Abstract
DNA replication generates sister chromatid pairs that are bound to one another until anaphase onset. The process, termed sister chromatid cohesion, requires the multisubunit cohesin complex that resides at centromeres and sites where genes converge. At the HMR mating-type locus of budding yeast, cohesin associates with a heterochromatin-like structure known as silent chromatin. In this report, we show that silent chromatin is necessary but not sufficient for cohesion of the replicating locus. A tRNA gene (tDNA) that delimits the silent chromatin domain is also required, as are subunits of the TFIIIB and RSC complexes that bind the gene. Non-tDNA boundary elements do not substitute for tDNAs in cohesion, suggesting that barrier activity is not responsible for the phenomenon. The results reveal an unexpected role for tDNAs and RNA polymerase III-associated proteins in establishment of sister chromatid cohesion.
Keywords: Sister chromatid cohesion, silent chromatin, transcriptional silencing, tDNA boundary/barrier element, cohesin, Sir, RNA polymerase III
The proliferation and development of all organisms requires high fidelity transmission of intact genomes between dividing cells. Sister chromatid cohesion is one of many processes that evolved to ensure proper chromosome segregation (Nasmyth 2002). DNA replication produces sister chromatids that are held together (cohesed) until mitosis. This ensures that kinetochores of each chromatid pair attach to microtubules from opposing poles of the mitotic spindle. When all kinetochores become properly attached (bioriented), the chromatid pairs separate synchronously with one full set of chromosomes migrating toward each pole.
Sister chromatid cohesion is mediated by a set of evolutionarily conserved proteins that form a protein complex known as cohesin (for reviews, see Nasmyth and Haering 2005; Dorsett 2006). The complex consists of two SMC subunits, Smc1 and Smc3, and two additional subunits, Scc3/Irr1 and Mcd1/Scc1. Cohesin loads onto chromatin late in G1 and becomes activated for cohesion in a replication-coupled process (Lengronne et al. 2006, and references therein). The complex is shaped like a ring with an inner diameter of ∼40 nm, large enough for a pair of 10-nm chromatin fibers (Gruber et al. 2003). Bound cohesin embraces DNA in a topological manner (Ivanov and Nasmyth 2005), and one popular model stipulates that both sister chromatids are encircled by a single cohesin ring. Variations on this theme have emerged (Milutinovich and Koshland 2003). Our work at the HMR locus, for example, indicates that cohesin binds topologically but not in a way that embraces both chromatids (Chang et al. 2005). At anaphase onset, programmed cleavage of Mcd1/Scc1 by the Esp1 site-specific protease triggers chromosome separation.
Cohesin accumulates at discrete sites on chromosomes (Blat and Kleckner 1999; Laloraya et al. 2000; Glynn et al. 2004; Lengronne et al. 2004). High-density binding occurs in regions surrounding centromeres to facilitate biorientation. The complex also contributes to post-replicative DNA repair by associating with domains that contain double-strand DNA breaks (Strom et al. 2004; Unal et al. 2004). The vast majority of remaining binding sites lie in intergenic regions between pairs of genes oriented toward one another. Active transcription influences the distribution of cohesin in these regions (Glynn et al. 2004; Lengronne et al. 2004). Thus, one theory holds that passage of RNA polymerase pushes cohesin to the ends of genes.
Cohesin also accumulates on large heterochromatic domains that contain few protein-encoding genes. Heterochromatin is a repressive structure that suppresses most transcription, as well as a variety of other DNA transactions (Grewal and Moazed 2003). In Schizosaccharomyces pombe, cohesin is maintained at pericentric heterochromatin by interacting with Swi6, a conserved heterochromatin protein (Bernard et al. 2001; Nonaka et al. 2002). In mutants lacking Swi6 or other heterochromatin features, cohesin is lost from pericentric heterochromatin and chromosomes lag on the elongating anaphase spindle, much like they do in cohesin mutants.
In the budding yeast Saccharomyces cerevisiae, cohesin associates with a heterochromatin-like structure, termed silent chromatin, which is found at telomeres and the transcriptionally repressed HMR and HML mating-type loci. At these locations, cis-acting elements termed silencers recruit a complex of silencing factors known as the Sir proteins (for review, see Rusché et al. 2003). Sir2 is an NAD-dependent histone deacetylase. Sir3 and Sir4 bind deacetylated histone tails. Iterative cycles of deacetylation by Sir2 and histone binding by Sir3/4 permit the complex to spread kilobases away from silencers. Binding of cohesin at HMR requires Sir3, Sir4, and the deacetylase activity of Sir2 (Chang et al. 2005). In sir mutants, cohesion of this locus and probably other silenced domains is lost.
Silent chromatin is restricted from spreading into adjacent domains of active chromatin by barrier elements (Valenzuela and Kamakaka 2006). A tRNA gene (tDNA) neighboring HMR is a principal component of the right-hand boundary of the silent chromatin domain (Donze and Kamakaka 2001). tDNAs within the pericentric repeat elements of S. pombe act similarly, serving as barriers to constrain pericentric heterochromatin (Noma et al. 2006; Scott et al. 2006). TFIIIC, an RNA polymerase III (RNA pol III) transcription factor, can form barriers independently of other RNA pol III factors in S. pombe. In S. cerevisiae, additional proteins of the RNA pol III transcriptional machinery are required (Donze and Kamakaka 2001). Intriguingly, the barrier activity of the HMR-proximal tDNA is compromised in smc1 and smc3 mutants (Donze et al. 1999). Bell and coworkers (Lau et al. 2002) showed that cohesin blocks not only silent chromatin spreading but the de novo establishment of silencing. A unifying interpretation of these findings is that cohesin inhibits heterochromatinization of euchromatic domains.
In this study we investigated the requirements for establishment of silent chromatin cohesion. Using a combination of fluorescence microscopy and site-specific recombination, we analyzed HMR alleles that replicate as extrachromosomal DNA circles. Our results identified essential roles for the HMR-proximal tDNA and components of the RNA pol III machinery in cohesion of silent chromatin.
Results
Silent chromatin is not sufficient for cohesion
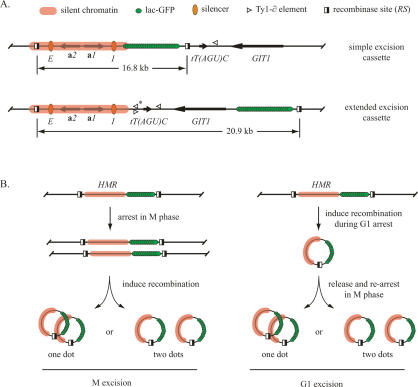
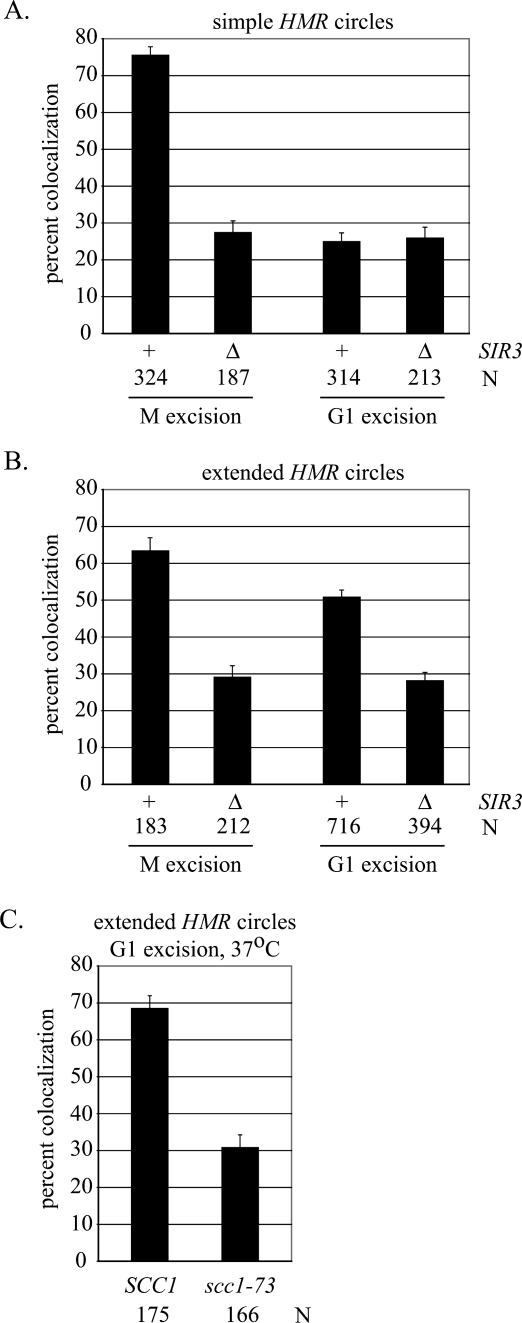
In previous work we modified HMR to monitor cohesion of the locus selectively (Chang et al. 2005). We integrated an array of lac operators near the I silencer and flanked the domain with target sites for the R site-specific recombinase. Inducible recombination uncoupled the construction (termed the simple excision cassette) (Fig. 1A) from the chromosome so that cohesion of HMR could be evaluated independently from cohesion of neighboring chromosomal domains. In this case, recombination uncoupled HMR from the neighboring tDNA. In cells expressing lac-GFP, excised HMR circles appeared as bright fluorescent dots. Excision during M-phase arrest (referred to as M excision) (Fig. 1B) produced a pair of dots that colocalized in 76% of wild-type cells and only 27% of sir3-null cells, in agreement with previous findings (Fig. 2A; Chang et al. 2005). The experiment demonstrates that silencing-dependent cohesion of HMR is maintained after unlinking the locus from neighboring chromosomal domains.
Figure 1.
Experimental design for the production of extrachromosomal HMR circles. (A) Organization of cassettes for excision of the native HMR locus, consisting of the a1 and a2 genes, as well as the E and I silencers (not drawn to scale). Relevant landmarks are defined graphically at the top of the figure. Endpoints of the silent chromatin domain were drawn arbitrarily. The right-hand RS site in the simple excision cassette disrupts a pair of overlapping Ty1 solo ∂ elements, entirely deleting one (*) that is present in the W303 but not S288C background. (B) Flow charts of the M and G1 excision protocols. Temporary G1 arrest was achieved with α-factor mating pheromone. M-phase arrest was achieved with microtubule destabilizing drugs (see Materials and Methods).
Figure 2.
Colocalization of extrachromosomal HMR circles produced by the M and G1 excision protocols. (A) Simple HMR circles were generated in strains RDY143 (SIR3) and RDY189 (Δsir3). N indicates number of cells examined. (B) Extended HMR circles were generated in strains RDY152 (SIR3) and RDY151 (Δsir3). (C) Cohesion of extended HMR circles requires MCD1/SCC1. Strains RDY152 (SCC1) and RDY213 (scc1-73) were subjected to the G1 excision protocol. Cells were grown at 24°C until 1 h after removal of α-factor, when the cultures were shifted to 37°C for 2 h. The temperature shift increases cohesion in the wild-type strain (cf. B and C).
In the present study we tested whether replication of the excised locus was sufficient to establish cohesion. To this end, HMR was uncoupled from the chromosome in G1 and colocalization was evaluated following a single round of DNA replication. G1 arrest was achieved with α-factor mating pheromone, which produced a uniform population of cells that contained one fluorescent dot per nucleus (data not shown). After galactose-induced recombination, the cultures were released from α-factor arrest and rearrested in the subsequent M phase with the microtubule-inhibiting drugs nocodazole and benomyl. Intact HMR silencers, both of which function as chromosomal origins of DNA replication, served to replicate the excised circle during the intervening S phase (Rivier and Rine 1992; Rivier et al. 1999). This is evident in Figure 2A, which shows that most cells emerging from the experimental protocol (hereafter referred to as the G1 excision protocol) contained pairs of dots that did not colocalize. Importantly, HMR circles in both wild-type and sir3-null strains displayed an equal deficit in colocalization, with only ∼25% of cells in each case containing single dots (Fig. 2A). The low values compare with those for M excision without Sir3 (Fig. 2A) or functional cohesin (Chang et al. 2005). Northern blot analysis confirmed that the a1 gene at HMR remained transcriptionally repressed in the wild-type strain, indicating that absence of cohesion was not due to an unexpected loss of silencing (Supplementary Fig. S1A). The results show that replication of silent chromatin alone is not sufficient to establish cohesion. Simple HMR circles lack an important feature required for the process.
Cohesion of HMR requires a neighboring chromatin domain
We hypothesized that establishment of cohesion at HMR requires a cis-acting element in the neighboring chromosomal DNA. Such an element would be linked to HMR during normal chromosomal replication (the M excision protocol) but unlinked when the locus replicates extrachromosomally (the G1 excision protocol). To test this hypothesis, we expanded the excision cassette to include additional neighboring chromosomal sequences. Specifically, the telomere-proximal recombinase site was moved downstream to an intergenic region ∼4 kb away. The new excision cassette (termed the extended excision cassette) (Fig. 1B) produced a larger ring that contained HMR, the GIT1 gene, a set of Ty1 retrotransposon long terminal repeats (solo ∂ elements), and the threonine tRNA gene that creates the right boundary of the silent chromatin domain [designated tT(AGU)C]. Following M excision, the pair of extended HMR circles colocalized in 63% of cells, a value that roughly parallels the result obtained for simple HMR circles (Fig. 2B). Deletion of SIR3 reduced colocalization of the extended circles to approximate background values (29%). These results indicate that pairing of the extended circles, like simple circles, relies on a Sir-dependent mechanism.
The extended HMR circles colocalized in 51% of cells following G1 excision. This represents a significant increase relative to simple circles produced by the same procedure (P > 0.001) (Fig. 2, cf. A and B). Moreover, in a sir3-null mutant, extended circles colocalized in only 28% cell, indicating that cohesion relied on silent chromatin. We repeated the G1 excision experiment with a conditional mutation in MCD1/SCC1 to test whether cohesin was responsible for the colocalization phenomenon. Cultures bearing newly formed circles were shifted from permissive to nonpermissive temperature following release from α-factor arrest. Figure 2C shows that the temperature shift eliminated colocalization in the cohesin mutant but not the wild-type strain. Taken together, these findings demonstrate that the extended circles contain elements that are both necessary and sufficient to establish cohesin-mediated cohesion of silent chromatin at HMR.
Cohesion of HMR requires the adjacent tRNA gene
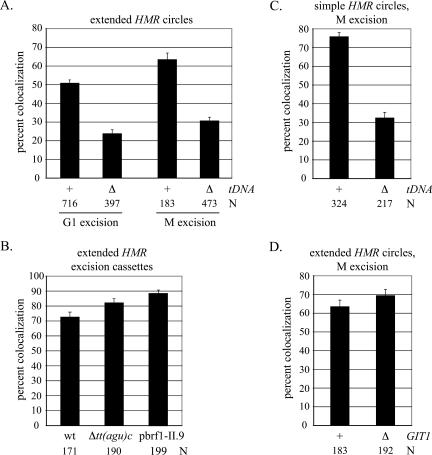
We made deletions in sequences unique to the extended excision cassette to identify the element(s) necessary for establishment of cohesion at HMR. tT(AGU)C was examined first because of the role of this gene as a silent chromatin barrier. A 100-base-pair (bp) fragment spanning the gene was replaced with a loxP site. The alteration reduced HMR pairing to background levels in the G1 excision protocol (24% colocalization) (Fig. 3A). Loss of the tDNA also disrupted pairing of the circles produced by M excision (31% colocalization) (Fig. 3A). The deletion had no effect on the unrecombined chromosomal arm, indicating that global cohesion was unperturbed (Fig. 3B). We conclude that tT(AGU)C promotes cohesion of the neighboring silent chromatin domain.
Figure 3.
tT(AGU)C establishes cohesion of HMR. (A) Colocalization of extended HMR circles produced by the G1 and M excision protocols using strains RDY152 (wt) and RDY180 [Δtt(agu)c∷loxP]. (B) Colocalization of the unexcised chromatids in strains RDY152 (wt), RDY180 [Δtt(agu)c∷loxP], and RDY209 (Δbrf1 with plasmid pbrf1-II.9, grown at 24°C). Cultures were supplemented with dextrose rather than galactose in the M excision protocol. (C) Colocalization of simple HMR circles requires tT(AGU)C. Circles were produced by the M excision protocol in strains RDY143 (wt) and RDY279 [Δtt(agu)c∷loxP]. (D) Colocalization of extended HMR circles does not require GIT1. Circles were produced by the M excision protocol using strains RDY152 (wt) and RDY226 (Δgit1∷klURA3).
The role of tT(AGU)C in cohesion was also examined in a strain carrying the simple excision cassette. In this construct, integration of the lac operator array displaces the tT(AGU)C from HMR by ∼14 kb. A TRP1 marker gene linked to the array expresses equally well in both wild-type and sir3 strains, indicating that silencing does not spread across the integrated DNA to the new distal tDNA position (Supplementary Fig. S2). Following the M excision protocol, HMR colocalizes in only 32% of the cells if the distal tDNA is deleted (cf. 76% in the wild type). This indicates that tT(AGU)C exerts its influence on cohesion even though it does not abut the silent chromatin domain and is not present on the excised DNA circle. Presumably the gene acts in cis to establish cohesion before HMR is uncoupled from the chromatin fiber by recombination.
The experiments above show that tT(AGU)C is necessary for cohesion of silent chromatin but is not necessarily sufficient. tT(AGU)C and the neighboring GIT1 gene are transcribed toward one another, like other convergent gene pairs where cohesin has been found (Glynn et al. 2004; Lengronne et al. 2004). It therefore seemed possible that convergent transcription of the tDNA and GIT1 was important for cohesion of HMR. To test this notion, we replaced the GIT1 ORF and 600 bp of upstream sequence with the URA3 gene from Kluyveromyces lactis (klURA3), orienting transcription of the new gene away from tT(AGU)C (see Fig. 6C, below). The results in Figure 3D show that the modified locus maintains cohesion following M excision (69% colocalization). We conclude that the role of tT(AGU)C in cohesion at HMR does not involve convergent transcription of the gene with GIT1. While undocumented transcription units cannot be ruled out, we note that large-scale transcriptome analysis did not identify nearby cDNAs that were oriented convergently with tT(AGU)C (Miura et al. 2006).
Figure 6.
Non-tDNA barrier elements do not support HMR cohesion. (A) Colocalization of HMR circles containing a CHA1p barrier following M excision. Strains RDY152 (wt), RDY180 [Δtt(agu)c∷loxP], and RDY206 [Δtt(agu)c∷CHA1p] were examined in rich media supplemented with 4 mM serine. (B) Colocalization of HMR circles bearing lexA operators following M excision. Strains RDY152 (wt) and RDY242 [Δtt(agu)c∷6lexOPs] were transformed with a plasmid expressing lexA (pLexA) or empty vector (pRS413). Plasmids were maintained by overnight growth in SC-trp,his + raffinose prior to replacing media with rich media containing raffinose and nocodazole. (C) Organization of the HMR region in RDY226 with GIT1 replaced by the K. lactis URA3 (klURA3). X marks the HMR-proximal tDNA that was substituted with heterologous sequences. (D) Barrier function of RDY226 (wt) and tDNA replacement strains RDY249 [Δtt(agu)c∷loxP], RDY251 [Δtt(agu)c∷CHA1p] and RDY263 [Δtt(agu)c∷6lexOPs]. Tenfold serial dilutions of each strain were spotted in rows on selective media containing or lacking 0.1% 5-FOA. All strains are prototrophic for tryptophan and grow on SC-trp media containing 4 mM serine. Plasmids pLexA and empty vector were maintained by SC-his selection.
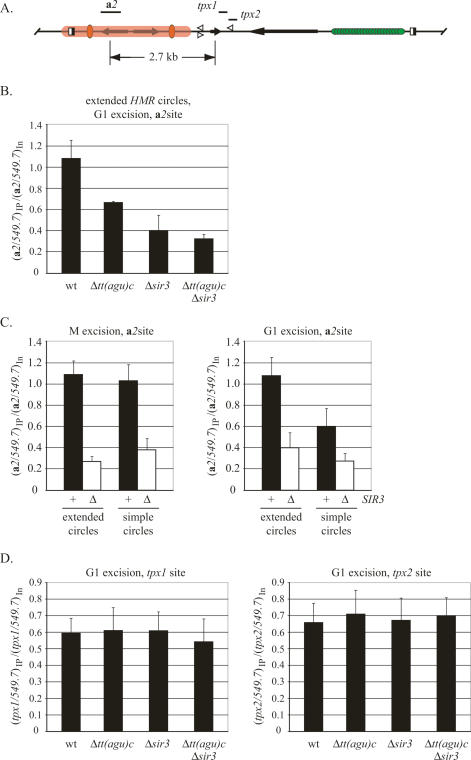
Cohesin binding at HMR
We used chromatin immunoprecipitation (ChIP) of TAP-tagged Mcd1/Scc1 to evaluate the role of tT(AGU)C in binding cohesin at HMR. Excised circles were first formed during G1 arrest in strains carrying extended excision cassettes. Cross-links were then generated with formaldehyde during the subsequent mitotic arrest. Association of Mcd1/Scc1 with the HMR a2 gene (Fig. 4A) was compared with a SIR-independent binding site on chromosome V (549.7). Figure 4B shows that the protein bound HMR a2 and that deleting SIR3 reduced binding (nearly threefold) to a level comparable with other well-characterized cohesin-free sites (534 on chromosome V and ACT1) (Chang et al. 2005; data not shown). Deleting the tDNA also reduced binding, albeit to an intermediate level. Deleting the tDNA from the sir3-null strain did not further diminish Mcd1/Scc1 binding. The results show that tT(AGU)C facilitates cohesin binding at HMR. In the absence of the tDNA, residual Mcd1/Scc1 either is bound in a nonproductive manner or is not present in sufficient quantity to establish cohesion.
Figure 4.
ChIP of Mcd1/Scc1-TAP. (A) Relative positions of the PCR-amplified sites. Primers are listed in Supplementary Table S2. (B) Mcd1/Scc1 binding at the a2 site of extended circles formed by G1 excision. Cross-links were generated during the subsequent M-phase arrest. The ratio of immunoprecipitated material (the specific site relative to the 549.7 control) was normalized to the same ratio of input material. The mean and standard deviation of three or more independent trials are presented (see Supplementary Fig. S3 for sample gels). Strains RDY177 (MCD1/SCC1-TAP), RDY179 (Δsir3 MCD1/SCC1-TAP), RDY181 [Δtt(agu)c∷loxP MCD1/SCC1-TAP], and RDY272 [Δtt(agu)c∷loxP Δsir3 MCD1/SCC1-TAP] were used. (C) Mcd1/Scc1 binding to a2 of simple and extended excision cassettes. Circles were formed by either the G1 or M excision protocols. Strains RDY178 (MCD1/SCC1-TAP) and RDY190 (Δsir3 MCD1/SCC1-TAP) were used to produce simple HMR circles. (D) Mcd1/Scc1-TAP binding to tT(AGU)C-proximal sites tpx1 and tpx2. Strains are listed in B.
We examined the binding of cohesin to simple HMR circles as well. When we uncoupled HMR from tT(AGU)C in M phase after establishment of cohesion, the level of Mcd1/Scc1 bound to HMR a2 compared with the level on extended circles (Fig. 4C). On the contrary, when we uncoupled HMR from tT(AGU)C in G1 (and cross-linked in the subsequent M phase), the simple circles associated with a reduced level of Mcd1/Scc1 (Fig. 4C). Notably, the amount of Mcd1/Scc1 on these circles compared with the amount on extended circles lacking the tDNA (Fig. 4, cf. B and C). Deleting SIR3 reduced Mcd1/Scc1 binding further in all cases. Collectively, the simple circle findings agree fully with the extended circle findings. tT(AGU)C must be present in cis during passage from G1 to M phase for cohesin to bind efficiently at HMR.
Lastly, we examined binding of Mcd1/Scc1 at sites near tT(AGU)C (designated tpx1 and tpx2 in Fig. 4A). Cohesin was previously shown to bind robustly to this region, which lies immediately downstream but adjacent to the tDNA (Laloraya et al. 2000). Cross-links were generated in M-phase-arrested cells that had undergone the G1 excision protocol. Only strains with extended excision cassettes were examined. Figure 4D shows that Mcd1/Scc1 bound the tDNA-proximal sites equally well in both wild type and a sir3 mutant, in agreement with earlier results (Kobayashi et al. 2004). More importantly, deletion of tT(AGU)C did not significantly reduce Mcd1/Scc1 binding in either strain. Like the residual cohesin at a2 described above, cohesin bound at this site does not support cohesion (Figs. 2, 3). The results indicate that cohesin binds to a tDNA-proximal region in a manner that requires neither the tDNA nor silent chromatin. Association with this site, like the residual cohesin at a2, is not sufficient for cohesion.
Cohesion of HMR requires the RNA pol III transcription machinery
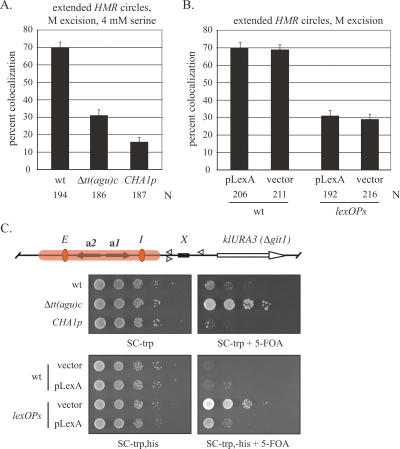
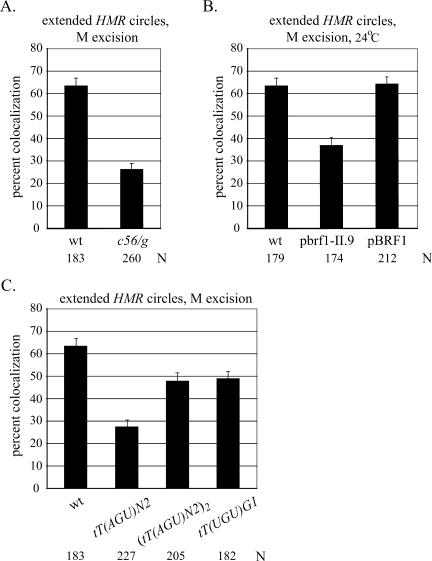
Select mutants were used to evaluate the role of the RNA pol III machinery in cohesion of HMR. RNA pol III transcription requires a hierarchical structure that utilizes two intragenic promoter elements at tDNAs, boxA and boxB (Schramm and Hernandez 2002). Transcription factor TFIIIC binds independently of other RNA pol III factors and recruits TFIIIB, which then recruits RNA pol III. A single point mutation in boxB (a c56/g transversion) prevents TFIIIC binding, and consequently blocks tDNA transcription and boundary function (Newman et al. 1983; Baker et al. 1986; Donze and Kamakaka 2001). When this single base-pair change was made in the extended excision cassette, colocalization of circles produced by the M excision protocol dropped to 26% (Fig. 5A). a1 transcripts from HMR could not be detected in this strain (Supplementary Fig. S1B). Therefore, the reduction in cohesion does not arise from an unexplained loss of silencing. These findings indicate that cohesion of HMR requires TFIIIC binding or some subsequent RNA pol III-related event.
Figure 5.
Cohesion of HMR requires RNA pol III machinery on tT(AGU)C. (A) Colocalization of extended HMR circles produced by the M excision protocol in strains RDY152 (wt) and RDY204 (c56/g). (B) Colocalization of extended HMR circles produced by the M excision protocol in strains RDY152 (wt), RDY209 (Δbrf1 with plasmid pbrf1-II.9), and RDY227 (Δbrf1 with plasmid pBRF1). Cultures were grown in nonselective rich media at 24°C. The difference between the values for the wild-type and mutant brf1 alleles (plasmids pBRF1 and pbrf1-II.9, respectively) is significant (P > 0.001). (C) Comparison of extended HMR circles with replacement tDNAs following M excision using strains RDY152 (wt), RDY231 [Δtdna∷tT(AGU)N2], RDY241 [Δtdna∷tT(AGU)N2]2}, and RDY205 [Δtdna∷tT(UGU)G1]. The difference between the values for the single and double tT(AGU)N2 replacement copies is significant (P > 0.001).
TFIIIB is composed of three polypeptides TBP, Bdp1, and Brf1. Select mutations in Brf1 block association of TBP, preventing assembly of TFIIIB on DNA (Andrau et al. 1999). We crossed one of these mutations (brf1-II.9) into our strain bearing the extended excision cassette. Previous work had shown that the conditional allele disrupted barrier function at HMR, even at the permissive temperature of 24°C (Donze and Kamakaka 2001). Figure 5B shows that colocalization of HMR circles formed by the M excision protocol dropped from 66.5% in the wild type to 37% in the mutant (Fig. 5B). This defect cannot be attributed to loss of silencing, which was found to be intact (Supplementary Fig. S1B). Colocalization was fully restored by reintroducing a plasmid-borne copy of the BRF1 gene, indicating that the cohesion defect was indeed due to mutation of the TFIIIB subunit. Furthermore, the defect appears to be localized near HMR since cohesion of the unrecombined chromosomal arm was not reduced (Fig. 4B). Taken together, these results indicate that recruitment of TFIIIB or a subsequent step in the RNA pol III transcription pathway is required for cohesion of HMR.
Heterologous tDNAs support cohesion of HMR
We entertained the possibility that tT(AGU)C-mediated cohesion of HMR was related to the boundary the gene creates. Therefore, we tested whether other tDNAs with barrier activity could substitute for tT(AGU)C. For this purpose, we utilized tT(UGU)G1 and twin copies of tT(AGU)N2, both of which block the spread of silencing when placed near HMR, and a single copy of tT(AGU)N2, which does not (Donze and Kamakaka 2001). Figure 5C shows that only the tDNA replacements with boundary-forming capacity supported colocalization of excised circles. In these cases, the Sir2 inhibitor splitomicin (Bedalov et al. 2001) reduced colocalization to background levels, indicating that the heterologous tDNAs also act through a silencing-dependent mechanism (Supplementary Fig. S4). The data show that cohesion at HMR can be established by tDNAs that create silent chromatin boundaries. These results, however, cannot distinguish whether cohesion relies on barrier activity per se or some upstream event, like transcription of the gene required to generate barrier activity.
Barrier function alone is not sufficient for cohesion of HMR
To further examine a possible relationship between silent chromatin barriers and cohesion, we analyzed barriers formed by sequences other than tDNAs. For this purpose, we replaced tT(AGU)C of the extended excision cassette with the serine-inducible CHA1 promoter (CHA1p) that normally resides on the right side of HML. We also replaced the tDNA with a series of binding sites for the bacterial lexA protein (six copies of the ColE1 operator, each containing two overlapping binding sites) (Ansari and Gartenberg 1997). Previous work revealed that inducing the CHA1 promoter or binding of lexA at high density blocks the spread of silencing (Donze and Kamakaka 2001; Bi et al. 2004). In this study we induced CHA1p with 4 mM serine. Figure 6A shows that circles containing the promoter colocalized in only 16% of the nuclei examined. Colocalization of wild-type and ΔtDNA circles, on the other hand, was not affected by the inducer (cf. Figs. 6A and 4A). HMR circles bearing lexA sites (lexOPs) were examined in strains that carried either a lexA expression plasmid or empty vector. In neither case was colocalization observed (Fig. 6B). Colocalization of wild-type circles was unaffected by the added plasmids. Collectively, these experiments show that neither CHA1p nor bound lexA can substitute for the role of a tDNA in silent chromatin cohesion.
To be certain that CHA1p and lexA create silencing barriers in our constructs, we integrated the klURA3 reporter gene ∼1.5 kb downstream from HMR at the GIT1 locus (Fig. 6C). Repression of the gene permits growth on 5-FOA, a drug that the klURA3 gene product converts to a toxic metabolite. Robust growth of the Δtt(agu)c∷loxP mutant relative to the wild-type strain demonstrates that this assay can measure variations in barrier activity over a 100-fold range (Fig. 6C). Similar results were found when comparing the c56/g mutant with wild type (data not shown). Growth of the CHA1-modified strain was completely blocked on media containing 5-FOA and serine, indicating that the heterologous promoter equals or exceeds the potency of the native tDNA barrier. Growth of strains expressing lexA was also hindered on 5-FOA, but only when lexOPs sites replaced the tDNA (Fig. 6C). Thus, lexA binding also creates a boundary at HMR. We conclude that a boundary between silenced and active chromatin domains is not sufficient for cohesion of HMR.
Cohesion of HMR requires Rsc2 but not Yta7 or Isw2
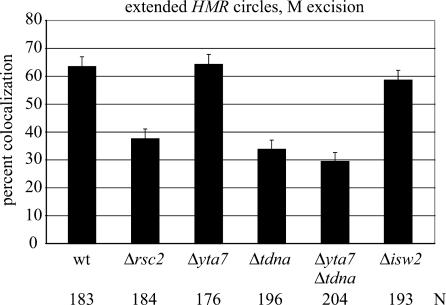
Rsc2 and Yta7 are bromodomain proteins that contribute to the natural silent chromatin barriers at HMR (Jambunathan et al. 2005; Tackett et al. 2005). Rsc2, as part of the RSC chromatin remodeling complex, binds tDNAs but does not appear to regulate their expression (Ng et al. 2002; Soutourina et al. 2006). Yta7 associates with silent chromatin boundaries as part of a Dpb4–chromatin remodeling complex (Tackett et al. 2005). The available evidence indicates that Yta7 and the tT(AGU)C function in distinct pathways at HMR. Deleting both elements causes a greater boundary defect than deleting either one alone (Jambunathan et al. 2005). In agreement with earlier work (Chang et al. 2005), we found that a RSC2 deletion impairs cohesion of extended HMR circles without causing derepression of the a1 gene (Fig. 7; Supplementary Fig. S1). Deletion of YTA7, on the other hand, had no impact (64% colocalization), and deletion of YTA7 from a tT(AGU)C-null strain did not exacerbate the colocalization defect. This result indicates that barrier function can be compromised without untoward effects on cohesion. The data lend weight to the idea that the tDNA promotes cohesion to HMR by means other than serving as a silent chromatin boundary.
Figure 7.
Influence of trans-acting factors on cohesion of HMR. Strains RDY176 (Δrsc2), RDY208 (Δyta7), and RDY225 (Δisw2) were used. The difference between values for the rsc2 and yta7 strains is significant (P > 0.001), whereas the differences between wild type, yta7, and isw2 is not.
Isw2 is the ATPase subunit of the yeast ISWI chromatin remodeling complexes that also bind tDNAs and modulate silent chromatin barrier activity at HMR (Gelbart et al. 2005; Oki and Kamakaka 2005; Tackett et al. 2005). The remodeler alters target site selection of Ty1 retrotransposons that integrate preferentially near tDNAs (Bachman et al. 2005). Here we find that deletion of ISW2 does not impair colocalization of extended HMR circles (Fig. 7). The result indicates that action of Isw2 at tT(AGU)C is not required for cohesion of the silent chromatin domain.
Discussion
tT(AGU)C establishes cohesion of the neighboring silent chromatin domain
Establishment of sister chromatid cohesion occurs during S phase and is thought to involve events at or near the replication fork. To investigate establishment of silent chromatin cohesion, we generated extrachromosomal HMR circles that replicated autonomously inside living cells. Despite remaining transcriptionally repressed, replicated circles failed to colocalize with one another unless the HMR-proximal tRNA gene tT(AGU)C was present in cis. Mutations in Brf1 and Rsc2, both subunits of complexes that associate with RNA pol III and bind the gene, attenuated the tDNA effect. Mutation of a critical residue in the tDNA promoter yielded similar consequences. Taken together, this work identifies tT(AGU)C and the associated RNA pol III machinery as a cohesion establishment complex at HMR.
tDNA-independent silent chromatin barriers do not establish cohesion
tT(AGU)C is distinguished by its ability to block silent chromatin from encroaching on the adjoining active chromosomal domain (Oki and Kamakaka 2005). The gene creates a discontinuity in arrayed nucleosomes that acts as a chain terminator to the propagation of chromatin-bound Sir proteins. The gene also abuts with one of the first documented cohesin-associated regions (CARC4) (Laloraya et al. 2000). Thus, we considered the possibility that the boundary between silent and nonsilent chromatin was responsible for cohesion at HMR. The failure of tDNA-independent boundaries to generate HMR cohesion, however, showed that barrier activity alone was not sufficient (Fig. 6). That two genes with documented roles in barrier activity at HMR, YTA7, and ISW2 had no measurable effect on cohesion of the locus reinforced these results (Fig. 7). Moreover, we found that tT(AGU)C mediated cohesion even when the tDNA was displaced from silent chromatin by an intervening lac operator array and active reporter gene (Fig. 3B). Collectively the evidence points to a role for tT(AGU)C in cohesion that is independent of the boundary it creates.
tT(AGU)C promotes binding of cohesin to the neighboring silent chromatin domain
Binding of cohesin throughout the HMR domain has been examined in a rigorous manner (at sites a1, 3′ untranslated region of a1, HMR-I in Chang et al. 2005, and sites tpx1, tpx2, and a2 above). At the a2 gene near the center of the silenced region, Mcd1/Scc1 binding diminished upon removal of the tDNA and fell to background levels in the absence of Sir3. At the tT(AGU)C-proximal sites tpx1 and tpx2 ∼2.7 kb away from a2, Mcd1/Scc1 binding was independent of both the tDNA and Sir3. Importantly, binding at these sites in strains lacking the tDNA (and the residual binding at a2) did not produce cohesion (Fig. 3). The results indicate that replication of cohesin-bound chromatin does not necessarily lead to cohesion. Maps of chromosomal cohesin-binding sites must be interpreted carefully since cohesin binding cannot be equated with cohesin function. Similar conclusions were reached when cohesin was found at locations where cohesin-independent mechanisms account for sister chromatid pairing (Lam et al. 2006; Shimada and Gasser 2007).
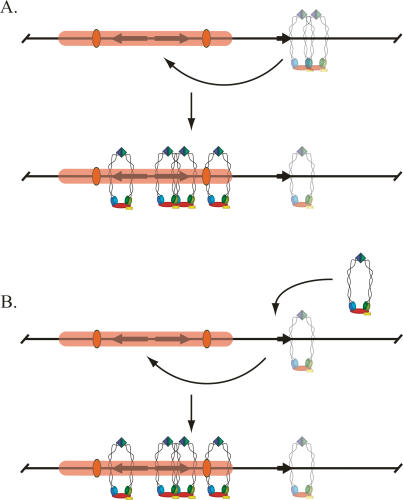
Our findings lead us to propose the existence of two pools of bound cohesin within the HMR domain: an active pool that associates with silent chromatin and participates in cohesion, and an inert pool that binds near the tDNA (and to a limited extent on silent chromatin). We view tT(AGU)C as an initiator element that promotes cohesion in one of two ways. In the first scenario, the gene functions by activating the inert pool. Alternatively, the gene loads and activates a second pool of cohesin de novo. In either case, activated cohesin could then migrate to silenced positions like a2, either by sliding along the chromatin fiber or by looping out the intervening DNA and transferring directly (Fig. 8).
Figure 8.
Models for the role of tT(AGU)C in cohesion of HMR. (A) tT(AGU)C activates a nonfunctional pool of cohesin on the adjacent tpx1 and tpx2 sites (represented as a translucent complex) that then migrates to the neighboring silenced chromosomal domain. (B) tT(AGU)C loads an active pool of cohesin, which then migrates to the adjacent silenced chromosomal domain. Silencing-dependent cohesin has thus far been detected on the a2 gene (Fig. 4), the a1 gene, and the HMR-I silencer (Chang et al. 2005).
A role for RNA pol III transcription complex in silent chromatin cohesion
How might the RNA pol III machinery participate? The simplest scenario is that Brf1 (or another pol III factor dependent on Brf1 for binding) directly recruits factors dedicated to cohesion. Precedent for such a scaffolding model comes from the targeting of yeast retrotransposons near pol III genes (Devine and Boeke 1996; Bachman et al. 2005). In the case of Ty3 viral-like particles of yeast, Brf1 and TBP alone are sufficient for targeting in vitro, presumably by interacting directly with the integrase (Yieh et al. 2002).
Cohesin might also be linked to RNA pol III transcription, via either the elongating polymerase or nascent RNA chain. A third possibility is that transcription causes secondary events that, in turn, promote cohesion. In this regard, we note that active tDNAs impede the movement of replication forks (Deshpande and Newlon 1996; Ivessa et al. 2003) and that two replisome-associated proteins responsible for the stalling, Tof1 and Csm3, are required for efficient sister chromatid cohesion (Mayer et al. 2004; Calzada et al. 2005; Tourriere et al. 2005).
A universal role for tDNAs in cohesion?
The ability of heterologous tDNAs to establish cohesion at HMR suggests that these and other tDNAs might function in cohesion at their endogenous locations, which distribute across every chromosome. We envision that other tDNAs are paired with secondary sites, like silent chromatin in the case of tT(AGU)C, and that these secondary sites capture activated cohesin. Individual tDNAs may serve dedicated functions. In S. pombe, for example, tDNAs at the boundaries of pericentric heterochromatin may establish cohesion for chromosome segregation, in addition to providing barrier activity (Noma et al. 2006). However, it is not likely that the RNA pol III acts alone in cohesin loading/activation. Cohesion of the unrecombined chromosomal arm, for example, persists in the brf1 mutant (Fig. 3). Moreover, large stretches of the genome, including the 80-kb domain surrounding HML, are devoid of all RNA pol III components (Harismendy et al. 2003), and minichromosomes lacking pol III transcription units establish cohesion efficiently (Chang et al. 2005).
Recent reports have highlighted roles for the RNA pol III pathway in spatial organization of genomes. In S. cerevisiae, tRNA gene families cluster near the nucleolus (Thompson et al. 2003). In S. pombe, TFIIIC concentrates in foci at the nucleolus and nuclear periphery where TFIIIC-bound sequences reside (Noma et al. 2006). It is tempting to speculate that the RNA pol III promoters embedded within the highly repetitive and dispersed Alu elements in mammalian chromosomes do the same (Deininger and Batzer 2002). Cohesin mediated by tDNAs may thus represent an additional layer of genome-wide chromosome organization.
Materials and methods
Strain and plasmid construction
Strains used in this study are listed in Supplementary Table S1. Those that contain the extended excision cassette were derived in several steps from the progenitor strain MRG2277, which has integrated copies of the lac-GFP (S65T) expression vector pGVH60 and the recombinase R-inducible expression vector pRINT, as well as a single RS target site upstream of HMR-E (at the SnaBI site). A second RS site and lacOP array were added with a single plasmid (pAFS52-RS-GIT1u), which integrates 1071 bp upstream of the GIT1 start codon. A cross between MRG2227 and strain CRC25 produced segregants RDY151 and RDY152. scc1-73 was introduced by crossing RDY152 with CRC83. PCR-mediated gene replacement (PMGR) was used to substitute tT(AGU)C with a loxP–URA3–loxP cassette from strain RRY5 to create RDY173. The module was then replaced using PMGR with templates that contain a single loxP site (strain RDY174), six ColE1 operators (plasmid pAA6), the CHA1 promoter (plasmid pDD560), the c56/g mutation (plasmid pDD450), or the tT(UGU)G1, tT(AGU)N2 or [tT(AGU)N2]2 tDNAs (plasmids pDD454, pDD451, and pDD589, respectively) (Ansari and Gartenberg 1997; Donze and Kamakaka 2001). The heterologous tDNA replacement fragments carried with them ∼250 bp of flanking DNA from their native chromosomal position. TAP tagging of MCD1/SCC1 was achieved by PMGR using strains from Chang et al. (2005) as templates. Various null mutants were obtained by PMGR using kanMX, natMX, or hphMX. The marker on the pRS plasmid bearing brf1-II.9 was swapped from LEU2 to URA3 by PMGR in strain DDY412. The new plasmid, as well as the brf1 genomic deletion, was crossed into RDY152 to generate RDY209. The pbrf1-II.9 plasmid in RDY209 was replaced with a plasmid bearing a wild-type copy of the gene (Conesa et al. 2005) to generate RDY227. PMGR was used to replace the GIT1 ORF and 624 bp upstream with the loxP–klURA3–loxP cassette of pUG72 (Güldener et al. 1996). Transcription of the integrated klURA3 gene was oriented in the opposite direction of GIT1. The TRP1 marker of the high-copy lexA expression vector pBTM116 was swapped to HIS3 using PMGR to generate pLexA. All strain modifications were confirmed by PCR and/or functional tests. Sequences of engineered loci are available on request.
Cell growth and microscopy
The M excision protocol was performed as described in Chang et al. (2005). For G1 excision, freshly streaked cells were grown to mid-log density in SC-trp media + 2% dextrose before diluting 1/200 into YPA (rich media) + raffinose for overnight growth. α-Factor was added (Cf = 20 nM) when cultures reached an OD of 0.2. Galactose (Cf = 2%) was added to induce excision 2.5 h later, when nearly all cells had adopted the “shmoo” morphology. Two hours after the addition of galactose, cells were collected by centrifugation, washed, and resuspended in rich media containing galactose (2%), nocodazole (10 μg/mL), and pronase E (100 μg/mL). Benomyl (Cf = 10 μg/mL) was added 1.5 h after resuspension, and cells were harvested 1.5 h later by centrifugation. Exceptions to this protocol are described in the figure legends. Paraformaldehyde fixation, mounting of cells on slides, fluorescence microscopy, and error analysis were as described in Chang et al. (2005).
ChIP
Cross-linking after M-phase excision utilized the standard cell growth protocol described above. The G1 excision protocol was modified by collecting cells 1.5 h after release from α-factor into media containing nocodazole. Immunoprecipitation procedures were as described in Chang et al. (2005) with the noted exceptions. PCR reactions were run in multiplex using oligo sets listed in Supplementary Table S2. Specificity of the ChIP reactions was confirmed with an additional set of primers that detected little immunoprecipitation of the cohesin-free ACT1 promoter (Lengronne et al. 2004; R.N. Dubey, unpubl.). Gels were stained with EtBr and destained in water before digital photography and quantization (Alpha Innotech, Inc). Individual bands were found to be nonsaturating and within the linear range.
Acknowledgments
We are grateful to David Donze, Hiten Madhani, and Giorgio Dieci for gifts of strains and plasmids. We thank Chuang-Rung Chang and Ching-Shyi Wu for technical advice. We thank David Donze, Abram Gabriel, Rohinton Kamakaka, and Ching-Shyi Wu for comments on the manuscript. This work was supported by a grant GM51402 from the NIH.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1583807
References
- Andrau J.C., Sentenac A., Werner M., Sentenac A., Werner M., Werner M. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 1999;288:511–520. doi: 10.1006/jmbi.1999.2724. [DOI] [PubMed] [Google Scholar]
- Ansari A., Gartenberg M.R., Gartenberg M.R. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol. 1997;17:7061–7068. doi: 10.1128/mcb.17.12.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman N., Gelbart M.E., Tsukiyama T., Boeke J.D., Gelbart M.E., Tsukiyama T., Boeke J.D., Tsukiyama T., Boeke J.D., Boeke J.D. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes & Dev. 2005;19:955–964. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R.E., Gabrielsen O., Hall B.D., Gabrielsen O., Hall B.D., Hall B.D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J. Biol. Chem. 1986;261:5275–5282. [PubMed] [Google Scholar]
- Bedalov A., Gatbonton T., Irvine W.P., Gottschling D.E., Simon J.A., Gatbonton T., Irvine W.P., Gottschling D.E., Simon J.A., Irvine W.P., Gottschling D.E., Simon J.A., Gottschling D.E., Simon J.A., Simon J.A. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Genier S., Javerzat J.P., Allshire R.C., Javerzat J.P., Allshire R.C., Allshire R.C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bi X., Yu Q., Sandmeier J.J., Zou Y., Yu Q., Sandmeier J.J., Zou Y., Sandmeier J.J., Zou Y., Zou Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 2004;24:2118–2131. doi: 10.1128/MCB.24.5.2118-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y., Kleckner N., Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Calzada A., Hodgson B., Kanemaki M., Bueno A., Labib K., Hodgson B., Kanemaki M., Bueno A., Labib K., Kanemaki M., Bueno A., Labib K., Bueno A., Labib K., Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes & Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.R., Wu C.S., Hom Y., Gartenberg M.R., Wu C.S., Hom Y., Gartenberg M.R., Hom Y., Gartenberg M.R., Gartenberg M.R. Targeting of cohesin by transcriptionally silent chromatin. Genes & Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa C., Ruotolo R., Soularue P., Simms T.A., Donze D., Sentenac A., Dieci G., Ruotolo R., Soularue P., Simms T.A., Donze D., Sentenac A., Dieci G., Soularue P., Simms T.A., Donze D., Sentenac A., Dieci G., Simms T.A., Donze D., Sentenac A., Dieci G., Donze D., Sentenac A., Dieci G., Sentenac A., Dieci G., Dieci G. Modulation of yeast genome expression in response to defective RNA polymerase III-dependent transcription. Mol. Cell. Biol. 2005;25:8631–8642. doi: 10.1128/MCB.25.19.8631-8642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A., Batzer M.A. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Deshpande A.M., Newlon C.S., Newlon C.S. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- Devine S.E., Boeke J.D., Boeke J.D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes & Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- Donze D., Kamakaka R.T., Kamakaka R.T. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams C.R., Rine J., Kamakaka R.T., Adams C.R., Rine J., Kamakaka R.T., Rine J., Kamakaka R.T., Kamakaka R.T. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2006;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M.E., Bachman N., Delrow J., Boeke J.D., Tsukiyama T., Bachman N., Delrow J., Boeke J.D., Tsukiyama T., Delrow J., Boeke J.D., Tsukiyama T., Boeke J.D., Tsukiyama T., Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes & Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E.F., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L., Koshland D.E., DeRisi J.L., Gerton J.L., DeRisi J.L., Gerton J.L., Gerton J.L. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I., Moazed D., Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Gruber S., Haering C.H., Nasmyth K., Haering C.H., Nasmyth K., Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H., Heck S., Fielder T., Beinhauer J., Hegemann J.H., Fielder T., Beinhauer J., Hegemann J.H., Beinhauer J., Hegemann J.H., Hegemann J.H. New efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O., Gendrel C.G., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O., Gendrel C.G., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O., Gidrol X., Sentenac A., Werner M., Lefebvre O., Sentenac A., Werner M., Lefebvre O., Werner M., Lefebvre O., Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D., Nasmyth K., Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A., Schnakenberg S.L., Zakian V.A., Zakian V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- Jambunathan N., Martinez A.W., Robert E.C., Agochukwu N.B., Ibos M.E., Dugas S.L., Donze D., Martinez A.W., Robert E.C., Agochukwu N.B., Ibos M.E., Dugas S.L., Donze D., Robert E.C., Agochukwu N.B., Ibos M.E., Dugas S.L., Donze D., Agochukwu N.B., Ibos M.E., Dugas S.L., Donze D., Ibos M.E., Dugas S.L., Donze D., Dugas S.L., Donze D., Donze D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR–tRNA boundary. Genetics. 2005;171:913–922. doi: 10.1534/genetics.105.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M., Horiuchi T., Tongaonkar P., Vu L., Nomura M., Tongaonkar P., Vu L., Nomura M., Vu L., Nomura M., Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- Laloraya S., Guacci V., Koshland D., Guacci V., Koshland D., Koshland D. Chromosomal address of the cohesion component Mcd1p. J. Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W.W., Peterson E.A., Yeung M., Lavoie B.D., Peterson E.A., Yeung M., Lavoie B.D., Yeung M., Lavoie B.D., Lavoie B.D. Condensin is required for chromosome arm cohesion during mitosis. Genes & Dev. 2006;20:2973–2984. doi: 10.1101/gad.1468806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A., Blitzblau H., Bell S.P., Blitzblau H., Bell S.P., Bell S.P. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes & Dev. 2002;16:2935–2945. doi: 10.1101/gad.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Itoh T., Watanabe Y., Shirahige K., Uhlmann F., Watanabe Y., Shirahige K., Uhlmann F., Shirahige K., Uhlmann F., Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K.-P., Shirahige K., Uhlmann F., McIntyre J., Katou Y., Kanoh Y., Hopfner K.-P., Shirahige K., Uhlmann F., Katou Y., Kanoh Y., Hopfner K.-P., Shirahige K., Uhlmann F., Kanoh Y., Hopfner K.-P., Shirahige K., Uhlmann F., Hopfner K.-P., Shirahige K., Uhlmann F., Shirahige K., Uhlmann F., Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Mayer M.L., Pot I., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Pot I., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W., Newitt R., Aebersold R., Boone C., Brown G.W., Aebersold R., Boone C., Brown G.W., Boone C., Brown G.W., Brown G.W., et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovich M., Koshland D.E., Koshland D.E. SMC complexes—Wrapped up in controversy. Science. 2003;300:1101–1102. doi: 10.1126/science.1084478. [DOI] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., Ito T., Kawaguchi N., Sese J., Toyoda A., Hattori M., Ito T., Sese J., Toyoda A., Hattori M., Ito T., Toyoda A., Hattori M., Ito T., Hattori M., Ito T., Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: The molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Newman A.J., Ogden R.C., Abelson J., Ogden R.C., Abelson J., Abelson J. tRNA gene transcription in yeast: Effects of specified base substitutions in the intragenic promoter. Cell. 1983;35:117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Robert F., Young R.A., Struhl K., Robert F., Young R.A., Struhl K., Young R.A., Struhl K., Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes & Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Cam H.P., Maraia R.J., Grewal S.I., Cam H.P., Maraia R.J., Grewal S.I., Maraia R.J., Grewal S.I., Grewal S.I. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Yamamoto M., Grewal S.I., Watanabe Y., Grewal S.I., Watanabe Y., Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Oki M., Kamakaka R.T., Kamakaka R.T. Barrier function at HMR. Mol. Cell. 2005;19:707–716. doi: 10.1016/j.molcel.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Rivier D.H., Rine J., Rine J. An origin of DNA replication and a transcription silencer require a common element. Science. 1992;256:659–663. doi: 10.1126/science.1585179. [DOI] [PubMed] [Google Scholar]
- Rivier D.H., Ekena J.L., Rine J., Ekena J.L., Rine J., Rine J. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics. 1999;151:521–529. doi: 10.1093/genetics/151.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché L.N., Kirchmaier A.L., Rine J., Kirchmaier A.L., Rine J., Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Schramm L., Hernandez N., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes & Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Scott K.C., Merrett S.L., Willard H.F., Merrett S.L., Willard H.F., Willard H.F. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- Shimada K., Gasser S.M., Gasser S.M. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell. 2007;128:85–99. doi: 10.1016/j.cell.2006.11.045. [DOI] [PubMed] [Google Scholar]
- Soutourina J., Bordas-Le Floch V., Gendrel G., Flores A., Dumay-Odelot H., Soularue P., Navarro F., Cairns B.R., Lefebvre O., Bordas-Le Floch V., Gendrel G., Flores A., Dumay-Odelot H., Soularue P., Navarro F., Cairns B.R., Lefebvre O., Gendrel G., Flores A., Dumay-Odelot H., Soularue P., Navarro F., Cairns B.R., Lefebvre O., Flores A., Dumay-Odelot H., Soularue P., Navarro F., Cairns B.R., Lefebvre O., Dumay-Odelot H., Soularue P., Navarro F., Cairns B.R., Lefebvre O., Soularue P., Navarro F., Cairns B.R., Lefebvre O., Navarro F., Cairns B.R., Lefebvre O., Cairns B.R., Lefebvre O., Lefebvre O., et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L., Lindroos H.B., Shirahige K., Sjogren C., Lindroos H.B., Shirahige K., Sjogren C., Shirahige K., Sjogren C., Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Tackett A.J., Dilworth D.J., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., Dilworth D.J., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., Aitchison J.D., Rout M.P., Chait B.T., Rout M.P., Chait B.T., Chait B.T. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., Engelke D.R., Haeusler R.A., Good P.D., Engelke D.R., Good P.D., Engelke D.R., Engelke D.R. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H., Versini G., Cordon-Preciado V., Alabert C., Pasero P., Versini G., Cordon-Preciado V., Alabert C., Pasero P., Cordon-Preciado V., Alabert C., Pasero P., Alabert C., Pasero P., Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J.E., Koshland D., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J.E., Koshland D., Sattler U., Shroff R., Lichten M., Haber J.E., Koshland D., Shroff R., Lichten M., Haber J.E., Koshland D., Lichten M., Haber J.E., Koshland D., Haber J.E., Koshland D., Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991– 1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Valenzuela L., Kamakaka R.T., Kamakaka R.T. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Yieh L., Hatzis H., Kassavetis G., Sandmeyer S.B., Hatzis H., Kassavetis G., Sandmeyer S.B., Kassavetis G., Sandmeyer S.B., Sandmeyer S.B. Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. J. Biol. Chem. 2002;277:25920–25928. doi: 10.1074/jbc.M202729200. [DOI] [PubMed] [Google Scholar]