Abstract
The T-box family transcription factor gene TBX20 acts in a conserved regulatory network, guiding heart formation and patterning in diverse species. Mouse Tbx20 is expressed in cardiac progenitor cells, differentiating cardiomyocytes, and developing valvular tissue, and its deletion or RNA interference–mediated knockdown is catastrophic for heart development. TBX20 interacts physically, functionally, and genetically with other cardiac transcription factors, including NKX2-5, GATA4, and TBX5, mutations of which cause congenital heart disease (CHD). Here, we report nonsense (Q195X) and missense (I152M) germline mutations within the T-box DNA-binding domain of human TBX20 that were associated with a family history of CHD and a complex spectrum of developmental anomalies, including defects in septation, chamber growth, and valvulogenesis. Biophysical characterization of wild-type and mutant proteins indicated how the missense mutation disrupts the structure and function of the TBX20 T-box. Dilated cardiomyopathy was a feature of the TBX20 mutant phenotype in humans and mice, suggesting that mutations in developmental transcription factors can provide a sensitized template for adult-onset heart disease. Our findings are the first to link TBX20 mutations to human pathology. They provide insights into how mutation of different genes in an interactive regulatory circuit lead to diverse clinical phenotypes, with implications for diagnosis, genetic screening, and patient follow-up.
Structural malformations of the heart (congenital heart disease [CHD]) are extremely common, present in nearly 1 in 100 live births and 1 in 10 stillborns. Treatment of CHD often involves highly invasive surgery in childhood, conferring a major economic burden on health resources and a life-long emotional burden for affected individuals and families. A subset of CHD is familial, and, in some cases, causative genes have been identified, most encoding cardiac transcription factors including NKX2-5 (MIM 600584), GATA4 (MIM 600576), and TBX5 (MIM 601620).1 These factors are part of a conserved regulatory network that controls cardiogenesis in species as diverse as man and insects.2,3 However, dominant mutations in cardiac developmental transcription factors account thus far for only a minority of familial cases and for few isolated cases of CHD.4,5 Therefore, a major imperative in this field remains to dissect cardiac developmental pathways in detail and to understand how mutations in genes encoding the various components of these pathways cause CHD at the genetic and mechanistic levels.
T-box transcription factors are characterized by the presence of a highly conserved, 180-aa, sequence-specific DNA-binding domain termed the “T-box.” These factors act as transcriptional activators and repressors and are known to function in a combinatorial and hierarchical fashion in many developmental processes.6 At least seven members of the T-box gene family are expressed in the developing heart in humans and vertebrate models.6 TBX1 (MIM 602054) is deleted in 22q11 deletion syndrome (MIM 188400 and 192430), the most common genetic deletion syndrome in humans, and has emerged as the leading candidate for causation of the complex cardiac and pharyngeal malformations that constitute the syndrome.7 TBX1 has multiple roles in pharyngeal development, including, in mouse, regulation of the Fgf8 gene (GenBank accession number NM_010205), involved in maintenance and growth of neural crest cells and an anterior heart progenitor population (the anterior second heart field) that contributes cardiomyocytes, smooth muscle, and endothelial cells to the outflow tract.6,8 Mutations in TBX5 cause the rare autosomal dominant Holt-Oram syndrome (MIM 142900), characterized by congenital forelimb and cardiac malformations, the latter including atrial septal defect (ASD), ventricular septal defect (VSD), tetralogy of Fallot, hypoplastic left heart, and conduction abnormalities.9,10 Tbx2 (GenBank accession number NM_009324), Tbx3 (GenBank accession numbers NM_198052 and NM_011535), and Tbx18 (GenBank accession number NM_023814) are involved in cardiac chamber and inflow-tract development, respectively, in mice,6,11 and, although TBX3 (MIM 601620) is mutated in ulnar-mammary syndrome,12 these genes have not thus far been implicated in CHD in humans.
TBX20 (MIM 606061) is an ancient member of the T-box superfamily related to TBX1, and the expression and function of the Tbx20 gene (GenBank accession number NM_020496) has recently been characterized in a number of models.6,13–15 In mice, Tbx20 is expressed in cardiac progenitor cells, as well as in the developing myocardium and endothelial cells associated with endocardial cushions, the precursor structures for the cardiac valves and the atrioventricular septum.16 Tbx20 carries strong transcriptional activation and repression domains, and it physically or genetically interacts with other cardiac developmental transcription factors, including Nkx2-5 (GenBank accession number NM_008700), Gata4 (GenBank accession number DQ436915), Gata5 (GenBank accession number NM_008093), and Tbx5 (GenBank accession number NM_011537).14,16 Loss of Tbx20 in mice is catastrophic for heart development. Homozygous mutants show a rudimentary heart that is poorly proliferative and lacks chamber myocardium and in which expression of the early transcription factor network is compromised.17–19 Tbx20 appears to directly repress another T-box gene, Tbx2, which is itself a repressor involved in allocation of chamber and nonchamber myocardium in the early heart tube. A partial knockdown of Tbx20 expression with RNA interference (RNAi) technology20 and analysis of Tbx20 function with use of a chick atrioventricular canal explant system21 have revealed later functions for Tbx20 in atrioventricular valve development. Adult heterozygous Tbx20-knockout mice show mild atrial septal abnormalities, including an increased prevalence of patent foramen ovale (PFO) and aneurysmal atrial septum primum, as well as mild dilated cardiomyopathy (DCM) and a genetic predisposition to frank ASD.17
The essential roles of Tbx20 in heart development and adult heart function in mice raise the possibility that mutations in human TBX20 (Ensembl Genome Browser [chromosome 7p14.2] accession number ENSG00000164532) contribute to CHD. We therefore screened 352 CHD-affected probands for TBX20 mutations and found one missense and one nonsense mutation in probands with a family history of CHD. Mutations lay within exons encoding the T-box DNA-binding domain, and we provide structural, functional, and biophysical evidence of their deleterious action. TBX20 mutations were associated with a complex spectrum of developmental and functional abnormalities, including defects in septation, valvulogenesis, and chamber growth and cardiomyopathy. The discovery of CHD mutations in an additional gene functioning in the conserved cardiac regulatory network highlights the importance of this network in development and evolution and as a molecular target in cardiac pathology.
Material and Methods
Patients and Clinical Details
Patients with CHD were unrelated individuals recruited without reference to family history during 2000–2006 from St. Vincent’s Hospital, Sydney Children’s Hospital, and The Children’s Hospital at Westmead, Sydney. Clinical evaluation by a cardiologist included medical history, 12-lead electrocardiography and transthoracic echocardiography, and/or transesophageal echocardiography (TEE). Diagnostic categorization of patients with CHD was made according to their most significant structural lesion. For example, a patient with an ASD and a left superior vena cava (SVC) would be categorized as having “ASD with other CHD” (see table 1). A patient with transposition of the great arteries and an ASD would be classified as having “other CHD,” since the transposition may represent more significant pathology. Ethnicity was determined by questionnaire. Informed written consent was obtained from all recruited patients. Study protocols were approved by the human research ethics committees of participating hospitals. The majority of white control individuals were unrelated anonymized individuals for whom atrial and ventricular septal status was undetermined. However, this group also included >100 “TEE controls,” who were unrelated individuals recruited from St. Vincent’s Hospital for whom TEE was performed for a number of indications and for whom ASD, VSD, and PFO were specifically excluded using intravenous saline contrast injection during the strain and release phases of the Valsalva maneuver. Mutation screening was also performed for a supplementary cohort of 90 probands with adult-onset familial DCM from St. Vincent’s Hospital or referred by collaborating physicians.
Table 1. .
Patient Cohort Details
| Phenotype(s) |
|||||
| ASD Onlya | ASD and Other CHDb | VSD Only | VSD and Other CHDc | Other CHDd | |
| No. of Subjects: | |||||
| Total | 151 | 24 | 41 | 22 | 115 |
| Male | 53 | 16 | 23 | 10 | 70 |
| With positive family historye | 20 | 5 | 4 | 2 | 8 |
| With AV conduction blockf | 5 | 3 | 0 | 1 | 0 |
| With atrial fibrillation | 8 | 0 | 0 | 0 | 0 |
| With LV dysfunction | 5g | 1h | 0 | 0 | 0 |
| Mean (range) age at enrollment, in years | 26 (0–79) | 12 (.2–62) | 6 (0–68) | 6 (0–59) | 4 (0–16) |
Three adults had mitral valve prolapse.
Including sinus venosus ASD (n=13; all others are secundum ASD); partial anomalous pulmonary venous connection (n=6); left SVC (n=2); valvular lesions (n=5), including one example of supravalvar mitral ring; and coarctation of the aorta (n=1).
Including ASD (n=7), left SVC (n=5), aortic valve abnormalities (n=5), coarctation of the aorta (n=4), double-chambered right ventricle (n=2), pulmonary stenosis (n=1), patent ductus arteriosus (n=1), and partial anomalous venous connection (n=1). One subject had mitral valve prolapse, and one had supravalvar mitral ring.
Including outflow tract lesions (n=75), atrioventricular septal defect and variants (n=18), functional single ventricle (n=17, including 2 with mitral valve atresia), heterotaxy (n=2), cor triatriatum (n=1), and Ebstein anomaly (n=1).
Positive family history was defined as at least one first-degree relative affected with CHD. Thirty-seven subjects were found to have syndromes known to be associated with CHD, including trisomy 21 (n=20) and 22q microdeletions (n=12). However, only two subjects with a positive family history were from this group.
First-degree or complete heart block. Complete and partial right bundle-branch block were not included in this group. Two subjects with ASD had left bundle-branch block.
All subjects were aged >55 years. Subjects had normal LV size and contractility but impaired diastolic relaxation (n=2) or impaired systolic function with (n=2) or without (n=1) LV dilation.
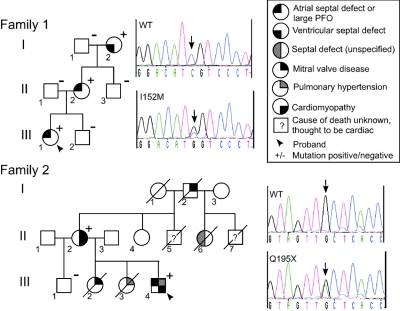
This patient (family 2, individual III:4) was positive for TBX20 mutation Q195X.
DNA Sequencing and Transcriptional Assays
TBX20 coding exons were amplified by PCR from 100 ng of leukocyte DNA, were purified with PCR Cleanup Plates (Millipore), and were sequenced using Big Dye Terminator v3.1 kit (Applied Biosystems) and ABI PRISM 3700 DNA Analyzer. Transfection assays and frog-embryo mRNA microinjection assays were performed as described elsewhere,16 except that, in the 293T-cell assay exploring Tbx20c function, Tbx20c plasmids were cotransfected with an expression plasmid encoding Sumo-1 (GenBank accession number NM_009460),22 which stimulated activity, although it is not known whether Tbx20 itself is sumoylated. Transcription data presented represent experiments performed in triplicate. Statistical analysis was performed using Student’s two-tailed t-test.
Molecular Modeling
A homology model of the T-box from mouse Tbx20 bound to DNA was produced using the program SWISS-MODEL, with use of the crystal structure of the T-box from human TBX3 (Protein Data Bank ID 1h6f)23 as template. The model structure was not subjected to energy minimization. Graphics were generated using PyMol and Adobe Illustrator CS2.
Purification of Tbx20 T-Boxes
Wild-type (WT) and I152M and Q195X Tbx20 proteins were prepared as glutathione S-transferase (GST) fusion proteins with use of a pGEX-4T-2 protein–expression vector in Escherichia coli BL21 (DE3) Rosetta cells (Novagen and Merck). Protein expression was induced by the addition of 0.4 mM isopropyl β-d-1-thiogalactopyranoside at OD600 of 0.6 and was continued at 22°C for 18 h. Cell pellets were resuspended in 20 mM 3-(N-Morpholino)propane sulphonic acid (MOPS) (pH 7.5), 150 mM NaCl, and 1 mM dithiothreitol (DTT) containing Complete Protease Inhibitors (Roche). Cells were lysed by sonication and then were treated with DNase I (Roche). Cell debris and inclusion bodies removed after centrifugation were solubilized in 8 M urea and were analyzed by SDS PAGE. DNA was precipitated from the lysate supernatant by addition of polyethyleneimine (0.1%), and fusion proteins were purified from the supernatant by affinity chromatography (Glutathione Sepharose 4B [Amersham Biosciences]) according to the manufacturer’s protocols. T-box domains were cleaved from GST on beads with thrombin and were further purified by cation-exchange chromatography on a UnoS.1 column (BioRad) running in 20 mM MOPS (pH 7.5), 1 mM DTT, and 10 μM ZnSO4, with a gradient of NaCl to remove any nucleic acids that remained associated with the protein throughout the affinity purification and cleavage steps. Q195X Tbx20 was unstable and formed inclusion bodies.
Biophysical Methods
Circular dichroism (CD) spectropolarimetry data were recorded on a Jasco J-720 spectropolarimeter equipped with a Neslab RTE-111 temperature controller. Far-UV CD spectra were collected at 20°C with a 1-mm cuvette, over the wavelength range 190–250 nm and with a speed of 20 nm/min, resolution of 0.5 nm, band width of 1 nm, and response time of 1 s. Final spectra were the average of three scans, corrected by subtracting a buffer-only spectrum. Protein concentration was estimated from A280, with use of a molar extinction coefficient of 21,430 M−1cm−1 (calculated from sequence data). Melting temperature (MT) was taken as the midpoint in the thermal denaturation curve, determined as the loss in secondary structure in the far-UV CD spectrum. Thermal data were collected at 215 nm, heating from 20°C to 80°C at 1°C per min, with a step size of 0.5°C, band width of 1 nm, and response time of 1 s. Protein concentration was 0.32 mg/ml for the far-UV spectra and 0.57 mg/ml for the thermal melt experiments, in 10 mM sodium phosphate and 150 mM NaF (pH 7.4). The MT was determined by fitting data to a sigmoidal function with use of the nonlinear least-squares fitter in MicroCal Origin. Surface plasmon-resonance analysis was performed on a Biacore 2000 SPR. A biotinylated double-stranded oligonucleotide corresponding to the T-half site16 was immobilized on a streptavidin-coated SA sensor chip (Biacore). Single-stranded oligonucleotides (5′-biotin-ctcttataggtgtgaaaaccgtg-3′ and 5′-cacggttttcacacctata-3′) were annealed before binding. The buffer used for all experiments was 50 mM MOPS, 150 mM NaCl, 1 mM DTT, 0.005% P20 surfactant, and 10 μM ZnSO4. The chip was pretreated according to the manufacturer’s instructions, with conditioning solution (3×100 μl injections at 50 μl/min with 50 mM NaOH and 1 M NaCl). Each biotinylated double-stranded oligonucleotide was diluted to 2 nM in 50 mM MOPS (pH 7.4), 500 mM NaCl, 1 mM DTT, 0.005% (v/v) P20 surfactant, and 10 μM ZnSO4 and was injected into one of the sensor-chip channels at a flow rate of 10 μl/min for 10 min, resulting in an immobilization level of ∼600 response units (RUs). The sensor chip was then washed with 50 mM MOPS, 150 mM NaCl, 1 mM DTT, 0.005% P20 surfactant, and 10 μM ZnSO4. Upstream, unmodified channel surfaces were used for reference subtraction. Kinetic measurements were performed at 20°C with a KINJECT protocol and a flow rate of 30 μl/min in the same buffer, with increasing protein concentrations across the range 0.1–10 μM. Ninety microliters of each protein concentration was injected and, at the end of the association phase, was replaced with continuous buffer flow, to monitor dissociation kinetics. WT and mutant protein samples were sampled alternately, zero-concentration samples were included for double referencing, and three cycles were performed. Data analysis was performed with the BIA evaluation software. For one-dimensional 1H nuclear magnetic resonance (NMR) spectra of WT and mutant Tbx20 T-box domains, proteins were prepared at 9.8 mg/ml (WT) and 1.3 mg/ml (I152M) in 50 mM MOPS (pH 7.5), 150 mM NaCl, 1 mM DTT, 10 μM ZnSO4 containing 10% D2O, and 20 μM dimethylsilapentane-5-sulfonic acid. Spectra were acquired at 293 K on a Bruker DRX-600 spectrometer and were processed using Topspin (Bruker).
TBX20 Gene
A TBX20 pseudogene covering exons 5 and 6 exists on human chromosome 12. This shows 98.4% homology to cognate regions on chromosome 7. Exon 5 and 6 primers used for mutation screening were specific to TBX20 on chromosome 7. Coding-exon PCR primers are available on request.
Results
We screened for mutations in TBX20 coding exons by direct DNA sequencing in 352 probands with CHD, 175 with ASD, 63 with VSD, and 115 with some other form of cardiac structural anomaly (table 1). Probands were recruited without reference to family history of CHD. However, 39 individuals (11%) had at least one first-degree relative with CHD. CHD in most subjects was diagnosed during the newborn period or during early childhood, with 23% diagnosed during adulthood. The majority of subjects were white (76%); the remainder were Asian (including Indian and Pakistani, 11%), Pacific Islander (Polynesian and Melanesian, 6%), Middle Eastern (5%), or Australian Aboriginal (2%). Unique TBX20 mutations within exons encoding the T-box DNA-binding domain were found in two white probands with ASD, each with a positive family history of CHD.
Family 1 carried the missense change TBX20 I152M (456C→G) (figs. 1, 2a, and 2b), which segregated with disease over 3 generations. The proband (III:1) had ASD, which was corrected surgically in early childhood. Her grandmother (I:2) had a small VSD, and her mother (II:2) had a large PFO with a permanent left-to-right blood shunt. Cardiac valves and left ventricular (LV) function were normal in all individuals. The I152M change was absent in >450 white controls.
Figure 1. .
Pedigrees of families with TBX20 mutations, with relevant sequence profiles of probands’ DNA and unaffected control individuals. The arrow under the sequence indicates the detected single-nucleotide change. In family 2, individual II:4, the only surviving unaffected member who was the descendent of an affected parent (and therefore a likely carrier), was not available for genotyping. For two deceased members (II:5 and II:7), there were no records available for the cause of death, although anecdotal evidence suggests a cardiac cause for both.
Figure 2. .
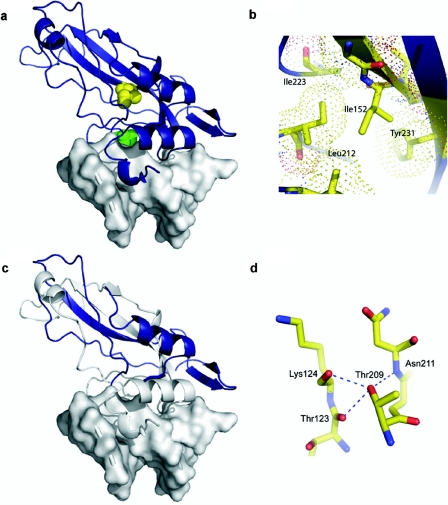
Structural impact of TBX20 mutations illustrated with a model of the TBX20 T-box (blue ribbon) bound to DNA (gray surface) on the basis of the x-ray crystal structure of the TBX3 domain.23 a, Space-filling representation of affected residues showing Ile152 (yellow) located in the core of the T-box and Thr209 (green) at the DNA-interaction face. b, Side chain of Ile152, packed within the hydrophobic core. The extra length and possibility of additional rotation within the side chain of methionine may disrupt the hydrophobic packing in this region and destabilize the structure. c, Q195X, which results in truncation of the TBX20 protein within the T-box. The region of the T-box expressed in the Q195X variant is shown in blue, with the remainder of the domain shown in gray. d, Thr209, involved in a stabilizing network of H bonds with residues that direct contact DNA. The loss of these H bonds would be expected to reduce stability of the chain in this region and have a significant effect on DNA binding.
Family 2 carried the change TBX20 Q195X (583C→T), which truncates TBX20 within the T-box DNA-binding domain16 (fig. 2c). Because of the ancient and conserved nature of the T-box fold and its established role as a sequence-specific DNA-binding domain,24 the truncated protein will most certainly lack DNA-binding ability and is likely to be functionally null. The change was present in the two living affected individuals in family 2 (fig. 1) but was absent in >300 white controls. Six members of the family, including four deceased members, had a significant cardiac history. For two additional deceased members, there were no records of the cause of death, but anecdotal evidence suggested a cardiac cause for both. The proband (III:4) had a small ASD and mild coarctation of the aorta. At age 31 years, he underwent percutaneous device closure of the ASD. He also had mild-to-moderate pulmonary hypertension diagnosed on cardiac catheterization at ages 7 mo and 6 years, although this resolved. Reduced LV function was also identified by echocardiography and cardiac catheterization in childhood, and this persisted into adulthood—at age 32 years, echocardiogram demonstrated a mildly dilated LV with mild global impairment of systolic function. Individual I:2 had unspecified mitral valve abnormalities necessitating surgical replacement. Individual II:2 had marked mitral valve prolapse with mild regurgitation, DCM, and apicolateral hypertrophy. Individual II:6 was scheduled for repair of a septal defect when she died in a vehicle accident during her 20s. Individual III:2 died at age 11 mo because of congenital mitral valve stenosis associated with a small LV and endocardial fibroelastosis. Individual III:3 died at age 7 years of right heart failure due to severe primary pulmonary hypertension. Cardiac catheterization of this individual revealed no structural lesion.
The presence of DCM in two individuals in family 2, as well as in Tbx20+/− mice in the absence of significant structural malformations,17 strongly suggests that heterozygosity for TBX20 loss-of-function mutations leads to a predisposition to adult-onset cardiomyopathy (see the “Discussion” section). However, we failed to detect any additional TBX20 mutations in 90 probands with familial DCM.
Among subjects with CHD, we detected one additional missense change, TBX20 T209I (626C→T), in a single white individual with ASD; this change was absent in >300 white controls. The proband’s family had a positive history of CHD (data not shown). However, the T209I change alone cannot account for the pathogenesis seen in this family, since one family member had ASD but was genotype negative. In addition, one member was genotype positive but phenotype negative, and the proband, in addition to having ASD, also had Klippel-Feil syndrome (MIM 118100), which can be associated with cardiac anomalies.
We introduced the observed changes in TBX20 into the mouse Tbx20 cDNA and compared transcriptional activity of the variant and WT Tbx20 proteins in cultured cell lines and in Xenopus laevis embryos after microinjection of synthetic mRNAs (fig. 3). In previous in vitro studies, the long isoform of Tbx20 (Tbx20a) was shown to have weak transcriptional activity when assayed alone, because of the dominant effects of its C-terminal trans-repression domain.16 The short Tbx20c isoform, which lacks this domain, has somewhat higher activity.16 However, a strong latent transcriptional activity in Tbx20 is revealed when Tbx20a is allowed to collaborate with interacting cardiac transcription factors Nkx2-5 and Gata4, and Tbx20a has strong dominant activities in the frog embryo overexpression assays.16
Figure 3. .
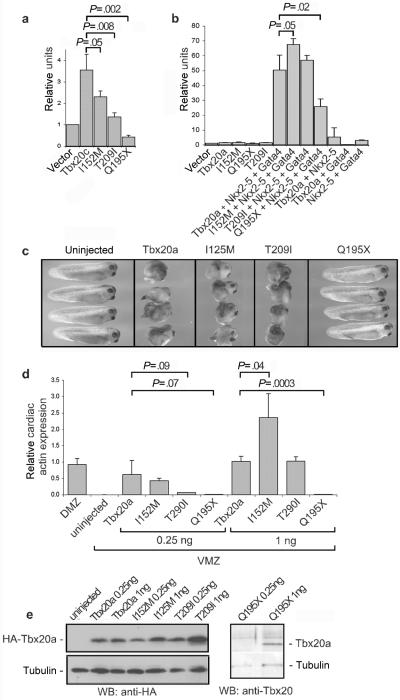
Functional analysis of TBX20 mutations. a, 293T cell–transfection assay measuring activation of the Nppa promoter in the presence of Tbx20c (short isoform lacking C-terminal trans activation and trans repression domains).16 b, COS cell–transfection assay measuring activation of the Nppa promoter in the presence of Tbx20a (full-length isoform), Nkx2-5, and Gata4, alone or in combination.16 Synergistic activation is seen only in the presence of all factors. c, Ability of WT and mutant Tbx20a proteins to disturb gastrulation movements in Xenopus laevis embryos after microinjection of respective mRNAs (1 ng) into fertilized eggs.16 WT protein likely disturbs gastrulation by dysregulating the function of endogenous T-box proteins, including brachyury and eomesodermin, and/or Gata factors, involved in mesoderm and endoderm formation. WT, I152M, and T209I are potent inhibitors, whereas Q195X is inactive. d, Comparison of the ability of Tbx20a WT and mutant proteins to activate expression of the endogenous Actc1 gene (encoding cardiac α-actin) in frog ventral-marginal-zone (VMZ) explants removed from gastrula embryos after microinjection of the indicated mRNAs into ventral cells of four-cell–stage embryos.16 Amounts of injected mRNA are indicated. Control tissue was the dorsal marginal zone (DMZ) that includes cardiac tissue. The histogram indicates normalized Actc1 levels, as determined by quantitative RT-PCR, with statistical significance (P) of indicated comparisons. e, Western blot (WB) showing Tbx20a protein expressed from injected mRNAs used in panel d, relative to levels of tubulin. Tbx20a proteins are linked to a C-terminal hemogluttinin (HA) epitope tag and are detected with anti-HA antibody, except for Tbx20a-Q195X, which lacks the tag and is detected with an anti-Tbx20 antibody.16
In experiments in which the Tbx20c vector was transfected into cultured cells, we also cotransfected a vector expressing Sumo-1,22 which enhanced activity of Tbx20c approximately twofold. As in previous studies,16 activity was assessed using a luciferase reporter plasmid driven by a proximal cis-regulatory element of the Nppa gene (GenBank accession number NM_008725), a known direct target of cardiac T-box factors.25 The activity of Tbx20c I152M, seen in family 1, was significantly reduced compared with WT Tbx20c (P=.05) (fig. 4a). However, activity was normal or slightly elevated compared with WT protein in the presence of Nkx2-5 and Gata4 (P=.05) (fig. 3b). In the mRNA-overexpression assay in whole Xenopus embryos,16 Tbx20a I152M was as potent as WT protein in disturbing gastrulation movements (fig. 3c), demonstrating its normal ability to interact with and disrupt the activity of endogenous T-box and/or Gata factors that regulate gastrulation.16 It also had a normal or elevated ability to induce ectopically the cardiomyocyte lineage in Xenopus gastrula ventral-marginal-zone explants, assessed by the activation of the endogenous cardiac α-actin gene (Actc1 [GenBank accession number NM_009608]) (P=.04) (fig. 3d and 3e). We conclude that Tbx20 I152M is functionally deficient when assayed in isolation, although this deficit can be masked in overexpression assays that depend on associations and/or synergies with other transcription factors (see the “Discussion” section).
Figure 4. .
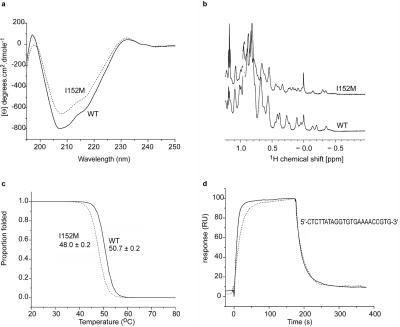
The I152M mutation, which affects the structure of the TBX20 T-box and its affinity for DNA. a, Far-UV CD spectra from WT (solid line) and I152M (dotted line). The spectra are broadly similar, although alterations to the two minima indicate some change to structure. b, 1H NMR spectra from WT and I152M T-boxes, which exhibit narrow lines and good resonance dispersion, confirming that the two domains are folded, with defined tertiary structure. The small amount of reordering within the hydrophobic core of the I152M domain is reflected in the differences in methyl resonances within the up-field region of the spectra (0.5 to −0.5 ppm). c, Thermal denaturation of Tbx20 WT and I152M T-boxes, followed as a function of secondary structure content, which demonstrates that the I152M mutation destabilizes the domain. The midpoint of thermal denaturation is reduced by ∼3°C, relative to the WT domain. d, Surface plasmon-resonance profiles of the binding of WT and I152M T-boxes to the T-half site DNA sequence, which indicate that the mutation reduces the on rate for the T site by fourfold but does not reduce the off rate.
To explore the functional deficit of TBX20 I152M further, we assessed the structural and biophysical consequences of the change in the context of the purified T-box domain. I152 is a highly conserved amino acid within the T-box family; the only substitution in Tbx20 orthologues is valine. We constructed a model of the mouse Tbx20 T-box (see the “Material and Methods” section) on the basis of the known crystal structure of the human TBX3 domain.23 In this model, the side chain of I152 is located in the domain’s core, packed against other hydrophobic residues (fig. 2a and 2b). Substitution of isoleucine for methionine in a similar context in other proteins is known to be destabilizing.26,27 We determined the far-UV CD spectra of bacterially expressed WT and mutant Tbx20 T-boxes. The WT protein displayed minima at 207 and 217 nm (fig. 4a), demonstrating a mainly β-sheet structure for the Tbx20 T-box, as in the determined crystallographic structures of other T-box domains.28,29 In the I152M spectrum, intensity differences in the two minima denoted some structural alteration. However, the local maximum at 232 nm, attributable to the packing of aromatic side chains distributed throughout the Tbx20 T-box, was unaffected in the I152M spectrum, suggesting that the overall structural change was relatively small.
We next determined the one-dimensional 1H NMR spectra and thermal stability profiles of WT and I152M T-boxes (fig. 4b). The narrow lines and good resonance dispersion in the NMR spectra indicated well-ordered tertiary structures, although differences in upfield-shifted methyl resonances confirmed that I152M induced some reordering of the hydrophobic core. The MT of the WT T-box was 51°C, similar to that of other DNA-binding domains.30,31 However, the I152M T-box had an MT of only 48°C (fig. 4c), indicating a significant degree of thermal instability and suggesting that, in vivo, TBX20 I152M populates an unfolded conformation more frequently than does the WT protein. We also explored the kinetics of DNA binding of WT and mutant T-boxes, using surface plasmon resonance. Comparative data showed that the I152M T-box bound to a high-affinity Tbx20 DNA-binding site (T site) with a fourfold lower affinity (±SD) than WT protein (Ka=0.42±0.005×106 M−1 vs. 1.58±0.02×10 M−1) (fig. 3d), an effect due entirely to a diminished DNA-binding on rate (ka=1.79±0.02×104 M−1s−1 vs. 6.96±0.07×104 M−1s−1; kd=0.0439±0.0003 s−1 vs. 0.0424±0.0002 s−1).
Tbx20 Q195X, seen in family 2, showed markedly reduced transcriptional activity for Nppa in transfection assays (fig. 3a and 3b) and lacked any ability to disrupt gastrulation movements or to induce the cardiac lineage in frog assays (fig. 3c–3e). Bacterially expressed protein was unstable.
The Tbx20 T209I change seen in the family 3 did not segregate with disease in the proband’s family. Nonetheless, it may be a deleterious allele. T209 is a highly conserved amino acid that lies in the T-box at the DNA interface (fig. 2a), and its polar side chains are involved in H-bonding networks that stabilize residues contacting DNA (fig. 2d). Tbx20 T209I showed reduced transcriptional activity when assayed alone, although not in the more complex assays (fig. 3a–3e), and the mutant T-box was unstable when expressed in bacteria, precipitating in inclusion bodies, strongly suggesting significant structural alteration.
Discussion
We have identified unique TBX20 mutations in two white CHD-affected families. Neither mutation was found in >300 whidete controls. The missense mutation, I152M (456C→G), occurred in a highly conserved amino acid in the T-box DNA-binding domain, and biophysical studies of the purified bacterially expressed T-box domain confirmed direct effects of this mutation on tertiary protein structure, thermal stability, and DNA binding. The most compelling evidence of the deleterious nature of the allele was the fourfold reduction in DNA-binding “on” rate. In vivo, the binary switch functions of signal-dependent developmental transcription factors are regulated acutely by opposing repressive activities and transcription factor degradation.32,33 The strongly reduced DNA-binding on rate is highly likely to have functional consequences for the timing of activation of cardiac developmental programs or their stability and efficacy. The compromised on rate is consistent with the observed structural instability of the Tbx20 T-box domain. When bound to DNA, the structure may be stabilized, and this could account for the lack of change in DNA-binding “off” rate. This mutation also segregated with cardiac septal pathology in family 1 over 3 generations, providing strong evidence of pathogenicity. Transcriptional activity of Tbx20 I152M was reduced by ∼40% when assayed in the context of the short Tbx20c isoform, although there was no change in overexpression assays that relied on synergistic interactions with other transcription factors. We conclude, therefore, that I152M has reduced function, although it is clearly not null. This is consistent with the presence of septal anomalies only in the three genotype-positive members of family 1. Although caution should be exercised in equating the severity of gene functional changes with severity of morphological or clinical phenotype, the atrial and ventricular septa do seem particularly vulnerable to genetic perturbation—they are the most common structural abnormalities of the heart and occur most prevalently in the absence of confounding malformations.
The other mutation detected, Q195X (583C→T), leads to the introduction of a stop codon within one of the exons encoding the T-box DNA-binding domain. This produces a TBX20 protein that is truncated within the T-box and that lacks the potent trans activation and trans repression domains present in the C terminus.16 Although we could not assess DNA binding of the truncated T-box directly because of the instability of the bacterially expressed protein, on the basis of our detailed understanding of T-box structure from crystallographic studies,28,29 the mutant protein will most certainly lack DNA-binding ability, and the functional assays confirm that it is severely compromised and most likely effectively null. Several deceased members of family 2 had diverse cardiac pathologies, and both of the affected members alive at the time of the study were genotype positive. Given the nature of the mutation, we can anticipate that the diverse spectrum of defects seen in this family is representative, at least in part, of TBX20 haploinsufficiency, although dominant-negative activities for the mutant protein cannot be discounted. The range of phenotypes is consistent with expression of Tbx20 in myocardium, as well as endocardial cushion tissue from which heart valves and atrioventricular septum are derived,16 and the catastrophic effects of Tbx20 knockdown in mice.17–20
It is noteworthy that members of family 2 were much more severely affected than were heterozygous Tbx20-null mice, which showed only hemodynamically insignificant malformations of the interatrial septum, an increase in PFO prevalence, and genetic susceptibility to ASD.17 A difference in phenotypic spectrum between human and mouse is also evident for NKX2-5 mutations,34–36 and our analysis of different mouse models of Nkx2-5 deficiency suggests that modifier genes subtly affecting the levels of NKX2-5 protein will have a significant influence on clinical outcome in humans.36–38
A prominent feature in patients with the Q195X mutation was mitral valve structural malformations, and it is noteworthy that both mitral valve stenosis and prolapse were observed in different members of family 2. Congenital mitral valve stenosis is a rare but serious malformation, generally associated with poor prognosis, whereas mitral valve prolapse is more common and is usually detected later in life. Indications for intervention in mitral valve prolapse relate more to the degree of mitral valve regurgitation than to the degree of the structural malformation itself. Whereas the respective etiologies of these types of mitral valve pathology are unknown, our data suggest that both can arise from loss of TBX20 function. Mouse Tbx20 is expressed strongly in cells of the endocardial cushions and, subsequently, the cardiac valves.16,17 Consistent with a functional role for TBX20 in this tissue, mouse embryos with a partial RNAi-mediated knockdown of Tbx20 expression show severely hypoplastic and/or immature atrioventricular valves.20 In chick endocardial-cushion explants, Tbx20 positively controls matrix metalloproteinase genes involved in endothelial-cell migration into the cushion matrix while repressing matrix genes involved in the latter stages of valve remodeling.21 Therefore, disturbance of a genetic program for atrioventricular valve induction and morphogenesis directly controlled by TBX20 is likely to underlie valve defects in TBX20 mutation–positive patients and mice.
DCM was present with structural CHD in two individuals with the Q195X mutation. Although one must be aware that this could reflect pathological decompensation after functional adaptation to structural defects, it is noteworthy that we found mild DCM without significant structural defects in adult Tbx20+/− mice,17 suggesting the alternative or additional explanation that the TBX20 mutation provides a sensitized developmental template for adult-onset DCM. Consistent with this notion, in patient II:2 (family 2), there was long-standing LV dilation and dysfunction that was disproportional to the severity of mitral valve regurgitation. Heart failure is also seen in some patients carrying NKX2-5 mutations, years after correction of structural CHD,34,39 and DCM is present even at fetal stages in a mouse model of Nkx2-5 deficiency.38 Careful consideration of the prevalence and age at onset of DCM in future studies of CHD-affected families, including TBX20 mutation–positive families, is warranted, since this has important implications for patient follow-up. Familial DCM shows a highly heterogeneous causation, with most mutations occurring in genes encoding myofilament, cytoskeletal, energy, and Ca2+-handling proteins.40 However, mutation of the transcriptional coactivator, EYA4 (MIM 603550), causes familial DCM and sensorineural hearing loss.41 A possible role for cardiac developmental transcription factor mutations in adult-onset DCM suggests that screening of patient DNA could be broadened to include such genes. However, TBX20 mutations are not a common cause of adult-onset cardiac dysfunction in the familial setting, since no additional TBX20 mutations were found after screening 90 probands with familial DCM. The participation of TBX20 mutations in idiopathic DCM remains to be explored.
The presence of primary pulmonary hypertension in one member (III:3) of family 2 is also of interest. Most primary pulmonary hypertension is idiopathic and, on histological evidence, is generally regarded to arise at the level of precapillary arterioles. Histochemical examination of the expression of β-galactosidase from the knockin Tbx20 lacZ allele in heterozygous mice revealed strong staining in the pulmonary venous system, although staining in the pulmonary arterial system was restricted to its proximal portion, in continuity with right ventricular myocardium (data not shown). This evidence raises the possibility that primary pulmonary hypertension in individual III:3 in family 2 is a comorbidity unrelated to the TBX20 mutation, although this should be monitored in future TBX20 mutation–positive families.
The third change detected in this study, T209I, did not segregate with pathology in the proband’s family. Whereas transcriptional defects and instability in bacteria hint at a deleterious function for this allele, its contribution to CHD is difficult to assess from analysis of this family.
T-box genes play critical roles in heart development.6 To our knowledge, our study establishes the first link between TBX20 mutation and human disease. Mutations were present in ∼0.6% (2 of 352) of CHD-affected patients screened, a prevalence similar to that for NKX2-5 mutations.4,5 However, among patients showing a family history of CHD in this study, ∼5% (2 of 39) carried a TBX20 mutation. If this prevalence is confirmed by additional studies, it is sufficient to warrant genetic screening for TBX20 mutations in CHD-affected families.
There is increasing awareness that the clinical CHD spectrum due to distinct mutations in the same gene—or mutations in different genes acting in a conserved pathway—can vary enormously within and among families.42,43 Conversely, defects in different cardiac developmental processes can give rise to similar forms of CHD.37,44,45 In TBX20 mutation–positive families, there is the additional complexity that TBX20 is essential for both myogenic and valvular development and that mutations are associated with myocardial dysfunction and DCM. Understanding the connections among heart developmental pathways, CHD, and cardiomyopathy remains a difficult challenge.1 An informed perspective in this area bears significantly on diagnosis, counseling, and long-term follow-up of CHD-affected families. The evident complexities in CHD causation and phenotypic manifestation may also significantly affect how genomewide screening efforts to discover mutations underlying the more common nonfamilial forms of CHD are designed and implemented.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council (NHMRC) (354400), National Heart Foundation of Australia (NHF), National Heart Lung and Blood Institute, National Institutes of Health (R01HL68885-01), Sylvia and Charles Viertel Charitable Foundation, St. Vincent’s Clinic Foundation, Rebecca Cooper Foundation, Sydney Heart Valve Bank, and Sydney Children’s Hospital Foundation. We thank participating family members; Olivia Baddeley and Haley Crotty, for data collection; and Owen Miller, Kevin Alford, David Amos, Terry Campbell, Gerald Carroll, Tim Carruthers, Mark Cooper, Richard Cranswick, Lloyd Davis, Deborah Hayes, Peter Hayes, Owen Jones, Dennis Kuchar, Lincoln Lee, Drew Mumford, Lynne Pressley, David Richmond, Shiva Roy, Neville Sammel, Jonathan Silberberg, Andrew Sindone, Charles Thorburn, and John Yiannikas, for patient referrals. E.K. was the recipient of an NHF scholarship, D.W. is an NHF Career Development Fellow, and M.S. is an NHMRC R. D. Wright Career Development Fellow.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Ensembl Genome Browser, http://www.ensembl.org/index.html (for TBX20 [accession number ENSG00000164532])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for Fgf8 [accession number NM_010205], Tbx2 [accession number NM_009324], Tbx3 [accession numbers NM_198052 and NM_011535]), Tbx18 [accession number NM_023814], Tbx20 [accession number NM_020496], Nkx2-5 [accession number NM_008700], Gata4 [accession number DQ436915], Gata5 [accession number NM_008093]), Tbx5 [accession number NM_011537], Sumo-1 [accession number NM_009460], Nppa [accession number NM_008725], and Actc1 [accession number NM_009608])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NKX2-5, GATA4, TBX5, TBX1, 22q11 deletion syndrome, Holt-Oram syndrome, TBX3, TBX20, Klippel-Feil syndrome, and EYA4)
- Protein Data Bank, http://www.rcsb.org/pdb/ (for human TBX3 [ID 1h6f])
- PyMol, http://pymol.sourceforge.net/ (for the PyMOL Molecular Graphics System)
- SWISS-MODEL, http://swissmodel.expasy.org/
References
- 1.Gruber PJ, Epstein JA (2004) Development gone awry: congenital heart disease. Circ Res 94:273–283 10.1161/01.RES.0000116144.43797.3B [DOI] [PubMed] [Google Scholar]
- 2.Harvey RP (1996) NK-2 homeobox genes and heart development. Dev Biol 178:203–216 10.1006/dbio.1996.0212 [DOI] [PubMed] [Google Scholar]
- 3.Cripps RM, Olson E (2002) Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol 246:14–28 10.1006/dbio.2002.0666 [DOI] [PubMed] [Google Scholar]
- 4.Elliott DA, Kirk E, Yeoh T, Chander S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, et al (2003) Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hyperplastic left heart syndrome. J Am Coll Cardiol 41:2072–2076 10.1016/S0735-1097(03)00420-0 [DOI] [PubMed] [Google Scholar]
- 5.McElhinney DB, Geiger E, Blinder J, Benson W, Goldmuntz E (2003) NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol 42:1650–1655 10.1016/j.jacc.2003.05.004 [DOI] [PubMed] [Google Scholar]
- 6.Stennard FA, Harvey RP (2005) T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development 132:4897–4910 10.1242/dev.02099 [DOI] [PubMed] [Google Scholar]
- 7.Yamagishi H, Srivastava D (2003) Unravelling the genetic and developmental mysteries of 22q11 deletion syndrome. Trends Mol Med 9:383–389 10.1016/S1471-4914(03)00141-2 [DOI] [PubMed] [Google Scholar]
- 8.Buckingham ME, Meilhac S, Zaffran S (2005) Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 6:826–835 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- 9.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al (1997) Mutations in human Tbx5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15:30–35 10.1038/ng0197-30 [DOI] [PubMed] [Google Scholar]
- 10.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis ARJ, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al (1997) Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 15:21–29 10.1038/ng0197-21 [DOI] [PubMed] [Google Scholar]
- 11.Christoffels VM, Mommersteeg MT, Trowe MO, Prall OWJ, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, et al (2006) Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res 98:1555–1563 10.1161/01.RES.0000227571.84189.65 [DOI] [PubMed] [Google Scholar]
- 12.Bamshad M, Lin RC, Law DJ, Watkins WS, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, et al (1997) Mutations in human TBX3 alter limb apocrine and genital development in ulnar-mammary syndrome. Nat Genet 16:311–315 10.1038/ng0797-311 [DOI] [PubMed] [Google Scholar]
- 13.Zaffran S, Reim I, Qian L, Lo PC, Bodmer R, Frasch M (2006) Cardioblast-intrinsic Tinman activity controls proper diversification and differention of myocardial cells in Drosophila. Development 133:4073–4083 10.1242/dev.02586 [DOI] [PubMed] [Google Scholar]
- 14.Brown DD, Martz SN, Binder O, Goetz SC, Price BMJ, Smith J, Conlon FL (2005) Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development 132:553–563 10.1242/dev.01596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szeto DP, Griffin KJ, Kimmelman DK (2002) hrT is required for cardiovascular development in zebrafish. Development 129:5093–5101 [DOI] [PubMed] [Google Scholar]
- 16.Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJP, Lai D, McDonald LPA, Niederreither K, Dolle P, Bruneau BG, et al (2003) Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4 and GATA5 in regulation of gene expression in the developing heart. Dev Biol 262:206–224 10.1016/S0012-1606(03)00385-3 [DOI] [PubMed] [Google Scholar]
- 17.Stennard FA, Costa MW, Lai D, Biben C, Furtado M, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, et al (2005) Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development 132:2451–2462 10.1242/dev.01799 [DOI] [PubMed] [Google Scholar]
- 18.Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S (2005) T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development 132:2475–2487 10.1242/dev.01832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A (2005) Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development 132:2697–2707 10.1242/dev.01854 [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi JJ, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsensein M, Georges R, Davidson L, Mo R, et al (2005) Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motorneuron development. Development 132:2463–2474 10.1242/dev.01827 [DOI] [PubMed] [Google Scholar]
- 21.Shelton EL, Yutzey KE (2007) Tbx20 regulation of endocardial cushion proliferation and extracellular matrix gene expression. Dev Biol 302:376–388 10.1016/j.ydbio.2006.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerscher O, Felberbaum R, Hochstasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22:159–180 10.1146/annurev.cellbio.22.010605.093503 [DOI] [PubMed] [Google Scholar]
- 23.Coll M, Seidman JG, Muller CW (2002) Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure 10:343–356 10.1016/S0969-2126(02)00722-0 [DOI] [PubMed] [Google Scholar]
- 24.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE (2005) T-box genes invertebrate development. Annu Rev Genet 39:219–239 10.1146/annurev.genet.39.073003.105925 [DOI] [PubMed] [Google Scholar]
- 25.Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM (2002) Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev 16:1234–1246 10.1101/gad.222902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gassner NC, Baase WA, Matthews BW (1996) A test of the “jigsaw puzzle” model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc Natl Acad Sci USA 93:12155–12158 10.1073/pnas.93.22.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohmura T, Ueda T, Hashimoto Y, Imoto T (2001) Tolerance of point substitutions of methionine for isoleucine in hen egg white lysozyme. Protein Eng 14:421–425 10.1093/protein/14.6.421 [DOI] [PubMed] [Google Scholar]
- 28.Provencher SW, Glockner J (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33–37 10.1021/bi00504a006 [DOI] [PubMed] [Google Scholar]
- 29.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC (1990) Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal Biochem 191:110–118 10.1016/0003-2697(90)90396-Q [DOI] [PubMed] [Google Scholar]
- 30.Bullock AN, Henckel J, DeDecker BS, Johnson CM, Nilkolova PV, Proctor MR, Lange DP, Fersht AR (1997) Thermodynamic stability of wild-type and mutant p53 core domain. Proc Natl Acad Sci USA 94:14338–14342 10.1073/pnas.94.26.14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T, Narhi LO, Wen J, Philo JS, Sitney K, Inoue J, Yamamoto T, Arakawa T (1998) Interactions between NFκB and its inhibitor iκB: biophysical characterization of a NFκB/iκB-α complex. J Protein Chem 17:757–763 10.1023/A:1020770000344 [DOI] [PubMed] [Google Scholar]
- 32.Barolo S, Posakony JW (2002) Three habits of highly effective signaling pathways: principles of transcriptional control by developing cell signalling. Genes Dev 16:1167–1181 10.1101/gad.976502 [DOI] [PubMed] [Google Scholar]
- 33.Kodadek T, Sikder D, Nalley K (2006) Keeping transcriptional activators under control. Cell 127:261–264 10.1016/j.cell.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 34.Schott J-J, Benson DW, Basson CT, Pease W, Silberach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG (1998) Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281:108–111 10.1126/science.281.5373.108 [DOI] [PubMed] [Google Scholar]
- 35.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kuperschmidt S, Roden DM, Schultheiss TM, O’Brien TX, et al (2004) Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest 113:1130–1137 10.1172/JCI200419846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, et al (2000) Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res 87:888–895 [DOI] [PubMed] [Google Scholar]
- 37.Prall OWJ, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al (2007) An Nkx2-5/Bmp2/Smad1 negative feedback loop controls second heart field progenitor specification and proliferation. Cell 128:947–959 10.1016/j.cell.2007.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elliott DA, Solloway MJ, Wise N, Biben C, Costa MW, Furtado M, Lange M, Dunwoodie SL, Harvey RP (2006) A tyrosine-rich domain within homeodomain transcription factor Nkx2-5 is an essential element in the early cardiac transcriptional regulatory machinery. Development 133:1311–1322 10.1242/dev.02305 [DOI] [PubMed] [Google Scholar]
- 39.Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, et al (1999) Mutations in the cardiac transcription factor Nkx2.5 affect diverse cardiac developmental pathways. J Clin Invest 104:1567–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fatkin D, Graham RM (2002) Molecular mechanisms of inherited cardiomyopathies. Physiol Rev 82:945–980 [DOI] [PubMed] [Google Scholar]
- 41.Schonberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, et al (2005) Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet 37:418–422 10.1038/ng1527 [DOI] [PubMed] [Google Scholar]
- 42.Benson DW, Sharkey A, Fatkin D, Lang P, Basson CT, McDonough B, Strauss AW, Seidman JG, Seidman CE (1998) Reduced penetrance, variable expressivity, and genetic heterogeneity of familial atrial septal defects. Circulation 97:2043–2048 [DOI] [PubMed] [Google Scholar]
- 43.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al (2003) GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424:443–447 10.1038/nature01827 [DOI] [PubMed] [Google Scholar]
- 44.Bajolle F, Zaffran S, Kelly RG, Hadchouel J, Bonnet D, Brown NA, Buckingham ME (2006) Rotation of the myocardial wall of the outflow tract is implicated in the normal positioning of the great arteries. Circ Res 98:421–428 10.1161/01.RES.0000202800.85341.6e [DOI] [PubMed] [Google Scholar]
- 45.Ward C, Stadt H, Hutson M, Kirby ML (2005) Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev Biol 284:72–83 10.1016/j.ydbio.2005.05.003 [DOI] [PubMed] [Google Scholar]