Abstract
Fifty-five epidemiologically linked Aspergillus fumigatus isolates obtained from six nosocomial outbreaks of invasive aspergillosis were subtyped by sequencing the polymorphic region of the gene encoding a putative cell surface protein, Afu3g08990 (denoted as CSP). Comparative sequence analysis showed that genetic diversity was generated in the coding region of this gene by both tandem repeats and point mutations. Each unique sequence in an outbreak cluster was assigned an arbitrary number or CSP sequence type. The CSP typing method was able to identify “clonal” and genotypically distinct A. fumigatus isolates, and the results of this method were concordant with those of another discriminatory genotyping technique, the Afut1 restriction fragment length polymorphism typing method. The novel single-locus sequence typing (CSP typing) strategy appears to be a simple, rapid, discriminatory tool that can be readily shared across laboratories. In addition, we found that A. fumigatus isolates substructured into multiple clades; interestingly, one clade consisted of isolates predominantly representing invasive clinical isolates recovered from cardiac transplant patients from two different outbreak situations. We also found that the A. fumigatus isolate Af293, whose genome has been sequenced, possesses a CSP gene structure that is substantially different from those of the other A. fumigatus strains studied here, highlighting the need for further taxonomic study.
Conidia of Aspergillus fumigatus become airborne easily, and subsequent inhalation of these infectious particles is believed to be the route of transmission for invasive aspergillosis. Accordingly, several nosocomial outbreaks of invasive aspergillosis have been reported with strong evidence suggesting that such infections are acquired from the environment of the hospital (10, 31, 35, 43). The Centers for Disease Control and Prevention (CDC) recommends that in the setting of a nosocomial aspergillosis outbreak and in the presence of continuing evidence of Aspergillus infection in the hospital population, an environmental assessment should be undertaken to determine and eliminate the source of infection (51). In such nosocomial outbreak investigations, Aspergillus strain typing methods can indicate the source and/or route of infection by determining whether epidemiologically related isolates are also genetically related. Once the source is identified, corrective measures can then be undertaken to eliminate the implicated source to contain the infection.
Several molecular methods have been evaluated for A. fumigatus strain typing, and these methods include randomly amplified polymorphic DNA typing (1), sequence-specific DNA primer analysis (32), polymorphic microsatellite markers (6, 7, 12), and analysis of hybridization profiles with a dispersed, repetitive DNA probe Afut1 (Afut1 restriction fragment length polymorphism [Afut1 RFLP]) (11, 36). Although a combination of typing methods appears to have more utility in strain typing, polymorphic microsatellite markers, and Afut1 methodologies used singly were found to be reproducible with a high discriminatory power (30) and appear to be more useful than other strain typing methods. However, these methods possess certain key shortcomings that restrict their utility in a clinical setting; most notably, they are not easily amenable to standardization across laboratories, and data collection requires specialized equipment and expertise to interpret sometimes difficult data sets. For these reasons, the former technologies remain largely inaccessible to many clinical microbiology laboratories. In contrast, DNA sequence-based typing methods are not plagued by these problems and, therefore, are being increasingly employed for routine identification of medically important fungi in clinical microbiology laboratories (18, 44). Comparative DNA sequencing methods such as those that determine sequence variation from several housekeeping genes (multilocus sequence typing [MLST]) have been elucidated for the routine typing of many different microbial species (13, 14, 33, 39). The MLST method has been particularly useful for strain discrimination in Candida spp. (9, 23, 52-54). and has been a powerful approach for A. fumigatus species identification (3-5, 20). However, the MLST method has not been suitable for A. fumigatus strain discrimination because of low levels of genetic diversity generated by the conserved protein-coding regions which results in an inability to differentiate strains (2, 47).
Recently, comparative genomic hybridization studies of A. fumigatus strains Af293, Af294, and Af71 have revealed that a number of genes, including those that encode enzymes and drug resistance transporters, were either absent or divergent in these A. fumigatus strains (37). Another study analyzing the entire A. fumigatus genome for open reading frames with coding tandem repeats found that several such genes were moderately to highly polymorphic in a test panel of A. fumigatus isolates (companion paper [31a]). The purpose of this study was to characterize the levels of polymorphism of one of these genes, the putative cell surface protein gene Afu3g08990 (hereby designated CSP) and to evaluate its discriminatory power on a panel of 55 epidemiologically linked A. fumigatus isolates from six different outbreak investigations. The results of our study show that CSP is a highly polymorphic marker that possesses sufficiently high levels of variability so as to successfully differentiate A. fumigatus strains.
MATERIALS AND METHODS
Aspergillus fumigatus strains.
Fifty-five isolates of epidemiologically linked A. fumigatus isolates were obtained from six CDC outbreak investigations and designated OB1 to OB6 (see Table 1). All of the isolates had available genotype data derived from Afut1 RFLP typing. Two of the six investigated outbreaks, OB2 and OB6, included isolates that were indistinguishable by Afut1 hybridization profile. Group OB2 included isolates that were previously recovered from the first reported case of person-to-person transmission of A. fumigatus; here the source of the outbreak was identified as the debridement and dressing of A. fumigatus-infected wounds from a patient in the same intensive care unit (43). Group OB6 included isolates recovered from a heart transplant unit in a Canadian hospital where the air vent duct was identified as the most likely source of the outbreak (unpublished data). Groups OB1, OB3, OB4, and OB5 did not yield a clear set of relationships based on Afut1 typing. Group OB1 isolates were obtained from a renal transplant unit in California (42), and group OB3 isolates were obtained from a cardiac transplant unit in New York. Isolates belonging to group OB4 were obtained from a surgical unit in a hospital in Colorado where the patients had undergone prosthetic heart valve replacement surgery (unpublished data). Cluster OB5 was comprised of isolates acquired from a poultry farm outbreak investigation in which A. fumigatus was recovered from the lungs of chickens and from chicken litter (unpublished data). The A. fumigatus isolates, ATCC 16903 and ATTC 42202 were also included in the analysis. Species identification of all 55 A. fumigatus isolates was confirmed by both morphological and molecular methods using sequence comparison of the β-tubulin region (5).
TABLE 1.
Sources, origins, and genotypes of A. fumigatus isolates obtained by Afut1 and CSP sequence analysis
| Isolate | Outbreak | Origin | Source | Yr of outbreak | Afut1 typea | CSP type(s)a,b |
|---|---|---|---|---|---|---|
| 648 | OB1 | California | Clinical | 2001 | 3 | 11 |
| 610 | OB1 | California | Environment | 2001 | 4 | 11 |
| 650 | OB1 | California | Environment | 2001 | 3 | 11 |
| 606 | OB1 | California | Clinical | 2001 | 7 | 12 |
| 607 | OB1 | California | Clinical | 2001 | 8 | 12 |
| 633 | OB1 | California | Clinical | 2001 | 9 | 12 |
| 683 | OB1 | California | Clinical | 2001 | 9 | 12 |
| 686 | OB1 | California | Clinical | 2001 | 9 | 12 |
| B5850 | OB2 | California | Clinical | 1998 | 37 | 23 |
| B5854c | OB2 | California | Clinical | 1998 | 37 | 23 |
| B5855 | OB2 | California | Clinical | 1998 | 37 | 23 |
| B5852c | OB2 | California | Clinical | 1998 | 37 | 23 |
| B5856c | OB2 | California | Clinical | 1998 | 37 | 23, 23 |
| B5868 | OB2 | California | Environment | 1998 | 37 | 23 |
| 38 | OB2 | California | Clinical | 1998 | NDe | 23 |
| B5863 | OB2 | California | Environment | 1998 | 37 | 23 |
| B5861 | OB2 | California | Environment | 1998 | ND | 21 |
| B5859 | OB2 | California | Clinical | 1998 | 38 | 24 |
| B5866 | OB2 | California | Environment | 1998 | 37 | 33, 33 |
| B5355 | OB3 | New York | Clinical | 1992 | 22 | 32 |
| B5360 | OB3 | New York | Environment | 1992 | 33 | 33 |
| B5357 | OB3 | New York | Environment | 1992 | 24 | 34 |
| B5361 | OB3 | New York | Clinical | 1992 | 32 | 31 |
| B5358 | OB3 | New York | Clinical | 1992 | ND | 31, 31 |
| B5450 | OB3 | New York | Clinical | 1992 | ND | 31 |
| 4339 | OB4 | Colorado | Clinical | 2005 | 73 | 41 |
| 4340 | OB4 | Colorado | Clinical | 2005 | 74 | 44, 44 |
| 3960c | OB4 | Colorado | Clinical | 2005 | 75 | 43, 43 |
| 3957c | OB4 | Colorado | Clinical | 2005 | 76 | 42 |
| 33d | OB5 | Georgia | Clinical | 2002 | 91 | 51 |
| 35d | OB5 | Georgia | Environment | 2002 | 92 | 52 |
| 34d | OB5 | Georgia | Clinical | 2002 | 93 | 53 |
| B6069 | OB6 | Canada | Clinical | 2002 | 41 | 61 |
| B6070 | OB6 | Canada | Clinical | 2002 | 41 | 61 |
| B6071 | OB6 | Canada | Clinical | 2002 | 41 | 61 |
| B6072 | OB6 | Canada | Clinical | 2002 | 41 | 61, 61 |
| B6073 | OB6 | Canada | Clinical | 2002 | 41 | 61 |
| B6075c | OB6 | Canada | Environment | 2002 | 41 | 61, 61 |
| B6077 | OB6 | Canada | Environment | 2002 | 41 | 61 |
| B6079 | OB6 | Canada | Environment | 2002 | 41 | 61, 61 |
| B6074 | OB6 | Canada | Clinical | 2002 | 41 | 61 |
| B6076 | OB6 | Canada | Environment | 2002 | 41 | 61 |
| B6078 | OB6 | Canada | Environment | 2002 | 41 | 61 |
| B6081 | OB6 | Canada | Environment | 2002 | 41 | 61, 61 |
| 97 | OB6 | Canada | Clinical | 2002 | 42 | 64 |
| 99c | OB6 | Canada | Clinical | 2002 | 42 | 64 |
| 101 | OB6 | Canada | Clinical | 2002 | 43 | 64 |
| 103c | OB6 | Canada | Clinical | 2002 | 44 | 65 |
| 105 | OB6 | Canada | Clinical | 2002 | 45 | 66 |
| B6083 | OB6 | Canada | Environment | 2002 | 46 | 62, 62 |
| B6084 | OB6 | Canada | Environment | 2002 | 46 | 62, 62 |
| B6082 | OB6 | Canada | Environment | 2002 | 46 | 62 |
| 127 | OB6 | Canada | Environment | 2002 | 46 | 62, 62 |
| 129 | OB6 | Canada | Environment | 2002 | 47 | 63 |
| 131 | OB6 | Canada | Environment | 2002 | 48 | 62 |
| ATCC 16903 | ATCC | 57 | 1 | |||
| ATCC 42202 | ATCC | 58 | 2 |
Afut1 and CSP type discrepancies are shown in bold type.
Some isolates were subjected to CSP typing twice. The second CSP type is the CSP type obtained after repeat DNA extraction and sequence analysis.
Isolates tested for reproducibility of the assay.
Isolates from an outbreak in a chicken house.
ND, not done.
PCR amplification and sequencing of polymorphic loci.
Fungal DNA was extracted and purified from these isolates as described previously (30). PCR primers for the CSP region were designed using the program GeneFisher (R. Giegerich, F. Meyer, and C. Schleiermacher, presented at the Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology) and were as follows: 5′-TTGGGTGGCATTGTGCCAA (forward) and 5′-GAGCATGACAACCCAGATACCA (reverse). PCR amplification was performed with 1 μl of DNA as the template in a total reaction mixture volume of 25 μl consisting of PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl); 0.2 mM each dATP, dGTP, dCTP, and dTTP; 1.2 to 1.6 mM MgSO4; 0.2 pmol of each primer; 1 U of Pfx polymerase (Invitrogen-BRL, Life Technologies, Carlsbad, CA); and 1× Pfx enhancer (Invitrogen). A total of 35 cycles of amplification were performed in a GeneAmp PCR system 9700 thermal cycler (PE-Applied Biosystems) after initial denaturation of DNA at 94°C for 5 min. Each cycle consisted of a denaturation step of 94°C for 15 s, an annealing step of 55°C for 30 s, and an extension step of 68°C for 30 s, and the last cycle was followed by a final extension at 68°C for 2 min. Products were visualized on a 1.2% agarose gel.
The resultant amplicons were purified using the reagents and protocols supplied by the manufacturer with the ExoSAP-IT PCR purification kit (USB Corporation, Cleveland, OH). The purified fragments were mixed with 4 μl of Big Dye (PE-Applied Biosystems) and 10 pmol of primer (same as the respective PCR primers), and a 10-μl reaction mixture was run on a thermal cycler at 96°C for 5 min, followed by 30 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Products were directly sequenced on an Applied Biosystems 3730 DNA analyzer in accordance with the protocols supplied by the manufacturer. Both strands were aligned, and the sequences were edited with the Sequencher version 4.7 software (Genecodes Inc., Ann Harbor, MI).
Identification and classification of CSP STs.
Sequences generated from the CSP locus were imported into the Bio-Edit sequence alignment program and manually inspected, and each unique CSP sequence (herein referred to as sequence type [ST]) was assigned a random numerical code for identification. ST assignment was done per outbreak to facilitate comparison with the archived Afut1 genotype data.
Evaluation of the CSP sequence typing scheme. (i) Reproducibility.
Reproducibility, defined as the ability to assign an identical type to the same isolate by a repeat assay, was assessed by sequence analysis of independent DNA preparations from randomly selected isolates. Twelve isolates—B6079, B6081, B6084, B6075, B6083, B6072, 127, B5358, B5866, B5856, 3960, and 4340—were tested twice for reproducibility.
(ii) In vitro stability.
In vitro stability of the hypervariable CSP locus was tested with the ATCC 16903 strain. Briefly, the isolate was subcultured on Sabouraud's dextrose agar plates for 20 passages; DNA was extracted from all 20 subcultures, and the CSP locus was sequenced.
(iii) Concordance.
Concordance between the CSP and Afut1 typing methods was calculated as described previously (46). Briefly, each isolate per outbreak was examined, and each isolate pair was classified as identical or different by the two genotyping methods. A two-by-two table was constructed for the two technique comparison and percent cell concordance was calculated. Percent concordance corresponds to the proportion of pairs for which the two methods are in agreement.
Phylogenetic analysis.
A phylogenetic analysis of all the CSP gene sequences was performed using the maximum-parsimony method. Statistical reliability of internal nodes was assessed using 1,000 bootstrap pseudoreplicates. The Neosartorya fischeri NRRL 181 CSP gene sequence (GenBank accession number XM_001263541) was included as the outgroup taxon in order to root the resultant trees. All phylogenetic analyses were conducted using the PHYLIP version 3.6 software package (16). Within this package, the program DNAPARS was used to conduct a heuristic search with 100 random sequence additions for the entire CSP gene fragment. The program DNAPENNY was used to conduct a branch-and-bound search reconsidering the input order of species for the analysis of the CSP repeat region (discussed below). For both heuristic and branch-and-bound searches, gaps were treated as a fifth character state.
Nucleotide distances were calculated using the computer program MEGA 3.1 (29). Estimates of the numbers of synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN) were calculated using the modified Nei-Gojobori method (56) with Jukes and Cantor (25) distances and the observed transition/transversion ratio. To test for the potential action of positive Darwinian selection across the entire CSP sequence and subregions within that sequence, we used Fisher's exact test to evaluate the difference in magnitude between the observed and potential numbers of synonymous and nonsynonymous substitutions (55). In order to evaluate the evidence for positive Darwinian selection acting on individual codons, the single-likelihood ancestral counting method (SLAC) (28) was used.
RESULTS
CSP locus as a polymorphic marker.
PCR product size differences were evident when the amplified products were visualized by agarose gel electrophoresis after ethidium bromide staining; the PCR amplicon size ranged from 550 bp to 700 bp in length. Sequence analysis of the PCR products showed that the CSP STs differed by numerous insertions (Fig. 1). In addition, all STs possessed a highly variable region consisting of 18-nucleotide tandem repeats. The number of repeats differed in strains, ranging from 7 to 13. When CSP STs were compared to the Afut1 genotypes (Table 1), we found that the two methods were comparable, with 100% concordance in groups OB2 to OB5. However, concordance values of only 62.5% and 91% were obtained when the two typing methods were used to compare groups OB1 and OB6. Specifically, Afut1 hybridization profiles revealed that isolates 606, 607, and 633 were closely related but distinct isolates, while the CSP typing method grouped all three isolates in a clonal cluster. Again, isolate 610 (group OB1) was closely related to isolates 648 and 650 by Afut1 analysis, but these isolates were indistinguishable by CSP typing. Similarly, in group OB6, 2 of 23 isolates (isolates 101 and 131) had distinct genotypes by the Afut1 method but identical sequences by the CSP typing method.
FIG. 1.
Alignment of the CSP repeat region. Representative CSP STs are shown as well as the N. fischeri ST. A dash indicates an insertion/deletion, and a dot indicates a nucleotide that is identical to the nucleotide in the top sequence.
Typeability, in vitro stability, and reproducibility of the CSP typing strategy.
The results of the CSP and Afut1 typing methods are presented in Table 1. All of the A. fumigatus isolates were amenable to both PCR amplification and sequencing of the CSP locus, rendering the CSP typing scheme 100% typeable. The typing technique was highly reproducible as shown by identical sequences generated after repeated DNA extraction and PCR amplification and sequencing of 12 randomly selected isolates (repeated at least two times). In vitro stability of the genomic region in which CSP resides was confirmed by examining the effects of multiple passages (20 generations) of strain ATCC 16903 on Sabouraud's dextrose agar plates. Results showed that there were no variations in the CSP STs analyzed from these passages.
Phylogenetic analysis.
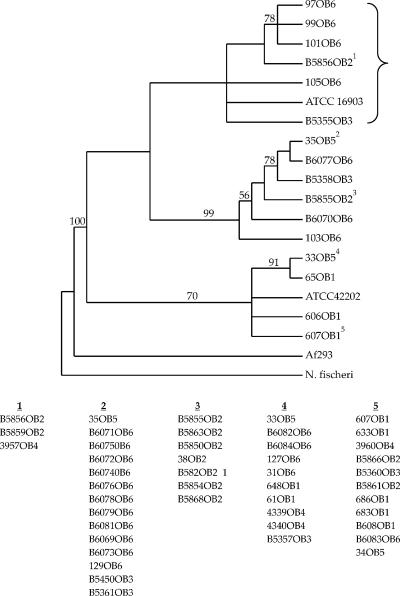
A phylogenetic analysis of the entire 558-bp CSP gene plus an additional 6 bp immediately upstream from the start codon resulted in 66,825 most parsimonious trees obtained using a heuristic search involving 100 random sequence additions. A strict consensus of these trees is presented in Fig. 2. Several clusters were characterized by moderate to high bootstrap values. All of the groups consisted of isolates originating from multiple outbreaks, suggesting that outbreak strains are widely distributed across North America. Isolate Af293, whose genome has been completely sequenced, clustered outside the outbreak strains and was found to be closer to N. fischeri (uncorrected percent divergence [p] of 0.106) than it was to the outbreak strains of A. fumigatus (range of p, 0.483 to 0.624) at the first 64 codon positions (192 bp) of the CSP gene alignment. However, among the remaining nucleotides (199 to 564), Af293 was nearly identical to the outbreak strains (p = 0 to 0.004). In contrast, N. fischeri was substantially different from Af293 and the other A. fumigatus strains (p = 0.066 to 0.071). It should also be pointed out that codon 28 is a stop codon among all outbreak strains and is followed by a putative alternative start codon at position 56. In addition, Af293 and N. fischeri lack codon 44 (each possesses a 3-bp deletion). As N. fischeri is the outgroup taxon for this analysis, it can be inferred that the presence of codon 44 among the outbreak strains represents an insertion at this site. Collectively, the above data indicate that Af293 may be from a lineage distinct from that of the outbreak strains studied here.
FIG. 2.
Phylogeny of unique A. fumigatus STs based on the 564-bp CSP gene fragment. N. fischeri is the outgroup taxon used to root the tree. The tree is a strict consensus of 66,825 most parsimonious trees that were 386 steps in length generated using a heuristic search. Numbers along branches represent bootstrap values. Only unique STs were used to construct the tree. Strains listed below the tree are grouped with other strains that possess identical CSP STs; the numbers above each grouping correspond to the numerical superscripts for each representative ST included in the phylogeny. Isolates showed within the brace were recovered exclusively from clinical samples.
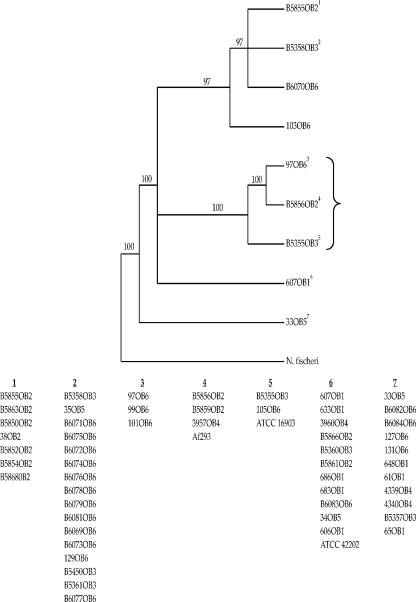
The level of nucleotide divergence observed across the entire CSP alignment (average p = 0.001; range = 0 to 0.005) was not substantially higher than what was observed for the repeat region alone (average p = 0.007; range = 0 to 0.042), although the range was greater. However, the repeat region is unusual in comparison to the flanking segments of the CSP gene due to numerous insertions and deletions in the former. Interestingly, these insertions and deletions appear to be highly phylogenetically informative. To verify this, we conducted a phylogenetic analysis of the tandem repeat region alone. A total of 15 most parsimonious trees were found using a branch-and-bound search. A strict consensus of these trees is shown in Fig. 3. The bootstrap support values for the clusters identified in this analysis were high. As in the case of the previous analysis, several multioutbreak clusters were found, although two outbreak-specific clusters (clusters 1 and 3) were identified. As in the case of the previous analysis, the clustering pattern suggests that outbreak strains are widely dispersed across North America. Also, strain Af293 did not cluster apart from the outbreak strains, as predicted based on the divergence values reported for the first 198 bp versus the remaining 366 bp, among which the tandem repeat region is found. Instead, Af293 was found to possess an identical CSP ST with cluster 4 strains, which include B5856OB2, B5859OB2, and 3957OB4 (Fig. 3). Finally, it should be pointed out that the bootstrap values associated with internal branches in Fig. 3 are much higher than what was observed in Fig. 2. This observation is consistent with our previous statement that insertions and deletions in the repeat region possess substantial phylogenetic signal.
FIG. 3.
Phylogeny of A. fumigatus strains based on the 159-bp 12-mer repeat region. N. fischeri is the outgroup taxon used to root the tree. The tree is a strict consensus of 15 most parsimonious trees that were 172 steps in length generated using a branch-and-bound search. Numbers along branches represent bootstrap values. Only unique STs were used to construct the tree; strains having identical CSP STs are listed below the tree as groups. The numbers above each grouping correspond to the numerical superscripts for each representative ST included in the phylogeny. Isolates recovered from clinical samples are shown within the brace.
Molecular evolutionary analysis of the repeat region.
Due to the unusually high number of insertion and deletions, we conducted analyses of positive Darwinian selection on the repeat region. Both methods (Fisher's exact test and the SLAC method) did not provide evidence of positive Darwinian selection. Interestingly, when we analyzed the number of distinct repeats (11) and compared them to each other, we found that dS was 0.311 and dN was 0.742, thus giving dN/dS of 2.4. However, the small number of observed changes between repeats (one synonymous change; eight nonsynonymous changes) and the small number of sites (four potentially synonymous sites; eight potentially nonsynonymous sites) requires the use of small sample test (Fisher's exact test), and as a result, the difference is not statistically significant (P = 0.59).
DISCUSSION
Over the last decade, sequence analysis of a single, highly variable gene has been successfully applied to strain differentiation among microorganisms that possess low levels of genetic variability and/or lack discernible population genetic structure. Notable cases include protein A (spa) and clumping factor (cflB) gene typing for Staphylococcus aureus (26, 27, 48) and flagellin A (flaA) gene typing for Campylobacter jejuni (34). The S. aureus spa typing method has been evaluated in numerous epidemiological studies and has been shown to be a valuable genotyping tool using well-validated strain panels (49). Similarly, we have demonstrated that the repeat region within the Afu3g08990 locus (CSP) is highly informative for subtyping, as shown through the presence of multiple, very well supported clusters inferred from our phylogenetic analysis (Fig. 3), and we recommend its use for A. fumigatus outbreak source tracking. In fact, results of the study demonstrate that the single-locus sequence typing strategy fulfills the basic tenets of an appropriate typing scheme (50), since it (i) assigned isolates to distinct subtypes; (ii) differentiated one subtype from another, at the same time not being so discriminatory as to assign a unique subtype to every isolate; and (iii) identified clusters of closely related isolates (“clonal complexes”).
The observation that CSP typing can group isolates in congruence with another well-established method, the Afut1 method, further validates the use of the CSP typing method as an alternate scheme for A. fumigatus strain typing. Although there was overall agreement between the two genotyping methods, there was discordance in two clusters, OB1 and OB6. In both of these outbreak clusters, isolates that were grouped as “clonal” by the CSP typing method were revealed as closely related isolates by Afut1 hybridization profiles. This could be a reflection of our conservative criterion for Afut1 genotyping whereby two isolates were considered different when the hybridization patterns differed by at least two bands. Nevertheless, this loss of resolution could be a drawback of any strategy that uses a single-locus scheme versus the Afut1 method, which reveals the genomic diversity of the organism. Thus, the CSP typing method alone may not have the resolving power of the highly discriminatory Afut1 method, but it has been demonstrated that a combination of genotyping methods yields better understanding of genetic relatedness than any one method used singly (30). Similarly, another single-locus sequence scheme (spa typing) was not able to resolve clonal groups of S. aureus isolates (27). In spite of this limitation, our study demonstrates for the first time the utility of a novel single-locus sequence typing scheme that can be used as a first-line strategy for rapid, easy to use and interpret method for A. fumigatus strain discrimination. Results of our study also demonstrated that the CSP gene is a stable and reproducible marker, making it an attractive genotyping tool.
Numerous studies have shown that the Afut1 RFLP and microsatellite-based methods are highly discriminatory techniques for A. fumigatus strain typing (8, 12, 30); however, both these methods have several disadvantages that include the need for specialized equipment and dedicated software, difficulty in data interpretation, and inability to share data between laboratories. In contrast, the only requirements of the proposed CSP-based typing scheme is the ability to perform PCR and have access to an automated sequencer; both this technology and expertise are now becoming available to many clinical microbiology laboratories. In addition, interpretation of the sequence information from a CSP typing scheme does not require sophisticated algorithms or dedicated software and thus can be seamlessly integrated into any laboratory. Since adequate typing information is obtained exclusively from a single locus, this typing method combines many of the advantages of a sequence-based system, such as MLST, while at the same time offering a more rapid and convenient system for outbreak investigations.
There are two findings from our study that carry important epidemiological implications for A. fumigatus. First, we found that CSP phylogenetic clusters consist of strains that have appeared in more than one outbreak (Fig. 3), indicating that CSP phylogenetic clusters are widely dispersed across North America. Second, we found that A. fumigatus outbreaks consist of strains from more than one phylogenetic cluster (Fig. 3), which is also consistent with a widespread geographic dispersal of clusters. Although these findings are not novel, our observation agrees with previous studies (45, 47) indicating that A. fumigatus strains are widespread across North America. However, our results differ from these other studies, as we detected substructuring (i.e., the presence of multiple, well-supported phylogenetic clusters) in spite of widespread geographic dispersal across North America.
What could explain this pattern of phylogenetic substructuring in the face of widespread dispersal? It could be that A. fumigatus is composed of cryptic lineages that are not detected with the more conserved genes typically characterized in MLST or other phylogenetic studies. However, the phylogenetic information in the CSP gene comes from the rapid polymorphism generated through insertions and deletions (and to a lesser extent, point mutations) found within the CSP repeat region. This pattern is reminiscent of what is seen in the case of merozoite surface repeat proteins of Plasmodium species (15, 17, 19), where the repeat regions have been shown to be immunogenic (40, 41), the ALS3 gene in the yeast Candida albicans (38), and the spherule outer wall glycoprotein (SOWgp) gene of Coccidioides, wherein the repeated elements appear to contribute to the virulence of this species both by functioning as an adhesin and by modulating the host immune response (21, 22). In a recent population-based study of the repetitive domain in the SOWgp gene using phylogenetic and genetic distance methods, it was shown that this gene evolves in Coccidioides by concerted evolution (24).
While the biological function of the Afu3g08990 gene (designated CSP in this study) is not known, data from Levdansky and colleagues (companion paper [31a]) show that the protein is expressed in the cell walls of both conidia and hyphae of A. fumigatus. Deletion of this gene in A. fumigatus resulted in phenotypic changes that included reduced adherence and premature fast germination (31a). In the present study, the phylogenetic tree constructed with the entire portion of the CSP locus revealed three distinct clades (Fig. 2). Notably, all isolates in one clade (shown within the brace in Fig. 2) were recovered from clinical samples and more than half these isolates (5/8 isolates) were invasive isolates causing infection in cardiac transplant patients. These isolates also grouped together in a clade (shown within the brace in Fig. 3) distinct from other clades in the phylogenetic tree constructed based on the 159-bp repeat region of the locus. It is tempting to speculate with this limited data that the phylogenetic lineages detected may correspond to antigenic lineages and not taxonomic lineages. However, this needs to be further explored with a more robust and larger set of isolates collected from both environmental and clinical origins. Nevertheless, we did find evidence to suggest that strain Af293 may represent a distinct lineage based on the phylogeny presented in Fig. 2 as well as the distinct structure of its CSP gene in comparison to that of other A. fumigatus strains (see Results). Interestingly, the sequence of the CSP gene from another strain of A. fumigatus, CEA10, that has been recently sequenced was found to be 100% similar to Af293 at the nucleotide level.
In conclusion, the present study demonstrates that the CSP region has a high degree of polymorphism, generating strain diversity patterns that distinguished epidemiologically linked strains and could therefore be a practical yet meaningful way to conduct epidemiological studies. Further, CSP typing appears to have significant advantages over many existing techniques in terms of cost, speed, ease of use and interpretation, and standardization, and it allows the creation of unambiguous data sets that can be readily exchanged between labs and be organized into global databases. Interestingly, this study also found evidence indicating that A. fumigatus is differentiated into distinct CSP phylogenetic lineages. While these may not represent distinct taxonomic lineages, our results suggest that strain Af293 may indeed be so. Because this strain's genome has been sequenced and is considered representative of A. fumigatus, the potential for taxonomic distinction between Af293 and the nosocomial strains studied here should be investigated further.
Acknowledgments
Sun T. Tay is the recipient of the HLCB scholarship from the University of Malaya, Malaysia.
All the medical centers are gratefully acknowledged for their generous contribution of A. fumigatus isolates used in this study. We thank Natalie Fedorova, The Institute for Genomic Research, Rockville, MD, for help with screening the A. fumigatus genome for polymorphic genes.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Aufauvre-Brown, A., J. Cohen, and D. W. Holden. 1992. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J. Clin. Microbiol. 30:2991-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain, J. M., A. Tavanti, A. D. Davidson, M. D. Jacobsen, D. Shaw, N. A. Gow, and F. C. Odds. 2007. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J. Clin. Microbiol. 45:1469-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee, S. A., J. Gribskov, M. Brandt, J. Ito, A. Fothergill, and K. A. Marr. 2005. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J. Clin. Microbiol. 43:5996-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balajee, S. A., J. L. Gribskov, E. Hanley, D. Nickle, and K. A. Marr. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balajee, S. A., D. Nickle, J. Varga, and K. A. Marr. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5:1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bart-Delabesse, E., C. Cordonnier, and S. Bretagne. 1999. Usefulness of genotyping with microsatellite markers to investigate hospital-acquired invasive aspergillosis. J. Hosp. Infect. 42:321-327. [DOI] [PubMed] [Google Scholar]
- 7.Bart-Delabesse, E., J. F. Humbert, E. Delabesse, and S. Bretagne. 1998. Microsatellite markers for typing Aspergillus fumigatus isolates. J. Clin. Microbiol. 36:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bart-Delabesse, E., J. Sarfati, J. P. Debeaupuis, W. van Leeuwen, A. van Belkum, S. Bretagne, and J. P. Latge. 2001. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates. J. Clin. Microbiol. 39:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bougnoux, M. E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretagne, S., E. Bart-Delabesse, J. Wechsler, M. Kuentz, N. Dhedin, and C. Cordonnier. 1997. Fatal primary cutaneous aspergillosis in a bone marrow transplant recipient: nosocomial acquisition in a laminar-air flow room. J. Hosp. Infect. 36:235-239. [DOI] [PubMed] [Google Scholar]
- 11.Chazalet, V., J. P. Debeaupuis, J. Sarfati, J. Lortholary, P. Ribaud, P. Shah, M. Cornet, H. Vu Thien, E. Gluckman, G. Brucker, and J. P. Latge. 1998. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J. Clin. Microbiol. 36:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Valk, H. A., J. F. Meis, I. M. Curfs, K. Muehlethaler, J. W. Mouton, and C. H. Klaassen. 2005. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J. Clin. Microbiol. 43:4112-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fakhr, M. K., L. K. Nolan, and C. M. Logue. 2005. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 43:2215-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felger, I., V. M. Marshal, J. C. Reeder, J. A. Hunt, C. S. Mgone, and H. P. Beck. 1997. Sequence diversity and molecular evolution of the merozoite surface antigen 2 of Plasmodium falciparum. J. Mol. Evol. 45:154-160. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle.
- 17.Ferreira, M. U., and D. L. Hartl. 2007. Plasmodium falciparum: worldwide sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-2 (MSP-2). Exp. Parasitol. 115:32-40. [DOI] [PubMed] [Google Scholar]
- 18.Hall, L., S. Wohlfiel, and G. D. Roberts. 2004. Experience with the MicroSeq D2 large-subunit ribosomal DNA sequencing kit for identification of filamentous fungi encountered in the clinical laboratory. J. Clin. Microbiol. 42:622-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann, E. H., R. S. Malafronte, S. L. Moraes-Avila, A. L. Osakabe, G. Wunderlich, A. M. Durham, P. E. Ribolla, H. A. del Portillo, and M. U. Ferreira. 2006. Origins of sequence diversity in the malaria vaccine candidate merozoite surface protein-2 (MSP-2) in Amazonian isolates of Plasmodium falciparum. Gene 376:224-230. [DOI] [PubMed] [Google Scholar]
- 20.Hong, S. B., S. J. Go, H. D. Shin, J. C. Frisvad, and R. A. Samson. 2005. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 97:1316-1329. [DOI] [PubMed] [Google Scholar]
- 21.Hung, C. Y., N. M. Ampel, L. Christian, K. R. Seshan, and G. T. Cole. 2000. A major cell surface antigen of Coccidioides immitis which elicits both humoral and cellular immune responses. Infect. Immun. 68:584-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung, C. Y., J. J. Yu, K. R. Seshan, U. Reichard, and G. T. Cole. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsen, M. D., N. A. Gow, M. C. Maiden, D. J. Shaw, and F. C. Odds. 2007. Strain typing and determination of population structure of Candida krusei by multilocus sequence typing. J. Clin. Microbiol. 45:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johannesson, H., J. P. Townsend, C. Y. Hung, G. T. Cole, and J. W. Taylor. 2005. Concerted evolution in the repeats of an immunomodulating cell surface protein, SOWgp, of the human pathogenic fungi Coccidioides immitis and C. posadasii. Genetics 171:109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jukes, T., and C. Cantor. 1969. Evolution of protein molecules. Academic Press, New York, NY.
- 26.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koreen, L., S. V. Ramaswamy, S. Naidich, I. V. Koreen, G. R. Graff, E. A. Graviss, and B. N. Kreiswirth. 2005. Comparative sequencing of the serine-aspartate repeat-encoding region of the clumping factor B gene (clfB) for resolution within clonal groups of Staphylococcus aureus. J. Clin. Microbiol. 43:3985-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosakovsky Pond, S. L., and S. D. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 30.Lasker, B. A. 2002. Evaluation of performance of four genotypic methods for studying the genetic epidemiology of Aspergillus fumigatus isolates. J. Clin. Microbiol. 40:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leenders, A., A. van Belkum, S. Janssen, S. de Marie, J. Kluytmans, J. Wielenga, B. Lowenberg, and H. Verbrugh. 1996. Molecular epidemiology of apparent outbreak of invasive aspergillosis in a hematology ward. J. Clin. Microbiol. 34:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Levdansky, E., J. Romano, Y. Shadkchan, H. Sharon, K. J. Verstrepen, G. R. Fink, and N. Osherov. 2007. Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryot. Cell 6:1380-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, D., P. F. Lehmann, B. H. Hamory, A. A. Padhye, E. Durry, R. W. Pinner, and B. A. Lasker. 1995. Comparison of three typing methods for clinical and environmental isolates of Aspergillus fumigatus. J. Clin. Microbiol. 33:1596-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz, P., J. Guinea, T. Pelaez, C. Duran, J. L. Blanco, and E. Bouza. 2004. Nosocomial invasive aspergillosis in a heart transplant patient acquired during a break in the HEPA air filtration system. Transpl. Infect. Dis. 6:50-54. [DOI] [PubMed] [Google Scholar]
- 36.Neuveglise, C., J. Sarfati, J. P. Latge, and S. Paris. 1996. Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Res. 24:1428-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 38.Oh, S. H., G. Cheng, J. A. Nuessen, R. Jajko, K. M. Yeater, X. Zhao, C. Pujol, D. R. Soll, and L. L. Hoyer. 2005. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151:673-681. [DOI] [PubMed] [Google Scholar]
- 39.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 40.Oliveira-Ferreira, J., L. R. Pratt-Riccio, M. Arruda, F. Santos, C. T. Ribeiro, A. C. Goldberg, and D. M. Banic. 2004. HLA class II and antibody responses to circumsporozoite protein repeats of P. vivax (VK210, VK247 and P. vivax-like) in individuals naturally exposed to malaria. Acta Trop. 92:63-69. [DOI] [PubMed] [Google Scholar]
- 41.Oliveira-Ferreira, J., E. Vargas-Serrato, J. W. Barnwell, A. Moreno, and M. R. Galinski. 2004. Immunogenicity of Plasmodium vivax merozoite surface protein-9 recombinant proteins expressed in E. coli. Vaccine 22:2023-2030. [DOI] [PubMed] [Google Scholar]
- 42.Panackal, A. A., A. Dahlman, K. T. Keil, C. L. Peterson, L. Mascola, S. Mirza, M. Phelan, B. A. Lasker, M. E. Brandt, J. Carpenter, M. Bell, D. W. Warnock, R. A. Hajjeh, and J. Morgan. 2003. Outbreak of invasive aspergillosis among renal transplant recipients. Transplantation 75:1050-1053. [DOI] [PubMed] [Google Scholar]
- 43.Pegues, D. A., B. A. Lasker, M. M. McNeil, P. M. Hamm, J. L. Lundal, and B. M. Kubak. 2002. Cluster of cases of invasive aspergillosis in a transplant intensive care unit: evidence of person-to-person airborne transmission. Clin. Infect. Dis. 34:412-416. [DOI] [PubMed] [Google Scholar]
- 44.Pounder, J. I., K. E. Simmon, C. A. Barton, S. L. Hohmann, M. E. Brandt, and C. A. Petti. 2007. Discovering potential pathogens among fungi identified as nonsporulating molds. J. Clin. Microbiol. 45:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pringle, A., D. M. Baker, J. L. Platt, J. P. Wares, J. P. Latge, and J. W. Taylor. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evol. Int. J. Org. Evol. 59:1886-1899. [PubMed] [Google Scholar]
- 46.Robinson, D. A., S. K. Hollingshead, J. M. Musser, A. J. Parkinson, D. E. Briles, and M. J. Crain. 1998. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J. Mol. Evol. 47:222-229. [DOI] [PubMed] [Google Scholar]
- 47.Rydholm, C., G. Szakacs, and F. Lutzoni. 2006. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot. Cell 5:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shopsin, B., and B. N. Kreiswirth. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 7:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soll, D. R. 2000. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 13:332-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tablan, O. C., L. J. Anderson, R. Besser, C. Bridges, and R. Hajjeh. 2004. Guidelines for preventing health-care-associated pneumonia, 2003: recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee. Morb. Mortal. Wkly. Rep. Recommend. Rep. 53:1-36. [PubMed] [Google Scholar]
- 52.Tavanti, A., A. D. Davidson, M. J. Fordyce, N. A. Gow, M. C. Maiden, and F. C. Odds. 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 43:5601-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tavanti, A., A. D. Davidson, N. A. Gow, M. C. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tavanti, A., A. D. Davidson, E. M. Johnson, M. C. Maiden, D. J. Shaw, N. A. Gow, and F. C. Odds. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 43:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, J., S. Kumar, and M. Nei. 1997. Small-sample tests of episodic adaptive evolution: a case study of primate lysozymes. Mol. Biol. Evol. 14:1335-1338. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, J., H. F. Rosenberg, and M. Nei. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]