Abstract
Giardia lamblia, an intestinal pathogen of mammals, including humans, is a significant cause of diarrheal disease around the world. Additionally, the parasite is found on a lineage which separated early from the main branch in eukaryotic evolution. The extent of genetic diversity among G. lamblia isolates is insufficiently understood, but this knowledge is a prerequisite to better understand the role of parasite variation in disease etiology and to examine the evolution of mechanisms of genetic exchange among eukaryotes. Intraisolate genetic variation in G. lamblia has never been estimated, and previous studies on interisolate genetic variation have included a limited sample of loci. Here we report a population genetics study of intra- and interisolate genetic diversity based on six coding and four noncoding regions from nine G. lamblia isolates. Our results indicate exceedingly low levels of genetic variation in two out of three G. lamblia groups that infect humans; this variation is sufficient to allow identification of isolate-specific markers. Low genetic diversity at both coding and noncoding regions, with an overall bias towards synonymous substitutions, was discovered. Surprisingly, we found a dichotomous haplotype structure in the third, more variable G. lamblia group, represented by a haplotype shared with one of the homogenous groups and an additional group-specific haplotype. We propose that the distinct patterns of genetic-variation distribution among lineages are a consequence of the presence of genetic exchange. More broadly, our findings have implications for the regulation of gene expression, as well as the mode of reproduction in the parasite.
Giardia lamblia, a member of the Diplomonadida, is a protozoan of interest for both evolutionary and medical reasons. Analyses of a majority of genes, including rRNA (10, 14, 46), genes involved in transcription (22) and translation (21), and genes of mitochondrial origin (2), have placed G. lamblia on one of the earliest diverging eukaryotic lineages; thus, studies of G. lamblia biology have shed light on early stages of eukaryotic evolution (11, 38, 41, 54). G. lamblia is responsible for causing approximately 1 billion cases of diarrheal disease annually worldwide (24, 59) and about 2.5 million annual infections in the United States (17). It is also a significant veterinary pathogen (56). The parasite has two life cycle stages: cysts that are responsible for the fecal-oral transmission of the parasite and trophozoites that establish the infection within the duodenum of the animal host.
To gain further insight into the evolution of mechanisms for genetic exchange and to better understand the role of parasite variation in disease etiology, we are interested in defining the extent of sequence diversity within G. lamblia populations. Due to the clonal population structure (5, 50-53) and the lack of a documented sexual stage of the parasite, G. lamblia has traditionally been considered to be an asexually reproducing organism. This conclusion has been challenged by the recent discovery of core meiotic machinery in G. lamblia's genome (39), but direct evidence of sexual recombination is still lacking. The genetic composition of G. lamblia has been shaped by lateral gene transfer from prokaryotic lineages, as evidenced by the presence of an archeal-type tRNA synthetase (11), as well as several bacterial metabolic enzymes (38). The presence of a double-stranded RNA virus that infects G. lamblia (57) suggests an additional mechanism for genetic exchange. Understanding the genetic structure of G. lamblia populations is complicated by the fact that G. lamblia is binucleated and functionally tetraploid (1, 3, 23, 62), with each nucleus being diploid (63). Unlike in binucleated ciliates, the two nuclei of G. lamblia appear to be equivalent based on all criteria tested to date (2, 8, 19, 25, 60, 63). It is unknown whether the two nuclei are functionally redundant or whether each serves a unique role in the cell. At the core of all of these issues is the question of genetic content of the two nuclei. The available evidence argues that the genetic contents of the two nuclei are distinct: every daughter cell inherits a copy of a left and a right nucleus during cell division (18, 44, 63), and the nuclear envelope retains its integrity during mitosis (7, 44). Thus, the expectation is that the sequence divergence between the two nuclei should be high, given that they contain isolated genomes that will independently evolve polymorphisms and experience substitutions.
The core mechanisms that would generate intraspecific variation, both between nuclei and among clones, remain unexplored, and the extent of such genetic variation is insufficiently understood. Previous studies of G. lamblia (6, 13, 30, 32, 34, 47, 55) have varied in their findings about genetic diversity of the parasite. Depending on the locus and isolate studied, the results ranged from a virtual lack of (6) to extensive (30) genetic variation, and it remains unclear at what level the variation exists (i.e., heterozygosity within an individual versus polymorphism within a population). Although some authors argued for close relatedness of G. lamblia isolates throughout the world (28), others emphasized that clonal lineages within G. lamblia are evolutionarily independent (30) and that while there is monophyly of G. lamblia, there is also extensive population substructure, so G. lamblia should be considered a species complex (4). Because previous works are based on either allozyme analysis or DNA sequence comparisons of a single locus, such opposing findings could be explained by biological reasons (e.g., locus-specific differences or distinct choices of isolates represented in different studies) and/or by methodological differences (e.g., allozyme analysis versus DNA sequencing or axenized versus field isolates).
In this paper, we report a study of genetic variation both within and across G. lamblia laboratory isolates (intra- and interisolate, respectively), with implications about the degree of genetic differentiation between the two nuclei. Our study encompasses 10 loci from nine isolates (preliminarily designated to represent three major G. lamblia groups, i.e., A-1, A-2, and B [36]), infecting both humans and other mammals. Throughout this work we focus on haplotypes, representing a set of single nucleotide polymorphisms (SNPs) at individual loci. We present data to indicate extremely low levels of genetic variation within and among G. lamblia group A-1 and A-2 populations. Further, we find greater sequence divergence and/or polymorphism within and among group B isolates and document the presence of divergent haplotypes within and among these isolates. Finally, considering the prediction that allelic diversity is high when there is a lack of genetic exchange, our study also allows us to consider different hypotheses about the mode of reproduction of G. lamblia.
MATERIALS AND METHODS
G. lamblia isolates and cell cultures.
We sampled nine G. lamblia isolates from diverse host and geographic origins representing the three medically relevant groups A-1, A-2, and B (2) (see Table S1 in the supplemental material). The Genome (Gen) isolate was used for the Giardia Genome Project (Marine Biological Laboratory, Woods Hole, MA) and was obtained originally by cloning a WB isolate (29). All other isolates (WB, CAT, Be-2, AB, N, GS, CM, and Be-1) were axenized in the laboratory, and their histories have been described previously (36). G. lamblia trophozoites were grown anaerobically for approximately 2 weeks in borosilicate tubes or polystyrene flasks in modified TYI-S-33 medium at 37°C (26), with the traditional phosphate buffer solution replaced by 0.024 M sodium bicarbonate. Giardia ardeae trophozoites were grown similarly, with the replacement of adult bovine serum with fetal bovine serum in the culture medium (15).
Amplification, cloning, and DNA sequencing.
Total genomic DNA was isolated from all G. lamblia isolates using DNA-STAT 60 (Tel Test, TX). Portions of coding regions (actin [621 bp], beta giardin [822 bp], chaperonin 60 [CPN60] [540 bp], [2Fe-2S] ferredoxin [402 bp], ribosomal protein L7a [RPL] [492 bp], and triosephosphate isomerase [TPI] [653 bp]) and full-length noncoding regions (the intergenic region between open reading frame [ORF] 10358 and ORF 10361, called intergenic region 1 [613 bp] [www.mbl.edu/Giardia]; the intergenic region between ORF 10019 and ORF 16475, called intergenic region 2 [762 bp] [www.mbl.edu/Giardia]; and the [2Fe-2S] ferredoxin introns [35 bp] and the RPL introns [109 bp]) were amplified by PCR using Pfu Turbo DNA polymerase (Stratagene) and locus-specific primers (see Table S2 in the supplemental material). Where appropriate, negative controls that did not contain a DNA template were used. Amplified gene products were cloned into a TOPO-TA pCR 4 vector and transfected into TOP10 Escherichia coli chemically competent cells (TOPO TA cloning kit for sequencing; Invitrogen).
For each locus, 20 clones per isolate were picked and sequenced on an ABI 3730XL capillary sequencer at the Marine Biological Laboratory. Low-quality sequences and truncated products were excluded from the analysis, yielding fewer than 20 sequences per locus per isolate in some instances (see Table S3 in the supplemental material). Detailed protocols for template isolation and sequencing can be found through the Marine Biological Laboratory's sequencing facility (http://jbpc.mbl.edu/SeqFacility/Pages). All loci were sequenced on both strands primed with M13 forward and M13 reverse oligonucleotides (Invitrogen).
Sequence analyses.
DNA sequences were trimmed manually to exclude the vector portion of the sequence, aligned initially using DNA Sequencher 4.1.4 (Gene Codes), and edited based on the visual inspection of contig alignments and individual sequence chromatographs. SNPs were identified in DnaSP 4.10.4 (43) as the number of segregating sites in the alignment. It should be noted that the sum of SNPs from each isolate of a group at a particular locus does not necessarily equal the total number of SNPs in that group at that particular locus but may also be higher or lower. For example, five SNPs were detected in an AB isolate and one SNP in an N isolate at the actin locus, while the total number of SNPs for group A-2 was five. This is because pooled SNP results (per group) depict not only intra- but also interisolate variation. Pooled SNP numbers lower than expected based on simple addition indicate the presence of shared polymorphisms, while higher pooled SNP numbers represent fixed differences among isolates.
Identification of haplotypes, estimation of π, θ, and Tajima's D, and analysis of fixed differences and shared polymorphisms were performed using DnaSP 4.10.4 (43). π, nucleotide diversity, is the average number of nucleotide substitutions between all unique pairwise comparisons of sequences (37, 40). θ, an estimate of heterozygosity, is estimated from the number of segregating sites in the alignment (40, 58). The major difference between the two measures of genetic variation is that π accounts for the frequency of polymorphisms, while θ does not. Tajima's D is a statistical test which represents the comparison between π and θ (40, 48). The null hypothesis of the test is that π and θ are approximately equal, which is expected under a neutral mutation-constant population size model.
Phylogenetic evolutionary analyses were conducted using MEGA version 3.1 (E. Kumar, K. Tamura, and M. Nei, 2004). Neighbor-joining trees were constructed using distances estimated under the Kimura two-parameter model, which includes both transitions and transversions (E. Kumar, K. Tamura, and M. Nei, 2004). Thanks to the availability of sequence data for a few close Giardia lamblia relatives in public databases, sequences from different parasite species were used as outgroups for the construction of different phylogenetic trees: Spironucleus barkhanus (GenBank accession number DX916114) was used at the actin locus, Giardia microti (GenBank accession number AY228649) at the TPI locus, and Giardia muris (GenBank accession number AY258618) at the beta giardin locus. Pairwise deletion of the gaps was executed in the analyses.
Nucleotide sequence accession numbers.
Sequences have been deposited in GenBank as population studies under the accession numbers EU014359 to EU014517. Sequences are organized hierarchically based on loci, isolate, and clone number.
RESULTS
Global patterns of nucleotide substitutions.
To identify SNPs, we examined 4,806 bp representing 10 loci, which included six genes, two introns, and two intergenic regions, from nine different G. lamblia laboratory isolates (Tables 1 and 2 and Table S1 in the supplemental material). The isolates used in the study were classified previously into groups A-1, A-2, and B (36). Given their varied histories, each laboratory isolate of parasites was treated as a population. Each locus was therefore amplified by PCR from genomic DNA, with resultant PCR products cloned and clones sequenced in order to sample multiple allelic variants within the populations. This method has been used traditionally for examining human immunodeficiency virus genetic variation within a single host (reviewed in reference 27). Because we sequenced approximately 20 clones per locus per isolate, the lowest allele frequency expected to be captured with this method is 1 in 20, compared to 1 in 2 with the traditional method of direct PCR sequencing. As described in subsequent sections, this more sensitive analysis resulted in the discovery of low-frequency haplotypes within the isolate populations.
TABLE 1.
SNPs and nucleotide diversity at coding regions in nine G. lamblia isolates
| Gene | Group | Isolate | No. of haplotypes | No. of SNPs | π (104)a | Tajima's Db |
|---|---|---|---|---|---|---|
| Actin | A-1 | WB | 2 | 1 | 1.6 | −1.16 (NS) |
| Gen | 1 | 0 | 0 | NA | ||
| CAT | 2 | 1 | 2 | −1.16 (NS) | ||
| Be-2 | 1 | 0 | 0 | NA | ||
| A-2 | AB | 4 | 5 | 9.6 | −1.78 (NS) | |
| N | 2 | 1 | 1.6 | −1.16 (NS) | ||
| B | GS | 3 | 2 | 3.6 | −1.51 (NS) | |
| CM | 3 | 26 | 44.6 | −2.48*** | ||
| Be-1 | 1 | 0 | 0 | NA | ||
| Beta giardin | A-1 | WB | 1 | 0 | 0 | NA |
| Gen | 1 | 0 | 0 | NA | ||
| CAT | 2 | 1 | 2.2 | −1.13 (NS) | ||
| Be-2 | 1 | 0 | 0 | NA | ||
| A-2 | AB | 1 | 0 | 0 | NA | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | 2 | 1 | 6.5 | 1.43 (NS) | |
| CM | 3 | 46 | 93.7 | −2.27*** | ||
| Be-1 | 2 | 39 | 119.5 | −1.88** | ||
| CPN60 | A-1 | WB | 2 | 1 | 3.1 | −1.14 (NS) |
| Gen | 2 | 1 | 2.7 | −1.16 (NS) | ||
| CAT | 5 | 4 | 9.8 | −1.84* | ||
| Be-2 | 3 | 2 | 4 | −1.51 (NS) | ||
| A-2 | AB | 2 | 1 | 2 | −1.17 (NS) | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | NDc | ND | ND | ND | |
| CM | ND | ND | ND | ND | ||
| Be-1 | 4 | 4 | 7.9 | −1.86* | ||
| [2Fe-2S] ferredoxin | A-1 | WB | 1 | 0 | 0 | NA |
| Gen | 1 | 0 | 0 | NA | ||
| CAT | 1 | 0 | 0 | NA | ||
| Be-2 | 4 | 3 | 12.2 | −1.35 (NS) | ||
| A-2 | AB | 2 | 2 | 5.3 | −1.51 (NS) | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | 9 | 9 | 29.5 | −2.03* | |
| CM | 10 | 17 | 43.1 | −2.42*** | ||
| Be-1 | 3 | 2 | 5 | −1.51 (NS) | ||
| RPL | A-1 | WB | 1 | 0 | 0 | NA |
| Gen | 2 | 1 | 4.2 | −0.56 (NS) | ||
| CAT | 1 | 0 | 0 | NA | ||
| Be-2 | 1 | 0 | 0 | NA | ||
| A-2 | AB | 2 | 1 | 2 | −1.16 (NS) | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | 6 | 58 | 142.7 | −2.39** | |
| CM | 2 | 1 | 2.2 | −1.16 (NS) | ||
| Be-1 | ND | ND | ND | ND | ||
| TPI | A-1 | WB | 1 | 0 | 0 | NA |
| Gen | 2 | 1 | 4.5 | −1.14 (NS) | ||
| CAT | 3 | 2 | 3 | −1.51 (NS) | ||
| Be-2 | 2 | 2 | 3.5 | −1.51 (NS) | ||
| A-2 | AB | 3 | 3 | 10.1 | −0.96 (NS) | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | 2 | 5 | 40.7 | 2.61** | |
| CM | 1 | 0 | 0 | NA | ||
| Be-1 | 3 | 128 | 781.2 | 1.62 (NS) |
π was estimated as described in Materials and Methods.
Tajima's D was calculated as described in Materials and Methods. NA, not applicable (in instances where D could not be calculated due to the lack of SNPs); NS, not significant. The asterisks indicate significance of Tajima's D test with the following confidence levels: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ND, not determined.
TABLE 2.
SNPs and nucleotide diversity at noncoding regions in nine G. lamblia isolates
| Region | Group | Isolate | No. of haplotypes | No. of SNPs | π (104)a | Tajima's Db |
|---|---|---|---|---|---|---|
| [2Fe-2S] ferredoxin intron | A-1 | WB | 1 | 0 | 0 | NA |
| Gen | 1 | 0 | 0 | NA | ||
| CAT | 1 | 0 | 0 | NA | ||
| Be-2 | 1 | 0 | 0 | NA | ||
| A-2 | AB | 1 | 0 | 0 | NA | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | 1 | 0 | 0 | NA | |
| CM | 2 | 1 | 28.6 | −1.16 (NS) | ||
| Be-1 | 1 | 0 | 0 | NA | ||
| RPL intron | A-1 | WB | 1 | 0 | 0 | NA |
| Gen | 1 | 0 | 0 | NA | ||
| CAT | 1 | 0 | 0 | NA | ||
| Be-2 | 1 | 0 | 0 | NA | ||
| A-2 | AB | 1 | 0 | 0 | NA | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | 2 | 17 | 165.7 | −2.39** | |
| CM | 1 | 0 | 0 | NA | ||
| Be-1 | NDc | ND | ND | ND | ||
| Intergenic 1 | A-1 | WB | 1 | 0 | 0 | NA |
| Gen | 1 | 0 | 0 | NA | ||
| CAT | 2 | 1 | 2 | −1.16 (NS) | ||
| Be-2 | 7 | 26 | 98.8 | −1.12 (NS) | ||
| A-2 | AB | 2 | 1 | 1.2 | −1.16 (NS) | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | 19 | 40 | 69.9 | −2.48*** | |
| CM | ND | ND | ND | ND | ||
| Be-1 | 1 | 0 | 0 | NA | ||
| Intergenic 2 | A-1 | WB | 2 | 1 | 1.7 | −1.16 (NS) |
| Gen | 2 | 1 | 3.3 | −0.40 (NS) | ||
| CAT | 3 | 2 | 6 | −0.65 (NS) | ||
| Be-2 | 2 | 1 | 1.7 | −1.16 (NS) | ||
| A-2 | AB | ND | ND | ND | ND | |
| N | 1 | 0 | 0 | NA | ||
| B | GS | ND | ND | ND | ND | |
| CM | ND | ND | ND | ND | ||
| Be-1 | 5 | 4 | 7.8 | −1.58 (NS) |
π was estimated as described in Materials and Methods.
Tajima's D was calculated as described in Materials and Methods. NA, not applicable (in instances where D could not be calculated due to the lack of SNPs); NS, not significant. The asterisks indicate significance of Tajima's D test with the following confidence levels: **, P < 0.01; ***, P < 0.001.
ND, not determined.
Examination of approximately 4,800 bp representing 10 loci over nine isolates yielded over 38,928 bp, and sequencing of approximately 20 clones per locus per isolate resulted in a grand total of 652,729 bp analyzed. We compared the sequences from each locus within and across the isolates to determine levels of both intra- and interisolate variation in G. lamblia populations. We discovered a total of 452 SNPs in our data set; thus, the density of SNPs was approximately one in 86 bp. A distinct difference in the degrees of polymorphism was apparent among groups, with one SNP per 403 bp found in group A-1 isolates, one SNP per 615 bp in group A-2 isolates, and one SNP in 26 bp in group B isolates. The higher degree of genetic diversity observed within and among group B isolates is mainly a consequence of the presence of two divergent haplotypes in this group and is discussed below in more depth. Of 452 total SNPs, 333 were detected in coding regions, while the remaining 119 were found in noncoding regions (see Table S3 in the supplemental material). Of 333 SNPs in coding regions, 81 were nonsynonymous SNPs (nsSNPs) and 252 were synonymous SNPs (sSNPs) (see Table S3 in the supplemental material), producing an nsSNP/sSNP ratio of approximately 0.32.
As an additional measure of genetic variation, we estimated nucleotide diversity, π, in our data set. The benefit of estimating this measure is that it depicts the level of genetic diversity detected by accounting for the frequency of polymorphisms. π estimates for 10 loci were examined across nine G. lamblia isolates and were further used to calculate the statistical test of neutrality, Tajima's D (Tables 1 and 2). Additionally, we discovered a total of insertions-deletions, but given their rarity and because they cannot be assumed to follow a single mutational model, we have limited our analysis to segregating sites as SNPs.
Variation in coding and noncoding regions across the three groups.
On average, SNPs occurred to similar extents in coding (one SNP in 88 bp) and noncoding (one SNP in 80 bp) regions. The genetic variability of coding regions mirrors that observed for the entire data set, with a significantly higher SNP density in group B isolates than in group A-1 and A-2 isolates (data not shown). Variation in intergenic regions was contributed primarily by the intergenic 1 region in the Be-2 isolate of group A-1 (26 SNPs) and the GS isolate of group B (40 SNPs) (Table 2 and Table S3 in the supplemental material), while the extent of polymorphism in the intergenic 2 region resembled that of coding regions (Table 2 and Table S3 in the supplemental material). The pattern of minimal levels of genetic diversity persisted across both introns analyzed (Table 2 and Table S3 in the supplemental material). In fact, there is a complete lack of intraisolate variation in intron regions (Table 2 and Table S3 in the supplemental material), with an exception of one SNP in the ferredoxin intron of the CM isolate and 17 SNPs at the RPL intron of the GS isolate, both from group B. The second case is reflective of the presence of two haplotypes at the RPL intron in the population of GS isolate sequences (see below), contributing to high GS intraisolate polymorphism.
Characterization of haplotypes and rare alleles.
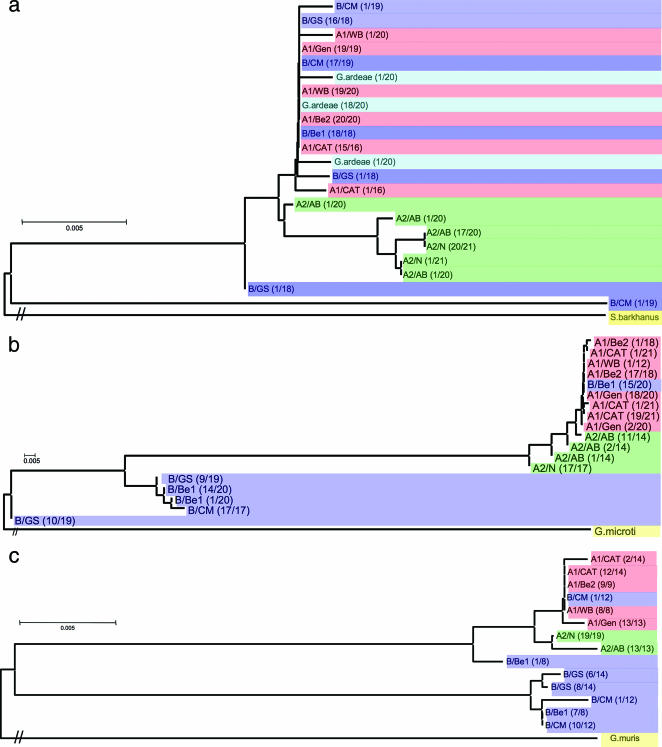
We used phylogenetic analyses to detect distinct haplotypes in G. lamblia populations and examine divergence among isolates, based on the distribution of SNPs in DNA sequences. At all loci examined, group A-1 isolate sequences formed a single cluster of haplotypes, as did the A-2 isolate sequences, thus forming two distinct clades (Fig. 1 and data not shown). In contrast, group B isolate sequences exhibited a more complex haplotype structure. At five loci (CPN60, ferredoxin gene, ferredoxin intron, intergenic 1 region, and intergenic 2 region), all group B sequences grouped with group A-1 sequences (data not shown). Thus, two main clades were detected at these five loci: A-1/B and A-2 (data not shown). However, at the remaining five loci (actin, RPL gene, RPL intron, TPI, and beta giardin) group B sequences exhibited a dichotomous structure, represented by two divergent haplotypes. A portion of sequences grouped with A-1 group sequences (as before), but additional sequences formed an independent long branch on the tree (a group B-specific clade) (Fig. 1 and data not shown). Thus, three main clades were detected at these five loci: A-1/B common haplotype, A-2 haplotype, and B-specific haplotype. The frequency of the group B-specific haplotype within the group B sequences was locus dependent. For example, the group B-specific haplotype was the prevalent one at the TPI and beta giardin loci (Fig. 1b and c and Table S3 in the supplemental material), while it was a rare haplotype at the actin locus (Fig. 1a and Table S3 in the supplemental material).
FIG. 1.
Neighbor-joining phylogenetic analyses of nine Giardia lamblia isolates at (a) actin, (b) TPI, and (c) beta giardin loci, carried out using the Kimura two-parameter model. Each unique sequence was used only once in the construction of the trees; thus, most branches represent multiple identical sequences. The haplotypes were named as follows: group designation/isolate designation. The frequency of each haplotype is shown in parentheses. Spironucleus barkhanus (GenBank accession number DX916114), Giardia microti (GenBank accession number AY228649), and Giardia muris (GenBank accession number AY258618) sequences were set as outgroups in the analyses of actin, TPI, and beta giardin loci, respectively. Different colors illustrate distinct groups: pink for group A-1, green for group A-2, blue for group B, light blue for G. ardeae, and yellow for the outgroup. For topology purposes, hash marks were used on the outgroup branches because of the extremely long branch lengths. Two major groupings are first apparent: A-1/B/A-2 and B. The first group is further subdivided into A-1/B and A-2. The trees show the presence of divergent haplotypes (A-1/B and B-specific haplotypes) in group B sequences. The group B-specific haplotype is the rare one at the actin locus but the prevalent one at the TPI and beta giardin loci.
The presence of two divergent haplotypes at some loci in group B isolates (Fig. 1 and data not shown) is clearly a major factor contributing to their higher levels of intra- and interisolate diversity (as noted before, one SNP in 403 bp in group A-1 isolates and one SNP in 615 bp in group A-2 isolates versus one SNP in 26 bp in group B isolates). Intraisolate and intragroup genetic variation in group A-1 isolates as well as group A-2 isolates was generally characterized by very few SNPs (Tables 1 and 2 and Table S3 in the supplemental material). Despite the fact that A-1 and A-2 isolates form separate clades (Fig. 1), interisolate and intergroup diversity among A-1 and A-2 isolates was typically characterized by a low number of SNPs (Table 3 and Table S3 in the supplemental material), representing definitive isolate and/or group markers (Table 4). Group B-specific haplotypes were, on the contrary, characterized by a significant number of polymorphic sites compared to group A-1 and A-2 sequences (Tables 1, 2, and 3 and Table S3 in the supplemental material). The presence of a group B-specific haplotype in an isolate is the source of higher genetic variation within that particular group B isolate, among that isolate and other group B isolates, and among that isolate and group A-1 and A-2 isolates. Thus, a majority of the variation detected was due to the differences among divergent haplotypes.
TABLE 3.
Distribution of SNPs among examined G. lamblia groups
| Group | No. of SNPs in region
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Actin | Beta giardin | CPN60 | Ferredoxin gene | RPL gene | TPI | Ferredoxin intron | RPL intron | Intergenic region 1 | Intergenic region 2 | |
| A-1 | 2 | 2 | 6 | 4 | 1 | 4 | 0 | 0 | 26 | 3 |
| A-2 | 5 | 2 | 1 | 2 | 1 | 3 | 0 | 0 | 1 | 0 |
| A | 7 | 6 | 10 | 10 | 6 | 8 | 0 | 4 | 37 | 25 |
| B | 27 | 47 | 4 | 29 | 59 | 130 | 1 | 17 | 40 | 4 |
| G. lamblia | 33 | 51 | 13 | 38 | 62 | 136 | 1 | 19 | 71 | 28 |
TABLE 4.
Pairwise comparisons of fixed differences between isolates at the TPI and beta giardin loci
| Locus | Isolate | No. of fixed differencesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WB | Gen | CAT | Be-2 | AB | N | GS | CM | Be-1 | ||
| TPI | WB | 0 | 0 | 0 | 1 | 3 | 124 | 127 | 0 | |
| Gen | 0 | 0 | 1 | 3 | 124 | 127 | 0 | |||
| CAT | 0 | 1 | 3 | 124 | 127 | 0 | ||||
| Be-2 | 1 | 3 | 124 | 127 | 0 | |||||
| AB | 0 | 122 | 125 | 0 | ||||||
| N | 123 | 126 | 1 | |||||||
| GS | 0 | 0 | ||||||||
| CM | 0 | |||||||||
| Be-1 | ||||||||||
| Beta giardin | WB | 1 | 0 | 0 | 4 | 2 | 44 | 0 | 5 | |
| Gen | 1 | 1 | 5 | 3 | 45 | 1 | 6 | |||
| CAT | 0 | 4 | 2 | 44 | 0 | 5 | ||||
| Be-2 | 4 | 2 | 44 | 0 | 5 | |||||
| AB | 2 | 44 | 2 | 6 | ||||||
| N | 44 | 1 | 5 | |||||||
| GS | 1 | 1 | ||||||||
| CM | 0 | |||||||||
| Be-1 | ||||||||||
Each value represents the number of SNPs fixed in the population of one isolate but not the other.
Last, where sequence data were available, Giardia ardeae, Giardia muris, Giardia microti, and/or Spironucleus barkhanus isolates were used as outgroups to construct phylogenies. The position of the A-1/B clade on the phylogenetic tree fell close to G. ardeae, while the other three species proved to be useful as the root of the phylogenetic tree.
Haplotype distribution and shared haplotypes in G. lamblia populations.
Sequence comparisons revealed that polymorphisms were often not shared between isolates: for example, the three SNPs discovered in the group A-2 AB isolate at the TPI locus were not the same as the two SNPs in the group A-1 WB isolate (Table 1 and data not shown). Thus, while examples of shared sequences are depicted in the common A-1/B clade at the actin, RPL gene, RPL intron, TPI, and beta giardin loci (Fig. 1), the majority of the polymorphisms were most often not shared among isolates from either within or between groups (Fig. 1 and data not shown).
Because these SNPs appeared to be isolate specific, we next performed pairwise interisolate comparisons to look for the number of fixed differences between each of the isolates at each locus to see whether the SNPs could be used to define isolates (Table 4 and Table S4 in the supplemental material). A fixed difference implies a nucleotide change that would be present in each sequence from one isolate but would not be present in the sequences of the other isolate. As examples, fixed differences at the TPI and beta giardin loci are shown in Table 4 and data from the TPI locus are further discussed. There was an absence of fixed differences among isolates within each of the three groups (A-1, A-2, and B). However, a comparison of either of the two A-2 isolates to any of the three A-1 isolates revealed fixed differences which were present in one group but not in the other (Table 4). These sequence changes, therefore, defined both isolates and groups. These findings are mirrored in the grouping of A-1 and A-2 group sequences into two separate clades (Fig. 1 and data not shown). Similarly, comparison of the group B isolates with isolates from the A-1 and A-2 groups revealed the presence of group B-specific markers (Table 4). While these substitutions were fixed in GS and CM isolates (thus representing isolate-specific markers) (Table 4), isolate Be-1 was comprised of a population of A-1/B haplotype sequences and group B-specific sequences (Fig. 1b); thus, the substitutions were not fixed in this population (Table 4). Therefore, our results confirmed the previous designation of group A-1 and A-2 isolates (36) and revealed a novel haplotype structure in group B isolates.
Estimation of Tajima's D to examine evolutionary history of the loci.
In order to learn about the mechanism responsible for maintaining the levels of observed genetic diversity, we calculated Tajima's D, a statistical test of neutrality and constant effective population size that compares π and θ (48) and whose sign can be informative about the evolutionary history of the locus in question (40). Tables 1 and 2 show Tajima's D values for 10 loci across nine G. lamblia isolates. In instances of a complete lack of genetic variation, it was impossible to calculate D values. In the majority of cases where variation was observed, calculated Tajima's D values were consistently negative across different loci, although in most cases the results of the test were not statistically significant.
DISCUSSION
General features of the genetic diversity in G. lamblia populations.
Our analysis of SNPs and other measures of genetic variation (π and θ) at 10 loci in G. lamblia populations reveals low levels of sequence diversity. Genetic variation was particularly low within an isolate (hence, most isolates were represented by a single haplotype), particularly in the case of isolates from groups A-1 and A-2. Levels of sequence diversity in interisolate comparisons varied by locus but revealed specific divergence patterns among the three groups (see below). We did not observe differences in the extent or pattern of genetic diversity between isolates, based either on their geographic origin (Table S1 in the supplemental material) or their host origin (Table S2 in the supplemental material), although we did not test for this directly.
The overall incidence of SNPs at 10 loci of G. lamblia was approximately 1 in 86 bp. However, the SNP rates in group A-1 and A-2 isolates (1 in 403 bp and 1 in 615 bp, respectively) were strikingly low compared to that in group B isolates (1 in 26 bp). This is exclusively due to the sequence variation between two divergent haplotypes within the group B isolates, one common with the A-1 group and one group B specific. We propose that the failure to detect a group B-specific haplotype at the remaining five loci is a not a consequence of their absence but rather of their low frequency in the population. It is important to note that our data apply only to the three major, and medically relevant, G. lamblia groups and that other studies have indicated higher levels of genetic variability in other species-specific G. lamblia groups (groups C through G) (31, 49). However, we note that G. ardeae sequences appeared more closely related to the A-1/B G. lamblia haplotype (Fig. 1), suggesting either that its previous designation as a separate species (15) might be questionable or that group B represents the most divergent distinct species.
Interestingly, low levels of genetic variation were detected not only in coding but also in noncoding regions, which are expected to have fewer functional constraints compared to coding regions. We propose two possible explanations: (i) due to G. lamblia's unusually short intergenic regions and introns, these regions might be influenced strongly by hitchhiking events, or (ii) due to the functional roles, such as splicing machinery requirements or roles in gene expression regulation, of noncoding regions, these regions might be conserved, as shown for Plasmodium falciparum (12).
Additional regions, such as microsatellites and variant-specific surface protein (VSP) genes, where one might expect to detect higher levels of genetic variation, were not analyzed in this study given that (i) microsatellites have not yet been described for Giardia and (ii) VSP gene repertoires differ across different Giardia groups (35).
Significance of rare haplotypes.
We investigated the significance of the haplotype distribution in the G. lamblia populations. As a preliminary step, to verify the presence of haplotypes detected a single time in our sampling set, we used SNP-specific primers to reamplify the haplotype from the isolate from which it was first identified and to further determine whether that haplotype is present in other isolates. This technique was used to confirm the presence of the rare actin haplotype in the group B CM isolate (group B-specific haplotype) (Fig. 1a) and also to indicate that it is a haplotype found across the group B isolates but not the group A-1 and A-2 isolates (data not shown).
Although a portion of discovered singletons might represent experimental errors, it is not plausible that they would significantly skew our results, given their rarity. This is because π is a frequency-dependent measure of variation, and so it will not be significantly impacted by low-frequency polymorphisms. Thus, we believe that overall estimates of extent and distribution of genetic variation within and among isolates can be stated with confidence.
Degree and distribution of genetic variation in G. lamblia populations with respect to other parasite populations.
Direct comparisons of the levels of sequence nucleotide polymorphisms in G. lamblia presented here to reports of levels in other parasitic protozoa are extremely challenging, given the different approaches. We report a density of SNPs in G. lamblia of ∼1 in 86 bp (1 in 403 bp in group A-1 isolates, 1 in 615 bp in group A-2 isolates, and 1 in 26 bp in group B isolates). Previous studies documented SNP incidence in a few parasite populations as follows: ∼1 in 526 bp in Plasmodium vivax (16), ∼1 in 917 bp in Plasmodium falciparum (33), and ∼1 in 175 bp in Entamoeba histolytica (9). Although the SNP incidence we found in group A-1 and group A-2 at first appears comparable to those seen in other parasites, this is not the case, given that the method used in our study is more sensitive and thus results in high SNP numbers compared to those from the other studies. We examined individual clones, as commonly applied for obtaining human immunodeficiency virus sequences from an individual host (reviewed in reference 27), rather than the more traditional method of sequencing a population of molecules from a PCR. Thus, the number of reported SNPs was recovered from a sample of approximately 20 sequences per locus per isolate of a cloned PCR product and not from direct sequencing of a PCR. The advantage of the method employed here lies in the fact that it allowed us to detect even rare alleles. Had we used the traditional experimental design, we would have seen our data structure collapsing, as A-1 and A-2 group sequences would most often be represented by a single sequence, and even the two divergent haplotypes in group B sequences would be detected only if they both had approximately the same frequencies. This was the case previously, when a few G. lamblia isolates were examined for SNPs at the TPI locus by use of a direct sequencing approach (6). Thus, simple comparisons of SNP rates across parasitic species cannot account for the differences in distinct methodologies employed.
There were slightly fewer synonymous changes (35 sSNPs versus 47 nsSNPs) (see Table S3 in the supplemental material) within and among A-1 and A-2 group isolates. However, substitutions defining the divergence between the group B-specific haplotype and A-1/B and A-2 haplotypes were heavily biased towards synonymous changes (217 sSNPs versus 34 nsSNPs) (see Table S3 in the supplemental material). An overall 0.32 ratio of nsSNPs/sSNPs found at six coding regions in G. lamblia populations would result in a high level of conservation of protein sequences compared to that found in P. falciparum (2.34 ratio of nsSNPs/sSNPs) (16), P. vivax (1.75 ratio of nsSNPs/sSNPs) (33), and even human populations (0.89 ratio of nsSNPs/sSNPs) (16). These data indicate a more stringent functional constraint in coding regions of G. lamblia, as was also shown recently for E. histolytica (9). In other words, although group B-specific sequences exhibited high divergence compared to the remainder of the data set (Tables 1 and 2 and Table S3 in the supplemental material), they showed a significant bias towards synonymous changes (Table S3 in the supplemental material), which suggests a strong purifying selection. In contrast, intraisolate variation in A-1 and A-2 groups (when captured) was depicted by a bias in nonsynonymous substitutions (Table S3 in the supplemental material).
Divergence between the two nuclei.
Determination of levels of intraisolate variation can also aid our understanding of sequence divergence between the two nuclei at the level of populations. Nearly complete sequence conservation within A-1 and A-2 group isolates, as well as the predominance of synonymous changes in group B isolates, suggests that the genetic contents of the two nuclei are virtually identical at the sequence level and that any protein products will have little to no variation. Thus, while the discovery of four alleles of the A6 VSP gene, only one of which is expressed at the cell surface at a time (61), argues that transcriptional regulation at the level of the two nuclei exists, the high level of sequence conservation suggests that gene expression may not require tight control at the nuclear level for most genes.
Mechanisms responsible for decreasing and/or maintaining low levels of genetic diversity in G. lamblia populations.
A negative value of Tajima's D (Tables 1 and 2) indicates an excess of rare variants in the population compared to the expectations under a model of neutral mutation, genetic drift, and population size equilibrium. Such an observation could be explained by the selective sweep hypothesis, as was proposed for Toxoplasma gondii (20), or, alternatively, could be a consequence of a bottleneck event. As our Tajima's D results consistently show negative values across most loci, and one would expect selection to act in a locus-dependent manner (40), a bottleneck event or recurrent bottleneck events might be a more plausible explanation for a lack of genetic diversity in G. lamblia populations. Although Tajima's D results were not statistically significant for group A-1 and A-2 isolates (Tables 1 and 2), indicating the possibility that the examined loci are effectively neutral, statistically significant negative Tajima's D values were found across all loci for group B isolates, providing support for the occurrence of a bottleneck event(s). Again, it is important to remember that, despite the fact that group B sequences exhibited the highest level of genetic diversity out of the three groups examined, this variation was contributed predominantly by the presence of two divergent haplotypes. Given that our study was performed on axenized isolates, the introduction of Giardia samples into the laboratory and subsequent expansion in culture could have constituted a bottleneck event.
It is important to recognize that an axenization-induced bottleneck event(s) (if any) could explain only the lack of intraisolate genetic diversity but could not account for the low interisolate variation we observed at many loci. Alternatively, a bottleneck event(s) in individual hosts during each infection could account for the lack of intraisolate variation. Three possible mechanisms could instead explain the low interisolate variation: (i) a recent and species-wide bottleneck event, (ii) a low mutation rate, and/or (iii) an active sexual cycle among isolates and groups in which recombination could lead to a homogenization of the genome. We propose that the presence of group A-1 and group B-specific haplotypes in group B populations is, in fact, a product of the genetic exchange, thus suggesting that a sexual cycle exists in the parasite.
Conclusions.
We believe that our study may prove useful in the future in understanding the etiology of the disease, since a diverse array of giardiasis manifestations, from asymptomatic infections to chronic diarrhea accompanied by malabsorption and requiring treatment, is seen in human patients. It is still unclear whether this is due to host or parasite effects or host-parasite interactions (reviewed in reference 42). For instance, studies of virulent and avirulent strains of E. histolytica have shown that the least virulent strain exhibited the most divergence, thus potentially linking genotypic variants to disease outcomes (45). Efforts to relate genotypic markers with various aspects of the infection could lead to promising means for an effective fight against the disease.
Supplementary Material
Acknowledgments
We are particularly grateful to Matthew Hamilton for his valuable ideas and comments during the course of this project and David Soria for help with construction of the phylogenetic trees. We thank Amanda Munson and Ernica Noel for assistance, Theodore Nash and Stanley Erlandsen for kindly providing us G. lamblia and G. ardeae isolates, and The Marine Biological Laboratory for sequencing efforts.
This work was supported by NIH grant 1R01AI/GM48922-01A1 to H.G.E. and a Georgetown pilot research grant to H.G.E. S.T. was supported in part by a Sigma Xi Grant-in-Aid of Research award and a Georgetown Graduate School dissertation fellowship award.
Footnotes
Published ahead of print on 8 June 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adam, R. D. 1992. Chromosome-size variation in Giardia lamblia: the role of rDNA repeats. Nucleic Acids Res. 20:3057-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam, R. D., T. E. Nash, and T. E. Wellems. 1988. The Giardia lamblia trophozoite contains sets of closely related chromosomes. Nucleic Acids Res. 16:4555-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, R. H., M. Adams, P. F. Boreham, G. Mayrhofer, and B. P. Meloni. 1989. Giardia intestinalis: electrophoretic evidence for a species complex. Int. J. Parasitol. 19:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Ayala, F. J. 1998. Is sex better? Parasites say “no.” Proc. Natl. Acad. Sci. USA 95:3346-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baruch, A. C., J. Isaac-Renton, and R. D. Adam. 1996. The molecular epidemiology of Giardia lamblia: a sequence-based approach. J. Infect. Dis. 174:233-236. [DOI] [PubMed] [Google Scholar]
- 7.Benchimol, M. 2004. Giardia lamblia: behavior of the nuclear envelope. Parasitol. Res. 94:254-264. [DOI] [PubMed] [Google Scholar]
- 8.Bernander, R., J. E. Palm, and S. G. Svard. 2001. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell. Microbiol. 3:55-62. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya, D., R. Haque, and U. Singh. 2005. Coding and noncoding genomic regions of Entamoeba histolytica have significantly different rates of sequence polymorphisms: implications for epidemiological studies. J. Clin. Microbiol. 43:4815-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boothroyd, J. C., A. Wang, D. A. Campbell, and C. C. Wang. 1987. An unusually compact ribosomal DNA repeat in the protozoan Giardia lamblia. Nucleic Acids Res. 15:4065-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunjun, S., C. Stathopoulos, D. Graham, B. Min, M. Kitabatake, A. L. Wang, C. C. Wang, C. P. Vivares, L. M. Weiss, and D. Soll. 2000. A dual-specificity aminoacyl-tRNA synthetase in the deep-rooted eukaryote Giardia lamblia. Proc. Natl. Acad. Sci. USA 97:12997-13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calderwood, M. S., L. Gannoun-Zaki, T. E. Wellems, and K. W. Deitsch. 2003. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 278:34125-34132. [DOI] [PubMed] [Google Scholar]
- 13.Cedillo-Rivera, R., J. M. Darby, J. A. Enciso-Moreno, G. Ortega-Pierres, and P. L. Ey. 2003. Genetic homogeneity of axenic isolates of Giardia intestinalis derived from acute and chronically infected individuals in Mexico. Parasitol. Res. 90:119-123. [DOI] [PubMed] [Google Scholar]
- 14.Edlind, T. D., and P. R. Chakraborty. 1987. Unusual ribosomal RNA of the intestinal parasite Giardia lamblia. Nucleic Acids Res. 15:7889-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erlandsen, S. L., W. J. Bemrick, C. L. Wells, D. E. Feely, L. Knudson, S. R. Campbell, H. van Keulen, and E. L. Jarroll. 1990. Axenic culture and characterization of Giardia ardeae from the great blue heron (Ardea herodias). J. Parasitol. 76:717-724. [PubMed] [Google Scholar]
- 16.Feng, X., J. M. Carlton, D. A. Joy, J. Mu, T. Furuya, B. B. Suh, Y. Wang, J. W. Barnwell, and X. Z. Su. 2003. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc. Natl. Acad. Sci. USA 100:8502-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furness, B. W., M. J. Beach, and J. M. Roberts. 2000. Giardiasis surveillance—United States, 1992-1997. Morb. Mortal. Wkly. Rep. 49:1-13. [PubMed] [Google Scholar]
- 18.Ghosh, S., M. Frisardi, R. Rogers, and J. Samuelson. 2001. How giardia swim and divide. Infect. Immun. 69:7866-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillin, F. D., D. S. Reiner, and J. M. McCaffery. 1996. Cell biology of the primitive eukaryote Giardia lamblia. Annu. Rev. Microbiol. 50:679-705. [DOI] [PubMed] [Google Scholar]
- 20.Grigg, M. E., S. Bonnefoy, A. B. Hehl, Y. Suzuki, and J. C. Boothroyd. 2001. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294:161-165. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto, T., Y. Nakamura, T. Kamaishi, F. Nakamura, J. Adachi, K. Okamoto, and M. Hasegawa. 1995. Phylogenetic place of mitochondrion-lacking protozoan, Giardia lamblia, inferred from amino acid sequences of elongation factor 2. Mol. Biol. Evol. 12:782-793. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto, T., Y. Nakamura, F. Nakamura, T. Shirakura, J. Adachi, N. Goto, K. Okamoto, and M. Hasegawa. 1994. Protein phylogeny gives a robust estimation for early divergences of eukaryotes: phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol. Biol. Evol. 11:65-71. [DOI] [PubMed] [Google Scholar]
- 23.Hou, G., S. M. Le Blancq, Y. E, H. Zhu, and M. G. Lee. 1995. Structure of a frequently rearranged rRNA-encoding chromosome in Giardia lamblia. Nucleic Acids Res. 23:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam, A. 1990. Giardiasis in developing countries, p. 235-266. In E. A. Myer (ed.), Giardiasis. Elsevier, Amsterdam, The Netherlands.
- 25.Kabnick, K. S., and D. A. Peattie. 1990. In situ analyses reveal that the two nuclei of Giardia lamblia are equivalent. J. Cell Sci. 95:353-360. [DOI] [PubMed] [Google Scholar]
- 26.Keister, D. B. 1983. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 77:487-488. [DOI] [PubMed] [Google Scholar]
- 27.Lemey, P., A. Rambaut, and O. G. Pybus. 2006. HIV evolutionary dynamics within and among hosts. AIDS Rev. 8:125-140. [PubMed] [Google Scholar]
- 28.Lu, S., J. Wen, J. Li, and F. Wang. 2002. DNA sequence analysis of the triose phosphate isomerase gene from isolates of Giardia lamblia. Chin. Med. J. (Engl. Ed.) 115:99-102. [PubMed] [Google Scholar]
- 29.McArthur, A. G., H. G. Morrison, J. E. Nixon, N. Q. Passamaneck, U. Kim, G. Hinkle, M. K. Crocker, M. E. Holder, R. Farr, C. I. Reich, G. E. Olsen, S. B. Aley, R. D. Adam, F. D. Gillin, and M. L. Sogin. 2000. The Giardia genome project database. FEMS Microbiol. Lett. 189:271-273. [DOI] [PubMed] [Google Scholar]
- 30.Meloni, B. P., A. J. Lymbery, and R. C. Thompson. 1995. Genetic characterization of isolates of Giardia duodenalis by enzyme electrophoresis: implications for reproductive biology, population structure, taxonomy, and epidemiology. J. Parasitol. 81:368-383. [PubMed] [Google Scholar]
- 31.Monis, P. T., R. H. Andrews, G. Mayrhofer, and P. L. Ey. 2003. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect. Genet. Evol. 3:29-38. [DOI] [PubMed] [Google Scholar]
- 32.Monis, P. T., G. Mayrhofer, R. H. Andrews, W. L. Homan, L. Limper, and P. L. Ey. 1996. Molecular genetic analysis of Giardia intestinalis isolates at the glutamate dehydrogenase locus. Parasitology 112:1-12. [DOI] [PubMed] [Google Scholar]
- 33.Mu, J., J. Duan, K. D. Makova, D. A. Joy, C. Q. Huynh, O. H. Branch, W. H. Li, and X. Z. Su. 2002. Chromosome-wide SNPs reveal an ancient origin for Plasmodium falciparum. Nature 418:323-326. [DOI] [PubMed] [Google Scholar]
- 34.Murtagh, J. J., Jr., M. R. Mowatt, C. M. Lee, F. J. Lee, K. Mishima, T. E. Nash, J. Moss, and M. Vaughan. 1992. Guanine nucleotide-binding proteins in the intestinal parasite Giardia lamblia. Isolation of a gene encoding an approximately 20-kDa ADP-ribosylation factor. J. Biol. Chem. 267:9654-9662. [PubMed] [Google Scholar]
- 35.Nash, T. E. 2002. Surface antigenic variation in Giardia lamblia. Mol. Microbiol. 45:585-590. [DOI] [PubMed] [Google Scholar]
- 36.Nash, T. E., T. McCutchan, D. Keister, J. B. Dame, J. D. Conrad, and F. D. Gillin. 1985. Restriction-endonuclease analysis of DNA from 15 Giardia isolates obtained from humans and animals. J. Infect. Dis. 152:64-73. [DOI] [PubMed] [Google Scholar]
- 37.Nei, M., and W.-H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nixon, J. E., A. Wang, J. Field, H. G. Morrison, A. G. McArthur, M. L. Sogin, B. J. Loftus, and J. Samuelson. 2002. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell 1:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramesh, M. A., S. B. Malik, and J. M. Logsdon, Jr. 2005. A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 15:185-191. [DOI] [PubMed] [Google Scholar]
- 40.Rand, D. 1996. Neutrality tests of molecular markers and the connection between DNA polymorphism, demography, and conservation biology. Conserv. Biol. 10:665-671. [Google Scholar]
- 41.Regoes, A., D. Zourmpanou, G. Leon-Avila, M. van der Giezen, J. Tovar, and A. B. Hehl. 2005. Protein import, replication, and inheritance of a vestigial mitochondrion. J. Biol. Chem. 280:30557-30563. [DOI] [PubMed] [Google Scholar]
- 42.Roxstrom-Lindquist, K., D. Palm, D. Reiner, E. Ringqvist, and S. G. Svard. 2006. Giardia immunity—an update. Trends Parasitol. 22:26-31. [DOI] [PubMed] [Google Scholar]
- 43.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 44.Sagolla, M. S., S. C. Dawson, J. J. Mancuso, and W. Z. Cande. 2006. Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite Giardia intestinalis. J. Cell Sci. 119:4889-4900. [DOI] [PubMed] [Google Scholar]
- 45.Shah, P. H., R. C. MacFarlane, D. Bhattacharya, J. C. Matese, J. Demeter, S. E. Stroup, and U. Singh. 2005. Comparative genomic hybridizations of Entamoeba strains reveal unique genetic fingerprints that correlate with virulence. Eukaryot. Cell 4:504-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sogin, M. L., J. H. Gunderson, H. J. Elwood, R. A. Alonso, and D. A. Peattie. 1989. Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science 243:75-77. [DOI] [PubMed] [Google Scholar]
- 47.Sulaiman, I. M., R. Fayer, C. Bern, R. H. Gilman, J. M. Trout, P. M. Schantz, P. Das, A. A. Lal, and L. Xiao. 2003. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 9:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, R. C., and B. P. Meloni. 1993. Molecular variation in Giardia. Acta Trop. 53:167-184. [DOI] [PubMed] [Google Scholar]
- 50.Tibayrenc, M. 1993. Entamoeba, giardia and toxoplasma: clones or cryptic species? Parasitol. Today 9:102-105. [DOI] [PubMed] [Google Scholar]
- 51.Tibayrenc, M., and F. J. Ayala. 1991. Towards a population genetics of microorganisms: the clonal theory of parasitic protozoa. Parasitol. Today 7:228-232. [DOI] [PubMed] [Google Scholar]
- 52.Tibayrenc, M., F. Kjellberg, J. Arnaud, B. Oury, S. F. Breniere, M. L. Darde, and F. J. Ayala. 1991. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc. Natl. Acad. Sci. USA 88:5129-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tibayrenc, M., F. Kjellberg, and F. J. Ayala. 1990. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc. Natl. Acad. Sci. USA 87:2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tovar, J., G. Leon-Avila, L. B. Sanchez, R. Sutak, J. Tachezy, M. van der Giezen, M. Hernandez, M. Muller, and J. M. Lucocq. 2003. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426:172-176. [DOI] [PubMed] [Google Scholar]
- 55.Upcroft, J. A., and P. Upcroft. 1994. Two distinct varieties of Giardia in a mixed infection from a single human patient. J. Eukaryot. Microbiol. 41:189-194. [DOI] [PubMed] [Google Scholar]
- 56.USDA. 1994. Cryptosporidium and Giardia in beef calves. Beef Cow/Calf Health and Productivity Audit, National Animal Health Monitoring System. USDA, Washington, DC.
- 57.Wang, A. L., H. M. Yang, K. A. Shen, and C. C. Wang. 1993. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc. Natl. Acad. Sci. USA 90:8595-8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watterson, G. A. 1975. On the number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7:256-276. [DOI] [PubMed] [Google Scholar]
- 59.WHO/UNICEF. 2000. Global water supply and sanitation assessment 2000 report. WHO/UNICEF Joint Monitoring Programme for Water Supply and Sanitation. WHO, Geneva, Switzerland.
- 60.Wiesehahn, G. P., E. L. Jarroll, D. G. Lindmark, E. A. Meyer, and L. M. Hallick. 1984. Giardia lamblia: autoradiographic analysis of nuclear replication. Exp. Parasitol. 58:94-100. [DOI] [PubMed] [Google Scholar]
- 61.Yang, Y., and R. D. Adam. 1994. Allele-specific expression of a variant-specific surface protein (VSP) of Giardia lamblia. Nucleic Acids Res. 22:2102-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, Y. M., Y. Ortega, C. Sterling, and R. D. Adam. 1994. Giardia lamblia trophozoites contain multiple alleles of a variant-specific surface protein gene with 105-base pair tandem repeats. Mol. Biochem. Parasitol. 68:267-276. [DOI] [PubMed] [Google Scholar]
- 63.Yu, L. Z., C. W. Birky, Jr., and R. D. Adam. 2002. The two nuclei of Giardia each have complete copies of the genome and are partitioned equationally at cytokinesis. Eukaryot. Cell 1:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.