Abstract
Previous work showed that the Aggregatibacter actinomycetemcomitans adhesin Aae demonstrated species specificity and tissue tropism to buccal epithelial cells (BECs) derived from humans and Old World primates, but a second, lower-affinity adhesin was noted. This study was designed to determine if Omp100 (also known as ApiA), a surface-expressed A. actinomycetemcomitans adhesin, is that second adhesin and if so to investigate its tissue tropism and species specificity. A targeted mutagenesis protocol was used to construct an isogenic apiA mutant and an aae apiA double mutant with wild-type A. actinomycetemcomitans. In addition, Escherichia coli strain DH5α was used to express apiA to further assess binding parameters. Results indicated that the apiA mutant strain showed significantly less binding to BECs than its parent strain (P ≤ 0.05). Further, binding mediated by ApiA was specific to BECs from humans and Old World primates, as seen in both wild-type A. actinomycetemcomitans and E. coli expressing ApiA (P ≤ 0.05). Pretreatment of wild-type A. actinomycetemcomitans cells with anti-ApiA antiserum reduced binding in a dose-dependent manner. The aae apiA double mutant completely abrogated A. actinomycetemcomitans binding to both human and Old World primate BECs. Taken together, these studies indicate that ApiA and Aae, in concert, modulate binding of A. actinomycetemcomitans to human BECs. Since the BEC is a prominent reservoir for A. actinomycetemcomitans, identification of this second adhesin could lead to important therapeutic strategies.
The gram-negative bacterium Aggregatibacter actinomycetemcomitans (formerly known as Actinobacillus actinomycetemcomitans) is considered the causative agent of localized aggressive periodontitis (LAP) (21, 32), a highly destructive form of periodontal disease that affects adolescents (32). Although A. actinomycetemcomitans is routinely recovered in high numbers from the subgingival plaque in patients with LAP, only 20% of healthy subjects display detectable levels of A. actinomycetemcomitans (32). In those individuals infected with A. actinomycetemcomitans, two lines of evidence suggest oral mucosal cell surfaces are prominent initial colonization sites as well as primary reservoirs for A. actinomycetemcomitans. First, A. actinomycetemcomitans has been isolated from the oral mucosa, of predentate children (16), and second, the host range in which A. actinomycetemcomitans is found in nature parallels the in vitro binding pattern seen when A. actinomycetemcomitans is tested with buccal epithelial cells (BECs) obtained from a variety of mammalian species (4).
In a recent publication our group demonstrated that Aae, an outer membrane protein, mediates attachment of A. actinomycetemcomitans (strain IDH781) to BECs from human and Old World primates, as opposed to BECs from other primates (7). Moreover, Escherichia coli expressing Aae demonstrated the same pattern of attachment, and thus binding was limited to BECs derived from humans and Old World primates. In a bacterial concentration-dependent BEC binding assay, results obtained from the aae mutant strain of A. actinomycetemcomitans and from E. coli expressing Aae indicated that the Aae adhesin had a high affinity for human BECs (7). It was also noted that aside from the Aae adhesin, a second, lower-affinity adhesin was present on the surface of A. actinomycetemcomitans that could also contribute to its binding to human BECs.
A. actinomycetemcomitans ApiA, also known as Omp100 (1), a member of the Yersinia adhesin (YadA) family of autotransporter adhesins, is a 100-kDa A. actinomycetemcomitans outer membrane protein adhesin that was shown to bind to epithelium (15). More recently, Li and colleagues demonstrated that ApiA expression resulted in binding to fibronectin and collagen as well as to epithelium (17). Although ApiA has been purported to be responsible for binding to different substrates, we decided to include apiA among a number of A. actinomycetemcomitans candidate adhesins to determine whether it could be the second BEC adhesin we observed.
The goals of this study were twofold: (i) to determine the gene responsible for this second A. actinomycetemcomitans adhesin, as noted in previous studies, and (ii) to determine the host range of this purported adhesin. If ApiA was identified as this second adhesin, an additional objective was to determine the effect of knocking out both apiA and aae on binding of A. actinomycetemcomitans to BECs derived from humans.
In this report we present evidence that ApiA exhibits a species-specific BEC binding phenotype similar to that of Aae and that a double knockout of apiA and aae completely abrogates the ability of A. actinomycetemcomitans to bind to human BECs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. actinomycetemcomitans strains were cultured in an A. actinomycetemcomitans growth medium (AAGM) that was composed of Trypticase soy broth supplemented with 6 g of yeast extract, 8 g of glucose, and 4 g of sodium bicarbonate in 1 liter deionized water and incubated at 37°C in 10% CO2. Strains harboring pMB7, pJAK16, or pJK654 were cultured in the AAGM containing 3 μg of chloramphenicol per ml to maintain harbored plasmids. Escherichia coli was cultured aerobically in Luria-Bertani broth. Chloramphenicol was supplemented in a concentration of 50 μg per ml to keep plasmids pVK43, pJAK16, and pJK654 stable in E. coli (Table 1).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source (reference) |
|---|---|---|
| A. actinomycetemcomitans strains | ||
| CU1000 | Clinical isolate (serotype f) | Fine et al., 1999 (6) |
| IDH781 | Clinical isolate (serotype d) | Haubek et al., 1995 (11) |
| IDH781N | Spontaneous Nalr variant of IDH781 | Fine et al., 2005 (7) |
| JK1046 | IDH781N aae::R6Kγori/KAN; Kmr | Fine et al., 2005 (7) |
| JK1047 | IDH781N flp-1::Tn903 kan; Kmr | Kaplan et al., 2004 (14) |
| JK1051 | IDH781N apia::R6Kγori/KAN; Kmr | This study |
| GJD1 | IDH781N aae::R6Kγori/KAN; apia::SPE; Kmr/Spr | This study |
| E. coli strains | ||
| TOP10 | Used for TOPO TA cloning | Invitrogen |
| DH5α | Used for functional expression of aae/apia | New England Biolabs |
| Plasmids | ||
| pCR2.1-TOPO | E. coli cloning vector; Kmr Apr | Invitrogen |
| pJAK16 | Broad-host-range expression vector; Cmr | Thomson et al., 1999 (28) |
| pJK654 | pJAK16 containing apia | This study |
| pVK43 | pJAK16 containing aae | This study |
| pMB7 | Used in facilitating transformation into A. actinomycetemcomitans | Rhodes et al., 2005 (22) |
| pGHM491 | Containing spectinomycin resistance gene | Gift from D. Figurski |
Nalr, nalidixic acid resistant; Apr, ampicillin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant.
Construction of apiA knockout strain.
We used a targeted-mutagenesis protocol to construct an isogenic mutant of IDH781N (7). To create apiA knockout in A. actinomycetemcomitans strain JK1051, a 2.0-kb kanamycin resistance transposon R6Kγori/KAN (Kmr) gene was inserted into the apiA open reading frame between bases 496 and 497 with the same promoter orientation as seen in the chromosome of IDH781N.
Genetic complementation of apiA mutation.
An ApiA expression plasmid was constructed by amplifying the apiA coding region from strain HK1651 (mapped to bp 602 to 1,489; GenBank accession no. AB064943) using a PCR primer pair (forward primer, CGCTGGATCCATAATGA AGA AAGTTTAGATGAC AT A TCAATTAT TTAAACACC; reverse primer, GACACTGCAGTTACCACTCAA AGTTTAAACC), which introduced a BamHI site 24 bp upstream from the apiA start codon and a PstI site immediately downstream from the apiA stop codon. The BamHI site and the PstI site are underlined in the forward primer and reverse primer, respectively. The PCR product was ligated into the BamHI/PstI sites of the broad-host-range plasmid pJAK16, which placed apiA under control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter (7). The resulting plasmid (named pJK654) was transformed into JK1051, and transformants were selected on agar containing 3 μg ml−1 of chloramphenicol.
Construction of apiA aae double-knockout strain.
This strain was constructed by first amplifying the entire apiA coding region and flanking regions by PCR (mapped to bp 87 to 1,797; accession no. AB064943). The forward primer (AATTCGTTGTCATAATGG) is located 516 bp upstream from the apiA start codon, and the reverse primer (GAATTCCCGGGCTGTGCC) is located 307 bp downstream from its stop codon. The PCR amplicon was ligated into the plasmid vector pCR2.1 using a TOPO cloning kit (Invitrogen). Next, the 1,033-bp spectinomycin resistance (Smr) gene from plasmid pGMH491 (provided by David Figurski) was inserted between the two internal ClaI sites in apiA (bp 850 and 1,230; accession no. AB064943) so that the Smr gene was transcribed in the same orientation as apiA. The resulting mutagenic plasmid (linearized with ScaI) was used to transform strain JK1046 (aae::Kmr) to spectinomycin resistance (20 μg ml−1), which resulted in strain GJD1 (aae::Kmr apiA::Smr).
Construction of E. coli strain expressing functional ApiA.
DH5α chemically competent cells (Invitrogen) were used to express functional ApiA. The plasmid pJK654 was transformed into DH5α, following the manufacturer's instructions. Transformation of pJAK16 (the empty plasmid) was used as a control. For either the epithelial attachment assay or Western blotting, clones of DH5α/pJK654 (apiA+) and DH5a/pJAK16 vector alone or pJAK16 apiA mutant were picked from Luria-Bertani plates supplemented with 50 μg/ml chloramphenicol and inoculated into Luria-Bertani broth containing 50 μg/ml chloramphenicol. After overnight culture at 37°C with shaking at 250 rpm, an aliquot from each cell culture was reinoculated into fresh Luria-Bertani broth at 37°C with shaking at 250 rpm for 3 h. IPTG was added to cell cultures at a final concentration of 1 mM for 3 h prior to in vitro studies.
Isolation of mammalian oral epithelium.
Human buccal and dorsal tongue epithelial cells were obtained from healthy adult volunteers. Collection of BECs from human volunteers was approved by the UMDNJ IRB committee. Collection of BECs from the mouths of animals was approved by the IACUC committees of University of Medicine and Dentistry of New Jersey, Rutgers University, and the IACUC committee of the New England Regional Primate Research Center. Collection methods have been described previously (5, 7). Briefly, BECs collected from the surface of the oral mucosa and/or the dorsal surface of the tongue were obtained by gentle but firm scraping of the mucosa with the flat end of a sterile wooden tongue blade. The blade with collected cells was suspended in 5 ml of phosphate-buffered saline (PBS) and subjected to vortex agitation to remove any residue of sloughed cells from the blade. Cells were then subjected to centrifugation at 100 × g for 3 min, washed, and resuspended in PBS, and cell counting was performed using a hemocytometer to obtain approximately 5 × 104 cells/ml. Cell membrane integrity was assayed by trypan blue exclusion. The presence of endogenous A. actinomycetemcomitans was checked both by culture and by PCR (5, 7). Only BECs that were intact and negative for the presence of A. actinomycetemcomitans were used in in vitro cell binding assays.
Oral epithelial cell binding assay.
The binding assay has been described previously (5, 7). Briefly, 200 μl of bacteria, either A. actinomycetemcomitans or E. coli, at a concentration of 5 × 108/ml was added to 200 μl of BECs contained in a 1.5-ml microcentrifuge tube to attain a ratio of 10,000 to 1 (A. actinomycetemcomitans to BEC). To determine A. actinomycetemcomitans binding, the tube containing the mixture of A. actinomycetemcomitans and BECs was rotated at 20 rpm at 37°C for 90 min. For E. coli binding, the tube was rotated at 20 rpm at 4°C overnight. Two hundred microliters of the mixture of bacteria and BECs was placed on top of 10 ml of 5% Ficoll 400 in PBS contained in a 15-ml centrifuge tube. The tube was centrifuged at 100 × g for 5 min to separate the unbound bacteria from the heavier BECs, which pelleted to the bottom of the tube. The supernatant was removed carefully by pipetting. The pellet was resuspended in an equal volume of PBS. Serial dilutions of the resuspension was plated on Trypticase soy agar plates for A. actinomycetemcomitans and on LB plates for E. coli to calculate numbers of bacterial cell CFU/BEC. Background controls included bacteria without BECs and BECs without bacteria added. Binding assays were performed in duplicate on at least three separate occasions. Intactness and viability of BECs were evaluated by light microscopy. More than 95% of the cells were intact and nonviable as determined by trypan blue exclusion.
Preparation of ApiA antiserum.
A chemically synthesized peptide with 15 amino acid residues (INENKKDIAINKANC) was selected as the ApiA antigen in order to generate antiserum from rabbits and was based on sequence analysis with blastp (NCBI) and PCGENE (Sigma-Genosys). The N-terminal 14 amino acid residues are located between the 55th and the 69th amino acid residues of the ApiA protein sequence (accession no. AB064943; NCBI). An extra cysteine was linked to the C-terminal end of the 14-mer peptide to facilitate the conjugation of keyhole limpet hemocyanin as the carrier protein. The keyhole limpet hemocyanin-conjugated peptide was used to inoculate New Zealand White rabbits, and the resulting serum was tested for immunoreactivity and specificity using the Western blotting and enzyme-linked immunosorbent assay methods.
Western blot analysis.
E. coli strain DH5α containing pJK654 (apiA+) or pJAK16 vector alone or pJAK16 apiA mutant was stimulated by addition of 1 mM IPTG for 3 h. In addition, JK1051 harboring pJK654 (apiA+) or pJAK16 vector alone or pJAK16 apiA mutant was treated with 1 mM IPTG for 3 h. Three ml of the bacteria containing approximately 3 × 109 cells were centrifuged at 1,500 × g for 5 min. The pellet was resuspended in 50 ml of distilled water, and 10 ml of 6× bacterial lysis buffer was added to the cell suspension to attain a 1× concentration of bacterial lysis buffer (50 mM Tris-Cl, pH 6.8, 100 mM dithiothreitol, 2% sodium dodecyl sulfate, 0.1% bromphenol blue, and 10% glycerol). After boiling for 5 min, the cell suspension was centrifuged at 15,000 × g for 5 min. The supernatant was loaded on a 10% sodium dodecyl sulfate-polyacrylamide gel for electrophoresis. Proteins in the gel were transferred to a 0. 2 mm nitrocellulose membrane (Millipore), and 5% bovine serum albumin was used to block nonspecific binding. ApiA was interrogated with 1:1,000-diluted ApiA antiserum. Alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) was used as the secondary antibody, and the reaction was developed by using the BCIP/NBT system (Bio-Rad).
Assay for blocking ApiA binding by ApiA antiserum.
Bacteria were prepared for the binding assay as follows. After two washes with PBS, bacterial strains were suspended in PBS and anti-ApiA antiserum was added to the bacterial suspension in a single dose of 1:25 (vol/vol). A dose-dependent anti-ApiA antiserum evaluation was performed using a series of doses ranging from 1:10 to 1:50 and 1:200. The mixture of bacteria and antiserum was rotated at 20 rpm for 1 h at 37°C, followed by centrifugation at 1,500 × g for 5 min. The supernatant containing unbound antibodies was removed, and the pellet was resuspended in PBS in preparation for the binding assay. Controls consisting of addition of preimmune serum at either 1:10 or 1:25 were prepared. The binding assay was performed as described previously, in this instance pretreating the bacteria to be tested with the preimmune sera and/or the active anti-ApiA antiserum.
Assay of autoaggregation.
The autoaggregation assay was modified from the procedures previously reported (3, 12, 13). Briefly, E. coli containing the apiA expression plasmids (pJK654), the aae expression plasmids (pVK43), or the empty vector alone (pJAK16) was cultured in preparation for the E. coli cell clumping and sedimentation assays. The cell clumping assay was performed as follows. After stimulation with IPTG, a 2.5-ml cell suspension was transferred to a 10-ml polypropylene tube. Cells were stained with ethidium bromide and excited with UV light to visually observe cell clumping. In the sedimentation assay, cells were suspended in a quartz cuvette (Perkin-Elmer) and monitored at a spectrum of 590 nm, and time-dependent sedimentation was recorded by noting the optical intensity with a UV-VIS spectrophotometer (UV mini 1240; Shimadzu).
A. actinomycetemcomitans colony phenotype and biofilm formation.
A. actinomycetemcomitans growth and colony morphology were examined as described previously (12, 13). Briefly, A. actinomycetemcomitans was grown on solid AAGM agar, and morphology was observed by means of transmitted light microscope. Colony morphology was recorded as rough textured or smooth textured. Biofilm formation was evaluated using a biofilm detachment assay (12, 13). Briefly, A. actinomycetemcomitans strains were adjusted to about 108 ml−1, inoculated into 96-well microtiter plates, and incubated at 37°C in 10% CO2 for 48 h. The plates were washed with PBS twice and stained with 200 μl crystal violet (Fisher) for 2 min. Plates were rinsed with tap water, and 200 μl of ethyl alcohol was added to each well, after which plates were scanned using a microplate reader (Bio-Rad) at 590 nm. Optical intensities were recorded and represented the biofilm formation that was found attached to the wells of the microtiter plate.
Statistical analysis.
Multiple group comparisons were assayed with one-way analysis of variance, followed by posthoc testing with the Student-Newman-Keuls test or the Bonferroni correction for selected pairwise comparisons. Significant differences were recorded only if they achieved a level of P values of <0.05.
RESULTS
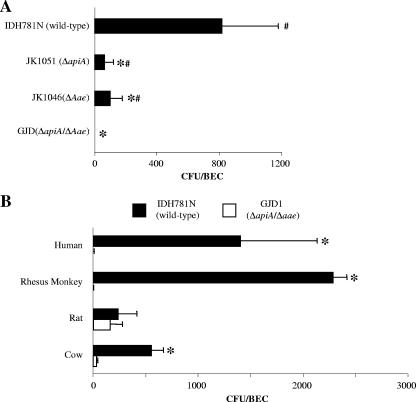
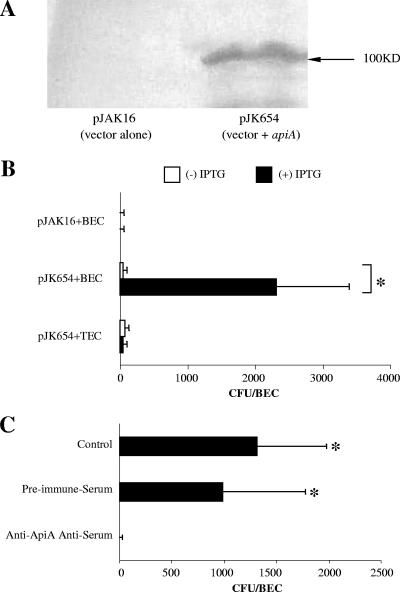
Previous studies showed that wild-type A. actinomycetemcomitans (IDH781) bound to BECs derived from humans, Old World primates, rats, and cows (7). Further, it was shown that the outer membrane adhesin Aae played a prominent role in attachment of A. actinomycetemcomitans to BECs obtained from humans and Old World primates (7). In this study, we set out to determine if a second outer membrane adhesin, ApiA, also known as Omp100, played a role in binding of A. actinomycetemcomitans to BECs obtained from humans and other mammals. Western blot analysis using rabbit antiserum developed to the ApiA peptide showed that ApiA was expressed in wild-type A. actinomycetemcomitans, strain IDH781N, but not in the apiA mutant strain JK1051 (Fig. 1A, left panel) and that a mutation in this adhesin affected binding to BECs (Fig. 1B and C). Further, complementation of the mutant strain with pJK654 (apiA+) restored the ability of JK1051 to express ApiA (Fig. 1A, right panel). The expressed adhesin migrated with an apparent molecular mass of 100 kDa (Fig. 1A).
FIG. 1.
A. actinomycetemcomitans binding to mammalian BECs as mediated by ApiA. (A) Western blot identification of ApiA expression in various strains. From left to right: wild-type A. actinomycetemcomitans IDH781N versus JK1051; JK1051 with pJAK16 (empty vector) versus pJK654 (complemented JK1051 with pJAK16 [apiA+]). Anti-ApiA antisera was used to detect ApiA expression. (B) Binding of wild-type IDH781N and the apiA mutant strain to BECs from various mammalian species. Mean number of bacterial cells per BEC was obtained from at least three assays; error bars indicate standard deviations. Solid bars show wild-type strain IDH781N; open bars show apiA mutant strain (the asterisk indicates a P value of <0.05). (C) Effect of anti-ApiA antisera and genetic complementation of apiA on binding to BECs. Antisera reduces binding in wild-type IDH781 (top two bars). Binding of apiA mutant JK1051 harboring plasmid pJAK16 (vector) or pJK654 (vector plus apiA) to human BECs showing no effect (middle bars). pJK654 complemented JK1051 and antisera to ApiA reduced binding (bottom two bars). Complemented strain showed a reduction in binding similar to that of the wild-type strain when treated with anti-ApiA antiserum (compare IDH781N and JK1051 to JK1051 and pJK654). #, P < 0.05, JK1051 harboring pJK654 versus JK1051 harboring pJAK16; *, P < 0.05, ApiA antiserum versus preimmune serum control. (D) Dose-dependent effects of anti-ApiA antisera on IDH781N binding to human BECs. Presented as a percentage of binding compared to results for IDH781N treated with the preimmune sera.
Binding experiments using wild-type IDH781N confirmed that A. actinomycetemcomitans bound to human, rhesus monkey, and rat BECs but did not attach to BECs derived from New World primates (Fig. 1B). Moreover, the IDH781N apiA mutant strain (JK1051) exhibited significantly reduced binding to human and rhesus monkey BECs (Fig. 1B; P < 0.05). On the other hand, there was no significant difference in binding of the apiA mutant strain JK1051 versus IDH781N to rat BECs. JK1051 cells harboring pJK654 (apiA-complemented strain) exhibited significantly more binding to human BECs (354 ± 104/BEC) than JK1051 cells harboring pJAK16 (the empty plasmid), which yielded a background of 21 ± 11/BEC (Fig. 1C; P < 0.05). These results suggested that pJK654 partially complemented the apiA mutation in JK1051. Pretreatment of A. actinomycetemcomitans with ApiA-specific antiserum prior to binding experiments significantly reduced BEC binding of the wild-type strain IDH781N cells as well as the complemented strain JK1051 cells harboring pJK654, as seen in Fig. 1C (P < 0.05). These results were further substantiated by demonstrating that these pretreatments reduced IDH781N binding to human BECs in a dose-dependent manner (Fig. 1D). However, pretreatment of wild-type A. actinomycetemcomitans with anti-ApiA antiserum did not result in total elimination of BEC binding (Fig. 1D). This finding was not unexpected, since the anti-ApiA antiserum does not cross-react with Aae, therefore enabling the pretreated IDH781N to continue to bind, since Aae, the other A. actinomycetemcomitans adhesin, also has specificity for BECs.
In experiments in which the A. actinomycetemcomitans apiA mutant strain JK1051 was complemented, we found that BEC binding was reduced compared to that for wild-type IDH781, suggesting that only partial complementation occurred (Fig. 1C). This reduced binding could be due to either amino acid sequence differences between IDH781 and HK1651 (the sequenced strain of A. actinomycetemcomitans used for our PCR primers) or chloramphenicol selection required to maintain the complementing plasmid. The HK1651 substitution has been done routinely in our laboratory in previous complementation studies with no incident (7, 14). On the other hand, choloramphenicol selection did in fact reduce BEC binding in the complemented strain, JK1051, containing pMB7, an empty plasmid with a chloramphenicol resistance gene, but had no effect on cell growth rate (data not shown).
An apiA aae double-mutant strain (named GJD1) was constructed and tested for binding to BECs and was found to completely abrogate binding to human and Old World primate BECs (Fig. 2A). In comparison to binding of GJD1 to human and Old World primate BECs, binding of GJD1 to rat BECs was slightly but not significantly reduced, while binding to BECs obtained from the mouths of cows was significantly reduced but not abrogated (Fig. 2B) (P < 0.05). As for the single mutations, the binding of JK1051 (apiA mutant strain) to human and Old World primate BECs, although significantly reduced, was still detectable. This was also true when the binding of JK1046 (aae mutant strain) was examined (Fig. 2A).
FIG. 2.
Binding of IDH781, apiA, aae, or apiA aae double mutant strain GJD1 to BECs. (A) Binding of wild-type strain IDH781N; apiA mutant JK1051; aae mutant JK1046; or apiA aae double mutant GJD1 to human BECs. Bars represent the mean; error bars represent the standard deviation. The asterisk indicates a P value of <0.05, versus IDH781; #, P < 0.05, versus GJD1. (B) Binding of wild-type strain IDH781N or the apiA aae double mutant GJD1 to BECs obtained from four mammalian species. *, P < 0.05, IDH781N versus GJD1.
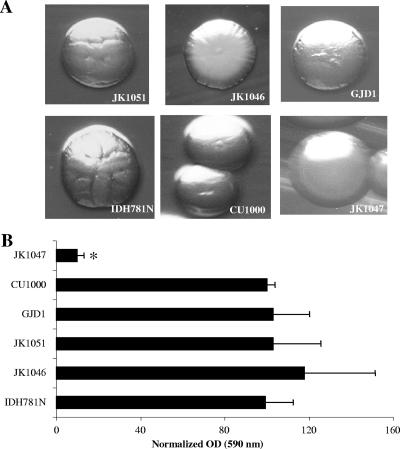
To determine the effect of these gene deletions on the A. actinomycetemcomitans physiology, we compared the growth rate, colonial morphology, and biofilm formation in the wild-type and deletion mutant strains. No differences were seen in terms of the growth rate characteristics of the strains tested (data not shown). IDH781N, CU1000, JK1051, JK1046, and GJD1 colonies all appear to possess the typical rough-bordered wild-type A. actinomycetemcomitans colony morphology. JK1047, an flp-1 knockout mutant, is an A. actinomycetemcomitans strain that has the smooth-colony morphology (6), and in these experiments it was used as a control to visualize the smooth phenotype (Fig. 3A —observe the smooth border in bottom right subpanel). To assess biofilm formation, a 96-well microtiter plate assay was used as described previously (7). A. actinomycetemcomitans strains IDH781N, JK1051, JK1046, GJD1, and CU1000 showed no differences in attachment to the abiotic surface of the polystyrene plate. JK1047 (flp-1 mutant) was used as a negative control and showed minimal attachment to the 96-well plate (12, 13) (see Fig. 3B). Taken together, these results indicate that apiA, aae, and the double mutant strain did not demonstrate changes in growth rate, colony morphology, and biofilm formation.
FIG. 3.
Phenotypic characterization of wild-type and mutant strains. (A) Colonial morphology of various strains of A. actinomycetemcomitans. Strains JK1051 (apiA), JK1046 (aae), GJD1 (apiA aae), IDH781N (wild type), CU1000 (wild type), and JK1047 (flp-1). (B) Biofilm formation of wild-type and mutant strains. Data are presented as normalized optical densities (OD) using IDH781N as the standard. Binding values are presented as a percentage of the IDH781N OD, which represents biofilm binding to polystyrene. Results for all strains are compared to the OD of wild-type strain IDH781N. Only the JK1047 (flp mutant) showed reduced biofilm binding.
To further investigate the role of ApiA, we expressed A. actinomycetemcomitans apiA in E. coli strain DH5α (Invitrogen) (Fig. 4). Western blot analysis of E. coli transformed with plasmid pJK654 displayed a specific band detected by anti-ApiA antisera which showed a molecular mass of 100 kDa (Fig. 4A). Further, DH5α/pJK654 cells pretreated with anti-ApiA antiserum exhibited significantly decreased binding to human BECs compared to cells pretreated with preimmune serum (Fig. 4C) (P < 0.05). Moreover, DH5α/pJK654 cells exhibited significantly increased binding to human BECs compared to the binding exhibited by DH5α/pJAK16 (empty plasmid) (Fig. 4B) (P < 0.05). In comparison, DH5α/pJK654 cells did not bind to human tongue epithelial cells.
FIG. 4.
Expression of ApiA in E. coli. (A) Western blot analysis of extracts from E. coli harboring pJAK16 (vector alone) or pJK654 (vector + apiA). Blots were probed with rabbit antiserum raised to ApiA. (B) Binding of E. coli cells harboring pJAK16 or pJK654 to human BECs and human tongue epithelial cells (TECs). E. coli cells were grown in medium containing 0 or 1 mM IPTG. The asterisk indicates a P value of <0.05, pJK654 (vector + apiA) versus pJAK16 (vector alone). (C) Binding of E. coli cells harboring pJK654 to human BECs. Bacterial cells were pretreated with preimmune serum or anti-ApiA antiserum prior to the binding assay. The asterisk indicates a P value of <0.05, versus the anti-ApiA antiserum pretreatment group.
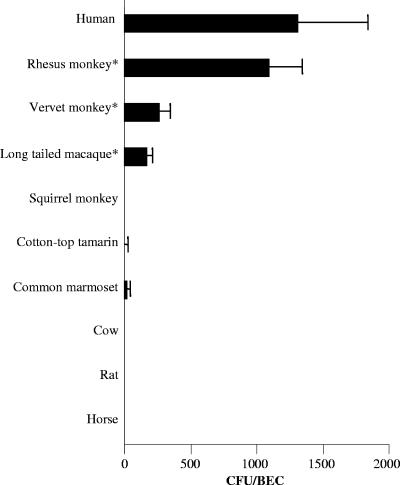
We also measured the binding of DH5α/pJK654 to BECs isolated from three Old World primates, three New World primates, and three nonprimate mammals (Fig. 5). Significantly more DH5α/pJK654 cells bound to BECs from Old World primates than to BECs from New World primates, although in one of three experiments, minimal binding was found in the common marmoset. DH5α/pJK654 cells did not bind at detectable levels to rat, cow, or horse BECs.
FIG. 5.
Binding of E. coli cells harboring ApiA expression plasmid pJK654 to BECs isolated from mammalian species. Bars represent means of triplicate assays, and error bars show standard deviations. E. coli bound only to Old World primate BECs; however, in one of three assays the common marmoset showed minimal binding.
Preliminary experiments demonstrated that E. coli expressing Aae initially bound to BECs at a concentration of 5 × 105 CFU/ml, whereas no binding of E. coli expressing ApiA was detected until E. coli expressing ApiA was added at a concentration of 5 × 106 CFU/ml (P < 0.05). These experiments demonstrated that the Aae adhesin permits initial binding at a lower concentration of cells added and suggest that ApiA has a lower affinity for human BECs than Aae; however, more experiments are required to confirm this suggestion (7). In contrast, with a concentration above 5 × 107 CFU/ml added, E. coli expressing ApiA bound more CFU per BEC than E. coli expressing Aae, implying that cell-to-cell autoaggregation could play a role in these results.
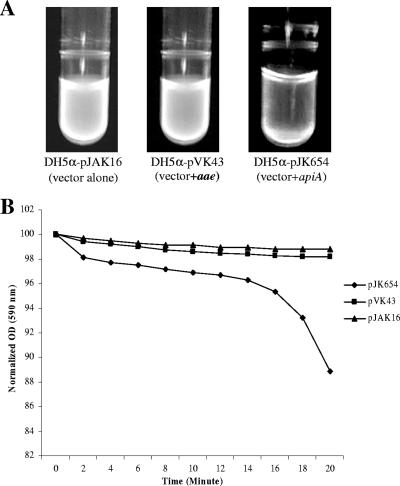
In an effort to better understand the ApiA adhesin phenotype in terms of its autoaggregating ability, we chose to examine ApiA expression in E. coli where other confounding attachment characteristics, as seen in wild-type A. actinomycetemcomitans, could be eliminated (Fig. 6). Experiments demonstrated that 1 h after a vigorous vortex agitation, DH5α/pJK654 cells formed autoaggregative clumps and left a relatively clear supernatant above the sedimented cells. Cell suspensions of DH5α/pJAK16 and DH5α/pVK43 (expressing Aae) appeared turbid and homogeneous in comparison to ApiA-expressing E. coli (Fig. 6A). When the cells were suspended in a quartz cuvette and monitored by light at 590 nm, time-dependent sedimentation was recorded for ApiA (Fig. 6B). Unlike the case with wild-type A. actinomycetemcomitans, autoaggregation by E. coli was easily dispersed by vortex agitation.
FIG. 6.
Autoaggregation of E. coli expressing ApiA and Aae. (A) E. coli strain harboring pJAK16 (vector), pJK654 (apiA+), or pVK43 (aae+) is seen in a visual autoaggregation assay. Cells were stained with ethidium bromide and excited by a UV light source. The cells harboring the apiA gene formed clumps that settled to the bottom of the tube, while the other strains remained in a homogeneous suspension. (B) Plot of the optical density of cell suspensions of E. coli expressing ApiA and Aae and an E. coli vector control. A 1-ml cell suspension was added to a quartz cuvette and monitored by light at 590 nm. The optical density (OD) was recorded at 2-min intervals. Data are presented as a percentage of the OD seen at zero time, where the cell suspension was homogeneous. Only the cells expressing ApiA (pJK654) sedimented.
DISCUSSION
The results obtained in these experiments are based on in vitro analysis of binding of A. actinomycetemcomitans to BECs derived from a range of mammalian species. The assay employed in this study has been used to evaluate binding of a wide variety of human pathogens, such as Neisseria meningitidis, E. coli, and pseudamonads, with human BECs as the target tissue (26, 29, 31). More recently the assay has been adapted for use with epithelium obtained from tissue sites better suited for evaluating interactions between bacteria and mammalian cells obtained from the urinary bladder (24) or vaginal epithelium (2). The binding assay used to study these interactions has several practical and research design advantages, one of which is that it can be used to evaluate adhesin/receptor relationships in intact but nonviable cells. Use of intact but dead cells provides a clear advantage when a distinction between bacterial attachment and invasion is required. This distinction is particularly true when a bacterium that attaches also has the ability to invade (18), since the process of internalization requires viable host cells (8). In these evaluations and in past studies using this assay, more than 95% of the BECs had normal intact morphology based on microscopic evaluation (7) and did not harbor A. actinomycetemcomitans or other pathogens (5) but were intact and permeable to trypan blue (data not shown). As a result, we are confident that this assay is capable of providing results that replicate mammalian/bacterial cell surface attachment, where adhesin/receptor interactions come into play (10). This conclusion is validated by the fact that the mammals that harbor A. actinomycetemcomitans in nature are the same mammals that demonstrate BEC binding of A. actinomycetemcomitans in vitro (4).
With that in mind, our findings indicate that ApiA and Aae are two prominent adhesins that mediate in vitro attachment of A. actinomycetemcomitans to BECs derived from humans and Old World primates. Evidence indicates that the primary reservoir for this organism is the oral cavity of humans (19), although A. actinomycetemcomitans has been isolated as an infectious contaminant from sites distant from the oral cavity in subacute bacterial endocarditis, pneumonia, brain abscesses, and kidney failure (30). In addition to humans and Old World primates, A. actinomycetemcomitans has also been isolated from the oral cavity of cows and rats, which taken together comprise the known natural host range for A. actinomycetemcomitans (4, 7). The results derived from this study suggest that both ApiA and Aae are required for efficient binding of A. actinomycetemcomitans to human BECs and confirm the findings of others that indicate that both Aae and ApiA function as outer membrane protein adhesins (1, 7, 17).
In contrast to data derived from human and monkey BECs, neither ApiA nor Aae appear to be the main adhesins responsible for the specific binding of A. actinomycetemcomitans to BECs derived from rats. Previously we demonstrated that an aae mutant had little if any effect on binding of A. actinomycetemcomitans to rat or cow BECs (7). In experiments reported in this article, the apiA mutant and the apiA aae double mutant reduced binding to rat BECs, but only minimally. Moreover, the apiA aae double mutant caused a significant decrease but did not abrogate binding to cow BECs. However, as mentioned, the double mutant did eliminate binding to BECs derived from humans and Old World primates. These results suggest that although the ApiA adhesin may have some effect on binding to cow BECs, other as yet unidentified adhesins may be present in cows as well as rats.
Furthermore, with respect to E. coli expression of ApiA and Aae, binding studies showed that a higher concentration of E. coli expressing ApiA is required to exhibit initial detectable binding compared to results for E. coli expressing Aae. This result could be due to differences in E. coli protein expression, or alternatively Aae expression may exhibit higher affinity for BECs. The latter hypothesis is supported by data that demonstrate that an A. actinomycetemcomitans Aae mutant requires a higher concentration of cells per BEC than its wild-type strain to achieve detectable levels of attachment, suggesting that Aae has a higher affinity for BECs, although more work needs to be done in this area (7). In contrast, at higher concentrations of bacterial cells added to BECs, the total numbers of CFU/BEC required were higher for E. coli expressing ApiA than for E. coli expressing Aae. These results suggest that autoaggregation was involved in E. coli expressing ApiA binding to BECs (17). The autoaggregative property of ApiA expressed in E. coli was further demonstrated by examining its ability to sediment as early as 20 min after vigorous vortex agitation (Fig. 6), a property that was not displayed in E. coli expressing Aae.
In conclusion, based on the data reported, we now can say convincingly that ApiA plays an important role in modulating A. actinomycetemcomitans binding to human and Old World primate BECs (7). Moreover, we are the first to report that a double knockout of aae and apiA completely abrogates the binding of A. actinomycetemcomitans to BECs from human and Old World primate BECs. If in fact, BECs provide A. actinomycetemcomitans with a protected domain in the oral cavity (23), then the data identifying the importance of the action of these two adhesins could have significant clinical and therapeutic implications.
Although ApiA and Aae are both autotransporter proteins, the surface-exposed domains of ApiA and Aae are not homologous. It is interesting to note that A. actinomycetemcomitans cells produce leukotoxin, a member of the RTX family of secreted toxins (9). As in the case of both ApiA and Aae, Leukotoxin interacts with PMNs and macrophages derived from humans and Old World primates but not with those derived from New World primates or other nonprimate mammals (27). It is clear that these surface-expressed proteins each have different host receptors. These findings suggest that the evolution of host specificity and host cell tropism in A. actinomycetemcomitans has occurred through a complicated process of natural selection resulting from multiple cell surface interactions (20, 25).
Acknowledgments
We extend our thanks Larry Katz of Rutgers and to Eva Ryden from UMDNJ for their help in obtaining buccal cells from the mammals used for this study. We also thank Galadriel Hovel-Miller and David Figurski from Columbia University for providing plasmid pGMH491. Kabilan Velliygounder is also acknowledged for his help with the earlier aspects of the study and for all his help during the study.
Our appreciation is extended to the National Institutes of Dental and Craniofacial Research for providing support in the form of grant no. DE-016306 to D.H.F.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 9 July 2007.
REFERENCES
- 1.Asakawa, R., H. Komatsuzawa, T. Kawai, S. Yamada, R. B. Goncalves, S. Izumi, T. Fujiwara, Y. Nakano, N. Suzuki, Y. Uchida, K. Ouhara, H. Shiba, M. A. Taubman, H. Kurihara, and M. Sugai. 2003. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 50:1125-1139. [DOI] [PubMed] [Google Scholar]
- 2.Boris, S., J. E. Suartez, F. Vasquez, and C. Barbes. 1998. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect. Immun. 66:1985-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Re, B., B. Sgorbati, M. Miglioli, and D. Palenzona. 2000. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 31:438-442. [DOI] [PubMed] [Google Scholar]
- 4.Eke, P. I., L. Braswell, R. Arnold, and M. Fritz. 1993. Sub-gingival microflora in Macaca mulatta species of rhesus monkey. J. Periodont. Res. 28:72-80. [DOI] [PubMed] [Google Scholar]
- 5.Fine, D. H., and D. Furgang. 2002. Lactoferrin iron levels affect attachment of Actinobacillus actinomycetemcomitans to buccal epithelial cells. J. Periodontol. 73:616-623. [DOI] [PubMed] [Google Scholar]
- 6.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 7.Fine, D. H., K. Velliyagounder, D. Furgang, and J. B. Kaplan. 2005. The Actinobacillus actinomycetemcomitans autotransporter adhesin Aae exhibits specificity for buccal epithelial cells from humans and old world primates. Infect. Immun. 73:1947-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay, B. B., F. Heffron, and S. Falkow. 1989. Epithelial cell surfaces induce Salmonella proteins required for bacterial adherence and invasion. Science 243:940-943. [DOI] [PubMed] [Google Scholar]
- 9.Frey, J., and P. Kuhnert. 2002. RTX toxins in Pasteurellaceae. Int. J. Med. Microbiol. 292:149-158. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons, R. J., and J. van Houte. 1971. Selective bacterial adherence to oral epithelial surfaces and its role as an ecological determinant. Infect. Immun. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haubek, D., K. Poulsen, S. Asikainen, and M. Kilian. 1995. Evidence for absence in northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 33:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan, J. B., M. F. Meyenhofer, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan, J. B., K. Velliyagounder, C. Ragunath, H. Rohde, D. Mack, J. K. Knobloch, and N. Ramasubbu. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186:8213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsuzawa, H., R. Asakawa, T. Kawai, K. Ochiai, T. Fujiwara, M. A. Taubman, M. Ohara, H. Kurihara, and M. Sugai. 2002. Identification of six major outer membrane 146 proteins from Actinobacillus actinomycetemcomitans. Gene 288:195-201. [DOI] [PubMed] [Google Scholar]
- 16.Lamell, C. W., A. L. Griffen, D. L. McClellan, and E. J. Leys. 2000. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J. Clin. Mirobiol. 38:1196-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, L., D. Matevski, M. Aspiras, R. P. Ellen, and G. Lepine. 2004. Two epithelial cell invasion-related loci of the oral pathogen Actinobacillus actinomycetemcomitans. Oral Microbiol. Immunol. 19:16-25. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, D. H., J. E. Lippmann, and P. M. Fives-Taylor. 1996. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect. Immun. 64:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller, H.-P., P. Eickholz, A. Heinecke, S. Pohl, R. F. Müller, and D. E. Lange. 1995. Simultaneous isolation of Actinobacillus actinomycetemcomitans from subgingival and extracrevicular locations of the mouth. J. Clin. Periodontol. 22:413-419. [DOI] [PubMed] [Google Scholar]
- 20.Nei, M., P. Xu, and G. Glazko. 2001. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc. Natl. Acad. Sci. USA 98:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nørskov-Lauritsen, N., and M. Kilian. 2006. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int. J. Syst. Evol. Microbiol. 56:2135-2146. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes, E. R., A. P. Tomaras, G. McGillivary, P. L. Connerly, and L. A. Actis. 2005. Genetic and functional analyses of the Actinobacillus actinomycetemcomitans AfeABCD siderophore-independent iron acquisition system. Infect. Immun. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis from subgingival and extracrevicular locations of the mouth. Infect. Immun. 71:2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakarya, S., G. Turncer Ertem, S. Oncu, I. Kocak, N. Erol, and S. Oncu. 2003. Eschericia coli bind to urinary bladder epithelium through nonspecific sialic acid mediated adherence. FEMS Immun. Med. Microbiol. 39:45-50. [DOI] [PubMed] [Google Scholar]
- 25.Schrago. C. G., and C. A. Russo. 2003. Timing the origin of New World monkeys. Mol. Biol. Evol. 20:1620-1625. [DOI] [PubMed] [Google Scholar]
- 26.Simpson, W. A., D. L. Hasty, and E. H. Beachy. 1985. Binding of fibronectin to human buccal epithelial cells inhibits the binding of type 1 fimbriated Eschericia coli. Infect. Immun. 48:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taichman, N. S., D. L. Simpson, S. Sakurada, M. Cranfield, J. DiRienzo, and J. Slots. 1987. Comparative studies on the biology of Actinobacillus actinomycetemcomitans leukotoxin in primates. Oral Microbiol. Immunol. 2:97-104. [DOI] [PubMed] [Google Scholar]
- 28.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trust, T. J., R. M. Gillespie, A. R. Bhatti, and L. A. White. 1983. Differences in the adhesive properties of Neisseria meningitidis for human buccal epithelial cells and erythrocytes. Infect. Immun. 41:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Winkelhoff, V. J., and J. Slots. 1999. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in nonoral infections. Periodontol. 2000 20:122-135. [DOI] [PubMed] [Google Scholar]
- 31.Woods, D. E., D. C. Straus, W. G. Johanson, V. K. Berry, and J. A. Bass. 1980. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect. Immun. 29:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]