Abstract
A family of HEAT-repeat containing ribosome synthesis factors was previously identified in Saccharomyces cerevisiae. We report the detailed characterization of two of these factors, Utp10 and Utp20, which were initially identified as components of the small subunit processome. Coprecipitation analyses confirmed the association of Utp10 and Utp20 with U3 snoRNA and the early pre-rRNA processing intermediates. Particularly strong association was seen with aberrant processing intermediates, which may help target these RNAs for degradation. Genetic depletion of either protein inhibited the early pre-rRNA processing steps in 18S rRNA maturation but had little effect on pre-rRNA transcription or synthesis of the 25S or 5.8S rRNAs. The absence of the poly(A) polymerase Trf5, a component of the TRAMP5 complex and exosome cofactor, led to stabilization of the aberrant 23S RNA in strains depleted of Utp10 or Utp20. In the case of Utp10, 20S pre-rRNA synthesis was also modestly increased by this loss of surveillance activity.
Keywords: yeast, RNA processing, RNA surveillance, ribosome synthesis, nucleolus, nuclear transport
INTRODUCTION
The eukaryotic ribosome is a large ribonucleoprotein (RNP) particle that assembles from small (40S) and large (60S) subunits during translation initiation. Biogenesis of the subunits is a highly complex process and mainly occurs in a specialized nuclear subcompartment, the nucleolus. However, late maturation steps take place in the nucleoplasm and in the cytoplasm, following nucleocytoplasmic export (for reviews, see Venema and Tollervey 1999; Lafontaine and Tollervey 2001; de la Cruz et al. 2003; Fromont-Racine et al. 2003). The 40S subunit is assembled around the 18S rRNA, whereas the 60S particle contains the 25S, 5.8S, and 5S rRNAs. Three rRNAs (18S, 25S, and 5.8S) are processed from a large precursor, the 35S pre-rRNA, transcribed by RNA polymerase I. Pre-rRNA processing involves a series of exonuclease and endonuclease steps that eliminate internal and external spacer sequences. Moreover, ∼100 nucleotide modifications are introduced, most of which are catalyzed by the small nucleolar RNPs (snoRNPs) (Kiss 2001; Bachellerie et al. 2002; Terns and Terns 2002).
In Saccharomyces cerevisiae, more than 180 nonribosomal RNAs and proteins have been shown to participate directly in post-transcriptional steps of ribosome biogenesis. The use of genetic and proteomic analyses allowed the prediction of several distinct RNP intermediates in the pathway of 40S and 60S ribosome subunit synthesis (for review, see de la Cruz et al. 2003; Fromont-Racine et al. 2003; Dez and Tollervey 2004). Among these, the small-subunit (SSU) processome, a large RNP complex containing the U3 snoRNA, is involved in the three early pre-rRNA cleavages at sites A0, A1, and A2 (Dragon et al. 2002; Bernstein and Baserga 2004; Osheim et al. 2004).
Here we characterize the defects in ribosome biogenesis in strains depleted of the ribosome synthesis factors Utp10 and Utp20, which were initially purified as components of the SSU processome (Dragon et al. 2002; Bernstein and Baserga 2004). Utp10, but not Utp20, was classed as a transcription-Utp (t-Utp), a group of ribosome synthesis factors that were reported to also be required for efficient pre-rRNA transcription by RNA polymerase I. Utp10 and Utp20 consist of repeated, predominantly α-helical motifs termed HEAT-repeats (Huntington, elongation A subunit, TOR) that are related to Armadillo-like repeats (Andrade et al. 2001). HEAT-repeat proteins perform many different functions in the cell and include the importin-β/karyopherin-β family of nucleo-cytoplasmic transport factors. By analogy with this family, we speculated that HEAT-repeat ribosome synthesis factors function in ribosomal subunit export (Dlakic and Tollervey 2004; Oeffinger et al. 2004; Dez et al. 2006). Consistent with this model, five HEAT-repeat ribosome synthesis factors (Noc2, Noc3, Noc4, Rrp12, and Sda1) were indeed found to be required for subunit export, while Noc1 was implicated in intranuclear movement of the preribosomes (Milkereit et al. 2001, 2003; Oeffinger et al. 2004; Dez et al. 2006). This does not, however, appear to be the case for Utp10 and Utp20, which are required for pre-rRNA processing on the pathway of 18S synthesis.
RESULTS
Structural modeling of Utp10 and Utp20
Yeast Utp10 and Utp20 were previously predicted to be HEAT-repeat proteins (Oeffinger et al. 2004). We used profile-profile searches (Soding 2005) to identify best structural matches for both proteins in the Protein Data Bank (PDB). Many significant matches were detected, and we chose Cand1 (Goldenberg et al. 2004) because it enabled us to model the largest part of both proteins. Both proteins are predicted to have extended, curved structures, with large surface areas available for potential interactions with other proteins and/or rRNA. Supplemental Figure S1 shows the model for Utp20 residues 1320–2442 (http://www.homepage.montana.edu/∼mdlakic/heat_Utp20p_suppl_FIG1.html).
Utp10 and Utp20 are components of 90S and 40S preribosomes
Previous analysis identified Utp10 and Utp20 as components of the SSU processome (Dragon et al. 2002; Bernstein and Baserga 2004). To confirm these results, we analyzed RNA species associated in vivo with these two proteins. A Utp20-TAP fusion was poorly functional, and Western analysis showed the fusion protein to be severely degraded (data not shown). The coprecipitation analyses were therefore performed using strains expressing N-terminal ProtA-tagged versions of Utp10 and Utp20 (see Table 1) under the control of a GAL10 promoter (Lafontaine and Tollervey 1996). Results obtained using Utp10-TAP were identical to the GAL∷ProtA-Utp10 construct (data not shown).
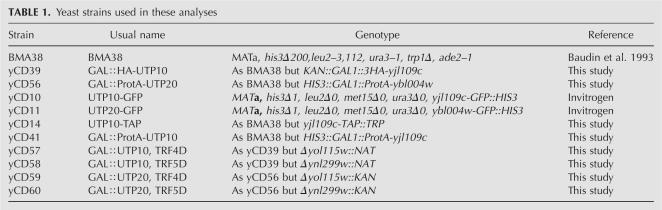
TABLE 1.
Yeast strains used in these analyses
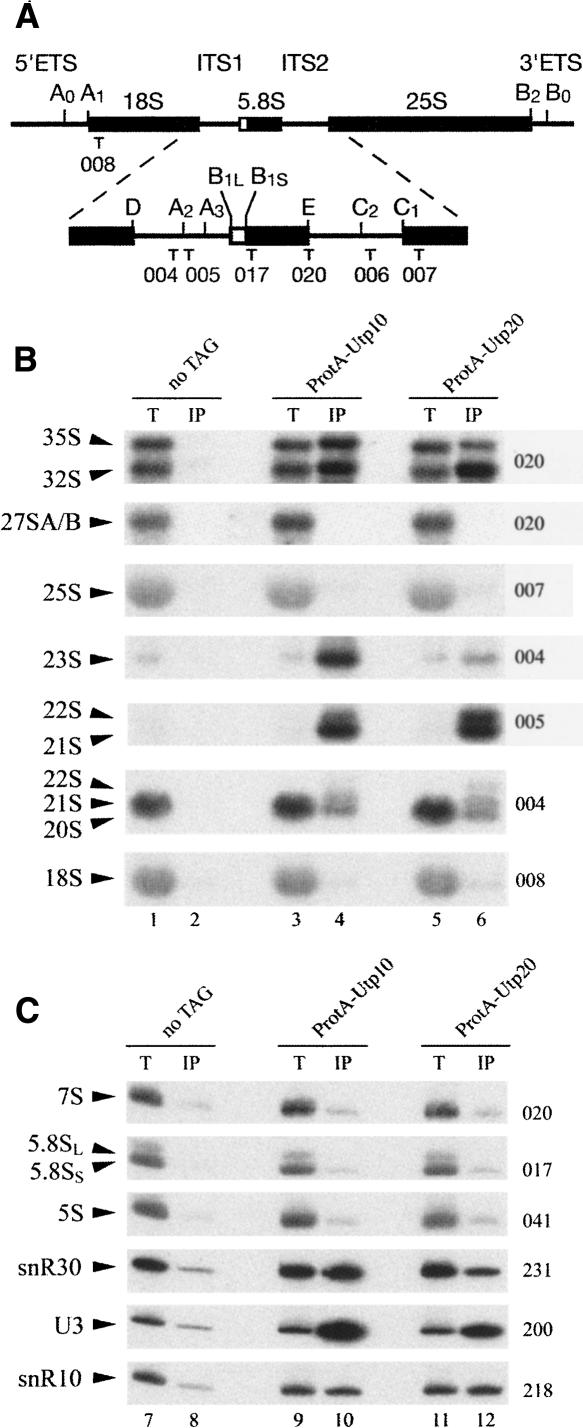
Growth of the ProtA-tagged strains on galactose medium was indistinguishable from the nontagged wild type, indicating that these fusion proteins were fully functional (data not shown). Following immunoprecipitation, bound RNAs were analyzed by Northern hybridization (Fig. 1) and compared with RNAs recovered in parallel from the nontagged control strain (no TAG lanes). Consistent with the association of Utp10 and Utp20 with the U3 processome, the U3 snoRNA was efficiently coprecipitated, as were snR10 and snR30 (Fig. 1C). The 35S and 32S pre-rRNAs also coprecipitated with both Utp10 and Utp20, confirming they are also part of the 90S preribosomal particles (Fig. 1B). However, whereas Utp10 and Utp20 precipitated the same amount of 32S pre-rRNA, the recovery of 35S pre-rRNA with Utp10 was higher than with Utp20. This observation suggests that Utp20 joins the 90S preribosomal particle later than Utp10, probably shortly before the cleavages at sites A0 and A1 that convert the 35S to 32S pre-rRNA. However, we cannot exclude the possibility that the differences in precipitation efficiencies reflect changes in accessibility of the TAP tag in different complexes. The aberrant 23S, 22S, and 21S rRNAs were surprisingly strongly coprecipitated with both Utp10 and Utp20. The late 20S pre-rRNA coprecipitated with Utp10 and Utp20, showing that the proteins stay associated with the pre-40S particle after A0, A1, and A2 cleavages. However, the 20S pre-rRNA precipitation efficiency was weaker than that for 35S and 32S pre-rRNA, presumably because much of the 20S pre-rRNA population is localized in the cytoplasm of the cells. In contrast, no coprecipitation was seen for pre-rRNAs on the pathway of 60S subunit synthesis, the 27SA/B and 7S pre-rRNAs, or any of the mature rRNA species.
FIGURE 1.
Utp10 and Utp20 are associated with early pre-rRNAs and snoRNAs. (A) The 35S pre-rRNA contains the sequences of the mature 18S, 5.8S, and 25S rRNAs, which are separated by internal transcribed spacers 1 and 2 (ITS1 and ITS2) and flanked by the 5′ and 3′ external transcribed spacers (5′ETS and 3′ETS). Locations of the different oligonucleotides used in this study are shown. (B, C) Northern analysis of rRNAs coprecipitated with Protein A–tagged versions of Utp10 and Utp20 (lanes 3–6, 9–12) or from extracts of cells lacking a tagged protein (lanes 1,2,7,8). Immunoprecipitation was performed on cell extracts using IgG-Sepharose. RNAs were extracted from the pellet after precipitation (lanes IP) or from total cell extract (lanes T) corresponding to 5% of the input for the immunoprecipitation reactions in panel B (1.2% denaturing agarose gel) or 10% of the input in the panel C (8% polyacrylamide/urea gel). Following separation, RNAs were transferred to a nylon membrane and hybridized with the anti-sense oligonucleotides indicated to the right of each panel.
Utp10 and Utp20 are required for pre-rRNA processing
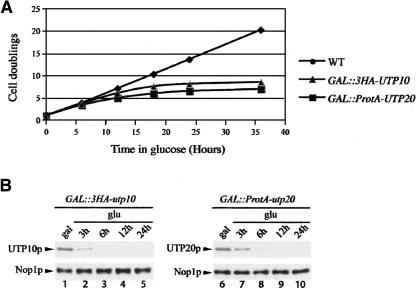
To define the role of Utp10 and Utp20 in ribosome biogenesis, we constructed N-terminal 3HA-utp10 and ProtA-utp20 fusions under the control of the glucose repressible GAL1 promoter, by one-step PCR (Lafontaine and Tollervey 1996; Longtine et al. 1998). On galactose containing YNB medium, the growth rates of the GAL∷HA-utp10, GAL∷ProtA-utp20 strains and otherwise isogenic wild type were almost identical (2 h). Twelve hours after transfer to the nonpermissive glucose medium, the growth rate of both strains was already substantially reduced with a doubling time of ∼5 h. Growth essentially ceased by 20 h after transfer (Fig. 2A). The kinetics of depletion of Utp10 and Utp20 were assessed by Western blotting (Fig. 2B), which showed that the abundance of 3HA-Utp10 was strongly reduced 3 h after transfer to glucose medium and became undetectable after 6 h. Utp20 depletion was slightly slower, with a low level of ProtA-Utp20 still detectable after 6 h in glucose media.
FIGURE 2.
Depletion of Utp10 and Utp20. (A) Growth rate of wild-type, GAL∷HA-UTP10, and GAL∷ProtA-UTP20 strains following a transfer from permissive galactose medium to nonpermissive glucose medium for the times indicated. Cells were maintained in exponential growth throughout the time course by addition of pre-warmed medium. (B) Western analysis of 3HA-Utp10 and ProtA-Utp20 depletion. Total protein was extracted at the times indicated and analyzed by Western blotting with antibodies that decorate the tags present on Utp10 and Utp20, or with anti-Nop1 as loading control. 3HA-Utp10 was decorated with rabbit anti-HA primary antibody, which was subsequently detected using anti-rabbit IgG linked to horseradish peroxidase. ProtA-Utp20 was detected with a peroxidase-antiperoxidase (PAP) complex.
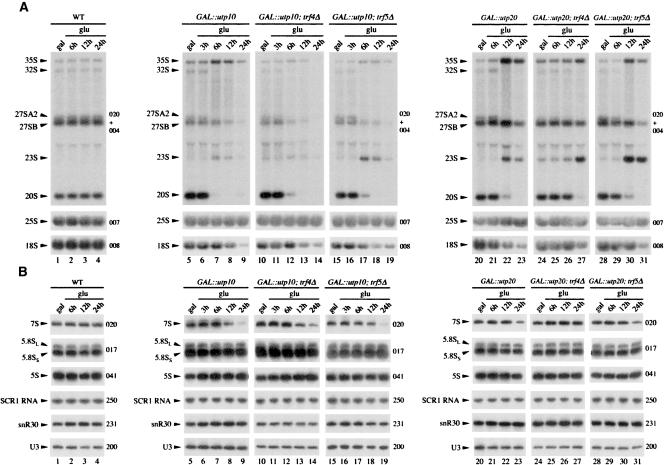
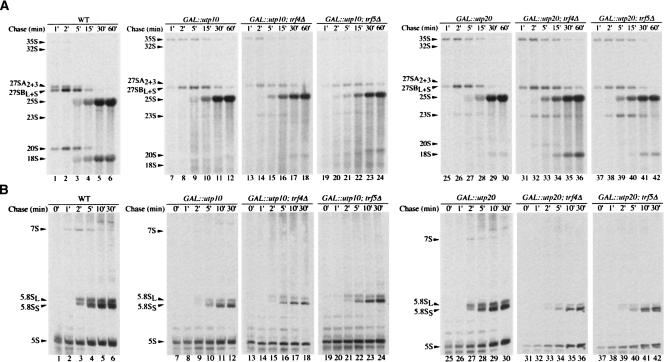
In order to determine the steps in pre-rRNA processing that require Utp10 and Utp20, we performed Northern analyses using GAL∷HA-utp10 (Fig. 3A,B, lanes 5–9), GAL∷ProtA-utp20 (Fig. 3A,B, lanes 20–23), and wild-type strains (Fig. 3A,B, lanes 1–4). Strains depleted of Utp10 or Utp20 showed defects in the pathway of 40S subunit synthesis. Levels of the 35S pre-rRNA increased, while the 32S, 27SA2, and 20S pre-rRNA were reduced, consistent with the inhibition of pre-rRNA cleavage at sites A0, A1, and A2. This conclusion was supported by the accumulation of the aberrant 23S RNA, which is produced by cleavage of the 35S pre-rRNA at site A3 in the absence of cleavage at sites A0, A1, and A2. Pre-rRNA processing defects were clearly visible following depletion of Utp10 for 6 h and after depletion of Utp20 for 12 h, consistent with the slower depletion of Utp20 seen in Western blots (Fig. 2B). At later time points, the mature 18S rRNA was depleted in the strains lacking Utp10 or Utp20, whereas levels of the 25S, 5.8S, and 5S rRNAs were stable. The levels of the 27SB and 7S pre-rRNA were also lower in both mutants, but this may largely reflect reduced synthesis as a consequence of growth inhibition. The ratio between the long and short forms of 5.8S rRNA was unaltered, indicating that the alternative pre-rRNA processing pathways that generate these rRNAs both remain active. Together these observations indicate that depletion of Utp10 or Utp20 leads to the inhibition of cleavage at sites A0, A1, and A2. Ongoing cleavage at A3 and maturation at B1L generates the 27SA3 and 27SBL pre-rRNAs, respectively, which can be matured to the 25S and 5.8S rRNAs in the absence of Utp10 or Utp20.
FIGURE 3.
18S rRNA synthesis is impaired in strains depleted of Utp10 and Utp20, and aberrant pre-rRNAs are stabilized in the absence of Trf5p. Cells were pregrown on galactose medium and transferred to glucose medium. Total RNA was extracted at the times indicated and analyzed by Northern blotting. (A) Northern hybridization of high molecular weight RNAs separated on an 1.2% agarose/glyoxal gel. (B) Northern hybridization of low molecular weight RNAs separated on an 8% polyacrylamide/8 M urea gel. Oligonucleotides used for Northern hybridizations are indicated on the right of each panel.
Pre-rRNA processing was also assessed by pulse-chase labeling, performed 6 h and 12 h, respectively, after transfer of the GAL∷HA-utp10 and GAL∷ProtA-utp20 strains to glucose medium (Fig. 4). Consistent with the Northern data, processing of the 35S pre-rRNA was delayed, whereas synthesis of 27SA and 20S pre-rRNA and 18S rRNA was greatly reduced, in the strains depleted of Utp10 (Fig. 4A, lanes 7–12) or Utp20 (Fig. 4A, lanes 25–30) compared with the wild type (Fig. 4A, lanes 1–6). The aberrant 23S RNA was visible but did not accumulate to high levels, consistent with previous reports that it is rapidly degraded (Allmang et al. 2000; Houseley and Tollervey 2006). Maturation of the 27SB pre-rRNA appeared to be slowed, but accumulation of mature 25S rRNA was not clearly reduced. These results confirmed that cleavage at A0, A1, and A2 is inhibited in cells lacking Utp10 and Utp20.
FIGURE 4.
Pulse-chase labeling of RNA. GAL∷HA-UTP10 and GAL∷ProtA-UTP20 strains were pre-grown on galactose medium and transferred to glucose medium for 6 h and 12 h, respectively. Cells were then pulse labeled with [3H] adenine for 2 min. After adding an excess of cold adenine, samples were collected at the indicated times, and RNAs were extracted, separated by gel electrophoresis, and transferred to a nylon membrane. Labeled RNAs were detected by fluorography. (A) High molecular weight RNAs separated on an 1.2% agarose/glyoxal gel. (B) Low molecular weight RNAs separated on an 8% polyacrylamide/8 M urea gel. Wild-type and GAL∷utp20 samples were each exposed for 24h, the GAL∷utp10 samples were exposed for 36 h.
In strains with fast-acting temperature-sensitive defects in pre-60S export, the nuclear-restricted 27S pre-rRNA and 25S rRNAs are degraded by the exosome following prior polyadenylation by the poly(A)polymerase Trf4 (Dez et al. 2006). We therefore assessed whether pre-rRNA degradation, particularly of the aberrant 23S RNA generated in the absence of Utp10 and Utp20, required either Trf4 or the homologous poly(A) polymerase Trf5 (Haracska et al. 2005; Egecioglu et al. 2006; Houseley and Tollervey 2006). GAL∷HA-utp10 and GAL∷ProtA-utp20 strains also carrying deletions of either TRF4 or TRF5 were created and analyzed (Figs. 3, 4). Northern analyses showed that the absence of Trf5 stabilized the 23S RNA in both Utp10 (Fig. 3A, lanes 15–19) and Utp20-depleted (Fig. 3A, lanes 28–31) strains, but the increase was much more marked following Utp10 depletion. In contrast, the absence of Trf4 did not clearly stabilize 23S RNA during Utp10 depletion (Fig. 3A, lanes 10–14). In the Utp20-depleted strain, the trf4Δ mutation delayed the appearance of the processing defects. However, the trf4Δ mutation, but not the trf5Δ mutation, significantly slows cell growth (data not shown), and we believe that this is responsible for the delayed onset of the phenotype. Pulse-chase analysis (Fig. 4A) confirmed that 23S is stabilized by the deletion of Trf5, and also revealed an increase in 20S synthesis, which appeared only at late chase time points. We speculate that stabilization of the 23S RNA in the absence of Trf5 allows a fraction to be matured to 20S in strains with reduced levels of Utp10. We did not, however, see any clear alteration in relative growth rates of the Utp10-depleted strain in the presence or absence of Trf5. It may be that the stabilized 20S is present in ribosomes that are defective due to loss of Utp10. So even though the pre-rRNA is partially rescued from degradation, it is probably still not functional.
Depletion of Utp10 and Utp20 leads to nuclear accumulation of Rps2-GFP but not of pre-RNA
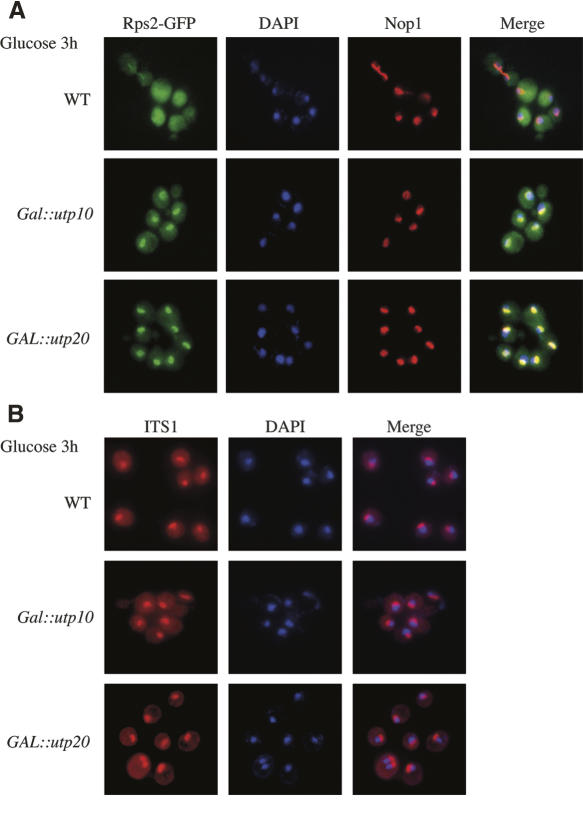
To determine whether Utp10 and Utp20 are required for nuclear export of the small ribosomal subunit, the localization of the 40S reporter construct Rps2-eGFP (Stage-Zimmermann et al. 2000) was analyzed in wild-type, GAL∷HA-utp10, and GAL∷ProtA-utp20 strains. Rps2-eGFP was found throughout the cell with no visible nuclear enrichment, during growth of all strains on galactose medium (data not shown) and following transfer of the wild-type strain to glucose (Fig. 5A). In the GAL∷HA-utp10 and GAL∷ProtA-utp20 strains shifted to glucose medium for 3 h, the Rps2-eGFP signal showed strong nuclear accumulation, with nucleolar enrichment (Fig. 5A). This phenotype was observed prior to the appearance of detectable pre-rRNA processing defects (Fig. 3), and no accumulation of the 60S reporter Rpl11b-eGFP was seen at this time (data not shown). At a later time point after the shift to glucose media, ∼10% of Utp10- or Utp20-depleted cells showed nuclear Rpl11b-eGFP accumulation (data not shown), probably reflecting the secondary consequence of slower pre-rRNA processing.
FIGURE 5.
Strains depleted of Utp10 or Utp20 did not show impaired export of 40S ribosomal subunits. (A) GAL∷HA-UTP10 and GAL∷ProtA-UTP20 strains expressing an Rps2-GFP fusion were pre-grown on galactose medium and transferred to glucose medium for 3 h. Cells were fixed, DAPI stained, and viewed by fluorescence microscopy. The nucleolus was visualized using an anti-Nop1 antibody, which was subsequently recognized by an Alexafluor 555 conjugated secondary antibody. (B) Pre-18S rRNA FISH with a probe complementary to the D–A2 segment of the ITS1 in wild-type, GAL∷HA-UTP10, and GAL∷ProtA-UTP20 strains grown for 3 h in glucose-containing medium.
To better determine the localization of the pre-40S particles, in situ hybridization was performed with a probe directed against the 5′ region of ITS1, which is present in the 20S pre-rRNA that is exported to the cytoplasm (Fig. 5B). Unexpectedly, this failed to confirm the pre-40S export defect. The levels of the nuclear and cytoplasmic signals obtained with the ITS1 probe were not clearly altered after depletion of Utp10 or Utp20 for by growth on glucose for 3 h. The basis of the nuclear accumulation of Rps2-GFP is currently unclear, but this does not appear to reflect pre-40S accumulation. We conclude that functional Utp10 and Utp20 are not directly required for efficient export of pre-40S ribosomal subunits from the nucleoplasm to the cytoplasm.
To follow the localization of Utp10 and Utp20, C-terminal fusions with GFP were constructed (see Materials and Methods). Utp10-GFP was strongly concentrated in a subnuclear region that appeared to form a cap on the nucleoplasm, identified by DAPI staining (Fig. 6A). Such a distribution is highly characteristic of localization to the yeast nucleolus. Utp20 was also enriched in the nucleolus, but additional distribution throughout the nucleoplasm and cytoplasm was seen (Fig. 6A).
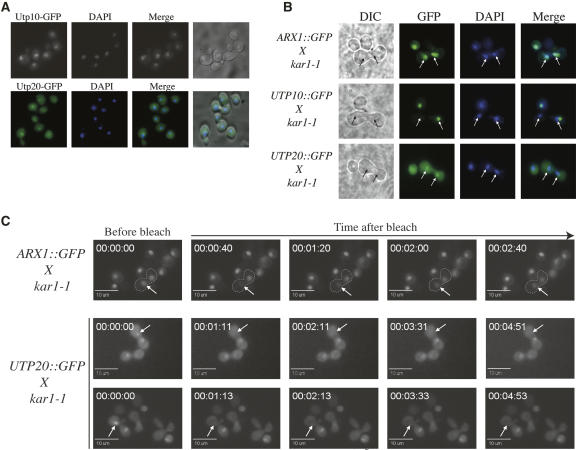
FIGURE 6.
Utp10 and Utp20 are concentrated in the nucleolus but do not shuttle between nuclei in a heterokaryon assay. (A) At steady state, Utp10-GFP localizes to the nucleolus, and Utp20-GFP is present in the whole cell but is enriched in the nucleolus. Cells were fixed, DAPI stained, and viewed by fluorescence microscopy. (B) Utp10-GFP does not shuttle from the nucleus to the cytoplasm. Strains expressing Utp10-GFP, Utp20-GFP, and Arx1-GFP were grown in YPD media, mated with a kar1-1 mutant strain, and incubated at 25°C until heterokaryons formed. Localization of the GFP-tagged proteins in heterokaryons is shown for Arx1-GFP (upper panel), Utp10-GFP (middle panel), and Utp20-GFP (lower panel). Cells are shown with DAPI-stained nucleoplasm. Arrows indicate the positions of nuclei in the heterokaryons. (C) Utp20-GFP does not rapidly shuttle from the nucleus to the cytoplasm. One nucleus from each heterokaryon expressing either Arx1-GFP or Utp20-GFP was bleached. The recovery of fluorescence of the bleached nuclei was followed during a time course after bleaching. Arrows indicate the position of the targeted nucleus.
To assess whether Utp10 and Utp20 accompany pre-40S particles from the nucleus through the NPC and out into the cytoplasm, heterokaryon experiments were performed (Fig. 6B,C). Strains expressing Utp10-GFP, Utp20-GFP, or the shuttling ribosome synthesis factor Arx1-GFP (Belaya et al. 2006) were crossed with a strain carrying kar1–1, which blocks nuclear fusion after mating, leading to heterokaryon formation (Vallen et al. 1992). As expected, Arx1p-GFP was found in both nuclei of the heterokaryons (Fig. 6B). In contrast, Utp10-GFP was consistently localized to only one nucleus in heterokaryons, indicating that this protein does not actively shuttle between the nuclei (Fig. 6B). Utp20-GFP was found in both nuclei of most heterokaryons examined, but the GFP intensity of the two nuclei often appeared uneven (data not shown; Fig. 6B).
To more clearly assess whether Utp20 actively shuttles between the nucleus and the cytoplasm, we used a modified heterokaryon assay utilizing a FRAP/FLIP photobleaching technique (White and Stelzer 1999; Belaya et al. 2006). In these experiments, one nucleus of the heterokaryon was completely bleached. Recovery of fluorescence in the targeted nucleus (fluorescence recovery after photobleaching; FRAP), as well as loss of fluorescence in the unbleached nucleus (fluorescence loss in photobleaching; FLIP), was followed in real time (Fig. 6C). As previously reported, bleaching of one nucleus in the heterokaryon expressing Arx1-GFP was followed by rapid recovery of the targeted nucleus, accompanied by loss of fluorescence in the nontargeted nucleus (Fig. 6C; Belaya et al. 2006). In contrast, bleaching of one nucleus in the heterokaryon expressing Utp20-GFP was followed by much slower recovery, indicating that Utp20-GFP does not rapidly shuttle between the nuclei.
These data show that a conventional heterokaryon assay, in which the proteins have at least an hour to equilibrate between the nuclei, led to a misleading conclusion on the shuttling behavior of Utp20-GFP. This emphasizes the importance of following protein shuttling over short time periods to assess the kinetics of the exchange.
DISCUSSION
Here we report the functional analysis of two members of the HEAT-repeat family of ribosome synthesis factors, Utp10 and Utp20. These proteins were identified as components of the SSU processome, and consistent with this, we report that genetic depletion of either protein specifically inhibits the early pre-rRNA processing steps that form the pathway of pre-40S subunit synthesis. In addition, Protein A–tagged forms of Utp10 and Utp20 coprecipitated with the 35S, 32S, and 20S pre-rRNAs, as well as several snoRNAs tested, including U3, but not with pre-rRNAs on the pathway of 60S subunit synthesis (27S and 7S pre-rRNA). This is consistent with specific association with early 90S and pre-40S particles. The efficiency of precipitation of 35S relative to later pre-rRNAs was greater with Utp10 than with Utp20. This differential association correlates with the fact that Utp10, but not Utp20, is classed as a t-Utp (Gallagher et al. 2004) or component of the UTP A complex (Krogan et al. 2004) and, as such, is predicted to assemble very early on the pre-rRNA transcript (Perez-Fernandez et al. 2007). However, the robust ongoing transcription in the Utp10-depleted cells revealed by metabolic labeling was surprising. The t-Utps, including Utp10, were reported to be required for pre-rRNA transcription by RNA polymerase I (Gallagher et al. 2004). However, following depletion of Utp10 for 6 h, a time point at which pre-rRNA processing was greatly impaired, pre-rRNA transcription was not strongly inhibited, calling this conclusion into question. Moreover, pre-rRNA transcription was not clearly differentially inhibited following depletion of Utp10 relative to Utp20, which is not a tUtp.
A striking feature of the coprecipitation data was the strong recovery of the aberrant 23S, 22S, and 21S RNAs with both Utp10 and Utp20. These RNAs are detectable in wild-type cells, but at very low levels, and were strongly enriched in the immunoprecipitates relative to the normal 35S, 32S, and 20S pre-rRNAs. The 21S and 20S RNAs are cleaved at the 5′ end of the 18S rRNA, showing that the 5′-ETS region is not required for the association of Utp10 or Utp20 with the pre-40S particles. These data suggest that the association of Utp10 and Utp20 with the, presumably defective, preribosomes that contain the 23S, 22S, and 21S RNAs is more stable than association with preribosomes that are maturing normally. It remains unclear how “normal” and “defective” preribosomes are distinguished by the nuclear surveillance machinery; however, one possibility is that this discrimination is primarily kinetic. The association of most processing factors with the pre-rRNA is expected normally to be very transient. However, any defect in pre-rRNA processing or ribosome assembly will slow maturation, potentially leading to prolonged association of processing factors with the defective preribosomes—regardless of the actual underlying defect. We speculate that retention of the pre-rRNA processing factors (or a subset of processing factors) on the pre-rRNA is sufficient to recruit the nuclear RNA surveillance machinery. The apparently very stable association of Utp10 and Utp20 with defective preribosomes might therefore be part of the mechanism that targets these particles to the degradation system.
The Trf/Air/Mtr4 polyadenylation (TRAMP) complexes are major nuclear cofactors for RNA degradation by the exosome complex of 3′-exoribonucleases (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Buhler et al. 2007). Two forms of the TRAMP complex have been identified in budding yeast, which differ in the presence of either Trf4 (in TRAMP4) or Trf5 (in TRAMP5). Trf4 and Trf5 are poly(A) polymerases that function, at least in part, by adding single-stranded adenosine tails to target RNAs, making them better substrates for 3′-degradation (Haracska et al. 2005; LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006; Houseley and Tollervey 2006). Mtr4 is a DExH box putative RNA helicase, which is shared between the TRAMP complexes. The remaining TRAMP component is either Air1 or Air2, two homologous zinc-knuckle proteins. At present, no functional distinctions have been detected between Air1 and Air2, and no phenotypes have been reported for air1Δ or air2Δ single mutant strains, suggesting that that they may be functionally redundant. If so, then functional differences between the TRAMP4 and TRAMP5 complexes are presumably due solely to distinctions between Trf4 and Trf5. The absence of Trf4 stabilized the 27SB pre-rRNA and 25S rRNA present in defective pre-60S particles in sda1-2 mutant strains (Dez et al. 2006). In contrast, we saw no convincing evidence for pre-rRNA stabilization in Utp10- or Utp20-depleted strains that also lacked Trf4. The aberrant 23S RNA was, however, stabilized by the absence of Trf5. Increased polyadenylation of 23S RNA in strains lacking the nuclear exosome component Rrp6 was also reported to be largely dependent on Trf5 (Houseley and Tollervey 2006). This would be consistent with the model that TRAMP5 acts early in the ribosome synthesis pathway during surveillance of 90S and pre-40S particles, whereas TRAMP4 functions in surveillance of later pre-60S subunits. It is likely that Trf4 and Trf5 share at least some common substrates since the double mutant shows synthetic lethality. However, the proteins are only ∼56% identical. They are highly homologous over the central polynucleotide polymerase domain and the C-terminal PAP-associated motif, but poorly conserved over their ∼150 amino acid N-terminal regions. It therefore seems likely that differential binding of recruitment factors to the N-terminal regions of Trf4 and Trf5 is responsible for differences in substrates specificity of the type we report here.
We also addressed the requirements for Utp10 and Utp20 in the export of ribosomal subunits to the cytoplasm. Fusions between GFP and the 40S subunit component Rps2 (Milkereit et al. 2003) and the 60S component Rpl11b (Stage-Zimmermann et al. 2000) were used as reporters for 40S and 60S subunit export. At least in some cases, such r-proteins were shown to truly reflect the localization of nuclear-accumulated preribosomes (Dez et al. 2006). Depletion of Utp10 or Utp20 did not lead to detectable nuclear accumulation of Rpl11b-GFP but did result in strong Rps2-GFP accumulation. A strong nuclear accumulation phenotype was visible in both the GAL∷utp10 and GAL∷utp20 strains 3 h after transfer to glucose medium. This is well before defects in pre-rRNA processing were detected. Unexpectedly, however, in situ hybridization with a probe directed against the 5′ region of ITS1, which is retained in the pre-40S particles that are exported to the cytoplasm, failed to confirm the nuclear accumulation of the pre-rRNA. We conclude that in these strains Rps2-GFP was not a faithful reporter of the localization of nuclear pre-40S ribosomes, and that Utp10 and Utp20 do not appear to play any direct role in 40S subunit export. The reason for the discrepancy between the data obtained with Rps2-GFP and the ITS1 probe is unclear. One possibility is that the pre-40S particles synthesized and exported in strains depleted of Utp10 and Utp20 are deficient in Rps2. Alternatively, degradation of pre-40S particles may release Rps2 either free or in partially disassembled preribosomal complexes, which may accumulate in the nucleolus.
To assess whether Utp10 and/or Utp20 accompany the pre-40S particles through the NPC and out into the cytoplasm, we initially used a conventional yeast heterokaryon assay. This showed that Utp10-GFP did not shuttle between nuclei in the heterokaryon, whereas the behavior of Utp20-GFP was consistent with shuttling. However, the use of a more sensitive assay based on photobleaching in heterokaryons (Belaya et al. 2006) allowed us to demonstrate that Utp20 does not shuttle at a substantial rate. In all cells examined Utp20-GFP gave a significant cytoplasmic signal, but the photobleaching experiment shows that this is due to leakage or inefficient nuclear import, rather than active exchange with the nuclear/nucleolar pool. These observations demonstrate that results of conventional heterokaryon assays should be interpreted with caution.
MATERIALS AND METHODS
Strains, media, plasmids, and cloning
A list of strains used is given in Table 1. Standard procedures were used for the propagation of yeast using YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or YNB medium (0.67% yeast nitrogen base, 0.5% (NH4)2SO4, and 2% glucose) supplemented with the required amino acids.
UTP10 and UTP20 GAL conditional mutants were constructed by one-step PCR using pFA6a-kanMX6-PGAL1–3HA (Longtine et al. 1998) and ptl27 (Lafontaine and Tollervey 1996), respectively. Transformants were selected for resistance to G418 or HIS prototrophy and screened by PCR and immunoblotting. trf4Δ and trf5Δ alleles were constructed using a one-step PCR strategy with pFA6a-natMX6 plasmid (Hentges et al. 2005), selected with nourseothricin (NAT), and verified by PCR. yCD14 and yCD41 strains were constructed using the classic TAP-Tag method and verified by PCR and Western-blot (note that yCD14 express only the Protein-A tag). yCD14 and yCD41 strains were transformed with pRpl11b-eGFP (Stage-Zimmermann et al. 2000) (kindly provided by Pam Silver, Harvard University) or pRps2-eGFP (Milkereit et al. 2003) (kindly provided by Ed Hurt, University of Heidelberg) to analyze the nuclear localization of (pre-)ribosomal subunits, or with a pADE2 vector for pulse chase analysis. Oligonucleotide sequences used to amplify the integrative cassettes are listed in Table 2.
TABLE 2.
Oligonucleotides used in these analyses

RNA extraction and Northern hybridization
RNA extraction and Northern hybridization were performed as previously described (Beltrame and Tollervey 1992). For high-molecular-weight RNA analysis, 5 μg of total of RNA were glyoxal denatured and resolved on a 1.2% agarose gel. Low-molecular-weight RNA products were resolved on 8% polyacrylamide/8.3 M urea gels.
Oligonucleotides used for Northern hybridizations are as follows:
004: 5′-CGGTTTTAATTGTCCTA;
005: 5′-ATGAAAACTCCACAGTG;
006: 5′-AGATTAGCCGCAGTTGG;
007: 5′-CTCCGCTTATTGATATGC;
008: 5′-CATGGCTTAATCTTTGAGAC;
017: 5′-GCGTTGTTCATCGATGC;
020: 5′-TGAGAAGGAAATGACGCT;
041: 5′-CTACTCGGTCAGGCTC;
200: 5′-UUAUGGGACUUGUU;
228: 5′-CATCCAGCTCAAGATCG;
403: 5′-ACCGTTTGGTCTACCCAAGTGAGAAGCCAAGACA; and
307: 5′-CACAGTTAACTGCGGTC.
Immunoprecipitations
Total cell extracts were prepared from strains that expressed ProtA-Utp10, ProtA-Utp20, or no tagged protein. Cells frozen in liquid nitrogen were broken in a mortar. Immunoprecipitations and analysis of coprecipitated RNAs were performed as previously described (Dez et al. 2004). Tot/IP ratios loaded were 1/20 for agarose gel and 1/10 for acrylamide gel.
Pulse chase analysis
Metabolic labeling of pre-rRNA was performed as previously described (Tollervey et al. 1993) with the following modifications. The strains were transformed with a plasmid containing the ADE2 gene. Strains were pregrown in synthetic galactose medium lacking adenine and transferred to glucose medium lacking adenine for 6 h (GAL∷HA-UTP10) or 12 h (GAL∷ProtA-UTP20). Cells with an OD600 of 0.4 were labeled with [8-3H] adenine (TRK343, Amersham) for 2 min followed by a chase of excess cold adenine. One-milliliter samples were collected at 1, 2, 5, 15, and 30 min following the addition of cold adenine, and cell pellets were frozen in liquid nitrogen. RNAs were then extracted and precipitated with ethanol.
Fluorescence microscopy
For immunofluorescence, cells were fixed in 3.7% paraformaldehyde at room temperature and spheroplasted using zymolase. Nop1 was detected with a mouse anti-Nop1 antibody (kindly provided by J. Aris, University of Florida) and a secondary goat anti-mouse antibody conjugated to Alexafluor 555 (Molecular Probes). DAPI was included in the mounting medium (Vectashield, Vector laboratories) to stain nuclear DNA. For RpS2-GFP localization assay, cells were pregrown in galactose medium and shifted to YPD media for 3 h before analysis. FISH experiments were performed as described previously (Leger-Sylvestre 2004) using Cy3-labeled ITS1 probe. Heterokaryon and FRAP analysis were performed as previously described (Belaya et al. 2006).
ACKNOWLEDGMENTS
We thank members of the Tollervey lab for help, advice, and discussion and Pam Silver and Ed Hurt for the pRpl11b-eGFP and pRps2-eGFP constructs. Many thanks to Emma Thomson and Martin Kos for critical reading of the manuscript. We also thank Philipp Milkereit and Isabelle Léger-Sylvestre for helpful discussion concerning the export assay. C.D. was the recipient of an EMBO long-term fellowship. This work was supported by the Wellcome Trust.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.609807.
REFERENCES
- Allmang, C., Mitchell, P., Petfalski, E., Tollervey, D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M.A., Petosa, C., O'Donoghue, S.I., Muller, C.W., Bork, P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.P., Cavaille, J., Huttenhofer, A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F., Cullin, C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae . Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaya, K., Tollervey, D., Kos, M. FLIPing heterokaryons to analyze nucleo-cytoplasmic shuttling of yeast proteins. RNA. 2006;12:921–930. doi: 10.1261/rna.2301806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame, M., Tollervey, D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, K.A., Baserga, S.J. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell. 2004;15:5038–5046. doi: 10.1091/mbc.E04-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler, M., Haas, W., Gygi, S.P., Moazed, D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- de la Cruz, J., Kressler, D., Linder, P. Ribosomal subunit assembly. In: Olson M.O.J., editor. The Nucleolus. Landes Bioscience; Georgetown, TX: 2003. pp. 262–290. [Google Scholar]
- Dez, C., Tollervey, D. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Dez, C., Froment, C., Noaillac-Depeyre, J., Monsarrat, B., Caizergues-Ferrer, M., Henry, Y. Npa1p, a component of very early Pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 2004;24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez, C., Houseley, J., Tollervey, D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae . EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic, M., Tollervey, D. The Noc proteins involved in ribosome synthesis and export contain divergent HEAT repeats. RNA. 2004;10:351–354. doi: 10.1261/rna.5184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu, D.E., Henras, A.K., Chanfreau, G.F. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine, M., Senger, B., Saveanu, C., Fasiolo, F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gallagher, J.E., Dunbar, D.A., Granneman, S., Mitchell, B.M., Osheim, Y., Beyer, A.L., Baserga, S.J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg, S.J., Cascio, T.C., Shumway, S.D., Garbutt, K.C., Liu, J., Xiong, Y., Zheng, N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Haracska, L., Johnson, R.E., Prakash, L., Prakash, S. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol. Cell. Biol. 2005;25:10183–10189. doi: 10.1128/MCB.25.22.10183-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges, P., Van Driessche, B., Tafforeau, L., Vandenhaute, J., Carr, A.M. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe . Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- Houseley, J., Tollervey, D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., Peng, W.T., Cagney, G., Robinson, M.D., Haw, R., Zhong, G., Guo, X., Zhang, X., Canadien, V., Richards, D.P., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;21:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D., Tollervey, D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3472. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D.L., Tollervey, D. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2001;2:514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre, I., Milkereit, P., Ferreira-Cerca, S., Saveanu, C., Rousselle, J.C., Choesmel, V., Guinefoleau, C., Gas, N., Gleizes, P.E. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. Embo J. 2004;23:2336–2347. doi: 10.1038/sj.emboj.7600252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Milkereit, P., Gadal, O., Podtelejnikov, A., Trumtel, S., Gas, N., Petfalski, E., Tollervey, D., Mann, M., Hurt, E., Tschochner, H. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Milkereit, P., Strauss, D., Bassler, J., Gadal, O., Kuhn, H., Schutz, S., Gas, N., Lechner, J., Hurt, E., Tschochner, H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- Oeffinger, M., Dlakic, M., Tollervey, D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes & Dev. 2004;18:196–209. doi: 10.1101/gad.285604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim, Y.N., French, S.L., Keck, K.M., Champion, E.A., Spasov, K., Dragon, F., Baserga, S.J., Beyer, A.L. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae . Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Perez-Fernandez, J., Roman, A., De Las Rivas, J., Bustelo, X.R., Dosil, M. The 90s pre-ribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007 doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann, T., Schmidt, U., Silver, P.A. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns, M.P., Terns, R.M. Small nucleolar RNAs: Versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D., Lehtonen, H., Jansen, R., Kern, H., Hurt, E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Vallen, E.A., Hiller, M.A., Scherson, T.Y., Rose, M.D. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J. Cell Biol. 1992;117:1277–1287. doi: 10.1083/jcb.117.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., Keller, W. A new yeast Poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J., Tollervey, D. Ribosome synthesis in Saccharomyces cerevisiae. . Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- White, J., Stelzer, E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- Wyers, F., Rougemaille, M., Badis, G., Rousselle, J.-C., Dufour, M.-E., Boulay, J., Régnault, B., Devaux, F., Namane, A., Séraphin, B., et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]