Abstract
Sperm-egg interaction is a crucial step in fertilization, yet the identity of most interacting sperm-egg proteins that mediate this process remains elusive. Rapid evolution of some fertilization proteins has been observed in a number of species, including evidence of positive selection in the evolution of components of the mammalian egg coat. The rapid evolution of the egg-coat proteins could strongly select for changes on the sperm receptor, to maintain the interaction. Here, we present evidence that positive selection has driven the evolution of PKDREJ, a candidate sperm receptor of mammalian egg-coat proteins. We sequenced PKDREJ from a panel of 14 primates, including humans, and conducted a comparative maximum-likelihood analysis of nucleotide changes and found evidence of positive selection. An additional panel of 48 humans was surveyed for nucleotide polymorphisms at the PKDREJ locus. The regions predicted to have been subject to adaptive evolution among primates show several amino acid polymorphisms within humans. The distribution of polymorphisms suggests that balancing selection may maintain diverse PKDREJ alleles in some populations. It remains unknown whether there are functional differences associated with these diverse alleles, but their existence could have consequences for human fertility.
Sperm-egg recognition is crucial for successful fertilization and therefore is key to reproduction.1 A priori, it could be expected that fertilization and the proteins that mediate it would be highly conserved. However, rapid evolution and striking divergence of numerous reproductive proteins between closely related species have been found in diverse taxa.2 Both sperm and egg gamete-recognition molecules exhibit adaptive evolution, suggesting some form of coevolutionary chase. This rapid, adaptive divergence could be the reason the process of sperm-egg recognition often exhibits species specificity.
The mammalian egg coat comprises at least three glycoproteins with zona pellucida (ZP) domains: ZP1, ZP2, and ZP3.3,4 The ZP3 protein was thought to be the primary inducer of the sperm acrosome reaction in mouse,5,6 although recent studies suggest that sperm recognize a three-dimensional structure that may require all three ZP proteins.3,7 Positive selection has been shown to promote the rapid divergence of ZP2 and ZP3.8,9 Two clusters of amino acids reported to be involved in the species-specific induction of the acrosomal reaction in mouse5,6 have evolved by positive selection, indicating that the selective pressure may be related to sperm-egg interaction.8,9 The identity of the sperm receptor that interacts with mammalian ZP proteins remains unknown and controversial; a minimum of 10 candidates have been proposed.10,11 If ZP3 and ZP2 egg proteins have undergone rapid evolution, sperm receptor or receptors for these proteins might also have experienced significant selective pressure to maintain the functional relationship with their cognate protein or protein complex. Such coevolution has been demonstrated for interacting sperm-egg recognition proteins in abalone.12 Hence, evidence of strong, positive selection on a candidate sperm surface provides an additional characteristic that might suggest its further study for a potential receptor role in mammalian fertilization.
We examined one of the sperm-receptor candidate proteins, PKDREJ, for evidence of rapid adaptive evolution among primates. This large, intronless gene encodes an ∼8-kb transcript in humans.13 Its sequence reveals a significant region of homology with members of the PKD gene family. Members of this gene family code for a number of membrane-bound proteins that form calcium ion channels and play important roles in cell-to-cell and cell-to-extracellular matrix interactions.14 The family includes the gene responsible for human polycystic kidney disease, polycystin-1,15 and for suREJ, a sea urchin protein important in fertilization.16 suREJ localizes to the plasma membrane over the acrosomal vesicle. Experimental evidence suggests that suREJ binds to the fucose sulfate polymers of the sea urchin egg jelly and initiates the acrosomal reaction by activating gated calcium channels. This is likely done through interactions among the two C-type lectins, the REJ domain, and the two forms of the polymer.17 All PKD family genes appear to contain a subunit of a nonspecific cation channel,13 the REJ domain followed by a GPS unit, a transmembrane region, the LHD/PLAT domain, and a variable number of transmembrane domains.
Four lines of indirect evidence support the idea that PKDREJ might function as a sperm surface receptor for the egg ZP. First, PKDREJ is a homologue to sea urchin REJ molecules demonstrated to be involved in sperm-egg interaction. In a phylogenetic analysis, PKDREJ groups with suREJ compared to the other mammalian PKD genes (W. J. Swanson and J. Gatesy, unpublished material). Second, the gene encoding PKDREJ shows testis-specific expression in both mouse18 and human,13 consistent with the expectation for a sperm protein involved in fertilization. Third, the PKDREJ protein localizes to the extracellular portion of the acrosomal region of spermatozoa.18 This is the region where sperm receptors for the ZP are expected to be located.18 Fourth, functional characterization of PKDREJ modulation of G-protein signaling is consistent with its potential role in the ZP-induced acrosome reaction.19 In the current study, we tested whether PKDREJ shows signs of rapid adaptive evolution, as has been observed in mammalian egg-coat proteins8,9 and other sperm and egg proteins.20
We determined the PKDREJ sequence from 13 nonhuman primates and 48 humans, to conduct statistical tests of inter- and intraspecific sequence variation. We found evidence that the divergence of PKDREJ has been promoted by adaptive evolution. Analysis of variation in the dN/dS ratio among sites pinpointed particular codons that show signs of positive selection, indicating that they may be important for the function of PKDREJ. By examination of variation both within and between species, we have documented intra- and interspecies diversifying selection. Our results suggest that PKDREJ shows evolutionary characteristics expected for a sperm receptor for the mammalian egg-coat ZP glycoproteins.13,18
Material and Methods
DNA Samples
A panel of genomic DNAs from 13 nonhuman primate species was used. Animals were chosen to be sufficiently diverged to test for the presence of positive selection but close enough to allow ease of sequence alignment. Identifiers in parentheses indicate the sample numbers from Coriell Cell Repositories' NIA Aging Cell Repository DNA Panel–Primate Panel: Phylogenetic PRP00001. Samples included chimpanzee (Pan troglodytes; NG06939), bonobo (Pan paniscus; NG05253), gorilla (Gorilla gorilla; NG05251), Sumatran orangutan (Pongo pygmaeus; NG12256), patas monkey (Erythrocebus patas; NG06116), Celebes crested macaque (Macaca nigra; NG07101), pigtailed macaque (M. nemestrina; NG08452), rhesus monkey (M. mulatta; NG07109), wooly monkey (Lagothrix lagotricha; NG05356), black-handed spider monkey (Ateles geofffroyi; NG05352), red-chested mustached tamarin (Saguinus labiatus; NG05308), common marmoset (Callithrix jacchus; NA07404), and ring-tailed lemur (Lemur catta; NG07099). To survey human polymorphisms for PKDREJ, we used DNAs from 48 individuals who comprised an African American panel of 25 individuals (NA17101–NA17140) and a CEPH European panel of 23 individuals (NA06990, NA07019, NA07348, NA07349, NA10830–NA10861, NA12547, NA12548, and NA12560) used by SeattleSNPs for nucleotide-variation discovery.
PCR and Sequencing
PKDREJ is a gene corresponding to the region 45030225–45037883 on chromosome 22 (GenBank accession number NM_006071; March 2006 assembly). Primers were designed using PRIMER3 version 0.221 and were based on the known human sequence. The primers and conditions for PCR and sequencing are available on request. Species-specific primers were designed from the sequence obtained from the closest known relative to the desired species. PCR products were diluted fivefold with deionized water, were cycle sequenced using BigDye version 3.1, were ethanol precipitated, and were analyzed on an ABI 3100 automated sequencer (Applied Biosystems).
Data Analysis
Primate PKDREJ sequences were imported into Sequencher 4.2 (Gene Codes) for manual assembly. A consensus sequence was created from multiple overlapping reads and was aligned with the human reference sequence from the UCSC Genome Browser. The 5′ 600-bp region of the PKDREJ coding region (∼300 aa) is a GC-rich region that could not be successfully amplified or sequenced for the majority of species or for the human panel. The exported consensus sequences were aligned by eye in Se-Al version 2.0 (Oxford Evolutionary Biology). A neighbor-joining tree was constructed using Kimura 2-parameter distances in MEGA3.22 A maximum-likelihood tree was produced from the 14 primate species sequences with use of DNAML in the Phylip version 3.5 package23 for use in PAML. We used an HKY+G model with empirical base frequencies, a transition:transversion ratio of 3.6, and a gamma-distribution-shape parameter of 0.1715. The model was chosen using hierarchical likelihood-ratio tests as implemented in ModelTest.24 Maximum likelihood–based methods25 were used to detect the presence of adaptive evolution on the amino acid sequence of PKDREJ. These tests were implemented using CODEML in the PAML package (v. 3.14). CODEML allows the use of models with dN/dS ratios that vary among sites.26
A likelihood-ratio test was used to examine the data for codons with dN/dS ratios significantly >1. This was done by comparing the likelihood of a null model, without selection, against the likelihood of the same model that includes an additional class of sites whose dN/dS ratio was free to vary. The neutral models included a model with a single dN/dS ratio averaged across all sites (M0), a model with a dN/dS class between 0 and 1 and a class with dN/dS=1 (M1), and a model with the dN/dS ratio that assumes a beta distribution limited to the interval 0,1 (M7). The selection models (M2 and M8) add one additional class of sites with dN/dS estimated from the data. To test for variation in the dN/dS ratio among sites, we also compared a model (M3) with three distinct classes of dN/dS to model 0. Significance was determined by comparing the negative of twice the log-likelihood difference (-2Δl) with the χ2 distribution, with degrees of freedom equal to the difference in the number of parameters estimated between the two nested models. A Bayesian analysis was used to calculate the posterior probabilities that sites with dN/dS>1 were influenced by positive selection.26 We used the new Bayes empirical Bayes approach, to have greater confidence in the prediction.27 Convergence was checked by repeating the analyses with different initial dN/dS values, and, in all cases, identical likelihoods and parameter estimates were obtained.
To test for correlations with mating systems, we analyzed for variation in the dN/dS ratio between lineages. We first compared a model with one dN/dS estimated for all lineages with a “free-ratio” model in which we estimated dN/dS for each lineage. We next compared a one-ratio model with a model in which each lineage was assigned a class on the basis of mating system, as described by Dorus et al.28 Significance was determined by comparing the negative of twice the log-likelihood difference (-2Δl) with the χ2 distribution, with degrees of freedom equal to the difference in the number of parameters estimated between the two nested models.
Chromatograms from the human panel were automatically base called, assembled, and scanned for SNPs with use of the Phred/Phrap/polyPhred programs (v. 14.0) and were visually inspected using Consed.29–32 The final consensus sequence was a single protein–coding region of 6,762 bp. Finished sequence data from the human panel were exported, and haplotypes were inferred using PHASE.33 The default-phase certainty parameters p=q=90% were used. Haplotypes were all well resolved. Two haplotypes per individual were obtained for this autosomal gene from the human panel of 48 individuals. DnaSP version 4.034 was used to estimate population genetic parameters and genetic distances and to perform tests of neutrality. The chimpanzee sequence that we determined experimentally was used as the outgroup. The protein architecture and domain identities were inferred using the SMART program.35 The human amino acid sequence and polymorphism data were then used in the programs SIFT36 and PolyPhen,37 to infer the possible consequences of amino acid changes on the function of the PKDREJ peptide.
Results
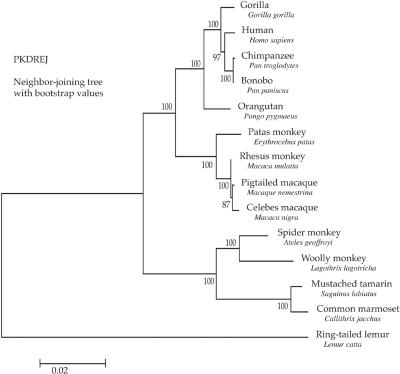
At least 5,867 of 6,762 bp of PKDREJ were amplified and sequenced for 14 primates. The first 895 bp contains a GC-rich region that was could not be PCR amplified. A portion of this region encodes the signal sequence that is cleaved off the mature protein, and the remainder is of unknown function. A neighbor-joining tree was constructed and resulted in a well-supported topology that agreed with the accepted phylogeny38,39 of humans, great apes, and Old and New World monkeys (fig. 1). A maximum-likelihood tree produced the same topology.
Figure 1. .
Neighbor-joining tree of the PKDREJ locus from the 14 primate species studied. Scale bar represents Kimura 2-parameter distances.
An analysis of the multispecies panel with use of CODEML indicates that positive selection has acted on PKDREJ (table 1). Model 3, which incorporates three categories of dN/dS values, was a significantly better fit to the data than was model 0, which estimates a single average dN/dS value over all sites. Whereas a comparison of model 3 and model 0 indicates a significant variation in dN/dS between sites, M3 is not a robust test of adaptive evolution. Thus, we compared more-general models that use a beta distribution (M7), which ascribes values of dN/dS between 0 and 1 across all sites with a model (M8) that includes an additional category with a class of sites that have dN/dS estimated from the data. This latter selection model M8 was a significantly better fit to the data than was the neutral M7 model whose dN/dS values are constrained between 0 and 1. This comparison is a robust test of positive selection.40 Approximately 3% of amino acids or 19 codons appear to have evolved by positive selection with an average dN/dS (ω) of 2.703, as estimated by model 8. To control for false-positive results due to ancestral recombination, we performed the analyses without the closely related species (we excluded chimpanzee, bonobo, gorilla, orangutan, and all but one macaque). The rationale for this is that these species are so closely related that there could still be sorting of ancestral polymorphisms with recombination, which could lead to false-positive results. The analysis of this smaller data set was completely consistent with the analysis of the broader data set containing all 14 primates, indicating positive selection.
Table 1. .
Maximum-Likelihood Estimates of Selection[Note]
| Model and Parameters | l | Positively Selected Sites |
| M0 (one ratio): | −14831.677 | None |
| dN/dS=.3914 | ||
| M1 (neutral): | −14743.041 | Not allowed |
| p0=.632 and ω0=.071 | ||
| p1=.368 and ω1=1.000 | ||
| M2 (selection): | −14739.552 | 314, 351, 1129, 1147, 1367, 1480, 1522, 345, 351, 387, 436, 528, and 547 |
| p0=.656 and ω0=.092 | ||
| p1=.326 and ω1=1.000 | ||
| p2=.018 and ω2=3.170 | ||
| M3 (discrete): | −14738.992 | 286, 312, 314, 345, 351, 387, 436, 528, 547, 629, 797, 870, 891, 953, 972, 1010, 1052, 1074, 1120, 1129, 1136, 1147, 1305, 1367, 1422, 1480, 1492, 1497, 1522, 1534, 1553, 1564, 1610, 1663, 1666, 1673, 1731, 2034, 2106, and 2209 |
| p0=.448 and ω0=.000 | ||
| p1=.497 and ω1=.623 | ||
| p2=.054 and ω2=2.398 | ||
| M7 (β): | −14744.192 | Not allowed |
| p=.103 and q=.145 | ||
| M8 (β and ω): | −14739.187 | 286, 312, 314, 345, 351, 547, 797, 870, 972, 1129, 1147, 1305, 1367, 1422, 1480, 1497, 1522, 1553, and 1666 |
| p0=.967, p=.229, and q=.402 | ||
| p1=.033 and ω=2.703 | ||
| M8a (β and fixed ω): | −14743.051 | Not allowed |
| p0=.646, p=7.695, and q=99.0 | ||
| p1=.355 and ω=1.000 |
Note.— p is the proportion of sites in each class, ω is the dN/dS ratio, and l is the log likelihood.
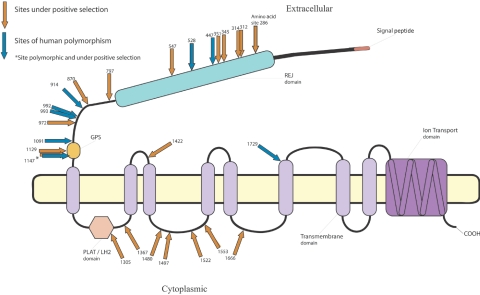
On the basis of predictions of protein architecture of PKDREJ,35 6 of the 19 sites predicted to have evolved by positive selection fall within the REJ module (fig. 2). Five sites fall in or between the GPS region and the REJ module. These sites are all predicted to be exposed to the extracellular environment and could therefore influence sperm-egg interaction(s) with other extracellular molecules. Seven sites fall on predicted low-complexity regions adjacent to the highly conserved and functionally important PLAT/LH2 domain that is potentially involved in mediating membrane attachment via binding to other proteins and transmembrane domains on the cytoplasmic side of the molecule. One additional site falls between transmembrane regions 2 and 3 in the extracellular side (fig. 2).
Figure 2. .
The PKDREJ protein with sites predicted to be subject to positive selection under model 8 (orange arrows) and amino acid polymorphic sites (blue arrows). The majority of polymorphic sites fall in the same domains as sites predicted to have evolved by positive selection.
To search for a correlation between the intensity of sperm competition and the amount of positive selection in all primate lineages studied (fig. 1), we compared dN/dS values along different branches with a neutral model that estimates a single dN/dS for all branches. For the branch-length model, a single dN/dS per branch is estimated on the basis of a set number of categories that we predetermined on the basis of the degree of sperm competition that might be expected, given the mating systems of the different species.28 In contrast with a study of the primate seminal-fluid proteins SEMG228 or SEMG1,41 no correlation was discovered between the amount of molecular evolution in a lineage of PKDREJ and the degree of sperm competition.
To survey polymorphisms within human populations, the PKDREJ gene was amplified and sequenced from a panel of 48 individuals (96 chromosomes). A total of 24 polymorphic sites were found in the human panel, 8 of which resulted in amino acid changes. All polymorphisms were diallelic. Two of the eight sites are located in the REJ module, and five more fall between the REJ module and the GPS site, corresponding to the regions that have several sites predicted to be under positive selection. One additional site falls at the end of transmembrane region 6. It is notable that all amino acid polymorphisms fall within parts of the protein that are predicted to be extracellular. The polymorphism at amino acid 447 (arginine→glycine) is at a site predicted by PAML to be under positive selection among species. The effect of the amino acid–changing polymorphisms was predicted using SIFT36 and PolyPhen.37 These programs compare the amino acid polymorphisms with amino acid variation in other members of the family, the location in the protein, and other factors, to predict whether the polymorphism would affect protein function (in particular, be deleterious). For genes under positive selection, we suggest it is possible to use SIFT and PolyPhen to detect polymorphisms that may have a functional consequence and therefore might be targets for positive selection. Results showed that two (R447G and R528E) of the eight nonsynonymous polymorphisms alter charge. PolyPhen reported changes to amino acid position 914 (L914P) as probably damaging to PKDREJ function, and SIFT reported changes to position 1147 (I1147M) as potentially affecting PKDREJ protein structure, indicating that these may have functional significance. Some of these derived alleles are at intermediate frequencies (table 2). In the case of amino acid 914, the nonsynonymous SNP is predicted to be possibly damaging, but the derived SNP occurs at a frequency of 0.63 overall and is similar in frequency in both populations. Slightly deleterious mutations are generally maintained at low frequencies in populations. Positive selection can act to increase the frequency of a mutation. The relatively high derived allele frequency, clustering of nonsynonymous SNPs by sites undergoing adaptive evolution, and indication of functional differences suggest that positive selection may be acting on these SNPs to alter protein function or specificity within humans as well as among primate species.
Table 2. .
SIFT and PolyPhen Prediction of Amino Acid Polymorphisms
| AA |
Prediction |
|||||
| Position | Ancestral | Derived | Frequency of Derived Allele (%) |
PolyPhen | SIFT | Location |
| 914 | P | L | 63 | Damaging | Tolerated | Extracellular |
| 1091 | N | S | 34 | Benign | Tolerated | Extracellular |
| 992 | T | P | 33 | Benign | Tolerated | Extracellular |
| 1147 | I | M | 15 | Benign | Damaging | GPS unit, extracellular |
| 993 | V | A | 8 | Benign | Tolerated | Extracellular |
| 528 | R | E | 3 | Benign | Tolerated | REJ module, extracellular |
| 1729 | V | I | 2 | Benign | Tolerated | Transmembrane, extracellular |
| 447 | R | G | 1 | Benign | Tolerated | REJ module, extracellular |
Twenty-six human haplotypes were inferred using PHASE.33 The most common haplotype appeared in 35% of the total population and in 63% of the European American population but in only 22% of the African American population. The European American population was dominated by a smaller number of haplotypes shared by a greater number of individuals. Among European Americans, there were four shared haplotypes among seven singletons, compared with eight shared haplotypes and 11 singletons in the African American population. The African American population had greater haplotype diversity (HD=0.904) compared with European Americans (HD=0.640). Nonsynonymous changes were largely shared by both populations, with only a single additional nonsynonymous change found exclusively in the African American population. The differences in diversity between these two population samples are primarily due to the abundance of singletons in the African American population, a pattern typical of that population.42 The difference in genetic makeup is reflected in the large FST=0.178 compared with the FST=0.157 human average42 and a significant χ2 value (P=.014) for genetic differentiation.
We compared the levels of polymorphism within humans with divergence between humans and chimpanzee or gorilla, using the McDonald-Kreitman (MK) test,43 which measures deviations from the expected ratio of nonsynonymous:synonymous polymorphisms to divergence. The MK test produced significant P values, such that neutrality can be rejected. We suggest two explanations for why neutrality is rejected. First, there could be an excess of synonymous polymorphisms within humans. An excess of synonymous polymorphisms could arise because of a balancing polymorphism at this locus. Alternatively, there could be a significant accumulation of nonsynonymous divergence since the human and chimpanzee lineages diverged. We favor the former idea, since there is additional evidence of balancing selection on the basis of analyses of the frequency spectrum (see below). The significant analyses of dN/dS suggests that balancing selection is most likely acting on nonsynonymous sites. Although the MK tests is generally considered to be robust to demographic effects,44,45 the test was run on subsamples of the human polymorphism data. Results from the European American sample proved to be nonsignificant, whereas results from the African American sample were highly significant.
Additional tests of neutrality on PKDREJ (Tajima’s D,46 Fu and Li’s D and F,47 and Fay and Wu’s H48) were performed on the total human sample and the two subsamples, with chimpanzee as the outgroup for Fay and Wu’s H (table 3). Significance was determined by coalescent simulations, with only Tajima’s D showing significance (P=.04) and for only the African American population, suggesting an excess of intermediate-frequency polymorphisms. Additionally, the value of Tajima’s D (D=1.5) for the African American sample was the second highest value seen in the SeattleSNPs database of 247 genes from the same African American samples, which places it in the 99th percentile of values (median Tajima’s D for the African American sample is −0.52). Thus, the magnitude and direction of Tajima’s D for PKDREJ is highly unusual for the African American population. The value of Tajima’s D for European Americans was 0.591, a typical value for this population based on comparisons with the SeattleSNPs database (median value is 0.379 for the CEPH population).
Table 3. .
Summary Statistics[Note]
| Sample Summary |
Parameter Estimate |
Test Statistic |
|||||||||||||
| Population | N | Synonymous | Nonsynonymous | Total | No. of Haplotypes | Φ | π | HD | k | Fs | FST | D | F | H | Tajima’s D |
| African American | 50 | 15 | 7 | 22 | 19 | 4.912 | 1×10−3 | .904 | 7.265 | −1.745 | … | .725 | 1.23638 | −.408 | 1.546 |
| European American | 46 |
10 |
6 |
16 |
11 |
3.641 | 6×10−4 | .64 | 4.254 | .253 | … | .3 | .457 | −2.381 | .529 |
| Total sample | 96 | 16 | 8 | 24 | 26 | … | 1×10−3 | .815 | 6.452 | −3.822 | .178 | −.135 | .43736 | −.928 | 1.135 |
Discussion
We compared PKDREJ sequences from 14 primates plus two population samples of humans. Multiple statistical tests reject equilibrium-neutral expectations and suggest that PKDREJ has evolved by positive selection in the primate lineage (table 1). Nineteen codons are identified that show signs of positive selection. These fall into several regions predicted to be important for function, including the REJ domain, the GPS domain, and a region between them. Of the 19 sites predicted to be subject to positive selection, 12 are in the predicted extracellular part of the molecule. An additional seven sites occur on the first three intracellular loops between transmembrane domains. These are low-complexity regions but contain a potential lipid-interaction site.49
We also surveyed PKDREJ for amino acid polymorphisms within humans. Several nonsynonymous polymorphic sites fell within or close to codons for which we predicted adaptive evolution. For example, 87% of human nonsynonymous polymorphisms fall within the REJ domain and the region before the GPS domain, the REJ module, and the extracellular region around the putative GPS domain representing ∼50% of the protein (fig. 2). All eight of the amino acid polymorphic sites are in predicted extracellular regions. The African American population appears to have an excess of intermediate-frequency variants compared with the expectations of an equilibrium-neutral model.
Although the function of PKDREJ remains unknown, it has been suggested as a candidate sperm receptor for the ZP.13,18,50 The rapid evolution of fertilization proteins is hypothesized to result from male-male competition in promiscuous mating systems in which sperm compete to fertilize the egg. Alternatively, there could be conflict between males and females, where the egg is selected to avoid fertilization by multiple sperm (which is usually fatal to the egg) but sperm are selected to fertilize rapidly.51 We were unable to document any correlation between rate of PKDREJ evolution and mating system, such as that reported for the primate seminal-fluid protein SEMG2.28 Whereas this result is consistent with female-male rather than male-male competition having driven the evolution of PKDREJ, the lack of correlation may instead be due to inaccuracies on the estimates of the degree of sperm competition. Additionally, sperm competition and sperm-egg conflict are not mutually exclusive, since intense sperm competition can select for the same characteristics that increase the likelihood of polyspermy and increased egg-sperm conflict.
Results of the MK test were significant, which we interpret as an excess of synonymous polymorphisms in human PKDREJ (table 4), although an excess of amino acid fixation between species may also contribute. Because the significance of the MK test result could be due, in part, to divergence in the chimpanzee outgroup, the MK test was repeated using gorilla as the outgroup. Results were again significant for the total population. The test was run on the two subpopulations, and results were found to be significant only for the African American sample. In all cases, the P value was diminished in relation to tests using the chimpanzee outgroup, suggesting that the choice of outgroup contributed to the significance of the result. A comparison of the neutrality index (NI) values (table 4), which provides a qualitative measure of the direction and extent of amino acid changes, were both <1, where a value of 1 indicates neutrality, >1 is purifying selection, and <1 is positive selection. The value of NI is closer to 1 when human is compared with gorilla than when human is compared with chimpanzee.
Table 4. .
MK Test: Chimpanzee/Gorilla Comparison[Note]
| Findings by Sample |
|||
| Comparison and Species | Total | European American | African American |
| Synonymous substitutions: | |||
| No. of polymorphic sites (human) | 16 | 10 | 15 |
| Fixed differences between species | |||
| Chimpanzee | 10 | 11 | 10 |
| Gorilla | 18 | 18 | 18 |
| Nonsynonymous substitutions: | |||
| No. of polymorphic sites (human) | 8 | 6 | 7 |
| No. of fixed differences between species: | |||
| Chimpanzee | 21 | 21 | 21 |
| Gorilla | 27 | 27 | 27 |
| NI: | |||
| Chimpanzee | .238 | .314 | .222 |
| Gorilla | .333 | .4 | .311 |
| Fisher’s exact test P (two tailed): | |||
| Chimpanzee | .015 | .122 | .013 |
| Gorilla | .045 | .151 | .039 |
Note.— Significant values are shown in bold.
Consistent with the finding of excess synonymous substitutions in the African American population by use of the MK test, analysis of the frequency spectrum also suggests that balancing selection acts on PKDREJ. For PKDREJ in the African American population, the value of Tajima’s D (1.5) was significantly positive and an extreme outlier compared with the values of 247 other loci from the SeattleSNPs data set. Positive values of Tajima’s D are due to an excess of alleles with intermediate-frequency variants.46 This pattern is seen under balancing selection as well as a narrow window of time during the recovery after population bottlenecks. Demographic changes affect all genes, whereas selection typically acts on a single locus. Large positive values of Tajima’s D are rare in the African American population, and other genes in the African American population show no evidence of a bottleneck. The distribution of polymorphisms and signs of selection on different human haplotypes in the African American population suggest that selection has driven the diversification of human reproductive alleles.
One of the striking features of the analysis presented here is the evidence of rapid, adaptive divergence of PKDREJ among primates, coupled with human polymorphism data indicating the presence of balancing selection. Thus, there is high divergence among species and high levels of variation within species. Such an observation is inconsistent with multiple complete selective sweeps and suggests variable selective pressures (e.g., balancing selection coupled with occasional selective sweeps) or perhaps a succession of partial selective sweeps. This pattern has been documented in a variety of other genes involved in reproduction. For example, both the Drosophila seminal-fluid protein Acp26Aa52–54 and the sea urchin sperm protein bindin55,56 show this pattern of high levels of differences both within and among species. Such consistent patterns across multiple taxonomic groups suggest a potentially fruitful area of population genetics for modeling the evolutionary processes that could lead to these patterns. For example, some models of sexual conflict predict diversification within species of genes involved in reproduction.57
Coevolution between sperm proteins and their egg receptors can result in the divergence of alleles. If a mutation in a sperm protein creates sufficient advantage in fertilizing eggs that display a particular variant of the egg-coat protein, it will be selected for despite the expense of being inferior at fertilizing eggs displaying other egg-coat alleles. This sperm protein may then become specialized, responding to changes in that egg protein while growing ever more divergent from other sperm alleles. Mismatch between mating types is more likely to occur between populations that infrequently exchange gametes, since there is a likely to be a trade-off between the benefit of increased effectiveness of fertilization and the odds of encountering a new egg-coat protein for which the sperm protein is an ineffective receptor.
The pattern of variability and departures from neutrality in the sequence evolution of PKDREJ strongly suggest positive selection on the evolution and diversification of PKDREJ. The putative structure of the PKDREJ protein and its testis-specific expression suggests that this gene is functionally analogous to the sea urchin sperm protein suREJ, which plays a role in fertilization. We have found evidence of adaptive changes in the evolution of PKDREJ among species as well as evidence of diversifying selection within species. There appears to have been selection for increased diversity in the number of haplotypes in the African American population sample. It is unknown whether there are functional differences between any of the haplotypes, but the existence of multiple alleles with functional differences could have consequences for human fertilization.
Acknowledgments
We thank two anonymous reviewers, members of the Swanson lab, John Gatesy, and Josh Akey for advice and discussion. Support was provided by National Institutes of Health grants HD42563 (to W.J.S.), HD41454 (to W.J.S.), HD38921 (to M.F.W.), and GM36431 (to C.F.A.) and National Science Foundation grants DEB-0213171 (to W.J.S.) and DEB-0410112 (to W.J.S.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PKDREJ [accession number NM_006071] and nonhuman primate PKDREJ sequences [accession numbers EF517278–EF517291])
- Oxford Evolutionary Biology, http://evolve.zoo.ox.ac.uk/software.html?id=seal (for Se-Al sequence alignment editor)
- SeattleSNPs, http://pga.gs.washington.edu/
References
- 1.Vacquier VD (1998) Evolution of gamete recognition proteins. Science 281:1995–1998 10.1126/science.281.5385.1995 [DOI] [PubMed] [Google Scholar]
- 2.Swanson WJ, Vacquier VD (2002) Rapid evolution of reproductive proteins. Nat Rev Genet 3:137–144 10.1038/nrg733 [DOI] [PubMed] [Google Scholar]
- 3.Dean J (2004) Reassessing the molecular biology of sperm-egg recognition with mouse genetics. Bioessays 26:29–38 10.1002/bies.10412 [DOI] [PubMed] [Google Scholar]
- 4.Wassarman PM (1999) Mammalian fertilization: molecular aspects of gamete adhesion, exocytosis, and fusion. Cell 96:175–183 10.1016/S0092-8674(00)80558-9 [DOI] [PubMed] [Google Scholar]
- 5.Kinloch RA, Sakai Y, Wassarman PM (1995) Mapping the mouse ZP3 combining site for sperm by exon swapping and site-directed mutagenesis. Proc Natl Acad Sci USA 92:263–267 10.1073/pnas.92.1.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Litscher ES, Wassarman PM (1998) Inactivation of the mouse sperm receptor, mZP3, by site-directed mutagenesis of individual serine residues located at the combining site for sperm. Proc Natl Acad Sci USA 95:6193–6197 10.1073/pnas.95.11.6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rankin TL, Coleman JS, Epifano O, Hoodbhoy T, Turner SG, Castle PE, Lee E, Gore-Langton R, Dean J (2003) Fertility and taxon-specific sperm binding persist after replacement of mouse sperm receptors with human homologs. Dev Cell 5:33–43 10.1016/S1534-5807(03)00195-3 [DOI] [PubMed] [Google Scholar]
- 8.Swanson WJ, Yang Z, Wolfner MF, Aquadro CF (2001) Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci USA 98:2509–2514 10.1073/pnas.051605998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansa SA, Lundrigan BL, Tucker PK (2003) Tests for positive selection on immune and reproductive genes in closely related species of the murine genus Mus. J Mol Evol 56:294–307 10.1007/s00239-002-2401-6 [DOI] [PubMed] [Google Scholar]
- 10.Wassarman PM, Jovine L, Litscher ES (2001) A profile of fertilization in mammals. Nat Cell Biol 3:E59–E64 10.1038/35055178 [DOI] [PubMed] [Google Scholar]
- 11.Wassarman PM, Jovine L, Litscher ES, Qi H, Williams Z (2004) Egg-sperm interactions at fertilization in mammals. Eur J Obstet Gynecol Reprod Biol Suppl 115:S57–S60 10.1016/j.ejogrb.2004.01.025 [DOI] [PubMed] [Google Scholar]
- 12.Galindo BE, Vacquier VD, Swanson WJ (2003) Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci USA 100:4639–4643 10.1073/pnas.0830022100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes J, Ward CJ, Aspinwall R, Butler R, Harris PC (1999) Identification of a human homologue of the sea urchin receptor for egg jelly: a polycystic kidney disease-like protein. Hum Mol Genet 8:543–549 10.1093/hmg/8.3.543 [DOI] [PubMed] [Google Scholar]
- 14.Gallagher AR, Hidaka S, Gretz N, Witzgall R (2002) Molecular basis of autosomal-dominant polycystic kidney disease. Cell Mol Life Sci 59:682–693 10.1007/s00018-002-8457-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millan JL, Gamble V, Harris PC (1995) The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet 10:151–160 10.1038/ng0695-151 [DOI] [PubMed] [Google Scholar]
- 16.Moy GW, Mendoza LM, Schulz JR, Swanson WJ, Glabe CG, Vacquier VD (1996) The sea urchin sperm receptor for egg jelly is a modular protein with extensive homology to the human polycystic kidney disease protein, PKD1. J Cell Biol 133:809–817 10.1083/jcb.133.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mengerink KJ, Moy GW, Vacquier VD (2002) suREJ3, a polycystin-1 protein, is cleaved at the GPS domain and localizes to the acrosomal region of sea urchin sperm. J Biol Chem 277:943–948 10.1074/jbc.M109673200 [DOI] [PubMed] [Google Scholar]
- 18.Butscheid Y, Chubanov V, Steger K, Meyer D, Dietrich A, Gudermann T (2006) Polycystic kidney disease and receptor for egg jelly is a plasma membrane protein of mouse sperm head. Mol Reprod Dev 73:350–360 10.1002/mrd.20410 [DOI] [PubMed] [Google Scholar]
- 19.Sutton KA, Jungnickel MK, Ward CJ, Harris PC, Florman HM (2006) Functional characterization of PKDREJ, a male germ cell-restricted polycystin. J Cell Physiol 209:493–500 10.1002/jcp.20755 [DOI] [PubMed] [Google Scholar]
- 20.Swanson WJ, Nielsen R, Yang Q (2003) Pervasive adaptive evolution in Mammalian fertilization proteins. Mol Biol Evol 20:18–20 [DOI] [PubMed] [Google Scholar]
- 21.Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J (2004) PHYLIP (Phylogeny Inference Package) release 3.6, Seattle [Google Scholar]
- 24.Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- 25.Yang Z (2000) Phylogenetic Analysis by Maximum Likelihood (PAML). release 3.1, London [Google Scholar]
- 26.Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Wong WS, Nielsen R (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118 10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- 28.Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT (2004) Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet 36:1326–1329 10.1038/ng1471 [DOI] [PubMed] [Google Scholar]
- 29.Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194 [PubMed] [Google Scholar]
- 30.Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- 31.Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- 32.Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 10.1093/nar/25.14.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- 35.Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95:5857–5864 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramensky V, Bork P, Sunyaev S (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30:3894–3900 10.1093/nar/gkf493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodman M (1999) The genomic record of humankind’s evolutionary roots. Am J Hum Genet 64:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman M, Bailey WJ, Hayasaka K, Stanhope MJ, Slightom J, Czelusniak J (1994) Molecular evidence on primate phylogeny from DNA sequences. Am J Phys Anthropol 94:3–24 10.1002/ajpa.1330940103 [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Nielsen R, Yang Z (2005) Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22:2472–2479 10.1093/molbev/msi237 [DOI] [PubMed] [Google Scholar]
- 41.Kingan SB, Tatar M, Rand DM (2003) Reduced polymorphism in the chimpanzee semen coagulating protein semenogelin I. J Mol Evol 57:159–169 10.1007/s00239-002-2463-0 [DOI] [PubMed] [Google Scholar]
- 42.Przeworski M, Hudson RR, Di Rienzo A (2000) Adjusting the focus on human variation. Trends Genet 16:296–302 10.1016/S0168-9525(00)02030-8 [DOI] [PubMed] [Google Scholar]
- 43.McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- 44.Nielsen R (2005) Molecular signatures of natural selection. Annu Rev Genet 39:197–218 10.1146/annurev.genet.39.073003.112420 [DOI] [PubMed] [Google Scholar]
- 45.McDonald JH (1996) Detecting non-neutral heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Mol Biol Evol 13:253–260 [DOI] [PubMed] [Google Scholar]
- 46.Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kierszenbaum AL (2004) Polycystins: what polycystic kidney disease tells us about sperm. Mol Reprod Dev 67:385–388 10.1002/mrd.20042 [DOI] [PubMed] [Google Scholar]
- 50.Mengerink KJ, Moy GW, Vacquier VD (2000) suREJ proteins: new signalling molecules in sea urchin spermatozoa. Zygote Suppl 8:S28–S30 [PubMed] [Google Scholar]
- 51.Frank SA (2000) Sperm competition and female avoidance of polyspermy mediated by sperm-egg biochemistry. Evol Ecol Res 2:613–625 [Google Scholar]
- 52.Tsaur SC, Ting CT, Wu CI (2001) Sex in Drosophila mauritiana: a very high level of amino acid polymorphism in a male reproductive protein gene, Acp26Aa. Mol Biol Evol 18:22–26 [DOI] [PubMed] [Google Scholar]
- 53.Herndon LA, Wolfner MF (1995) A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc Natl Acad Sci USA 92:10114–10118 10.1073/pnas.92.22.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aguade M, Miyashita N, Langley CH (1992) Polymorphism and divergence in the Mst26A male accessory gland gene region in Drosophila. Genetics 132:755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Metz EC, Palumbi SR (1996) Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol Biol Evol 13:397–406 [DOI] [PubMed] [Google Scholar]
- 56.Vacquier VD, Moy GW (1977) Isolation of bindin: the protein responsible for adhesion of sperm to sea urchin eggs. Proc Natl Acad Sci USA 74:2456–2460 10.1073/pnas.74.6.2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gavrilets S, Waxman D (2002) Sympatric speciation by sexual conflict. Proc Natl Acad Sci USA 99:10533–10538 10.1073/pnas.152011499 [DOI] [PMC free article] [PubMed] [Google Scholar]