Abstract
Measles virus continues to cause morbidity and mortality despite the existence of a safe and efficacious vaccine. Measles is associated with induction of both a long-lived protective immune response and immunosuppression. To gain insight into immunological changes during measles virus infection, we examined gene expression in blood mononuclear cells from children with acute measles and children in the convalescent phase compared to uninfected control children. There were 13 significantly upregulated and 206 downregulated genes. Upregulated genes included the immune regulatory molecules interleukin 1β (IL-1β), CIAS-1, tumor necrosis factor alpha, PDE4B, PTGS2, IL-8, CXCL2, CCL4, ICAM-1, CD83, GOS-2, IER3 (IEX-1), and TNFAIP3 (A20). Plasma levels of IL-1β and IL-8 were elevated during measles virus infection. Downregulated genes mainly involved three gene ontology biological processes, transcription, signal transduction, and the immune response, and included IL-16 and cell surface receptors IL-4R, IL-6R, IL-7R, IL-27RA, CCR2, and CCR7. Most mRNAs had not returned to control values 1 month after discharge, consistent with prolonged immune response abnormalities during measles virus infection.
Measles virus (MV) is remarkable for the robust immune response it elicits in the face of a generalized immune suppression that lasts several weeks following acute infection (10). After infection of the respiratory epithelium, MV is transported to the regional lymph nodes, probably by dendritic cells or macrophages (9). After replication in lymphoid tissue, there is systemic spread to multiple organs including skin, spleen, and liver. Infection induces a robust immune response beginning at the time of the rash with production of immunoglobulin M antibody to the MV nucleoprotein and induction of cellular immune responses. Antibodies are made to most MV proteins, with immunoglobulin G1 as the predominant isotype (9). The cellular immune response is responsible for the characteristic measles rash and is important for viral clearance (33, 48). Patients with agammaglobulinemia develop a rash and recover, while those with defects in cellular immunity may not (8, 23).
During the rash and recovery, there is evidence of immune activation that includes lymphoproliferation, an increase in plasma levels of soluble CD4 (sCD4), sCD8, soluble interleukin 2 receptor (IL-2R), β2-microglobulin, neopterin, sFas, sFasL, and soluble tumor necrosis factor receptor, and increased surface expression of Fas, CD25, and CD38 on circulating lymphocytes (10, 11, 37). Cytokines are also increased in plasma during the immune response to MV. Gamma interferon (IFN-γ) is detected early and then returns to baseline after the rash appears (26, 29). IL-2, IL-4, IL-10, and IL-13 are elevated during acute infection, and increased levels of IL-4 and IL-10 persist for several weeks after recovery (26). IL-12 levels are lower than those of the controls, indicating that some cytokine responses are downregulated by MV infection (1, 26, 29, 34). During acute disease and recovery, lymphoproliferative responses to mitogens and delayed type hypersensitivity responses to recall antigens are repressed, and there is an increased susceptibility to other infections (9, 13).
To better characterize MV-induced immunological changes at a transcriptional level, microarrays were used to globally survey mRNA levels in peripheral blood mononuclear cells (PBMCs) during acute measles and convalescence.
MATERIALS AND METHODS
Subjects and samples.
Peripheral blood samples for microarray and plasma analysis were collected at a single time from three control children and at study entry, hospital discharge, and 1-month follow-up from five children hospitalized with measles at the University Teaching Hospital in Lusaka, Zambia (Table 1). The median numbers of days after rash onset at entry, discharge, and follow-up were 4, 8, and 40, respectively. Children with available PBMCs with high-quality RNA from all three time points were selected for microarray analysis from a larger prospective study of measles (24). Plasma samples from an additional 8 control children and 19 children with measles were used for analysis of plasma levels of cytokines and chemokines. All were confirmed to have measles and to be negative for human immunodeficiency virus type 1 (HIV-1) infection (24).
TABLE 1.
Clinical characteristics of patients and controls
| Subject | Type | Age (mo) | Sex | No. of days after onset of rash ata:
|
||
|---|---|---|---|---|---|---|
| Entry | Discharge | Follow-up | ||||
| 187 | Control | 26 | Female | NA | NA | NA |
| 188 | Control | 15 | Male | NA | NA | NA |
| 194 | Control | 9 | Male | NA | NA | NA |
| 33 | Patient | 21 | Male | 3 | 8 | 61 |
| 41 | Patient | 79 | Female | 2 | 4 | 36 |
| 43 | Patient | 9 | Male | 7 | 11 | 41 |
| 538 | Patient | 11 | Female | 4 | 9 | 39 |
| 536 | Patient | 8 | Male | 4 | 6 | 40 |
NA, not applicable.
White blood cell (WBC) and differential counts were performed manually. Percentages of CD4+ and CD8+ lymphocytes were determined by flow cytometry as previously described (37). Plasma was separated and stored at −80°C. PBMCs were isolated on Ficoll-Hypaque gradients, washed, and stored in RNAzol or RNA-Stat (Tel-Test, Inc., Friendswood, TX) at −80°C. All studies were performed with protocols approved by the Committee for Human Research of the Johns Hopkins Bloomberg School of Public Health and the Ethics Committee of the University of Zambia.
GeneChip assays.
RNAs from PBMCs of five children with measles and three control children (Table 1) were isolated following the manufacturer's instructions and used for microarray studies. RNA quality was determined by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100 (Agilent, Palo Alto, CA). For each sample, 100 ng of total RNA was processed according to the amplification protocol from the Affymetrix GeneChip Eukaryotic Small Sample Target Labeling Technical Note, version 1 (Affymetrix, Santa Clara, CA). Two cycles of amplification, each with a cDNA synthesis step followed by an in vitro transcription reaction, were used to generate cRNA. The second in vitro transcription synthesis incorporated biotinylated ribonucleotides to produce labeled antisense cRNA targets for hybridization. Fifteen micrograms of cRNA was fragmented by metal-induced hydrolysis in fragmentation buffer (250 mM Tris acetate, pH 8.1, 150 mM magnesium acetate, 500 mM potassium acetate) at 94°C for 35 min. Hybridization cocktails were prepared as recommended for arrays of “Standard” format and applied to Affymetrix Human Genome U133A GeneChips. Hybridization was performed at 45°C for 16 h at 60 rpm in the Affymetrix rotisserie hybridization oven. The signal amplification protocol for washing and staining of eukaryotic targets was performed in an automated fluidics station (Affymetrix FS450) as described in the Affymetrix Technical Manual, Revision 3. The arrays were transferred to the GCS3000 laser scanner (Affymetrix) and scanned at an emission wavelength of 570 nm at 2.5-mm resolution. Intensity of hybridization for each probe pair was computed by GCOS 1.1 software.
Exploratory data analysis was performed on the samples using R v2.2.0 and the add-on package affy v1.5.8 (14) (http://www.R-project.org). Background correction, normalization, and summarization were performed using Robust MultiArray Analysis (14). Significance testing was performed with Significance Analysis of Microarrays (SAM) v2.21 to compare control samples with those obtained from children with measles at entry, discharge, and follow-up (46). A gene was considered significant if it had a change in expression of >2.0-fold and a false discovery rate (FDR) of <15% at the 90th percentile. SAM allows the investigator to choose the FDR, and with these parameters, no more than 15% of the genes identified as significant would be false discoveries. A more stringent FDR did not significantly alter the results. The genes were annotated with the 20 June 2005 Affymetrix release and grouped into gene ontologies (GO) with Onto-Express using the default settings (6). Only groups with corrected P < 0.05 and including five or more genes were considered in the analysis.
Cytokine and chemokine assays.
Enzyme immunoassays (EIAs) for IL-1β, IL-8, CCL4, and CXCL2 were performed on stored plasma according to the manufacturers' protocols (R&D Systems and IBL-America, Minneapolis, MN). Plasma levels of IL-16 were previously determined by EIA (25).
Microarray data accession number.
The microarray data are available at GEO under accession number GSE5808.
RESULTS
Leukocytes.
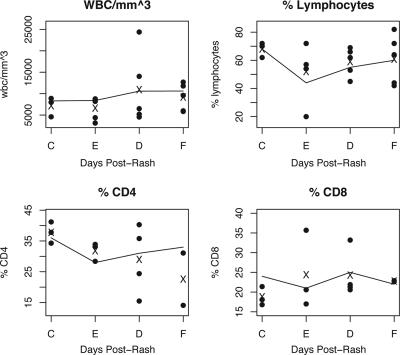
WBC and differential counts were available for most children at each time point, and the percentages of CD4+ and CD8+ lymphocytes were determined on a subset of the samples (Fig. 1). WBC counts and the percentage of lymphocytes, CD4+, and CD8+ T lymphocytes closely resembled those reported for the larger study from which these samples were drawn (Fig. 1) (24). WBC counts were slightly elevated at discharge and follow-up with a lower lymphocyte percentage at entry and generally lower percentages of CD4+ T cells compared to controls.
FIG. 1.
WBC composition. The individual counts for each patient are shown with filled circles, and the mean values for the samples in this study are displayed with an “X.” The means for the larger cohort are shown with a line connecting the sampling times. Data are shown for controls (C) and for children with measles at entry (E; days 2 to 7), discharge (D; days 4 to 11), and follow-up (F; days 36 to 61).
Gene expression.
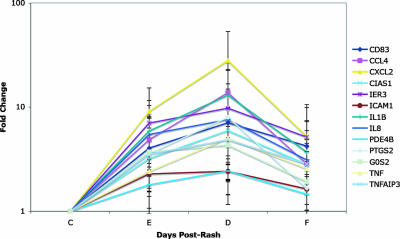
Microarray analysis was performed on mRNAs from PBMCs obtained from children with measles (see Table S1 in the supplemental material). There were no significantly regulated genes at entry, 224 at discharge, and 32 at follow-up. The results were pooled and duplicate genes were averaged, leaving 13 upregulated genes and 206 downregulated genes. Similar results were obtained by comparing controls versus days 2 to 4, 6 to 11, and 36 to 61. The significant groups with more than five genes for the GO biological processes annotation were identified. The only upregulated groups were signal transduction and apoptosis, which overlapped significantly. Upregulated genes included the cytokines IL-1β and tumor necrosis factor alpha (TNF-α), chemokines CCL4, CXCL2, and IL-8, cell surface molecules ICAM-1 and CD83, and regulatory proteins CIAS-1 (NALP3), IER3 (IEX-1), PDE4B, PRGS2, GOS2, and TNFAIP3 (A20) (Fig. 2). These mRNAs increased at entry, peaked at discharge, and declined but remained above the control levels at follow-up.
FIG. 2.
Upregulated genes. The mean and standard deviation of the change in expression (n-fold) for each gene is shown. CIAS1, cold autoinflammatory syndrome 1 (NALP3); IER3, immediate early response 3 (IEX-1); ICAM1, intercellular adhesion molecule 1; PDE4B, phosphodiesterase 4B; PTGS2, prostaglandin-endoperoxide synthase 2; GOS2, putative lymphocyte G0/G1 switch gene; TNFAIP3, tumor necrosis factor α-induced protein 3 (A20); C, control; E, entry; D, discharge; F, follow-up.
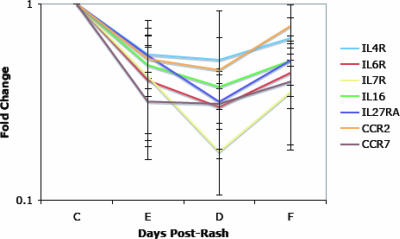
The downregulated gene classifications fell into three broad GO biological process groups: transcription, signal transduction, and the immune response (Table 2). These gene expression levels were decreased at entry, and for most, the lowest expression levels were at discharge. There was a tendency to recover by follow-up, although not back to control levels (Fig. 3). The biological process group for transcription had the largest number of genes and included four classifications of downregulated genes: regulation of transcription, DNA-dependent (e.g., Sp3 transcription factor [Sp3], GATA binding protein 3 [GATA3], and lymphoid enhancer-binding factor 1 [LEF1]); transcription (e.g., BTB and CNC homology 1, basic leucine zipper transcription factor 2 [Bach2], Kruppel-like factor 2 [KLF2], and zinc finger protein 161 [ZNF161]); regulation of transcription from RNA polymerase II promoter (e.g., transcription factor 4 [TCF4], Kruppel-like factor 12 [KLF12], and Spi-B transcription factor [SPIB]); and nuclear mRNA splicing, via spliceosome (e.g., splicing factor 3a, subunit 2 and 3b, subunit 1 [SF3A2 and SF3B1] and RNA binding motif protein 14 [RBM14]). The largest gene groups in these classifications were zinc finger proteins (n = 10) and transcription factors (n = 10). The signal transduction biological process group included the classifications signal transduction, protein amino acid phosphorylation, and cell surface receptor-linked signal transduction. There were nine receptor genes and eight kinases in this group. The immune response biological process group included the classifications immune response, antimicrobial humoral response, and cellular defense response. Genes within these classifications included the cytokines IL-16 and TNFSF3 (LTβ) and the cytokine and chemokine receptors IL-4R, IL-6R, IL-7R, IL-27RA, CCR2, and CCR7 (Fig. 3).
TABLE 2.
Significantly downregulated genes grouped by gene ontology (GO) biological process as analyzed using Onto-Express software
| GO IDc | Function name | Unique inputa | Corrected Pb |
|---|---|---|---|
| 6355 | Regulation of transcription, DNA dependent | 35 | 0.0019 |
| 6350 | Transcription | 30 | 0.0012 |
| 7165 | Signal transduction | 24 | 0.038 |
| 6468 | Protein amino acid phosphorylation | 13 | 0.031 |
| 6955 | Immune response | 12 | 0.004 |
| 6357 | Regulation of transcription from RNA polymerase II promoter | 11 | 0.0025 |
| 398 | Nuclear mRNA splicing, via spliceosome | 7 | 0.018 |
| 7166 | Cell surface receptor-linked signal transduction | 6 | 0.019 |
| 19735 | Antimicrobial humoral response (sensu Vertebrata) | 6 | 0.0023 |
| 6968 | Cellular defense response | 5 | 0.0058 |
Number of genes in each group.
Probability that a finding is not due to chance after correcting for multiple comparisons.
ID, identification.
FIG. 3.
Examples of downregulated immune response genes. The mean and standard deviation of the change in expression (n-fold) for each gene is shown. IL27RA, IL-27 receptor alpha chain (WSX-1); C, control; E, entry; D, discharge; F, follow-up.
Plasma cytokine and chemokine levels.
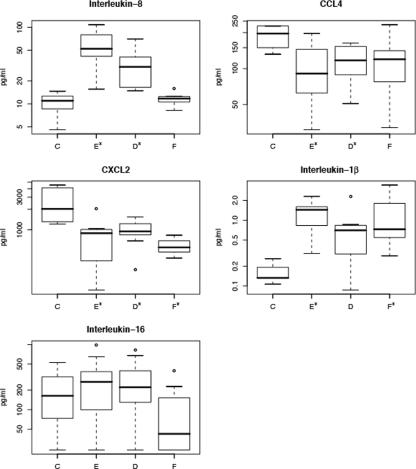
To determine whether changes in mRNA expression levels resulted in altered plasma levels of cytokines or chemokines, EIAs were performed. IL-1β and IL-8 levels were significantly increased during acute measles (Fig. 4). IL-8 levels returned to baseline at follow-up, while IL-1β levels remained elevated. CXCL2 and CCL4 were both higher in controls than during acute infection. Plasma levels of IL-16 were lower at follow-up than at earlier times during infection and than in controls.
FIG. 4.
Plasma levels of chemokines and cytokines. Box plots of plasma protein levels of IL-8, CCL4, CXCL2, IL-1β, and IL-16, as measured by EIA, are shown. Time points with P < 0.05 relative to the control level are indicated by an *. IL-16 plasma levels were previously reported for this cohort (25).
DISCUSSION
PBMCs are the most accessible tissue for analysis of gene expression during infectious disease in humans, but only a few infections have been studied. Examination of changes in gene expression in PBMCs from children with measles at hospital entry, discharge, and 1-month follow-up identified 13 upregulated genes and 206 downregulated genes. The upregulated genes tended to be genes regulated by NF-κB rather than by IFN and were consistent with an ongoing immune response. The downregulated genes were involved in cellular signaling and transcription and had not returned to control levels at follow-up. These gene expression patterns are consistent with, and help to explain, the prolonged alteration of lymphocyte responses characteristic of measles.
Remarkably, IFN-responsive genes were not upregulated and may have been actively suppressed. The function of one upregulated gene, TNFAIP3 (A20), is to downregulate the activity of IRF-3, a key transcription factor for induction of IFN (19, 38). This pattern is in contrast to the mRNAs increased by in vitro MV infection of dendritic cells and PBMCs that were predominantly IFN and IFN-responsive genes (3, 50). Likewise, PBMCs from influenza virus-infected humans and smallpox virus-infected monkeys showed significant upregulation of immune modulators induced by IFN (35, 36). However, PBMCs from patients with severe acute respiratory syndrome showed a similar pattern to measles with several upregulated genes from the innate immune system, but no IFN-induced genes (35). This suggests that gene expression in PBMCs during the phase of measles studied reflects primarily the immune response to infection, not infection per se. Because measles is not recognized until the rash appears as virus clearance is initiated 10 to 14 days after infection, and changes in gene expression are rapid and tightly controlled (3, 4, 50), it is possible that an IFN response would have been detected earlier in infection.
Although changes in gene expression were not statistically significant until discharge, the patterns of expression of both up- and downregulated genes were established at entry. The lack of statistical significance is probably related to the greater variability at this time of rapid change related to initiation of the immune response. In addition, the mRNAs for several proteins that are significantly increased or decreased in plasma during measles virus infection (e.g., IL-2, IL-4, IL-10, and IL-12) (1, 26, 29, 34) were not identified as regulated. This could be because PBMCs are not the main source of these cytokines, because regulation is not at the level of transcription, or because the criteria used for gene identification missed biologically relevant changes in PBMC gene expression.
Among the upregulated genes were proinflammatory cytokines IL-1β and TNF-α. IL-1β mRNA and protein production are also upregulated in MV-infected monocyte-derived cells and cultured PBMCs taken from patients during the convalescent phase of infection (18, 49). Production of biologically active IL-1β is only partially regulated at the transcriptional level, as the mRNA is translated as a leaderless 35-kDa precursor protein that must be processed by caspase-1 to the active secreted form. IL-1β processing is regulated within the “inflammasome,” a complex of the proteins CIAS-1 (NALP3), Cardinal, ASC, and pro-IL-1β (28, 44). CIAS-1 mRNA was also upregulated during measles virus infection and IL-1β protein was increased in the plasma of the children studied, documenting the biologic relevance of the increased expression of these genes. A previous study did not detect IL-1β in serum, but the assay may have lacked sensitivity (29).
Cell surface molecules that were upregulated included ICAM-1 and CD83. ICAM-1 is induced on activated lymphocytes and monocytes and enhances both antigen presentation and vascular adhesion (16). CD83 is a marker of mature dendritic cells and regulates the development of cellular immune responses (17, 41).
There were several chemokines significantly upregulated during measles virus infection, specifically CCL4 (MIP-1β), CXCL2 (GRO-β), and CXCL8 (IL-8). CCL4 is produced at sites of interaction between antigen-specific dendritic cells and CD4+ T cells and attracts naïve CD8+ T cells that express CCR5 (5, 22, 32). CXCL2 and IL-8 are structurally related GRO family chemokines that are produced by macrophages (42). Both IL-8 and CXCL2 bind to CXCR2, but IL-8 also binds to CXCR1 expressed on a subset of effector CD8 T cells (12, 45). CXCL2 may contribute to lung injury during viral infection (20). Infection of epithelial cells with a number of respiratory viruses, including MV, increases expression of IL-8 in vitro (40). IL-8 is increased in the plasma of monkeys early after MV infection (40) and, in this study, was increased in children with measles during the rash. However, CCL4 and CXCL2 plasma levels were lower during acute infection, suggesting that the changes seen in PBMCs may have local effects that were not reflected in plasma.
Expression of many more genes was decreased than increased, and these decreased mRNAs encoded proteins involved in signaling and regulation of transcription. In contrast, in lipopolysaccharide-challenged humans, the downregulated genes were involved in respiration, protein synthesis, and degradation (4), and in smallpox virus-infected monkeys, the downregulated genes were T and B cell markers and reflected lymphopenia. Acute measles changes were most similar to those induced by chronic HIV-1 infection in humans, where genes associated with signal transduction, immune response, and transcription were also largely downregulated (27). Simian immunodeficiency virus-infected macaques sampled at 3 and 7 weeks after infection had two downregulated zinc finger proteins similar to measles but otherwise showed quite different responses with few downregulated genes (47). Although the cell composition of the PBMC compartment is changing during measles virus infection, we do not think that this had a significant effect on the results, because the largest alterations in cell composition occurred at entry. This fact may have contributed to the variability and lack of significantly regulated genes at this time.
Among the downregulated genes was the CD4 chemotactic cytokine IL-16, and this was reflected in decreased IL-16 plasma levels at follow-up (7, 25). Several receptors for cytokines and chemokines were downregulated: IL-4R, IL-6R, IL-7R, IL-27RA, CCR2, and CCR7. CCR2, the receptor for MCP1 (CCL2), MCP3 (CCL7), and MCP5 (CCL12), is highly expressed on monocytes and is downregulated when monocytes differentiate into macrophages (21, 31). Both TLR2 and TLR4 agonists downregulate CCR2, and the MV H protein can activate TLR2 (2, 30). CCR7 is important for dendritic cell and T cell homing to lymph nodes and is upregulated early after MV infection of dendritic cells (50) but is absent on effector memory cells (15), which express receptors necessary for migration to inflamed tissues. This decrease is consistent with an ongoing T cell effector response during the rash phase of measles (9, 39). Also consistent with an effector memory response is the increase in phosphodiesterase-4 (PDE4B) that is expressed at high levels in memory CD4+ T cells, where it regulates degradation of cyclic AMP, a mediator of inflammatory responses (43).
In summary, the gene expression patterns during the rash and convalescent phase of measles reflect ongoing immune responses and extensive downregulation of signaling and transcription pathways that are likely to underlie the inability of lymphocytes to respond to stimuli, such as tuberculin, with a delayed type hypersensitivity response or to mitogens with proliferation.
Supplementary Material
Acknowledgments
We thank Anne Jedlicka and Margaret V. Mintz for help with the microarray processing; Giovanni Parmigiani for help with statistical analysis; Mwaka Monze and Francis Kasolo for facilitating research at the Virology Laboratory and the University Teaching Hospital; clinical staff for work with patient recruitment; Felicity Cutts and the London School of Hygiene and Tropical Medicine for helping to establish the cohort; and the Japan International Cooperation Agency for generously allowing the use of laboratory facilities in Zambia.
This work was supported by grants R01 AI-23047 and T32 ES-07141 from the National Institutes of Health and by the Bill and Melinda Gates Foundation. The microarray core facility was supported by the Johns Hopkins Malaria Research Institute.
Footnotes
Published ahead of print on 30 May 2007.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Atabani, S., A. Byrnes, A. Jaye, I. Kidd, A. Magnusen, H. Whittle, and C. Karp. 2001. Natural measles causes prolonged suppression of interleukin-12 production. J. Infect. Dis. 1841-9. [DOI] [PubMed] [Google Scholar]
- 2.Bieback, K., E. Lien, I. M. Klagge, E. Avota, J. Schneider-Schaulies, W. P. Duprex, H. Wagner, C. J. Kirschning, V. ter Meulen, and S. Schneider-Schaulies. 2002. Hemagglutinin protein of wild-type measles virus activates Toll-like receptor 2 signaling. J. Virol. 768729-8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolt, G., K. Berg, and M. Blixenkrone-Moller. 2002. Measles virus-induced modulation of host-cell gene expression. J. Gen. Virol. 831157-1165. [DOI] [PubMed] [Google Scholar]
- 4.Calvano, S. E., W. Xiao, D. R. Richards, R. M. Felciano, H. V. Baker, R. J. Cho, R. O. Chen, B. H. Brownstein, J. P. Cobb, S. K. Tschoeke, C. Miller-Graziano, L. L. Moldawer, M. N. Mindrinos, R. W. Davis, R. G. Tompkins, and S. F. Lowry. 2005. A network-based analysis of systemic inflammation in humans. Nature 4371032-1037. [DOI] [PubMed] [Google Scholar]
- 5.Castellino, F., A. Y. Huang, G. Altan-Bonnet, S. Stoll, C. Scheinecker, and R. N. Germain. 2006. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 440890-895. [DOI] [PubMed] [Google Scholar]
- 6.Draghici, S., P. Khatri, P. Bhavsar, A. Shah, S. A. Krawetz, and M. A. Tainsky. 2003. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 313775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass, W. G., R. T. Sarisky, and A. M. Vecchio. 2006. Not-so-sweet sixteen: the role of IL-16 in infectious and immune-mediated inflammatory diseases. J. Interferon Cytokine Res. 26511-520. [DOI] [PubMed] [Google Scholar]
- 8.Good, R. A., and S. J. Zak. 1956. Disturbances in gammaglobulin synthesis as “experiments of nature.” Pediatrics 18109-149. [PubMed] [Google Scholar]
- 9.Griffin, D. E. 2007. Measles virus, p. 1551-1586. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 10.Griffin, D. E., B. J. Ward, and L. M. Esolen. 1994. Pathogenesis of measles virus infection: an hypothesis for altered immune responses. J. Infect. Dis. 170S24-S31. [DOI] [PubMed] [Google Scholar]
- 11.Griffin, D. E., B. J. Ward, E. Jauregui, R. T. Johnson, and A. Vaisberg. 1992. Immune activation during measles: beta-2-microglobulin in plasma and cerebrospinal fluid in complicated and uncomplicated disease. J. Infect. Dis. 1661170-1173. [DOI] [PubMed] [Google Scholar]
- 12.Hess, C., T. K. Means, P. Autissier, T. Woodberry, M. Altfeld, M. M. Addo, N. Frahm, C. Brander, B. D. Walker, and A. D. Luster. 2004. IL-8 responsiveness defines a subset of CD8 T cells poised to kill. Blood 1043463-3471. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch, R. L., D. E. Griffin, R. T. Johnson, S. J. Cooper, I. Lindo de Soriano, S. Roedenbeck, and A. Vaisberg. 1984. Cellular immune responses during complicated and uncomplicated measles virus infections of man. Clin. Immunol. Immunopathol. 311-12. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry, R. A., L. Gautier, and L. M. Cope. 2003. An R package for analyses of affymatrix oligonucleotide arrays, p. 102-119. In G. Parmigiani, E. S. Garrett, R. A. Irizarry, and S. I. Zeger (ed.), The analysis of gene expression data: methods and software. Springer-Verlag, New York, NY.
- 15.Langenkamp, A., K. Nagata, K. Murphy, L. Wu, A. Lanzavecchia, and F. Sallusto. 2003. Kinetics and expression patterns of chemokine receptors in human CD4+ T lymphocytes primed by myeloid or plasmacytoid dendritic cells. Eur. J. Immunol. 33474-482. [DOI] [PubMed] [Google Scholar]
- 16.Lebedeva, T., M. L. Dustin, and Y. Sykulev. 2005. ICAM-1 co-stimulates target cells to facilitate antigen presentation. Curr. Opin. Immunol. 17251-258. [DOI] [PubMed] [Google Scholar]
- 17.Lechmann, M., S. Berchtold, J. Hauber, and A. Steinkasserer. 2002. CD83 on dendritic cells: more than just a marker for maturation. Trends Immunol. 23273-275. [DOI] [PubMed] [Google Scholar]
- 18.Leopardi, R., R. Vainionpaa, M. Hurme, P. Siljander, and A. A. Salmi. 1992. Measles virus infection enhances IL-1β but reduces tumor necrosis factor-α expression in human monocytes. J. Immunol. 1492397-2401. [PubMed] [Google Scholar]
- 19.Lin, R., L. Yang, P. Nakhaei, Q. Sun, E. Sharif-Askari, I. Julkunen, and J. Hiscott. 2006. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J. Biol. Chem. 2812095-2103. [DOI] [PubMed] [Google Scholar]
- 20.Londhe, V. A., J. A. Belperio, M. P. Keane, M. D. Burdick, Y. Y. Xue, and R. M. Strieter. 2005. CXCR2 is critical for dsRNA-induced lung injury: relevance to viral lung infection. J. Inflamm. (London) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luther, S. A., and J. G. Cyster. 2001. Chemokines as regulators of T cell differentiation. Nat. Immunol. 2102-107. [DOI] [PubMed] [Google Scholar]
- 22.Maurer, M., and E. von Stebut. 2004. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 361882-1886. [DOI] [PubMed] [Google Scholar]
- 23.Moss, W. J., F. Cutts, and D. E. Griffin. 1999. Implications of the HIV epidemic for control and eradication of measles. Clin. Infect. Dis. 29106-112. [DOI] [PubMed] [Google Scholar]
- 24.Moss, W. J., M. Monze, J. J. Ryon, T. C. Quinn, D. E. Griffin, and F. Cutts. 2002. Prospective study of measles in hospitalized, human immunodeficiency virus (HIV)-infected and HIV-uninfected children in Zambia. Clin. Infect. Dis. 35189-196. [DOI] [PubMed] [Google Scholar]
- 25.Moss, W. J., J. J. Ryon, M. Monze, F. Cutts, T. C. Quinn, and D. E. Griffin. 2002. Suppression of human immunodeficiency virus replication during acute measles. J. Infect. Dis. 1851035-1042. [DOI] [PubMed] [Google Scholar]
- 26.Moss, W. J., J. J. Ryon, M. Monze, and D. E. Griffin. 2002. Differential regulation of interleukin (IL)-4, IL-5, and IL-10 during measles in Zambian children. J. Infect. Dis. 186879-887. [DOI] [PubMed] [Google Scholar]
- 27.Ockenhouse, C. F., W. B. Bernstein, Z. Wang, and M. T. Vahey. 2005. Functional genomic relationships in HIV-1 disease revealed by gene-expression profiling of primary human peripheral blood mononuclear cells. J. Infect. Dis. 1912064-2074. [DOI] [PubMed] [Google Scholar]
- 28.Ogura, Y., F. S. Sutterwala, and R. A. Flavell. 2006. The inflammasome: first line of the immune response to cell stress. Cell 126659-662. [DOI] [PubMed] [Google Scholar]
- 29.Ohga, S., C. Miyazaki, K. Okada, K. Akazawa, and K. Ueda. 1992. The inflammatory cytokines in measles: correlation between serum interferon-gamma levels and lymphocyte subpopulations. Eur. J. Pediatr. 151492-496. [DOI] [PubMed] [Google Scholar]
- 30.Parker, L. C., M. K. Whyte, S. N. Vogel, S. K. Dower, and I. Sabroe. 2004. Toll-like receptor (TLR)2 and TLR4 agonists regulate CCR expression in human monocytic cells. J. Immunol. 1724977-4986. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, R. J., M. Lutz, and B. Premack. 2005. Differential signaling mechanisms regulate expression of CC chemokine receptor-2 during monocyte maturation. J. Inflamm. (London) 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piqueras, B., J. Connolly, H. Freitas, A. K. Palucka, and J. Banchereau. 2006. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood 1072613-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polack, F. P., P. G. Auwaerter, S.-H. Lee, H. C. Nousari, A. Valsamakis, K. M. Leiferman, A. Diwan, R. J. Adams, and D. E. Griffin. 1999. Production of atypical measles in rhesus macaques: evidence for disease mediated by immune complex formation and eosinophils in the presence of fusion-inhibiting antibody. Nat. Med. 5629-634. [DOI] [PubMed] [Google Scholar]
- 34.Polack, F. P., S. J. Hoffman, W. J. Moss, and D. E. Griffin. 2002. Altered synthesis of interleukin-12 and type 1 and type 2 cytokines in rhesus macaques during measles and atypical measles. J. Infect. Dis. 18513-19. [DOI] [PubMed] [Google Scholar]
- 35.Reghunathan, R., M. Jayapal, L. Y. Hsu, H. H. Chng, D. Tai, B. P. Leung, and A. J. Melendez. 2005. Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunol. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubins, K. H., L. E. Hensley, P. B. Jahrling, A. R. Whitney, T. W. Geisbert, J. W. Huggins, A. Owen, J. W. LeDuc, P. O. Brown, and D. A. Relman. 2004. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc. Natl. Acad. Sci. USA 10115190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryon, J. J., W. J. Moss, M. Monze, and D. Griffin. 2002. Functional and phenotypic changes in circulating lymphocytes from hospitalized Zambian children with measles. Clin. Diagn. Lab Immunol. 9994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitoh, T., M. Yamamoto, M. Miyagishi, K. Taira, M. Nakanishi, T. Fujita, S. Akira, N. Yamamoto, and S. Yamaoka. 2005. A20 is a negative regulator of IFN regulatory factor 3 signaling. J. Immunol. 1741507-1512. [DOI] [PubMed] [Google Scholar]
- 39.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401708-712. [DOI] [PubMed] [Google Scholar]
- 40.Sato, H., R. Miura, and C. Kai. 2005. Measles virus infection induces interleukin-8 release in human pulmonary epithelial cells. Comp. Immunol. Microbiol. Infect. Dis. 28311-320. [DOI] [PubMed] [Google Scholar]
- 41.Scholler, N., M. Hayden-Ledbetter, A. Dahlin, I. Hellstrom, K. E. Hellstrom, and J. A. Ledbetter. 2002. Cutting edge: CD83 regulates the development of cellular immunity. J. Immunol. 1682599-2602. [DOI] [PubMed] [Google Scholar]
- 42.Smith, D. F., E. Galkina, K. Ley, and Y. Huo. 2005. GRO family chemokines are specialized for monocyte arrest from flow. Am. J. Physiol. Heart Circ. Physiol. 289H1976-H1984. [DOI] [PubMed] [Google Scholar]
- 43.Sun, Y., L. Li, F. Lau, J. A. Beavo, and E. A. Clark. 2000. Infection of CD4+ memory T cells by HIV-1 requires expression of phosphodiesterase 4. J. Immunol. 1651755-1761. [DOI] [PubMed] [Google Scholar]
- 44.Sutterwala, F. S., Y. Ogura, M. Szczepanik, M. Lara-Tejero, G. S. Lichtenberger, E. P. Grant, J. Bertin, A. J. Coyle, J. E. Galan, P. W. Askenase, and R. A. Flavell. 2006. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity 24317-327. [DOI] [PubMed] [Google Scholar]
- 45.Takata, H., H. Tomiyama, M. Fujiwara, N. Kobayashi, and M. Takiguchi. 2004. Cutting edge: expression of chemokine receptor CXCR1 on human effector CD8+ T cells. J. Immunol. 1732231-2235. [DOI] [PubMed] [Google Scholar]
- 46.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vahey, M. T., M. E. Nau, M. Taubman, J. Yalley-Ogunro, P. Silvera, and M. G. Lewis. 2003. Patterns of gene expression in peripheral blood mononuclear cells of rhesus macaques infected with SIVmac251 and exhibiting differential rates of disease progression. AIDS Res. Hum. Retrovir. 19369-387. [DOI] [PubMed] [Google Scholar]
- 48.van Binnendijk, R. S., M. C. M. Poelen, K. C. Kuijpers, A. D. M. E. Osterhaus, and F. G. C. M. Uytdehaag. 1990. The predominance of CD8+ T cells after infection with measles virus suggests a role for CD8+ class I MHC-restricted cytotoxic T lymphocytes (CTL) in recovery from measles. Clonal analyses of human CD8+ class I MHC-restricted CTL. J. Immunol. 1442394-2399. [PubMed] [Google Scholar]
- 49.Ward, B. J., R. T. Johnson, A. Vaisberg, E. Jauregui, and D. E. Griffin. 1991. Cytokine production in vitro and the lymphoproliferative defect of natural measles virus infection. Clin. Immunol. Immunopathol. 61236-248. [DOI] [PubMed] [Google Scholar]
- 50.Zilliox, M. J., G. Parmigiani, and D. E. Griffin. 2006. Gene expression patterns in dendritic cells infected with measles virus compared with other pathogens. Proc. Natl. Acad. Sci. USA 1033363-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.