Abstract
Human immunodeficiency virus type 1 (HIV-1) infection of individuals carrying the two alleles of the CCR5Δ32 mutation (CCR5−/−) has rarely been reported, but how the virus overcomes the CCR5Δ32 protective effect in these cases has not been delineated. We have investigated this in 6 infected (HIV+) and 25 HIV− CCR5−/− individuals. CD4+ T lymphocytes isolated from HIV− CCR5−/− peripheral blood mononuclear cells (PBMCs) showed lower levels of CXCR4 expression that correlated with lower X4 Env-mediated fusion. Endogenous CCR5Δ32 protein was detected in all HIV− CCR5−/− PBMC samples (n = 25) but not in four of six unrelated HIV+ CCR5−/− PBMC samples. Low levels were detected in another two HIV+ CCR5−/− PBMC samples. The expression of adenovirus 5 (Ad5)-encoded CCR5Δ32 protein restored the protective effect in PBMCs from three HIV+ CCR5−/− individuals but failed to restore the protective effect in PBMCs isolated from another three HIV+ CCR5−/− individuals. In the latter samples, pulse-chase analyses demonstrated the disappearance of endogenous Ad5-encoded CCR5Δ32 protein and the accumulation of Ad5-encoded CCR5 during the chase periods. PBMCs isolated from CCR5−/− individuals showed resistance to primary X4 but were readily infected by a lab-adapted X4 strain. Low levels of Ad5-encoded CCR5Δ32 protein conferred resistance to primary X4 but not to lab-adapted X4 virus. These data provide strong support for the hypothesis that the CCR5Δ32 protein actively confers resistance to HIV-1 in vivo and suggest that the loss or reduction of CCR5Δ32 protein expression may account for HIV-1 infection of CCR5−/− individuals. The results also suggest that other cellular or virally induced factors may be involved in the stability of CCR5Δ32 protein.
Infection by human immunodeficiency virus type 1 (HIV-1) occurs by the binding of the viral envelope protein gp120 to two proteins on the surfaces of target cells, CD4 and a coreceptor. The coreceptor is almost always a chemokine receptor, typically either CCR5 or CXCR4 (4, 14, 17-20). CCR5 and CXCR4 belong to a large family of seven transmembrane G-protein-coupled receptors. These receptors are characterized by the presence of seven transmembrane domains, four extracellular domains, and four intracellular domains. HIV-1 strains are classified (8) into three major groups based on their coreceptor usage: R5 (CCR5-tropic), X4 (CXCR4-tropic), and R5/X4 (able to use either CCR5 or CXCR4).
A naturally occurring 32-bp deletion in the human CCR5 gene is highly associated with resistance to HIV-1 infection (11, 16, 18, 19, 40). CCR5Δ32 encodes a truncated protein that is not detected on the cell surface and, therefore, is not functional as a coreceptor (11, 16, 18, 19, 40). CCR5Δ32 is common among Caucasians (∼10% allele frequency in North America) but is absent in native African and Asian populations (11, 16, 18, 40). Approximately 1% of Caucasians are homozygous for the mutant CCR5Δ32 allele (30).
The majority of individuals carrying the two alleles of the CCR5Δ32 mutation (CCR5−/−) are highly protected against HIV-1 infection (reviewed in reference 34). Individuals who are heterozygous for the mutant allele (CCR5+/−) are not protected against infection, but once infected, their progression to AIDS is delayed (11, 16, 18, 31, 38, 40), indicating that partial resistance can occur in the presence of a single copy of CCR5Δ32. In rare cases, CCR5Δ32 homozygosity was associated with HIV-1 infection (6, 9, 23, 29, 32, 35, 37, 38) but, in these cases, the mechanism of infection has not been defined. In most cases, exclusive use of CXCR4 by virus isolates or the presence of env sequences typical of X4 viruses was observed. The isolation of dual-tropic (R5/X4) HIV-1 from infected individuals homozygous for the CCR5Δ32 allele has also been reported (23, 24, 33).
Our previous work suggested that HIV resistance in CCR5Δ32 homozygotes may result from both genetic loss of CCR5 on the cell surface and active downregulation of CXCR4 expression by the mutant CCR5Δ32 protein. We and others have demonstrated that the CCR5Δ32 protein may form heterodimers with wild-type CCR5 and CXCR4 which are retained in the endoplasmic reticulum and result in reduced cell surface expression of the coreceptors (2, 7, 12, 39). We have previously demonstrated the absence of detectable CCR5Δ32 protein in two infected CCR5−/− individuals (2). Here, we have examined the mechanism of the failure of the CCR5Δ32 protective genotype in six HIV-infected CCR5−/− individuals. In particular, we have examined expression of the CCR5Δ32 and CXCR4 proteins in six HIV+ CCR5−/− individuals and analyzed the effect of providing intracellular recombinant CCR5Δ32 protein on HIV-1 Env-mediated fusion and virus infection. The results suggest that the expression and stability of the CCR5Δ32 protein are critical for resistance to HIV-1 infection.
MATERIALS AND METHODS
Cells and viruses.
All the cell lines used in the present study were purchased from the American Type Culture Collection (Rockville, MD). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Quality Biologicals, Gaithersburg, MD) containing 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 2 mM l-glutamine, and antibiotics. Peripheral blood mononuclear cells (PBMCs) from all donors were activated with phytohemagglutinin (PHA) (10 μg/ml) (Sigma Chemicals, St. Louis) for 3 days before any experiment. All HIV viruses were obtained from the AIDS Reagent Program, NIAID.
PBMC samples.
A list of the PBMC samples utilized in the present study is summarized in Table 1. The clinical profile of patient 1 has been described previously (9, 33). Patient 2 and patient 1 are two CCR5−/− individuals. Patient 2 is uninfected, while patient 1 is infected and the mode of infection appears to be via homosexual intercourse. The three HIV+ SEROCO samples are all from CCR5−/− homozygous individuals who are homosexuals. Two of the SEROCO samples have not been described previously. SEROCO sample 2 (SEROCO 2) has been previously reported (38). The Shep 164 sample has been previously documented (37) and examined in our previous study (2). The Multicenter AIDS Cohort Study (MACS)-infected CCR5−/− homozygous sample (identification no. 20705) has been described previously (23). The other HIV− CCR5−/−, CCR5+/−, and CCR5+/+ samples were obtained from MACS.
TABLE 1.
List of PBMC samples used in this studya
| PBMC sample | CCR5 genotype | HIV status | Route of transmission | Virus isolated | CD4b counts | Reference |
|---|---|---|---|---|---|---|
| Patient 1 | −/− | HIV+ | Homosexual activity | R5/X4 | 440 | 33 |
| Shep 164 | −/− | HIV+ | Homosexual activity | X4 | 221 | 37 |
| SEROCO 1 | −/− | HIV+ | Homosexual activity | ND | 170 | This study |
| SEROCO 2 | −/− | HIV+ | Homosexual activity | X4 | 60 | 38 |
| SEROCO 3 | −/− | HIV+ | Homosexual activity | ND | 486 | This study |
| MACS 1 | −/− | HIV+ | Homosexual activity | R5/X4 | 525 | 23 |
| MACS 2-26 | −/− | HIV− | NA | NA | NA | MACS |
| MACS 27-47 | +/− | HIV− | NA | NA | NA | MACS |
| Patient 2 | −/− | HIV− | NA | NA | 950 | 33 |
ND, not determined; NA, not applicable.
CD4 counts at the time of sample collection.
FACS analysis.
Cells were washed twice in fluorescence-activated cell sorter (FACS) buffer (supplemented with 0.5% FBS and 0.02% sodium azide), resuspended in 100 μl FACS buffer at 107/ml, and incubated with a 1:200 dilution of monoclonal antibodies (purchased from Pharmingen, Inc., San Diego, CA) raised against the different receptors (CD4, CCR5, and CXCR4) at 4°C for 30 min. The cells were first stained with phycoerythrin (PE)-conjugated anti-CCR5 (catalog no. 556042), washed, restained with fluorescein isothiocyanate (FITC)-conjugated anti-CD4 (catalog no. 555346), washed, and finally restained with allophycocyanin (APC)-labeled anti-CXCR4 monoclonal antibodies (catalog no. 555976). Finally, cells were washed twice with phosphate-buffered saline (PBS), resuspended in 500 μl ice-cold FACS buffer, and analyzed in a FACScan cytometer (Becton Dickinson, San Jose, CA).
HIV-1 Env-mediated cell fusion.
The basic features of the fusion assay were developed by using the HIV-1 glycoprotein (Env)-CD4 interaction of two different populations of cells, one expressing CD4 and the other expressing the HIV-Env (3). Target PBMCs (endogenous CD4) were infected with vCB-21R (encoding LacZ under T7 promoter) or Ad5pT7-lacZ. HeLa cells coinfected with vTF7-3 and one of the HIV-1 Envs served as effector cells. After mixing of the effectors and target cell populations and incubation at 37°C for 2.5 h, fusion specificity was measured by β-galactosidase production in a colorimetric lysate assay as described previously (3).
Western blotting.
Cell lines or PBMCs either uninfected or infected with adenovirus type 5 (Ad5)/Δ32 were subjected to immunoblotting to verify the expression of either endogenous or Ad5-encoded CCR5Δ32 protein. Cell lysates were prepared, fractionated on 12.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels, and immunoblotted onto polyvinylidene difluoride membranes (Millipore). After blocking, the membranes were reacted with the Δ32-specific antibodies at 1/100 dilution (2), washed, and incubated with horseradish peroxidase-conjugated secondary antibodies. Protein bands were detected by the addition of substrate.
Pulse-chase analysis.
Pulse-chase experiments were performed with CCR5−/− PBMCs to analyze the expression of endogenous and Ad5-encoded CCR5Δ32 protein. For an analysis of expressed proteins, the CCR5−/− PBMCs were infected with Ad5CCR5 or Ad5 CCR5Δ32 at 30 PFU/cell and incubated for 48 h to allow expression of the recombinant proteins. At the appropriate times after infection, supernatant medium was removed and replaced, sequentially, with methionine-free/cysteine-free DMEM without radiolabel for 30 min and methionine-free/cysteine-free DMEM containing 200 μCi of [35S]methionine and 200 μCi of [35S]cysteine (Amersham Corp.) for 30 min and cells were washed three times with complete DMEM and then reincubated with standard methionine- and cysteine-containing DMEM without radiolabel for 1 or 2 h. Following the chase period of 1 or 2 h, the cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (0.15 M sodium chloride, 1% [wt/vol] deoxycholic acid, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS, 0.01 M Tris-hydrochloride, pH 7.4, 1 mM methionine, 1 mM cysteine, and 10 μg/ml aprotinin containing 0.1% bovine serum albumin [BSA]). The cell lysates were cleared from cell debris by centrifugation and then used for immunoprecipitation with the appropriate antibody. The CCR5 protein was immunoprecipitated using CTC-6 monoclonal antibodies purchased from R&D. The CCR5Δ32 protein was immunoprecipitated by using a previously described polyclonal antibody raised against the last 31 frameshift amino acids in the CCR5Δ32 protein (2).
Following incubation with the antibodies for 2 h on ice, the resulting immune complexes were reacted with protein A-Sepharose beads (Pharmacia) at 4°C for 1 h and then washed three times with RIPA buffer containing 0.1% BSA and one last time with RIPA buffer without BSA. The immunoprecipitated protein samples were suspended in electrophoresis sample buffer and heated for 1 min at 90°C before analysis by SDS-polyacrylamide gel electrophoresis. The gel was then dried and exposed to an X-ray film.
Productive HIV-1 infection assays.
PHA-activated PBMCs isolated from individuals with different CCR5 genotypes were infected with either Ad5, Ad5/Δ32, or Ad5/CCR5 virus vectors at a multiplicity of infection (MOI) of 50 PFU/cell. Infected PBMCs were incubated for 2 days to allow expression of recombinant proteins (CCR5 or CCR5Δ32 protein) and then infected with either IIIB (X4) or Ba-L (R5). The HIV-1 virus was adsorbed for 2 h, and cells were washed three times with PBS and maintained in RPMI 1640 supplemented with 10% FBS and PHA plus interleukin-2. Culture fluid (50 μl) was harvested after cell resuspension every 3 days and replaced with fresh medium. The amount of p24 antigen in the cell-containing supernatants was measured using an enzyme-linked immunosorbent assay (ELISA) kit purchased from DuPont (Wilmington, DE).
RESULTS
CXCR4 surface expression correlates with X4 Env-mediated cell fusion.
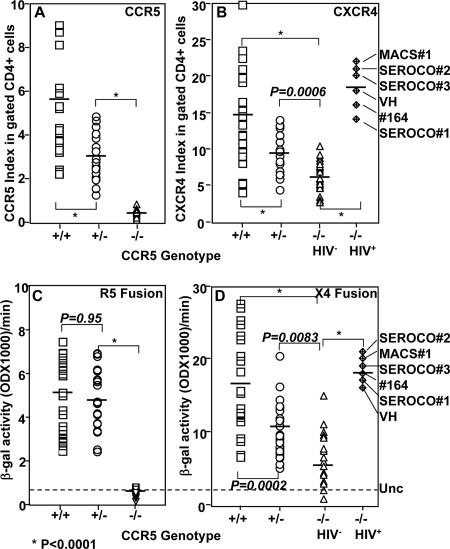
To analyze cell surface expression of the HIV coreceptors in PBMCs, we performed three-color staining (FITC-CD4, PE-CCR5, and APC-CXCR4) using PHA-activated PBMCs isolated from individuals with different CCR5 genotypes. Individuals homozygous for the wild-type CCR5 are referred to as CCR5+/+, those homozygous for the CCR5Δ32 deletion as CCR5−/− to indicate the lack of CCR5 expression, and those heterozygous for CCR5 are designated CCR5+/− to indicate the presence of one allele of wild-type CCR5. We measured the mean fluorescence intensity (MFI) of CXCR4 and CCR5 expression in the CD4+ gated population. Cell surface expression was evaluated as the MFI of CCR5 or CXCR4 expression divided by the MFI value obtained with the corresponding isotype-matched control. The results, expressed as the CCR5 or CXCR4 index, demonstrate efficient expression of CCR5 in CCR5+/+ and CCR5+/− PBMCs and a lack of CCR5 expression in CCR5−/− samples (Fig. 1A). Consistent with the previous data in the literature (reviewed in reference 1), we found that CCR5 expression levels in heterozygous PBMCs were lower than those in PBMCs homozygous for wild-type CCR5. The average value (n = 25) of the CCR5 index was significantly lower in CCR5+/− than in CCR5+/+ samples (Fig. 1A). The CCR5 index calculated for CCR5−/− PBMCs served as a negative control for the background staining in this three-color staining (Fig. 1A).
FIG. 1.
Analysis of coreceptor expression and Env-mediated fusion of PHA-activated PBMCs isolated from CCR5+/+ (+/+), CCR5+/− (+/−), or CCR5−/− (−/−) individuals (n = 25/genotype). The PBMC samples were washed twice with PBS and then sequentially stained with PE-conjugated anti-CCR5 monoclonal antibody (3A9) or its isotype-matched control; washed and then stained with FITC-conjugated anti-CD4 monoclonal antibody or its isotype-matched control; and washed and then stained with APC-labeled anti-CXCR4 monoclonal antibody or its isotype-matched control. The MFIs of CCR5 and CXCR4 expression were measured by gating on the CD4+ population (A and B). The coreceptor expression index represents the MFI of the coreceptor staining divided by the MFI value obtained with the isotype control. (C and D) HIV-1 Env-mediated fusion with PBMC samples stained for the analysis shown in panels A and B. PHA-activated PBMCs expressing T7-lacZ were mixed with Env-expressing cells containing T7 RNA polymerase and incubated for 2.5 h at 37°C. The ability of the PBMCs to undergo cell fusion with R5 (C) or X4 (D) Env was quantified by measuring β-galactosidase (β-gal) produced. The broken line indicates the average background value obtained with the uncleaved Env (Unc) control. The indicated P values were calculated using the Student t test. The identities of the HIV+ CCR5−/− samples are indicated on the right side.
The CXCR4 index calculated for the different samples indicated that all samples tested positive for CXCR4 expression. However, the calculated CXCR4 indexes for CCR5−/− and CCR5+/− samples were significantly lower than those observed with the CCR5+/+ PBMCs (Fig. 1B). The CXCR4 index for the six infected CCR5−/− individuals was relatively higher than that for uninfected CCR5−/− or CCR5+/− PBMCs (Fig. 1B).
To determine whether the coreceptor expression index correlates with the susceptibility of PBMCs to Env-mediated fusion, we utilized a cell fusion assay that has been described previously (3). The fusion assay represents the first step in viral infection, where the envelope glycoprotein of the virus interacts with the host cell receptor/coreceptor, resulting in membrane fusion and viral entry. This assay is dependent on cell fusion between two cell populations, one expressing CD4/coreceptor and T7 RNA polymerase, the other expressing HIV-1 Env and lacZ under the T7 promoter; cell fusion activates the production of β-galactosidase. A significant R5 Env-mediated fusion with CCR5−/− PBMCs was never observed in our assays (Fig. 1C). In contrast, CCR5+/+ and CCR5+/− PBMCs were similarly susceptible to R5 Env-mediated fusion (Fig. 1C) but were significantly more susceptible to X4 Env-mediated cell fusion than were CCR5−/− samples (Fig. 1D). All the HIV+ CCR5−/− samples showed higher X4 fusion values than did uninfected CCR5−/− samples (Fig. 1D). These experiments demonstrated that the potency of X4 but not R5 Env-mediated fusion correlated with the coreceptor expression index.
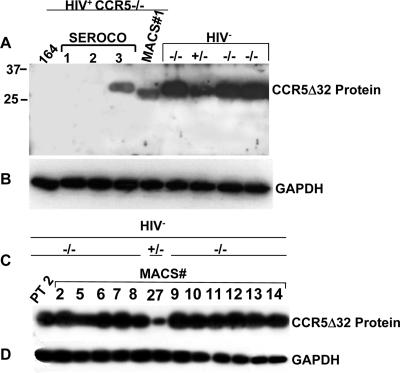
Expression levels of endogenous CCR5Δ32 protein in CCR5−/− PBMCs correlate with resistance to HIV-1.
To investigate the potential reasons for the rare loss of the protective effect by the CCR5Δ32 mutation, we examined endogenous expression of CCR5Δ32 protein in PBMCs from six HIV+ CCR5Δ32 homozygous individuals, using HIV− CCR5Δ32 homozygous PBMCs as controls. We first confirmed the HIV status of these individuals by amplification of Gag sequences and verified the CCR5 genotype by PCR analysis of genomic DNA (data not shown). To examine CCR5Δ32 protein expression, we used a polyclonal antiserum raised against the 31 frameshift amino acid residues found in CCR5Δ32 but not in CCR5 (2). High-level expression of the CCR5Δ32 protein was detected in PBMCs from all HIV− CCR5−/− individuals, and lower levels of expression were detected in two CCR5+/− individuals tested (Fig. 2A and C). We examined 25 HIV− CCR5−/− samples (results for 15 are shown in Fig. 2, and results for 10 other samples are not shown) and detected high levels of CCR5Δ32 protein. In contrast, the CCR5Δ32 protein was not detectable in PBMCs from three HIV+ CCR5−/− individuals and was expressed at reduced levels in PBMCs from two HIV+ CCR5−/− individuals (Fig. 2A). We previously reported the absence of the CCR5Δ32 protein in another unrelated individual (2). Taken together, the number of HIV+ CCR5−/− PBMC samples in which the CCR5Δ32 protein could not be detected is now four. To determine whether the absence of CCR5Δ32 protein in these infected individuals was due to the quality of the loaded samples, the same blot was stripped and reprobed with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Fig. 2B and D). These experiments demonstrated that all uninfected CCR5−/− samples express endogenous CCR5Δ32 protein and suggest that reduced CCR5Δ32 protein expression or a lack of CCR5Δ32 protein expression in HIV+ CCR5−/− individuals might explain the loss of the protective effect.
FIG. 2.
Expression analysis of CCR5Δ32 protein in infected and uninfected CCR5−/− individuals. (A) Immunodetection of native CCR5Δ32 protein in PBMCs isolated from individuals homozygous (−/−) and heterozygous (+/−) for the CCR5Δ32 mutant allele. The Western blot was probed with antibodies generated against the novel frameshift amino acids that specifically detect CCR5Δ32 protein. (B) The blot in panel A was stripped and reprobed with monoclonal antibodies to GAPDH to control for gel loading. (C) Immunodetection (same procedure as described for panel A) of native CCR5Δ32 protein in 11 CCR5−/− and 1 (+/−) PBMC samples obtained from the MACS. (D) The blot in panel C was stripped and reprobed with GAPDH antibodies. The numbers above each lane represent MACS sample numbers. PT 2, patient 2.
Rescue of the CCR5Δ32 protective effect in HIV+ CCR5−/− PBMCs.
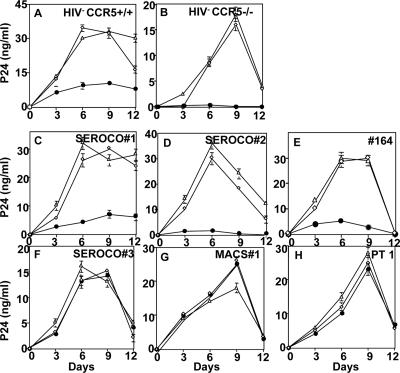
To test whether the absence or the reduced levels of endogenous CCR5Δ32 protein in HIV+ CCR5−/− PBMCs could be rescued by the expression of recombinant CCR5Δ32 protein, we performed a series of experiments using Ad5-encoded CCR5Δ32 protein (2). We hypothesized that the expression of recombinant CCR5Δ32 protein may render these PBMCs resistant to IIIB infection. We used the X4 IIIB infection as a model since, in our hands, all CCR5−/− PBMCs showed more efficient productive infection with this lab-adapted strain of HIV-1.
As a negative control for the CCR5Δ32 effect, we used a recombinant Ad5 encoding the homologous wild-type CCR5 protein. To control the effects of the Ad5-encoded proteins, we included cells infected with wild-type Ad5. The results show that the expression of Ad5-encoded CCR5Δ32 protein but not CCR5 protein in CCR5+/+ PBMCs conferred resistance to R5 (data not shown) as well as X4 (IIIB) strains of HIV-1 (Fig. 3A). Moreover, we found that the expression of CCR5 in HIV− samples restored R5 infection, indicating that CCR5 is the major determinant of R5 infection (data not shown). The HIV− CCR5−/− PBMC sample expressing either endogenous or Ad5-encoded CCR5Δ32 (Ad5/D32) but not wild-type CCR5 (Ad5/CCR5) protein showed efficient resistance to X4 (IIIB) infection (Fig. 3B).
FIG. 3.
Rescue of the protective effect of CCR5Δ32 by Ad5-encoded CCR5Δ32 protein. The indicated PBMC samples (A to H) were infected with either Ad5 (⋄), Ad5/CCR5 (▵), or Ad5/Δ32 (•) at 50 pfu/cell for each virus, incubated for 2 days to allow protein expression, and then infected with the X4 IIIB. Culture fluids were harvested after cell suspension every 3 days and replaced with fresh medium. The amount of p24 antigen in the cell-containing supernatants was measured using an ELISA kit purchased from DuPont. The zidovudine control infection resulted in p24 values below 1 ng/ml (data not shown). Panels C to H show HIV+ CCR5−/− PBMCs. Error bars indicate standard deviations. PT 1, patient 1.
The restoration of the CCR5Δ32 protective effect by the Ad5-encoded CCR5Δ32 protein was accomplished in three HIV+ CCR5−/− PBMCs. Significant resistance to X4 infection was observed in these samples expressing Ad5-encoded CCR5Δ32 protein (Fig. 3C, D, and E). In contrast, this protective effect (resistance to X4) was not observed in the other three HIV+ CCR5−/− PBMCs (Fig. 3F, G, and H). Reverse transcription-PCR analysis demonstrated similar efficiencies of CCR5Δ32 RNA transcription by the Ad5 vector in all PBMC samples (data not shown). The results demonstrate that the expression of recombinant CCR5Δ32 protein in some HIV+ CCR5−/− PBMCs can restore the protective effect.
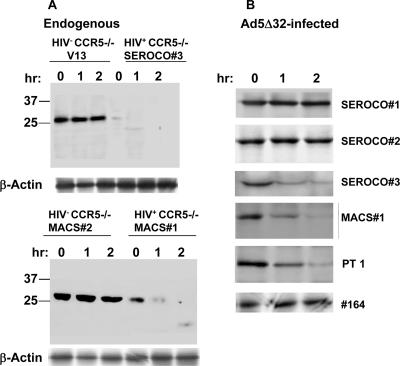
Pulse-chase analysis of CCR5Δ32 protein expressed in CCR5−/− PBMCs.
Since the CCR5Δ32 effect could not be restored in some HIV+ CCR5−/− PBMCs, we decided to perform pulse-chase analysis of the CCR5Δ32 protein in all HIV− CCR5−/− samples. In order to analyze the stability of CCR5Δ32 protein, we first performed pulse-chase experiments on endogenous protein expressed in some CCR5−/− PBMC samples. The number of cells limited our pulse-chase analysis of the endogenous protein to those samples that had larger numbers of cells available. The results demonstrate the accumulation of endogenous CCR5Δ32 protein in HIV− CCR5−/− PBMCs (V13 and MACS 2) (Fig. 4A) and the disappearance of a faint protein band detected in CCR5−/− HIV+ PBMCs (SEROCO 3 and MACS 1) (Fig. 4A). To further analyze CCR5Δ32 protein stability, we infected the PBMC samples with Ad5/Δ32 and performed the same pulse-chase experiments. The results revealed that the Ad5-encoded CCR5Δ32 protein accumulated in PBMCs isolated from some HIV+ CCR5−/− PBMCs and diminished in three other HIV+ CCR5−/− samples during the chase period (Fig. 4B). The results suggest that instability of the CCR5Δ32 protein in PBMCs isolated from three HIV+ CCR5−/− individuals might explain the loss of the CCR5Δ32 protective effect. This identifies two different categories of HIV+ CCR5−/− individuals, one where the protective effect can be restored due to the stability of the CCR5Δ32 protein and another where the inability to restore the protective effect correlated with the instability of the CCR5Δ32 protein.
FIG. 4.
Pulse-chase analysis of CCR5Δ32 protein stability in CCR5−/− PBMCs. PHA-stimulated cells were pulse labeled with [35S]methionine and [35S]cysteine for 30 min in methionine- and cysteine-free medium DMEM containing 200 μCi of [35S]methionine and 200 μCi of [35S]cysteine per milliliter. Following the pulse, the cells were washed with FBS and reincubated in complete DMEM without radioactive label for 1 and 2 h before lysis and immunoprecipitation with rabbit antisera directed against the unique 31 amino acids of the CCR5Δ32 protein. The immunoprecipitated protein samples were analyzed on a 12.5% SDS-polyacrylamide gel. The dried gel was exposed to X-ray film and developed after 24 h. PT 1, patient 1; hr, hour.
Expression of Ad5-encoded CCR5 protein in HIV+ CCR5−/− PBMCs restored R5 Env-mediated fusion.
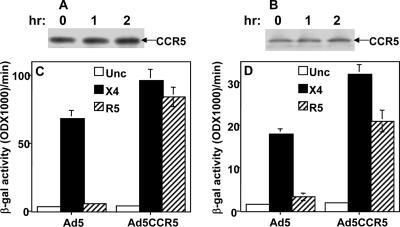
In order to confirm that the failure to restore the CCR5Δ32 protein protective effect is due to intracellular protein instability, we examined the intracellular stability of expressed CCR5 (Ad5 encoded) in the two HIV+ CCR5−/− PBMC samples. The PBMC samples were coinfected with Ad5CCR5 and the reporter Ad5pT7-lacZ and used in pulse-chase and Env-mediated fusion assays. The pulse-chase analysis of Ad5-encoded CCR5 in these PBMC samples demonstrated the accumulation of recombinant CCR5 protein after 2 h of chase (Fig. 5A and B). To confirm that the recombinant CCR5 protein is functional as a fusion cofactor, we examined the PBMC samples in the Env-mediated fusion assay. The results demonstrate an efficient restoration of the ability of R5 Env-expressing cells to fuse with these HIV+ CCR5−/− PBMC samples. Both samples expressing the Ad5-encoded CCR5 showed efficient R5 and X4 Env fusion (Fig. 5C and D). These results demonstrated intracellular stability of the Ad5-encoded CCR5 protein efficiently and the restoration of R5 Env-mediated fusion in both PBMC samples. These data support our hypothesis that intracellular instability of CCR5Δ32 protein in some HIV+ CCR5−/− PBMCs might explain the loss of the protective effect.
FIG. 5.
Expression and stability of recombinant CCR5 restored R5 Env-mediated fusion with HIV+ CCR5−/− PBMC samples. (A and B) Pulse-chase analysis of CCR5 protein encoded by Ad5CCR5. The cells were coinfected with Ad5CCR5 and Ad5pT7-lacZ and used for pulse labeling. Cell lysates were prepared and immunoprecipitated with CTC-6 monoclonal antibodies to CCR5 (R&D). (C and D) HIV-1 Env-mediated fusion with two HIV+ CCR5−/− PBMC samples expressing Ad5-encoded CCR5. The PBMCs were coinfected with Ad5CCR5 and Ad5pT7-lacZ reporter. The infected cells were incubated for 48 h and then mixed with HeLa cells expressing T7 RNA polymerase and one of the indicated HIV-1 Envs. Unc is a negative Env control that has a mutation at the cleavage site and, therefore, does not promote cell fusion. The results were evaluated by measuring the β-galactosidase (β-gal) produced as a result of Env-mediated cell fusion. The PBMC samples used were MACS 1 (A and C) and SEROCO 3 (B and D). hr, hour. Error bars indicate standard deviations.
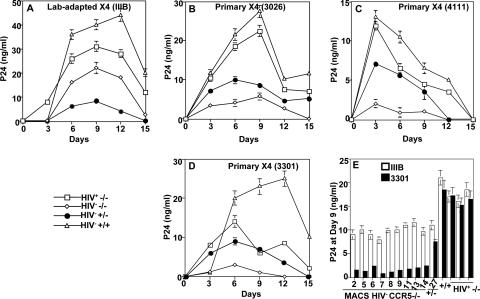
PBMCs from CCR5Δ32 homozygotes show resistance to infection by primary X4 strains.
To analyze the susceptibility of CCR5−/− PBMCs to X4 infection, we compared productive HIV-1 infection of the lab-adapted IIIB strain to that of three primary X4 strains. The infection profiles show that PBMCs isolated from CCR5+/+ and HIV+ CCR5−/− individuals produced higher amounts of p24 upon X4 infection (Fig. 6A to D). HIV− CCR5−/− PBMCs showed significant p24 production with lab-adapted IIIB (Fig. 6A) but resisted productive infection by the three primary X4 isolates (Fig. 6B, C, and D). These results were reproduced with nine other CCR5−/− PBMCs from the MACS cohort (Fig. 6E). Interestingly, all HIV+ CCR5−/− PBMC samples produced significantly higher amounts of p24 than did HIV− CCR5−/− PBMCs during the course of infection with the primary X4 isolates (Fig. 6B to E). These results suggest that the susceptibility of CCR5−/− PBMCs to X4 is dependent on the HIV-1 strain used in the infection assay.
FIG. 6.
Infection kinetics of PBMCs with lab-adapted IIIB (X4) and primary X4 (isolate 4111, 3301, or 3026). Infections were performed at 10 ng/ml of the indicated X4 isolate. Culture fluid was harvested after cell resuspension every 3 days and replaced with fresh medium (A, B, C, and D). The amount of p24 antigen in the cell-containing supernatants was measured using an ELISA kit purchased from DuPont. The symbols with labeling indicate the genotype and HIV status of the PBMC sample. The MACS identity number of each PBMC sample is shown. Panel E shows the results of productive infection of the indicated PMBC samples at day 9. Error bars indicate standard deviations.
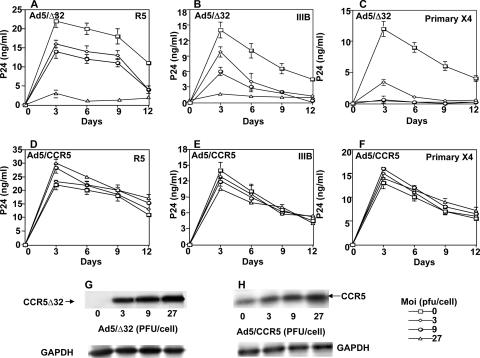
Lower levels of Ad5-encoded CCR5Δ32 protein induce resistance to primary X4.
To determine whether CCR5Δ32 protein levels correlate with the susceptibility to HIV-1, we carried out a series of functional experiments using recombinant CCR5Δ32 protein encoded by an adenovirus system to provide increasing levels of the protein. The replication-defective recombinant vectors designated Ad5/Δ32 and Ad5/CCR5 were used to infect CCR5+/+ PBMCs at increasing MOIs. We have previously demonstrated that the CCR5 protein encoded by Ad5/CCR5 is a functional HIV coreceptor (2). The HIV infection assays demonstrated an increased inhibitory effect that correlated with increased expression of intracellular CCR5Δ32 protein. Both R5 (Fig. 7A) and X4 infection (Fig. 7B and C) showed reduced susceptibilities that correlated with the increased Ad5/Δ32 virus input and increased CCR5Δ32 protein expression. Resistance to primary X4 was consistently observed with cells expressing less CCR5Δ32 protein (3 PFU) (Fig. 7C) compared to IIIB infection (27 PFU) (Fig. 7B). In contrast, increased Ad5/CCR5 virus input and CCR5 protein expression had no significant effect on either R5 (Fig. 7D) or X4 infection (Fig. 7E and F). The increased levels of CCR5Δ32 and CCR5 proteins encoded by the adenoviral vectors were confirmed by Western blot analysis of protein extracts of infected PBMCs (Fig. 7G and H). These results demonstrate that expression levels of CCR5Δ32 protein correlate with the level of resistance to HIV-1 infection and suggest that infection by primary and lab-adapted X4 strains may require different coreceptor levels.
FIG. 7.
Lower expression levels of Ad5-encoded CCR5Δ32 protein induce resistance to primary X4. Affinity-purified CD4+ T lymphocytes isolated from CCR5+/+ PBMCs were used uninfected (0) or infected with increasing MOIs (3 to 27) of the indicated recombinant Ad5 viruses, incubated for 48 h to allow expression of the recombinant proteins, washed with FBS, and then infected with Ba-L (R5) (A and D), IIIB (B and E), or primary X4 (C and F). The amount of p24 antigen in the cell-containing supernatants was measured using ELISA. The zidovudine control infection resulted in p24 values below 1 ng/ml (not shown). The error bars represent replicates of the same experiment. The results shown represent one of three independent experiments using samples from three different donors. A sample of each Ad5 infection at the indicated MOI was analyzed by Western blotting to verify expression of the recombinant proteins (E and F). The CCR5 blots were performed at the same day the cells were infected with HIV-1 (day 0). The primary X4 used in these experiments was isolate 4111.
DISCUSSION
Our previous studies suggested that resistance to HIV-1 infection in CCR5Δ32 homozygotes may result from both genetic loss of CCR5 on the cell surface and active downregulation of CXCR4 expression by the mutant CCR5Δ32 protein (2). We have shown that the truncated CCR5Δ32 protein can act as a negative regulator of wild-type CCR5 and CXCR4. The dominant negative activity of the mutant CCR5Δ32 protein correlated with its ability to reduce cell surface expression of the major HIV coreceptors and to form heterodimeric complexes with CCR5 and CXCR4 (2). The present study was designed to investigate the mechanism of failure of the protective effect observed in some rare cases of individuals homozygous for the CCR5Δ32 mutant allele. The restoration of R5 Env-mediated fusion and HIV-1 infection by Ad5-encoded CCR5 in CCR5−/− PBMCs demonstrated that CCR5 is the major coreceptor on PBMCs utilized by R5 isolates. Restoration of the protective effect in three HIV+ CCR5−/− PBMC samples suggested that the expression and stability of the truncated CCR5Δ32 protein is a critical determinant of resistance to HIV-1 in vivo.
The baseline CXCR4 level was high in all six infected homozygous individuals examined. The significantly higher CXCR4 levels in these infected CCR5−/− individuals may have been a consequence of the absence or reduced expression of CCR5Δ32 protein. We have previously demonstrated that CCR5Δ32 protein physically associates with CXCR4 and reduces its availability at the cell surface (2). It is possible that the absence of CCR5Δ32 protein allows more CXCR4 molecules to traffic to the cell surface. It is also possible that Tat produced by the infecting viruses upregulated CXCR4 in all these HIV+ samples. Selective upregulation of CXCR4 expression by Tat has previously been reported (21).
Multiple factors might be involved in the observed absence or reduced levels of CCR5Δ32 protein in HIV+ CCR5−/− patients. Modulation of the protein expression might have been a consequence of HIV-1 infection. Two of these patients had been infected with R5/X4 strains (23, 33). It has been previously demonstrated that CCR5 protein expression was eliminated by chronic infection with R5 viruses (13). It is possible that a similar mechanism is responsible for the absence or reduced levels of CCR5Δ32 protein in these two samples (MACS 1 and patient 1) that harbored R5/X4 strains. However, this may not be the case in the samples that harbored X4 strains (SEROCO 2 and Shep 164). The pure X4 strains signaling via CXCR4 may alter CCR5Δ32 protein turnover. Previous studies have demonstrated that both R5 and X4 envelopes induce factors relevant to viral replication, including the expression of genes involved in cell proliferation, cell cycle, transcription factors, and mitogen-activated protein kinases (15). Cicala et al. have also demonstrated that within each of these categories, R5 and X4 envelopes induced different subsets of genes. These differentially induced genes may influence the expression levels of cellular proteins, including CCR5Δ32 protein (15).
Posttranslational modification of the CCR5Δ32 protein may also contribute to its stability in some infected individuals. The observed faster migration of the CCR5Δ32 protein of the MACS 1 sample might be due to different protein modification in this PBMC sample. We have sequenced the cDNA clones encoding the CCR5Δ32 protein from all PBMC samples and did not detect any difference from the reported wild-type amino acid sequence (data not shown). It is also possible that some other genetic determinants impact CCR5Δ32 protein expression, stability, or degradation. For example, polymorphisms in the promoter region of CCR5 may affect gene expression and result in the downmodulation of protein expression. We have detected some of these polymorphisms in the promoter regions (data not shown); however, their potential effect on gene expression has yet to be determined.
The ability to restore the CCR5Δ32 protein protective effect was associated with stable expression and accumulation during the chase period. It is not clear why the Ad5-encoded CCR5Δ32 protein expressed in some HIV+ CCR5−/− PBMCs also disappeared during the chase period. It is possible that an unknown cellular factor(s) may contribute to the stability of the CCR5Δ32 protein. Previous studies demonstrated that CCR5 is palmitoylated at its carboxyl-terminal tail and that palmitoylation is critical for receptor trafficking and efficient activation of intracellular signaling pathways (10, 36). It has been demonstrated that interfering with this normal lipid modification promotes rapid proteolytic degradation of CCR5 and decreases its proper surface expression (36). The situation may be different for the truncated CCR5Δ32 protein that lacks the carboxyl-terminal tail present in CCR5 and instead contains 31 frameshift residues. Future work focusing on the potential posttranslational modifications of CCR5Δ32 protein and its potential interaction with other cellular proteins may provide important insight regarding the intracellular activity and stability of this biologically important protein.
It has previously been demonstrated that an optimal number of coreceptor molecules are required for efficient HIV-1 infection (28). Other studies have suggested that susceptibility to HIV-1 infection can be modulated by coreceptor expression levels (22, 25). For example, Hladik et al. reported that exposed seronegative individuals with a CCR5Δ32 heterozygous genotype are resistant to sexual transmission due to significant reduction in their CCR5 expression (25). Another independent study reported that low copy numbers of the CCL3L1 (MIP-1αP) gene are associated with enhanced HIV/AIDS susceptibility (22). CCL3L1 is a potent HIV-suppressive chemokine and a ligand for CCR5. Gonzalez et al. demonstrated that an increasing CCL3L1 copy number was positively associated with CCL3/CCL3L1 secretion and negatively associated with the proportion of CD4+ T cells that express CCR5 (22). Two different mechanisms have been proposed for chemokine inhibition of HIV-1 infection, steric blockade and ligand-mediated receptor internalization, both of which result in reduced availability of the coreceptor at the cell surface (5).
Our previous studies proposed intracellular interaction of CCR5Δ32 protein with CCR5 and CXCR4 to explain the resistance of CCR5−/− cells to HIV-1 (2). The present study demonstrated that increased stable expression levels of CCR5Δ32 protein correlate with enhanced resistance to HIV-1 infection. Higher levels of expressed CCR5Δ32 protein have always correlated with the detection of lower cell surface staining of both coreceptors (data not shown). Significantly higher levels of Ad5-encoded CCR5Δ32 protein were required to induce resistance to the lab-adapted IIIB isolate. The more efficient resistance to primary X4 by the Ad5-encoded CCR5Δ32 protein may reflect different requirements of receptor/coreceptor density. The optimal numbers of receptor/coreceptor molecules required for efficient HIV-1 infection are not known. Previous in vitro studies using HeLa-CD4 cell clones have demonstrated that R5 and X4 viral strains can have differential requirements for CD4 and coreceptor levels (26, 27). The studies by Kozak et al. implied that the stoichiometry of receptor/coreceptor might play an important role in susceptibility to HIV-1 infection (27). The results of this study suggest that CCR5Δ32 protein expression in CCR5−/− individuals causes the downmodulation of CXCR4 to levels that might result in an unfavorable stoichiometry of the molecules involved in HIV-1 entry. An analysis of the different requirements by primary versus lab-adapted HIV-1 strains will provide important insight into the mechanism of viral entry and the design of entry inhibitors.
In summary, our findings highlight the complexity of the CCR5Δ32 protective effect, in which many cellular molecules, including CCR5 and CXCR4, could be involved. The instability of CCR5Δ32 protein in PBMCs from some CCR5−/− individuals highlights the important role of this protein as an HIV suppressive factor. Comprehensive analyses of the novel antiviral activity of CCR5Δ32 protein will be important to elucidate its mechanism of action. Ultimately, such studies not only will lead to a better understanding of the host cell interaction of HIV-1 but also might support the design of novel drugs that mimic CCR5Δ32 protein activity.
Acknowledgments
This study was supported by NIH grant A152019-01A1 to G.A. Q.J. was supported by a scholarship from the Chinese Scholarship Council, Beijing, China.
We thank Margaret Bauer for critical comments. We thank the MACS Cohort for providing the PBMC samples from all CCR5 genotypes. We thank Hassan Naif for providing PBMCs from patients 1 and 2 and H. W. Sheppard and S. P. Buchbinder for the Shep 164. Human PBMC samples were obtained under Indiana University exempt IRB study no. 4.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Agrawal, L., X. Lu, Q. Jin, and G. Alkhatib. 2006. Anti-HIV therapy: current and future directions. Curr. Pharm. Des. 12:2031-2055. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, L., X. Lu, J. Qingwen, Z. VanHorn-Ali, V. Nicolescue, D. McDermott, P. M. Murphy, and G. Alkhatib. 2004. Role for CCR5Δ32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells. J. Virol. 78:2277-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib, G., C. C. Broder, and E. A. Berger. 1996. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J. Virol. 70:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 5.Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology 234:340-348. [DOI] [PubMed] [Google Scholar]
- 6.Balotta, C., P. Bagnarelli, M. Violin, A. L. Ridolfo, D. Zhou, A. Berlusconi, S. Corvasce, M. Corbellino, M. Clementi, M. Clerici, M. Moroni, and M. Galli. 1997. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. AIDS 11:F67-F71. [DOI] [PubMed] [Google Scholar]
- 7.Benkirane, M., D. Y. Jin, R. F. Chun, R. A. Koup, and K. T. Jeang. 1997. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5Δ32. J. Biol. Chem. 272:30603-30606. [DOI] [PubMed] [Google Scholar]
- 8.Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 9.Biti, R., R. Ffrench, J. Young, B. Bennetts, G. Stewart, and T. Liang. 1997. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat. Med. 3:252-253. [DOI] [PubMed] [Google Scholar]
- 10.Blanpain, C., V. Wittamer, J. M. Vanderwinden, A. Boom, B. Renneboog, B. Lee, E. Le Poul, L. El Asmar, C. Govaerts, G. Vassart, R. W. Doms, and M. Parmentier. 2001. Palmitoylation of CCR5 is critical for receptor trafficking and efficient activation of intracellular signaling pathways. J. Biol. Chem. 276:23795-23804. [DOI] [PubMed] [Google Scholar]
- 11.Braciak, T. A., K. Bacon, Z. Xing, D. J. Torry, F. L. Graham, T. J. Schall, C. D. Richards, K. Croitoru, and J. Gauldie. 1996. Overexpression of RANTES using a recombinant adenovirus vector induces the tissue-directed recruitment of monocytes to the lung. J. Immunol. 157:5076-5084. [PubMed] [Google Scholar]
- 12.Chelli, M., and M. Alizon. 2001. Determinants of the trans-dominant negative effect of truncated forms of the CCR5 chemokine receptor. J. Biol. Chem. 276:46975-46982. [DOI] [PubMed] [Google Scholar]
- 13.Chenine, A. L., Q. Sattentau, and M. Moulard. 2000. Selective HIV-1-induced downmodulation of CD4 and coreceptors. Arch. Virol. 145:455-471. [DOI] [PubMed] [Google Scholar]
- 14.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 15.Cicala, C., J. Arthos, E. Martinelli, N. Censoplano, C. C. Cruz, E. Chung, S. M. Selig, D. Van Ryk, J. Yang, S. Jagannatha, T. W. Chun, P. Ren, R. A. Lempicki, and A. S. Fauci. 2006. R5 and X4 HIV envelopes induce distinct gene expression profiles in primary peripheral blood mononuclear cells. Proc. Natl. Acad. Sci. USA 103:3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. (Erratum, 274:1069.) [DOI] [PubMed] [Google Scholar]
- 17.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 18.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 19.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. X. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 20.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 21.Gibellini, D., M. C. Re, F. Vitone, N. Rizzo, C. Maldini, M. La Placa, and G. Zauli. 2003. Selective up-regulation of functional CXCR4 expression in erythroid cells by HIV-1 Tat protein. Clin. Exp. Immunol. 131:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez, E., H. Kulkarni, H. Bolivar, A. Mangano, R. Sanchez, G. Catano, R. J. Nibbs, B. I. Freedman, M. P. Quinones, M. J. Bamshad, K. K. Murthy, B. H. Rovin, W. Bradley, R. A. Clark, S. A. Anderson, R. J. O'Connell, B. K. Agan, S. S. Ahuja, R. Bologna, L. Sen, M. J. Dolan, and S. K. Ahuja. 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434-1440. [DOI] [PubMed] [Google Scholar]
- 23.Gorry, P. R., C. Zhang, S. Wu, K. Kunstman, E. Trachtenberg, J. Phair, S. Wolinsky, and D. Gabuzda. 2002. Persistence of dual-tropic HIV-1 in an individual homozygous for the CCR5D32 allele. Lancet 359:1832-1834. [DOI] [PubMed] [Google Scholar]
- 24.Gray, L., M. J. Churchill, N. Keane, J. Sterjovski, A. M. Ellett, D. F. Purcell, P. Poumbourios, C. Kol, B. Wang, N. K. Saksena, S. L. Wesselingh, P. Price, M. French, D. Gabuzda, and P. R. Gorry. 2006. Genetic and functional analysis of R5X4 human immunodeficiency virus type 1 envelope glycoproteins derived from two individuals homozygous for the CCR5Δ32 allele. J. Virol. 80:3684-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hladik, F., H. Liu, E. Speelmon, D. Livingston-Rosanoff, S. Wilson, P. Sakchalathorn, Y. Hwangbo, B. Greene, T. Zhu, and M. J. McElrath. 2005. Combined effect of CCR5-Δ32 heterozygosity and the CCR5 promoter polymorphism −2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J. Virol. 79:11677-11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabat, D., S. L. Kozak, K. Wehrly, and B. Chesebro. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 68:2570-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak, S. L., E. J. Platt, N. Madani, F. E. Ferro, Jr., K. Peden, and D. Kabat. 1997. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J. Virol. 71:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuipers, H., C. Workman, W. Dyer, A. Geczy, J. Sullivan, and R. Oelrichs. 1999. An HIV-1-infected individual homozygous for the CCR-5 Δ32 allele and the SDF-1 3′A allele. AIDS 13:433-434. [DOI] [PubMed] [Google Scholar]
- 30.Martinson, J. J., N. H. Chapman, D. C. Rees, Y. T. Liu, and J. B. Clegg. 1997. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet. 16:100-103. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, L., M. Magierowska, J. B. Hubert, C. Rouzioux, C. Deveau, F. Sanson, P. Debre, J. F. Delfraissy, I. Theodorou, et al. 1997. Early protective effect of CCR-5 Δ32 heterozygosity on HIV-1 disease progression: relationship with viral load. AIDS 11:F73-F78. [DOI] [PubMed] [Google Scholar]
- 32.Michael, N. L., J. A. Nelson, V. N. KewalRamani, G. Chang, S. J. O'Brien, J. R. Mascola, B. Volsky, M. Louder, G. C. White II, D. R. Littman, R. Swanstrom, and T. R. O'Brien. 1998. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J. Virol. 72:6040-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naif, H. M., A. L. Cunningham, M. Alali, S. Li, N. Nasr, M. M. Buhler, D. Schols, E. de Clercq, and G. Stewart. 2002. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Δ32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J. Virol. 76:3114-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, T. R., C. Winkler, M. Dean, J. A. Nelson, M. Carrington, N. L. Michael, and G. C. White II. 1997. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet 349:1219. [DOI] [PubMed] [Google Scholar]
- 36.Percherancier, Y., T. Planchenault, A. Valenzuela-Fernandez, J. L. Virelizier, F. Arenzana-Seisdedos, and F. Bachelerie. 2001. Palmitoylation-dependent control of degradation, life span, and membrane expression of the CCR5 receptor. J. Biol. Chem. 276:31936-31944. [DOI] [PubMed] [Google Scholar]
- 37.Sheppard, H. W., C. Celum, N. L. Michael, S. O'Brien, M. Dean, M. Carrington, D. Dondero, and S. P. Buchbinder. 2002. HIV-1 infection in individuals with the CCR5-Δ32/Δ32 genotype: acquisition of syncytium-inducing virus at seroconversion. J. Acquir. Immune Defic. Syndr. 29:307-313. [DOI] [PubMed] [Google Scholar]
- 38.Theodorou, I., L. Meyer, M. Magierowska, C. Katlama, C. Rouzioux, et. al. 1997. HIV-1 infection in an individual homozygous for CCR5 delta 32. Lancet 349:1219-1220. [PubMed] [Google Scholar]
- 39.Venkatesan, S., A. Petrovic, D. I. Van Ryk, M. Locati, D. Weissman, and P. M. Murphy. 2001. Reduced cell surface expression of CCR5 in CCR5δ32 heterozygotes is mediated by gene dosage, not by receptor sequestration. J. Biol. Chem. 16:16. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerman, P. A., A. Buckler-White, G. Alkhatib, T. Spalding, J. Kubofcik, C. Combadiere, D. Weissman, O. Cohen, A. Rubbert, G. Lam, M. Vaccarezza, P. E. Kennedy, V. Kumaraswami, J. V. Giorgi, R. Detels, J. Hunter, M. Chopek, E. A. Berger, A. S. Fauci, T. B. Nutman, and P. M. Murphy. 1997. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies of populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol. Med. 3:23-36. [PMC free article] [PubMed] [Google Scholar]