 14 | 12.1 | ORS307 | 0–18.0 | 2.8 | 0.7 | 5.6 | No | |
| 15 | 50.5 | ORS7 | 35.1–57.1 | 2.0 | 1.61 | 4.3 | No | |
| 17 | 4.0 | ORS565 | 0–10.0 | 3.8 | −0.14 | 5.5 | No | |
| No. ray flowers | 5 | 19.6 | ORS505 | 8.5–29.6 | 1.9 | −0.2 | 6.7 | No |
| 8 | 32.4 | ORS147 | 23.5–41.8 | 0.8 | 1.77 | 3.1 | No | |
| 10 | 13.8 | ORS534 | 4.0–22.9 | 1.4 | 0.04 | 3.6 | No | |
| 12 | 72.3 | HT466 | 65.7–72.8 | 1.9 | 0.28 | 4.3 | No | |
| 15 | 57.1 | ORS687 | 48.5–58.2 | 2.8 | 0.23 | 13.1 | No | |
| 17 | 26.8 | ORS735 | 10.0–33.7 | 1.2 | 0.92 | 4.6 | No | |
| No. selfed seeds | 1 | 14.4 | HT39 | 8.0–30.3 | 48.7 | −0.5 | 6.6 | No |
| 8 | 15.5 | ZVG34 | 6.2–29.5 | 44.2 | 0.69 | 5.4 | No | |
| 12 | 72.3 | HT466 | 65.7–72.8 | 60.4 | 0.06 | 6.8 | No | |
| 17 | 18.0 | ORS735 | 10.0–33.7 | 37.3 | 1.08 | 7.2 | Yes | |
| Achene weight | 1 | 6.6 | HT1018 | 2.0–18.3 | 1.6 | 0.70 | 8.6 | No |
| 8 | 35.2 | ORS456 | 19.5–35.8 | 1.7 | 0 | 7.6 | No | |
| 9 | 19.0 | HT294 | 6.0–35.8 | 1.3 | 0.37 | 4.2 | Yes | |
| 10 | 15.8 | ORS437 | 9.8–18.9 | 2.6 | 0.36 | 19.0 | Yes | |
| Shatteringg | 4a | 0 | HT298 | 0–4.0 | −0.1 | 0.26 | 10.7 | n/a |
| 4b | 33.4 | ORS674 | 32.6–41.4 | 0.1 | 0.04 | 6.4 | n/a | |
| 10 | 15.8 | ORS437 | 7.8–20.1 | −0.1 | 0.38 | 9.0 | n/a | |
| Seed germination | 12 | 72.3 | HT466 | 71.4–72.8 | −10.0 | −1.31 | 17.3 | n/a |
| 15 | 57.1 | ORS687 | 48.5–58.2 | −20.3 | 0.16 | 17.8 | n/a |
When multiple QTL for a single trait occurred on the same linkage group, a letter was used to uniquely identify them.
Absolute position from the top of the linkage group (in centimorgans).
Refers to the region flanking each QTL peak in which the LOD score declines by one.
Refers to the additive effect (a) of the Hopi allele. Underlined values indicate instances in which the allelic effects are in the wrong direction. See text for details.
Refers to the dominance ratio (d/a) of the Hopi allele.
Percentage of phenotypic variation explained by each QTL using CIM. PVE values for QTL with effects in the direction of the wild phenotype are underlined.
Indicates whether or not a given QTL was detected in the previous cultivated × wild sunflower QTL analysis (Burke et al. 2002). The determination of overlap between studies was based on the one-LOD confidence intervals. Note that the previous analysis of shattering was based on a different (indirect) measure and that seed germination has not been previously analyzed.
Figure 1.—
Results of the CIM analysis for the 16 linkage groups on which QTL were detected. QTL positions are indicated by bars alongside each linkage group. The length of each bar is equal to the one-LOD support interval for that QTL. Loci at which the crop allele had the expected effect are indicated by a filled bar, whereas those at which the crop allele conferred a wild-like phenotype are represented by unfilled bars.
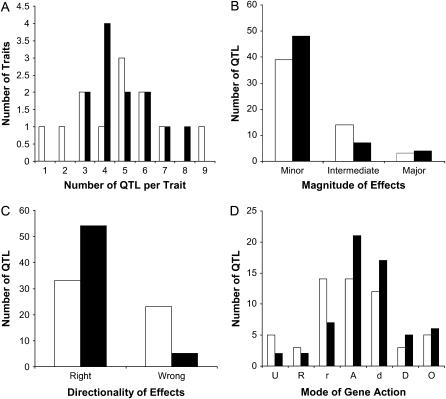
Figure 2.—
Comparison of the number of QTL (A) per trait, (B) magnitude of effects, (C) directionality of effects, and (D) mode of gene action between a previous study based on an elite × wild sunflower mapping population (open bars) (Burke et al. 2002) and the present study, which was based on a primitive × wild mapping population (closed bars). The following thresholds were used to classify the mode of gene action for each QTL: underdominant ≤ −1.25 < recessive ≤ −0.75 < partially recessive ≤ −0.25 < additive ≤ 0.25 < partially dominant ≤ 0.75 < dominant < 1.25 ≤ overdominant.
As previously documented, there was a paucity of QTL of major effect. Individual QTL explained 2.5–59.0% of the observed phenotypic variance for a particular trait, but only four QTL had PVE ≥25% (Figure 2B). Three of these major-effect QTL co-occur on the bottom of LG15 and influence days to flower, plant height, and number of leaves along the main stem. The only other QTL of major effect is located on LG10 and influences the number of heads produced. Contrary to previous findings, the majority of domestication-related QTL identified here (56 of 61) had effects in the expected direction (Figure 2C). That is, the Hopi allele produced a more crop-like phenotype, and the wild allele produced a more wild-like phenotype. The exceptions were QTL for shattering on LG4, number of heads produced on LG6, days to flower and number of leaves along the main stem on LG7, and leaf size on LG16. All five QTL with effects in “wrong” direction were minor, explaining from 2.5 to 6.4% of the phenotypic variance. In terms of the mode of gene action, the Hopi allele at each locus exhibited a dominance ratio (d/a) ranging from −2.19 to 11.47 with a mean of 0.38 (Table 2; Figure 2D).
Results of the search for epistasis amongst significant QTL using MIM are presented in Table 3. There were 30 significant pairwise interactions across the 13 traits. For the most part, these interactions were minor, and their phenotypic effects were mixed. Indeed, 28 of 30 interactions had an effect of <5%, and half were in the expected direction, whereas the other half were in the wrong direction. The two exceptions were interactions for branching and seed germination. In the former case, there was a dominant × additive interaction with an effect of 9.3% between QTL on LG13 and LG17, whereas the latter case had an additive × additive interaction with an effect of 11.5%. In both cases, the interactions acted in the expected direction, with Hopi/Hopi genotypes producing an even more Hopi-like phenotype.
TABLE 3.
Summary of significant interactions amongst individually significant QTL
| Trait | Linkage groups | Type of interactiona | Phenotypic effectb | Effect (%) |
|---|---|---|---|---|
| Height | 6 × 15 | A × A | 12.5 | 2.2 |
| 9 × 15 | D × A | 17.8 | 1.3 | |
| 9 × 15 | D × D | −22.9 | 1.0 | |
| No. main stem leaves | 6 × 15 | A × A | 2.4 | 3.8 |
| 7 × 9 | A × A | −1.2 | 1.3 | |
| Leaf size | 5 × 8 | A × A | −46.8 | 1.6 |
| 5 × 14 | D × D | 100.2 | 2.2 | |
| 10 × 14 | D × A | 70.5 | 1.2 | |
| No. branches | 10 × 17 | D × A | −1.5 | 0.8 |
| 13 × 17 | D × A | −5.0 | 9.3 | |
| 13 × 17 | D × D | 3.0 | −1.1 | |
| 16 × 17 | A × A | 1.4 | 3.7 | |
| No. heads | 8 × 16 | A × A | 0.8 | 0.8 |
| 8 × 17 | D × A | −1.4 | 2.2 | |
| 13a × 17 | D × A | −1.2 | 1.1 | |
| Disk diameter | 1 × 8 | A × A | −1.9 | 0.4 |
| 1 × 14 | A × A | −2.3 | 1.0 | |
| 1 × 17 | A × A | −2.9 | 2.1 | |
| 6 × 14 | D × A | 2.7 | 1.1 | |
| 6 × 15 | A × A | 2.3 | 1.8 | |
| 6 × 17 | A × A | 2.3 | 1.3 | |
| 8 × 14 | A × A | −2.7 | 1.8 | |
| 15 × 17 | A × A | −2.2 | 0.3 | |
| No. ray flowers | 8 × 15 | D × A | 2.1 | 1.8 |
| 10 × 12 | D × A | −2.0 | 1.9 | |
| 10 × 15 | D × A | −1.9 | 1.5 | |
| No. selfed seeds | 1 × 17 | A × A | 38.5 | 0.3 |
| 8 × 17 | A × A | 42.2 | 1.0 | |
| Achene weight | 1 × 8 | A × A | −2.3 | 2.0 |
| Seed germination | 12 × 15 | A × A | −14.3 | 11.5 |
A × A = additive × additive; A × D = additive × dominant; D × A = dominant × additive; D × D = dominant × dominant.
Underlined values indicate an interaction with effects in the wrong direction.
DISCUSSION
QTL numbers and magnitudes of effect:
In general terms, the results of this study confirm that sunflower is an exception to the rule in that its domestication involved changes at a large number of loci, each of relatively small effect (Figure 2, A and B). Indeed, we analyzed 13 traits and identified a total of 61 QTL, only 4 of which had PVE > 25%. In contrast, the domestication of crops such as maize (Doebley and Stec 1991, 1993), rice (Xiao et al. 1998; Xiong et al. 1999), and beans (Koinange et al. 1996) were all driven by relatively major changes at a much smaller number of loci. This observed lack of major QTL suggests that the transition from wild to domesticated sunflower was relatively smooth with few major phenotypic leaps.
Gene action and interaction:
In terms of the predominant mode of gene action, our results mirror those of Burke et al. (2002) and stand in stark contrast to the view that domestication is generally driven by recessive genetic changes (e.g., Ladizinsky 1985; Lester 1989). In fact, inspection of Figure 2D reveals a preponderance of nonrecessive QTL, suggesting that selection during domestication likely resulted in a rapid phenotypic response, as most of these QTL would have been at least partially visible to selection, even when rare. These findings are in accord with previous QTL results from other taxa, including tomato (Paterson et al. 1991) and maize (Doebley et al. 1994).
With regard to the role of gene interaction in domestication, our results provide somewhat limited evidence of epistasis. When combined with the overall lack of epistasis documented by Burke et al. (2002), these results suggest that neither the initial domestication of sunflower nor its subsequent improvement relied heavily upon the fixation of favorably interacting gene complexes. While MIM detected significant QTL × QTL interactions for 10 of the 13 traits, the vast majority of these interactions had effects of ≤5% (Table 3). The exceptions to this were an interaction between two branching QTL located on LG13 and LG17 and an interaction between the two seed dormancy QTL on LG12 and LG15. This latter case, which involves a synergistic additive × additive interaction between two QTL of intermediate effect (PVE = 17.3 and 17.8%, respectively), is particularly noteworthy because seed dormancy was not previously analyzed by Burke et al. (2002). Gandhi et al. (2005) did, however, map QTL related to seed dormancy in a different elite × wild cross and identified a QTL of intermediate effect in the same region of LG15, as well as two other QTL that were not recovered here; no significant epistatic interactions were detected among those QTL.
QTL concordance:
Despite the foregoing similarities between our results and those of Burke et al. (2002), a direct comparison of QTL locations reveals a relatively low level of concordance. Indeed, comparing the 59 QTL identified in this study (ignoring the two seed dormancy QTL because this trait was not previously analyzed) to the 56 QTL previously identified for this same suite of traits reveals only 15 cases in which QTL for the same trait mapped to the same linkage group in both studies. Twelve of these cases involved QTL with overlapping one-LOD support intervals, suggesting that the same QTL was detected in both crosses, 2 showed clear evidence of nonoverlap, and in one case the degree of overlap could not be determined because of a paucity of shared markers (LG5). This relatively low rate of correspondence between studies is likely due to a combination of factors, including differences between the parents used in each cross, QTL × environment interactions, and difficulties associated with reliably detecting QTL of small effect (Beavis 1994).
Because sunflower is an annual plant, it was impossible to use the same wild individual in both the present and previous crosses. Moreover, wild sunflower is an obligate outcrosser that exhibits high levels of heterozygosity (e.g., Ivanov 1975; Fernandez-Martinez and Knowles 1978; Tang and Knapp 2003; Harter et al. 2004). Thus, in an attempt to minimize problems with intra-taxon polymorphism and maintain continuity with previous work, the wild parent for the present cross was drawn from the same population that Burke et al. (2002) utilized. Although variation is evident in the wild for all of the traits in question, the phenotypic differences between wild and cultivated sunflower are largely consistent across environments. Despite this, it is still possible that some of the differences between the two studies resulted from allelic variation between the wild parents used in the two studies.
A more likely explanation is that a sizable fraction of the differences result from the cultivar parents used in the two studies having very different evolutionary histories; in fact, this was the primary motivation of the present study. The cultivar parent utilized by Burke et al. (2002) was a highly improved, elite oilseed line that has subsequently been found to bear the signature of post-domestication selective sweeps, presumably due to selection on oil-related characters (Burke et al. 2005). In contrast, the cultivar parent used in the present study is a primitive Native American landrace. The results presented herein should, therefore, provide a much more accurate picture of the genetic changes necessary to transform wild into domesticated sunflower, as they are largely free from the confounding effects of improvement subsequent to the initial domestication event. In this context, it is worth noting that LG6 has previously been shown to a harbor a large cluster of QTL that mostly have effects in the wrong direction (Burke et al. 2002). Subsequent work has suggested that at least some of these QTL arose as a byproduct of selection during sunflower improvement (Burke et al. 2005), and our results are fully consistent with this hypothesis. Indeed, only a subset of the QTL that were initially identified were recovered in the present analysis, and all but one of the QTL on this linkage group now have effects in the expected direction.
Another key difference between the cultivars utilized in these two studies is that they are adapted to relatively different habitats. As such, some of the QTL that have been identified in just one population are likely to reflect differences in local adaptation. Most notable in this context is flowering time (and associated traits such as height and number of stem leaves) in the Hopi × wild cross analyzed here. The Hopi landrace exhibits late flowering, presumably as an adaptation to the extremely long growing season of the desert southwest. Our results indicate that this flowering time difference is conditioned by a major QTL at the bottom of LG15, which, as one might expect, was not present in the previous cross.
As noted above, other factors that could account for the relatively low level of QTL concordance are QTL × environment interactions and the difficulties associated with reliably detecting QTL of small effect. With regard to QTL × environment interactions, it has previously been shown that individual QTL can vary in their degree of environmental sensitivity, with some QTL being robust across environments, while others can be detected only under certain conditions (e.g., Paterson et al. 1991, 2003). Thus, even though both populations were greenhouse grown, and the traits of interest are reasonably robust across environments, it is conceivable that some fraction of the QTL were detected in one study but not the other because of differences in growing conditions.
Regarding the issue of detectability, it is well known that QTL of minor effect suffer a higher false-negative rate as compared to QTL of major effect (Beavis 1994). Indeed, Doebley and Stec (1993) found much higher agreement in QTL locations in a comparison between two teosinte × maize populations for QTL of major effect (81% concordance for QTL with r2 ≥ 20%) as compared to QTL of intermediate or minor effect (55 and 28% concordance for QTL with 10% ≤ r2 ≤ 20% and QTL with r2 < 10%, respectively). Given the typically small effect sizes associated with QTL identified in both the present and previous analyses, the relatively low QTL concordance is therefore not surprising. Consistent with this idea is the fact that the handful of major QTL identified by Burke et al. (2002), including QTL for flowering time and the number of stem leaves on LG6 and the number of selfed seeds on LG17, were all recovered in the present study. The key difference is that the estimated effect sizes for all of these QTL were much lower in the present study. In the case of flowering time and the number of stem leaves, the reduced PVE in the Hopi × wild cross is likely due to an overall increase in phenotypic variance for these traits within the mapping population (our unpublished data) due to the adaptation of the Hopi sunflower to the long growing seasons of the desert southwest (see above). In the case of selfed seed production, the prior identification of two QTL at the bottom of LG17 (Burke et al. 2002) has subsequently been shown to be an artifact of inconsistent locus ordering; this region is now believed to harbor the S locus (Gandhi et al. 2005). Values reported in Figure 2 from the earlier study have been adjusted to account for the reordering of these markers. The low PVE associated with this locus in the present analysis is potentially an artifact of extreme segregation distortion in this region (all four markers on this linkage group deviate significantly from the expected segregation ratios, with all P < 0.001).
Conversely, the four QTL of major effect identified in the present study had not been previously identified. This result is not surprising for the QTL related to flowering time that are located near the bottom of LG15, as they are likely a byproduct of adaptation of the Hopi landrace to local growing conditions. However, in the one remaining case (number of heads produced; LG10), this is somewhat unexpected. Indeed, this QTL explains 28% of the segregating phenotypic variance and colocalizes with the B locus, which is known to influence apical branching (Tang et al. 2006). In fact, the region harboring the B locus is known to have manifold effects, influencing not only plant architecture but also achene/seed morphology. In the present analysis, this QTL is embedded within a larger cluster of loci that influence apical dominance as well as leaf and disc morphology, seed weight, and shattering. In fact, Burke et al. (2002) also found QTL related to seed size in this vicinity, but none related to branching. One possible explanation for this is that, despite their simple inheritance in crosses between cultivars, branching-related traits in wild × cultivar crosses are thought to be genetically complex (Burke et al. 2002) and may well be influenced by genetic background.
QTL directionality and evidence of selection:
Perhaps the greatest departure between our results and those of Burke et al. (2002) relates to the directionality of QTL effects (Figure 2C). As previously noted, nearly one-third of all QTL identified by Burke et al. (2002) had effects in the wrong direction, with the wild allele producing a more crop-like phenotype and vice versa. As noted above, however, a subsequent analysis of improvement-related traits (i.e., seed oil content and composition) combined with a population genetic scan for selection suggested that this result was due to post-domestication selection and breeding (Burke et al. 2005). The low frequency of wrong-way QTL in the present study (only 5 of 61 QTL had such effects) is fully consistent with this hypothesis. While data on QTL directionality can be used to statistically test for past directional selection (Orr 1998), the power of this approach is limited by QTL numbers. Thus, following the methods of Rieseberg et al. (2002), we pooled our data across traits and tested for selection on the domestication syndrome as a whole. In this case, the results were highly significant (P < 0.001), providing clear evidence that sunflower domestication was driven by consistent directional selection on a wide variety of traits.
Conclusions:
Our results confirm that the domestication of sunflower was driven by selection on a large number of loci, most of which had small to moderate phenotypic effects. However, the underlying cause of this departure from the typical genetic architecture of domestication remains a mystery. For example, while sunflower is an ancient polyploid (Adams and Wendel 2005; Sossey-Alaoui et al. 1998), and thus potentially exhibits high levels of genetic redundancy across the genome, this sort of redundancy alone cannot be the explanation. Indeed, virtually all major crops have experienced large-scale genome duplication at some point in their evolutionary history. Another possibility is that the population bottleneck leading to domesticated sunflower was less severe than that which occurred during the evolution of other crop lineages, resulting in a relatively large effective population size during domestication. This, in turn, would allow for selection to target mutations of minor effect more efficiently in sunflower than in other crop lineages. While the available data indicate that sunflower suffered a similar loss in genetic diversity during domestication as compared to other crop plants (e.g., Liu and Burke 2006)—a fact that argues against the idea that sunflower experienced a relatively mild bottleneck—a better understanding of the dynamics of the sunflower domestication bottleneck awaits more rigorous analysis. Ultimately, it may be that Doebley and Stec (1991) had it right; evolution under domestication may simply be an opportunistic process that makes use of whatever genetic variation happens to be available. Whether or not this lack of suitable mutations of major effect in sunflower reflects some sort of fundamental genomic constraint remains an open question.
Acknowledgments
We thank Tamara Berthel, Chris Buckner, Mark Chapman, Michael Foster, Meghan Jennings, Natasha Sherman, Ben Stephens, and Jessica Wenlzer for assistance in the greenhouse and/or lab, and Dörthe Dräger, Adam Heesacker, Steve Knapp, and Shunxue Tang for technical assistance with genotyping and access to unpublished marker data. We also thank Jim Birchler, Mark Chapman, Aizhong Liu, Natasha Sherman, and an anonymous reviewer for comments on a previous version of this manuscript. This work was supported by grants from the United States Department of Agriculture Cooperative State Research, Education and Extension Service National Research Initiative (CSREES-NRI) Plant Genome Program (award 03-35300-13104 to J.M.B.) and the National Science Foundation (award DBI-0332411 to J.M.B.).
References
- Adams, K. L., and J. F. Wendel, 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Basten, C. J., B. S. Weir and Z.-B. Zeng, 1994. Zmap-a QTL cartographer, pp. 65–66 in Proceeding of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, edited by C. Smith, J. S. Gavora, B. Benkel, J. Chesnais, W. Fairfull et al. Organizing Committee, 5th World Congress on Genetics Applied to Livestock Production. Guelph, Ontario, Canada.
- Basten, C. J., B. S. Weir and Z.-B. Zeng, 2004. QTL Cartographer, Version 1.17. Department of Statistics, North Carolina State University, Raleigh, NC.
- Beavis, W. D., 1994. The power and deceit of QTL experiments: lessons from comparative QTL studies, pp. 250–266 in 49th Annual Corn and Sorghum Research Conference. American Seed Trade Association, Washington, DC.
- Bost, B., D. De Vienne, F. Hospital, L. Moreau and C. Dillmann, 2001. Genetic and nongenetic bases for the L-shaped distribution of quantitative trait loci effects. Genetics 157: 1773–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, H. D., K. G. Otto, B. E. Frewen, J. K. Mckay and D. W. Schemske, 1998. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149: 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. M., S. Tang, S. J. Knapp and L. H. Rieseberg, 2002. Genetic analysis of sunflower domestication. Genetics 161: 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. M., S. J. Knapp and L. H. Rieseberg, 2005. Genetic consequences of selection during the evolution of cultivated sunflower. Genetics 171: 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1991. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1993. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics 134: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Stec, J. Wendel and M. Edwards, 1990. Genetic and morphological analysis of a maize-teosinte F2 population: implications for the origin of maize. Proc. Natl. Acad. Sci. USA 87: 9888–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Bacigalupo and A. Stec, 1994. Inheritance of kernel weight in two maize-teosinte hybrid populations: implications for crop evolution. J. Hered. 85: 191–195. [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez, J., and P. F. Knowles, 1978. Inheritance of self-incompatibility in wild sunflower, pp. 484–489 in 8th International Sunflower Conference. International Sunflower Association, Paris.
- Fishman, L., A. J. Kelly and J. H. Willis, 2002. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution 56: 2138–2155. [DOI] [PubMed] [Google Scholar]
- Gandhi, S. D., A. F. Heesacker, C. A. Freeman, J. Argyris, K. Bradford et al., 2005. The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor. Appl. Genet. 111: 619–629. [DOI] [PubMed] [Google Scholar]
- Harlan, J. R., 1992. Crops & Man. American Society of Agronomy, Madison, WI.
- Harter, A. V., K. A. Gardner, D. Falush, D. L. Lentz, R. A. Bye et al., 2004. Origin of extant domesticated sunflowers in eastern North America. Nature 430: 201–205. [DOI] [PubMed] [Google Scholar]
- Ivanov, I. G., 1975. Study on compatibility and incompatibility display in crossing selfed sunflower lines. Rastenievud Nauki 12: 36–40. [Google Scholar]
- Kao, C. H., and Z.-B. Zeng, 1997. General formulas for obtaining the MLEs and the asymptotic variance-covariance matrix in mapping quantitative trait loci when using the EM algorithm. Biometrics 53: 653–665. [PubMed] [Google Scholar]
- Kao, C. H., Z. B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinange, E. M. K., S. P. Singh and P. Gepts, 1996. Genetic control of the domestication syndrome in common bean. Crop Sci. 36: 1037–1045. [Google Scholar]
- Ladizinsky, G., 1985. Founder effect in crop-plant evolution. Econ. Bot. 39: 191–199. [Google Scholar]
- Lai, Z., K. Livingstone, Y. Zou, S. A. Church, S. J. Knapp et al., 2005. Identification and mapping of SNPs from ESTs in sunflower. Theor. Appl. Genet. 111: 1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lester, R. N., 1989. Evolution under domestication involving disturbance of genic balance. Euphytica 44: 125–132. [Google Scholar]
- Lincoln, S. E., and E. S. Lander, 1992. Systematic detection of errors in genetic linkage data. Genomics 14: 604–610. [DOI] [PubMed] [Google Scholar]
- Liu, A. Z., and J. M. Burke, 2006. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics 173: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1998. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., S. Damon, J. D. Hewitt, D. Zamir, H. D. Rabinowitch et al., 1991. Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127: 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., Y. Saranga, M. Menz, C. X. Jiang and R. J. Wright, 2003. QTL analysis of genotype × environment interactions affecting cotton fiber quality. Theor. Appl. Genet. 106: 384–396. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., A. Widmer, A. M. Arntz and J. M. Burke, 2002. Directional selection is the primary cause of phenotypic diversification. Proc. Natl. Acad. Sci. USA 99: 12242–12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra, J., 2005. Quantitative trait loci and the study of plant domestication. Genetica 123: 197–204. [DOI] [PubMed] [Google Scholar]
- Sanguinetti, C. J., E. D. Neto and A. J. G. Simpson, 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17: 914–921. [PubMed] [Google Scholar]
- Schuelke, M., 2000. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18: 233–234. [DOI] [PubMed] [Google Scholar]
- Slabaugh, M. B., G. M. Huestis, J. Leonard, J. L. Holloway, C. Rosato et al., 1997. Sequence-based genetic markers for genes and gene families: single-strand conformational polymorphisms for the fatty acid synthesis genes of Cuphea. Theor. Appl. Genet. 94: 400–408. [Google Scholar]
- Sossey-Alaoui, K., H. Serieys, M. Tersac, P. Lambert, E. Schilling et al., 1998. Evidence for several genomes in Helianthus. Theor. Appl. Genet. 97: 422–430. [Google Scholar]
- Stuber, C. W., R. H. Moll, M. M. Goodman, H. E. Schaffer and B. S. Weir, 1980. Allozyme frequency changes associated with selection for increased grain-yield in maize (Zea mays L.). Genetics 95: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., and S. J. Knapp, 2003. Microsatellites uncover extraordinary diversity in native American land races and wild populations of cultivated sunflower. Theor. Appl. Genet. 106: 990–1003. [DOI] [PubMed] [Google Scholar]
- Tang, S., J. K. Yu, M. B. Slabaugh, D. K. Shintani and S. J. Knapp, 2002. Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105: 1124–1136. [DOI] [PubMed] [Google Scholar]
- Tang, S., V. K. Kishore and S. J. Knapp, 2003. PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor. Appl. Genet. 107: 6–19. [DOI] [PubMed] [Google Scholar]
- Tang, S., A. Leon, W. C. Bridges and S. J. Knapp, 2006. Quantitative trait loci for genetically correlated seed traits are tightly linked to branching and pericarp pigment loci in sunflower. Crop Sci. 46: 721–734. [Google Scholar]
- Wang, R.-L., A. Stec, J. Hey, L. Lukens and J. Doebley, 1999. The limits of selection during maize domestication. Nature 398: 236–239. [DOI] [PubMed] [Google Scholar]
- Wills, D. M., and J. M. Burke, 2006. Chloroplast DNA variation confirms a single origin of domesticated sunflower (Helianthus annuus L.). J. Hered. 97: 403–408. [DOI] [PubMed] [Google Scholar]
- Wills, D. M., M. L. Hester, A. Z. Liu and J. M. Burke, 2005. Chloroplast SSR polymorphisms in the Compositae and the mode of organellar inheritance in Helianthus annuus. Theor. Appl. Genet. 110: 941–947. [DOI] [PubMed] [Google Scholar]
- Xiao, J. H., J. M. Li, S. Grandillo, S. N. Ahn, L. P. Yuan et al., 1998. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150: 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L. X., K. D. Liu, X. K. Dai, C. G. Xu and Q. F. Zhang, 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251. [Google Scholar]
- Yu, J. K., S. Tang, M. B. Slabaugh, A. Heesacker, G. Cole et al., 2003. Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43: 367–387. [Google Scholar]
- Zeng, Z.-B., 1993. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z.-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]