Abstract
Coupled translation, first described in the M2 gene of pneumovirus respiratory syncytial virus (RSV), is an alternative mechanism of translational initiation in which the ribosomes which translate the first (M2-1) open reading frame (ORF) move a short distance upstream after termination and reinitiate translation from a second (M2-2) overlapping ORF. Here, we show that the same mechanism occurs in two closely related viruses, avian pneumovirus (APV) and pneumonia virus of mice (PVM), although with markedly different efficiencies. To identify the reasons for the variation in efficiency of coupled expression between RSV and APV, we used chimeric M2-1 genes containing different lengths of the M2-1 ORF from each virus. An essential component allowing coupled expression in the chimeras was a segment from the RSV M2-1 coding region containing a high degree of secondary structure. Additional sequences at the 5′ end of the RSV M2-1 ORF also promoted coupled translation when the region with high levels of secondary structure was present. These data indicate that at least two distant parts of the mRNA transcript, together with a suitable overlapping region, are involved in the coupling process. Replacement of the last 102 nucleotides of the RSV M2-1 ORF with the equivalent APV sequence showed identical levels of coupled translation. Thus, the overlapping region can direct the ribosome back onto the start codon of the second ORF while the upstream coding sequence of the M2-1 ORF determines the levels of coupled expression.
The evolutionary pressures that have constrained viruses that use RNA as genetic material to maintain small genomes have resulted in the exploitation of novel strategies to maximize the number of encoded proteins. While many of these strategies involve transcriptional regulation, an increasing number of translational regulatory mechanisms have been identified. Viruses depend solely on their host to provide the ribosomal components necessary to synthesize their proteins, and thus any translational regulatory mechanism utilized by the virus must be compatible with the host system. Analysis of virus translation has revealed novel features, such as internal ribosome entry sites in picornaviruses, which had not previously been appreciated and which may play a role in control of protein expression in uninfected host cells (3, 13). Most eukaryotic transcripts contain a single open reading frame (ORF) and initiate translation by the scanning mechanism, as reviewed by Kapp and Lorsch (15). For those transcripts expressed in eukaryotes, or from the viruses that infect them, which contain more than one ORF, the second ORF can be accessed by scanning ribosomes if the sequence context of the 5′ proximal AUG codon is nonoptimal. Ribosomes may then scan further down the mRNA by leaky scanning to the next initiation codon, where, if it is in a favorable sequence context, translation of a second ORF can begin (4, 7, 16). However, access to a second ORF only occurs frequently if the initiation codons of both the first and second ORFs are near each other in the mRNA sequence. Ribosomes may also access internal ORFs using alternative strategies, including internal ribosome entry sites, ribosome shunting, and a coupled translation termination/initiation process, all of which are dependent on sequences and structures present within the mRNA molecule that direct ribosomal initiation at the desired location (2, 12, 14, 17, 20, 22).
Coupled translation initiation/termination was first described for the human respiratory syncytial virus (RSV) M2 mRNA transcript, and subsequently a similar process was reported in two caliciviruses (2, 18, 19). RSV is a member of the subfamily Pneumovirinae, which contains two genera, the pneumoviruses (including RSV and pneumonia virus of mice [PVM]) and the metapneumoviruses (MPVs; including avian pneumovirus [APV] and human MPV [hMPV]), all of which cause acute respiratory infections in their hosts. All M2 transcripts contain two ORFs, M2-1 and M2-2, which overlap in the same arrangement (8) (Fig. 1). Expression of the RSV M2-2 ORF occurs via an unusual coupled translation event in which termination of translation of the M2-1 ORF is required before translation of the M2-2 ORF can be initiated. In RSV strain A2, initiation occurs at each of three possible in-frame AUG initiation codons in vivo (2, 11). We have previously shown that various regions of the RSV M2-1 ORF play a role in coupled translation. Surprisingly, the most important is not the region of overlap between the two coupled ORFs but a region approximately 150 nucleotides upstream of the overlap region which contains a very stable structural element required in the translational coupling process (11). Mutations which alter the predicted structure of this region significantly reduce the level of coupled translation.
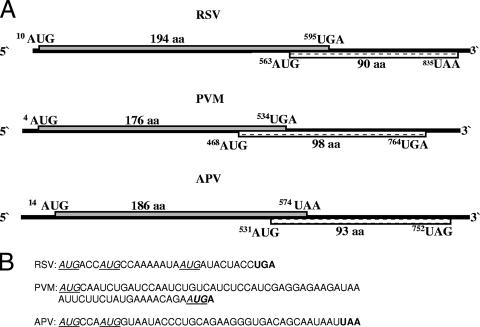
FIG. 1.
(A) Organization of selected pneumovirus M2 transcripts of RSV strain A2, PVM strain 15, and APV strain CVL/14, adapted from reference 8. The superscripts refer to the nucleotide positions of the first residues of the start and stop codons of the ORFs in the mRNAs. (B) Sequence of the overlap in the M2 transcript of RSV, PVM, and APV ORFs. Start codons for the second ORFs are underlined and italicized, and the stop codons of the M2-1 ORFs are shown in boldface.
Western blot analysis demonstrated that the PVM M2-2 protein was present in infected cells but failed to detect the equivalent protein in APV-infected cells (1). There is no conservation of sequence in the overlap regions in the M2 ORF of pneumoviruses nor of the coding region of the M2-2 ORF. However, the consistency of the organization of the ORFs of the M2 mRNA suggests that a similar coupled translation process is likely to be used to access the M2-2 ORF in other pneumoviruses. This raises the possibility that the sequence differences between the viruses may lead to differential expression of the second ORF, representing an additional level of translational control directed by the sequence, and possibly structure, of the upstream region. Here we describe analysis of further sequence requirements for coupled translation of the RSV M2-2 ORF. In addition, we demonstrate that the translation of the M2-2 ORF in PVM and APV also uses a coupled translation process but that in these viruses the level of expression is lower than that seen with RSV and is dependent on the sequences upstream of the overlap region where the coupling process occurs. These data indicate that the mRNA sequences in the overlap and, most importantly, upstream regions of the M2 mRNA interact to determine the efficiency of the translational coupling process.
MATERIALS AND METHODS
Reporter gene assays.
The standard reporter gene assay used 250 ng of plasmid DNA to transfect subconfluent HEp2 cells in each well of a 3.8-cm2 well. The HEp2 cells were preinfected with a vaccinia virus strain, vTF7-3, expressing the T7 polymerase at a multiplicity of infection (MOI) of 1 PFU per cell (10). After 48 h, cellular proteins were extracted in 150 μl of chloramphenicol acetyltransferase (CAT) lysis buffer (Roche). In order to obtain a detectable level of CAT with the APV constructs, the reporter assay was modified. Here, 2 μg of plasmid DNA was transfected into 9.6-cm2 well plates preinfected with vTF7-3 at an MOI of 5 PFU per cell. Proteins were extracted as before, CAT protein expression was measured as described previously (2), and data were plotted taking the appropriate positive control as 100%, with errors plotted as standard deviations (SDs) of the mean. Further statistical analysis was carried using the Student's t test.
For cloning, all PCRs were carried out using Pfu DNA polymerase (MBI Fermentas) and the sequences of the amplified fragments were verified. The primers used in this study are listed in Table 1.
TABLE 1.
Primers used in this study
| Name | Sequence |
|---|---|
| PVM M2-1F | TTCCCGGGGCAAATATGAGTGTGAGACCTTGCAAA |
| PVM M2-1R | GGTCTAGAATCAYTCTGTTTTCATAGAAGAATTTAT |
| 3′ CATSphI | GCTGTTAAGCATGCTTACGCCCCGCCCTGCCACTC |
| PVMqcstartF | GACTGCCAGTAGGAGTACTCTGCAATCTGATCCAATC |
| PVMqcstartR | GACAGATTGGATCAGATTGCAGAGTACTCCTACTGGC |
| PVMqcstopF | CTTCTATGAAAACAGAATGGTTCTATAGAAAAAAATCACTGGATAT |
| PVMqcstopR | ATATCCAGTGATTTTTTTCTATAGAACCATTCTGTTTTCATAGAAG |
| PVMqcstart2F | AATTCTTCTATGAAAACAGACTGATTCTAGAGAAAAAAATCACTGG |
| PVMqcstart2R | CCAGTGATTTTTTTCTCTAGAATCAGTCTGTTTTCATAGAAGAATT |
| APVAvaI | TTCCCGGGGCAAATATGTCTAGGCGAAATCCCTGC |
| APVstart1 | GGAGATCTTTAATTATTGCTGTCACCCTTCTGCAGGGTATTACCATTGGCGTTGTG |
| APVstart2 | GGAGATCTTTAATTATTGCTGTCACCCTTCTGCAGGGTATTACCGTTGGC |
| APVstop | GGAGATCTTTGATTATTGCTGTCACCCTT |
| APVwild | GGAGATCTTTAATTATTGCTGTCACCCTT |
| APVqcstopF | CTGGATATACCACCGTTGCTATATCCCAATGGCATCG |
| APVqcstopR | CGATGCCATTGGGATATAGCAACGGTGGTATATCCAG |
| APV M2-2 R | CCTCTAGATTATATGAGGTATATATAGCTATAAAC |
| APV M2-2 F | TTCCCGGGGCAAATATGCCAATGGTAATACCCTGC |
PVM M2-1-CAT constructs.
The PVM M2-1 ORF was amplified from the plasmid pM2-1PVM (a kind gift from Lindsay Thorpe and which contains the complete M2 ORF of PVM cloned in pBlueScribe) using primers PVM M2-1F and PVM M2-1R. The CAT ORF was amplified using primers 5′ CAT (2) and 3′ CATSphI and was ligated to the PVM M2-1 PCR product and cloned into pBlueScribe (Stratagene) digested with AvaI and SphI to generate plasmids pPVMWC and pPVMmut2. pPVMmut1 and pPVMmut3 were generated using quick change mutagenesis with the primers PVMqcstartF and PVMqcstartR on plasmids pPVMWC and pPVMmut2, respectively. Quick change mutagenesis was used to generate pPVMstop, using primers PVMqcstopF and PVMqcstopR on plasmid pPVMWC. Plasmids pPVMmut4 and pPVMmut5 were generated using primers PVMqcstart2F and PVMqcstart2R with the plasmids pPVMmut2 and pPVMmut3 as templates, respectively.
APV M2-1-CAT constructs.
The APV M2-1 ORF and the overlapping sequence were amplified from pAPVM2-6.9.00 (a kind gift from Nicole Edworthy and which contains the entire APV M2 gene), using the primer APVAvaI and an appropriate APV reverse primer (APVwild, APVstart1, APVstart2, or APVstop) and cloned into pBlueScribe, generating constructs without a mutation or constructions with mutations in the first M2-2 start codon, second M2-2 start codon, or M2-1 stop codon, respectively. The CAT ORF was amplified as previously described (2) and cloned into the downstream of the APV M2-1 stop codon via an XbaI restriction enzyme site, generating pAPVWC (no mutations), pAPVmut1 (first start mutation), pAPVmut2 (second start mutation), and pAPVStop1 (M2-1 stop codon mutation). The mutant pAPVmut3 was constructed by removing the remaining start codon in pAPVmut1 by mutagenesis using the primers APVAvaI and APVstart2. Finally, pAPVStop2 was generated using quick change mutagenesis with primers APVqcstopF and APVqcstopR on the pAPVStop1 template.
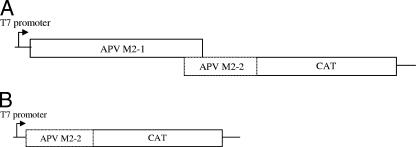
Full-length APV M2 gene construct.
The full-length APV M2-1 and M2-2 ORFs were amplified using the primers APVAvaI and APVM2-2R from plasmid pAPVM2-6.9.00. The primer APVM2-2R removes 4 nucleotides from the 3′ terminus of the M2-2 ORF so that the CAT ORF is in frame when cloned into the XbaI site present in the new construct, generating pAPVFLWC (see Fig. 4A). As a control, the APV M2-2 ORF alone was fused to the CAT gene ORF using the primers APVM2-2F and APVM2-2R as described above (plasmid pM2-2CAT Fig. 4B).
FIG. 4.
Schematic representation of (A) chimeric APV M2-2-CAT protein (plasmid pAPVM2-2CAT) and (B) chimeric RSV/APV M2-1 with overlapping CAT ORFs made to evaluate coupled translation in full-length APV M2 reporter constructs.
Chimeric M2-1-CAT constructs.
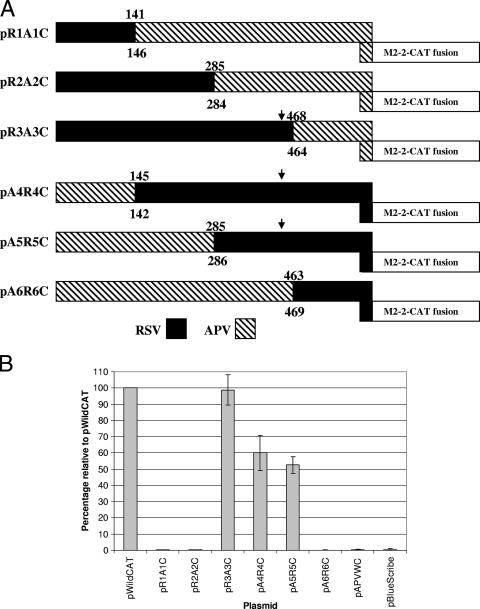
Chimeras were generated using PCR as part of another study (H. Stokes et al., unpublished data). Chimera pR1A1 contains residues 1 to 44 from RSV and 45 to 186 from APV M2-1 proteins. Chimera pR2A2 contains residues 1 to 92 from RSV and 92 to 186 from the APV M2-1 protein. Chimera pR3A3 contains residues 1 to 153 from RSV and 152 to 186 from the APV M2-1 protein. Chimera pA4R4 contains residues 1 to 44 from APV and 45 to 194 from the RSV M2-1 protein. Chimera pA5R5 contains residues 1 to 91 from APV and 93 to 194 from the RSV M2-1 protein, and chimera pA6R6 contains residues 1 to 151 from APV and 154 to 194 from the RSV M2-1 protein. Primers M2END (2) and APVwild were used to amplify and clone the M2-1 chimera from plasmids pR1A1, pR2A2 and pR3A3 into pBlueScribe. Primers APVAvaI and M2WILD (2) were used to amplify and clone the M2-1 chimera from pA4R4, pA5R5, and pA6R6. The CAT gene ORF was cloned into the XbaI site downstream of the chimeric M2-1 stop codon as described above, generating plasmids shown in Fig. 5, where the nomenclature to distinguish the reporter constructs from the parental plasmids is the addition of C (for CAT) following the name of the parental chimera.
FIG. 5.
(A) Schematic representation of the RSV-APV M2-1 chimeric proteins. The numbers indicate the nucleotide positions of the junction regions of the chimeric genes (above RSV and below APV). The position of the 3′ end of the region identified as being important for coupled expression in the RSV M2 gene (11) is indicated with an arrow, when present. The M2-2 ORF fused to the CAT gene ORF is also shown. (B) Levels of CAT protein expression of each mutant relative to the RSV pWildCAT positive control.
RESULTS
Coupled translation of the PVM M2 mRNA.
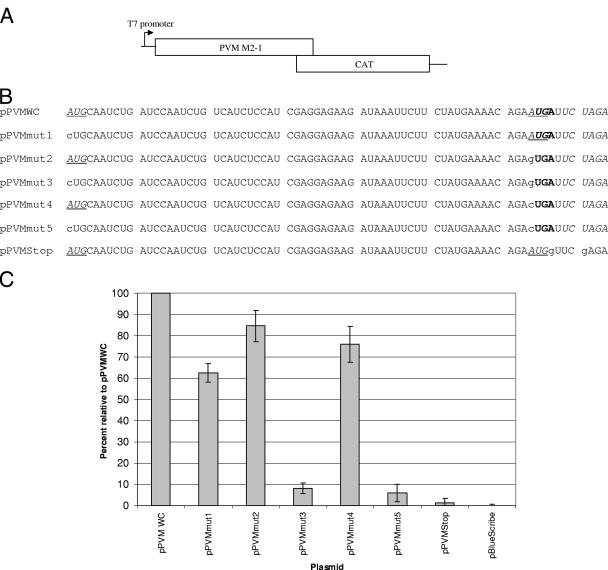
To determine whether the two ORFs in the PVM M2 mRNA were translationally coupled, we used a strategy similar to that used to demonstrate translational coupling in RSV. The PVM M2-1 upstream ORF was not altered in any way, but the M2-2 ORF downstream of the M2-1 ORF stop codon was replaced with the gene expressing the bacterial enzyme CAT, whose product can easily be detected and quantified by enzyme-linked immunosorbent assay (ELISA) and which was placed in frame with the putative M2-2 ORF translation initiation codon(s). This plasmid construct was designated pPVMWC (Fig. 2). To determine which of the two possible initiation codons was used for translation of the M2-2 ORF, each of the start codons was individually mutated with start 1 removed in pPVMmut1 and start 2 removed in pPVMmut2, or in combination with pPVMmut3 lacking either of the putative start codons. Two additional mutants, pPVMmut4 and pPVMmut5, were generated in which the second potential initiation codon was converted to CUG instead of the GUG (that could possibly act as a start codon) in pPVMmut2 and pPVMmut3. Finally, plasmid pPVMstop was constructed in which the M2-1 ORF stop codon and an additional in-frame stop codon 3 nucleotides downstream in the CAT ORF were mutated, extending the M2-1 ORF by 138 nucleotides and so increasing the distance between the site of translational termination of the M2-1 ORF and the initiation codons for the M2-2 ORF. The sequences of the mutated plasmids are shown in Fig. 2B.
FIG. 2.
(A) Schematic representation of the reporter gene constructs made to evaluate coupled translation in PVM. Transcription from the plasmids were under the control of the bacteriophage T7 promoter, as indicated. (B) Sequence alignment of the overlap regions in the PVM M2-1/CAT constructs. The CAT ORF is cloned in frame using the XbaI site (italics) to the M2-2 start codon(s) within the overlap. Positions of the M2-2 ORF start codons are underlined and in italics, and the positions of the M2-1 ORF stop codons are shown in boldface. Mutated residues are in lowercase. (C) Levels of CAT protein expression of each mutant relative to the pPVMWC positive control.
The levels of expression of the CAT protein fused with the beginning of the M2-2 protein for the various constructs are shown in Fig. 2C. Removal of either of the two potential initiation codons results in a measurable decrease in CAT protein expression suggesting both start codons function in unison for maximal expression (Fig. 2C). Expression was significantly lower when the first start codon was removed than when the second start codon, which overlaps with the termination codon of ORF 1, was removed (pPVMmut1 to pPVMmut2, P = <0.001; and pPVMmut1 to pPVMmut4, P < 0.03). Removal of both start codons, in both pPVMmut3 and pPVMmut5, reduced CAT expression to extremely low levels. Removal of both potential start codons did not completely eliminate reporter gene expression, as was reported for the equivalent RSV constructs (2). These data show that, as for RSV, the translation of the M2-2 ORF can be initiated from either of the 5′ proximal AUG initiation codons, although there is a preference for the first initiation codon. Using the CAT ELISA system to measure directly the levels of protein synthesized in transfected cells showed that translation from the PVM M2 gene second ORF was 6.3-fold lower (SD, ±2.2) than seen for RSV when comparing the constructs in which the overlap regions were not mutated (pPVMWC here and pWildCAT for RSV) (2). The most significant observation is that seen with the construct pPVMStop. In this construct, the stop codon for the upstream M2-1 ORF has been relocated 138 nucleotides downstream of its original position by insertion of a single nucleotide immediately following the AUG start 2 sequence to disrupt the natural UGA stop coding of the M2-1 ORF. The insertion of a G residue at this position increases the match of start 2 to the optimal Kozak sequence and should, in principle increase translation initiation at this position if it occurs by a conventional scanning process. However, when the M2-1 ORF is extended the two M2-2 ORF translation initiation codons are not utilized and expression of CAT protein from the M2-2 ORF falls to almost zero (Fig. 2C). This is exactly the same situation as that seen for the RSV M2-2 ORF and indicates that, as in the RSV situation, the PVM M2-2 protein is synthesized using a coupled translation process in which the translation of the M2-2 ORF is only initiated using ribosomes that have terminated translation of the upstream M2-1 ORF. The position of the point of termination of translation of the upstream ORF is critical for the coupled translation process, and extension of the distance between the points of termination and initiation by 138 nucleotides is sufficient to disrupt the process.
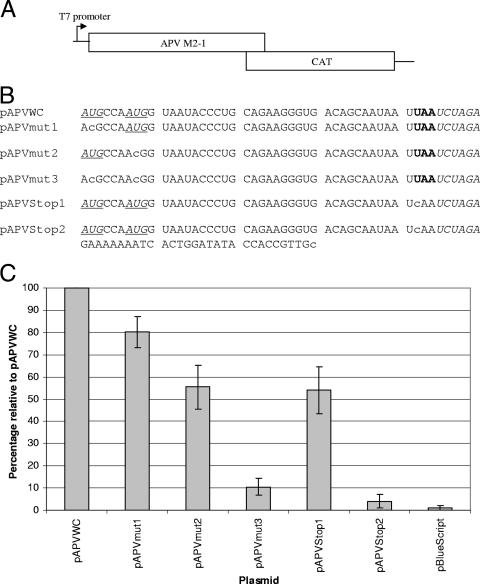
Coupled translation of the APV M2 mRNA.
Initial attempts to detect a product of the APV M2-2 gene using antibodies were unsuccessful (1). It is possible that low antibody avidity or low-level expression contributed to the failure to detect the M2-2 protein. Expression of the APV M2-2 gene in a reverse genetics system indicated that the gene product inhibits genome RNA synthesis, as is seen for the RSV M2-2 protein (N. Edworthy and A. Easton, unpublished data). These data suggest that the APV M2-2 gene is translated into a functional product but that the level of protein produced may be very low. To test if the coupled translation mechanism described for the RSV and PVM M2 mRNAs also occurs in APV M2 mRNA, the same approach was used. The APV M2 gene was cloned, and the M2-2 ORF was replaced with the CAT gene (Fig. 3A). The sequences of the various constructs in the region of gene overlap are shown in Fig. 3B. The APV M2-2 ORF contains two potential AUG initiation codons which lie upstream of the stop codon for the M2-1 ORF (Fig. 1B). Each of these was mutated in separate constructs (pAPVmut1 and pAPVmut2), and both were removed (pAPVmut3). In addition, a mutant (pAPVStop1) was generated in which the stop codon for the M2-1 ORF was mutated (UAA→CAA [Fig. 3B]), moving the point of termination of the M2-1 ORF 36 nucleotides further downstream to an in-frame stop codon in the CAT ORF. A second mutant in which this stop codon was also mutated, relocating the stop codon 57 nucleotides downstream from its original location (pAPVStop2) was also constructed. Initial attempts to detect reporter gene expression were unsuccessful with the standard protocol used for the equivalent RSV and PVM plasmids. In order to detect CAT protein expression from the APV constructs, it was necessary to increase the number of transfected cells twofold and the amount of plasmid DNA sixfold while infecting the transfected cells with a higher MOI of the recombinant vaccinia virus necessary to generate T7 RNA polymerase required to direct transcription from the plasmid DNA. Overall, these modifications increase the gene dosage of both reporter plasmid and T7 RNA polymerase gene, thus increasing expression. This allowed expression from the APV M2-2 ORF to be detected and quantified (Fig. 3C). Comparison with the level of CAT expression from the RSV M2 reporter plasmids pWildCAT described previously (2) showed coupled expression of the second ORF in the APV M2 reporter was approximately 67-fold lower (SD, ±16). This suggests that the failure to detect the protein previously was due to the very low level of expression as originally proposed (1).
FIG. 3.
(A) Schematic representation of the reporter gene constructs made to evaluate coupled translation in APV. (B) Sequence alignment of the overlap regions in the APV M2-1/CAT constructs. The CAT ORF is cloned in frame using the XbaI site (italics) to the M2-2 start codon(s) within the overlap. Positions of the M2-2 ORF start codons are underlined and in italics, and the positions of the M2-1 ORF stop codons are shown in boldface. Mutated residues are in lowercase. Additional sequences downstream of the XbaI site in pAPVStop2 are shown with the additional mutation removing the in-frame stop codon highlighted. (C) Levels of CAT protein expression of each mutant relative to the pAPVWC positive control.
Analysis of expression of the APV M2-2 ORF indicated that both putative start codons are functional (Fig. 3C). Deletion of either reduces expression by approximately 20% for the first initiation codon and approximately 45% for the second initiation codon. Deletion of both potential initiation codons dramatically reduced the level of reporter gene expression to background, although as with RSV (1) and PVM (pPVMstop [Fig. 2]), expression was not reduced completely to zero in the assay. Mutagenesis of the M2-1 ORF stop codon in the construct pAPVStop1 to relocate it 36 nucleotides downstream reduced the level of CAT reporter gene expression to approximately 55% of that seen with the “parental” pAPVWC construct. When this further stop codon was mutated, creating construct pAPVStop2 CAT, with the M2-1 translational termination site now 57 nucleotides downstream from its original location, gene expression dropped to 4% of control levels, similar to the situation seen with RSV (2). The distance-dependent translation indicates that the expression of the APV M2-2 ORF is coupled to the translation of the M2-1 ORF since in pAPVStop1 and pAPVStop2 the M2-2 translation initiation codons are not altered and the only effect of the mutations is to alter position of the termination of translation of the M2-1 ORF, which lies downstream of the M2-2 ORF initiation codons.
Role of additional sequences in coupled translation.
It has been shown that the RSV M2 transcript sequences upstream of the region of overlap between the M2-1 and M2-2 ORFs and lying within the M2-1 protein ORF are required for coupled translation to occur (11). The data show that the very low level of coupled translation in the APV M2 gene is due to the absence of sequence/structural elements within the M2-1 ORF that promote coupling to the levels seen in RSV. However, a less likely possibility is that in the APV M2 mRNA the sequence/structure which promotes coupled translation is present in the M2-2 ORF and hence is absent from the construct used to analyze the process due to its replacement with the CAT gene ORF. To investigate this, the CAT ORF was fused in frame at the 3′ terminus of M2-2 ORF, generating an M2-2/CAT fusion protein (pAPVFLWC in Fig. 4A), with all of the M2-2 ORF present. As a control, the M2-2/CAT chimeric ORF was also cloned immediately downstream of the T7 promoter to ensure that the ELISA detection system was not affected (pAPVM2-2CAT in Fig. 4B). Using the standard reporter assay method, no CAT protein could be detected using pAPVFLWC while the M2-2/CAT chimeric protein expressed from pAPVM2-2CAT was detected. This indicates that sequences within the APV M2-2 ORF do not promote coupled translation and that the very low levels of coupled translation that we have detected reflect the situation in vivo.
To further address the question of why coupled expression in APV was much lower than that measured in RSV and PVM, we utilized a series of chimeric M2-1 protein constructs that had been generated for other studies (Stokes et al., unpublished). The overlap region between the 3′ end of the M2-1 ORF and the beginning of the M2-2 ORF was retained and fused with the CAT ORF to generate reporter protein constructs analogous to those in pWildCAT (for the RSV M2 gene) and pAPVWC (Fig. 5). In this way, it was possible to measure the level of coupled translation in the constructs which contained different portions of either the RSV or APV M2-1 ORF. Constructs made were named pR1A1C, pR2A2C, pR3A3C, pA4R4C, pA5R5C, and pA6R6C. A diagrammatic representation of the chimeras indicating the nucleotides present from each of the two virus M2 mRNAs is shown in Fig. 5A. Previously, we identified key segments in the RSV M2-1 ORF as playing a vital role in coupled translation. The frequency of the coupled translation was reduced by progressively deleting portions of the M2-1 ORF, with some regions having a more important role. In particular, a region approximately 150 nucleotides upstream of the overlap region, encoding amino acid residues 143 to 149 of the RSV M2-1 protein and containing a high degree of secondary structure, plays a critical role in the coupling process (11). Three of the chimeric constructs, pR3A3C, pA4R4C, and pA5R5C, contained this critical region of the RSV M2-1 ORF, as indicated in Fig. 5A. Analysis of the levels of CAT protein expression by the chimeric constructs indicated that coupled translation only occurred in those containing the critical RSV sequence (Fig. 5B). This confirmed that this region was able to direct coupled translation, even when combined with the overlap region from a different virus, and that it is functional irrespective of the sequences which lie immediately downstream of it, as long as an overlap region capable of permitting coupled translation is present. In addition, the data for constructs pR1A1C, pR2A2C, and pA6R6C also showed that the APV M2-1 ORF does not contain any sequences which are capable of enhancing coupled translation. This is likely to be the reason for the low levels of expression from the APV M2-2 ORF. Replacement of the overlap region of the APV M2 mRNA with that from RSV in construct pA6R6C, when tested in the more sensitive reporter assay, gave a level of expression from the second ORF 1,300-fold lower than seen with the RSV wild-type sequence (data not shown). These data indicate that the RSV overlap region alone is not capable of efficiently directing the coupling process, as previously reported (11).
DISCUSSION
All viruses rely on the infected cell to provide the necessary components for translation of mRNA, and the study of translational regulation in virus systems has identified novel processes that have subsequently been shown to be used by the host to control protein expression of specific genes. Coupled translation is a novel mechanism that has, to date, been detected in human RSV M2 mRNA and rabbit hemorrhagic disease virus VP10 mRNA (2, 19). The process occurs on mRNAs which contain two overlapping reading frames in which the second ORF is accessed by the ribosomes that first translate the upstream ORF. This requires the presence of sequences upstream of the ORF overlap regions to function efficiently, and only low levels of coupled translation are detected in the absence of these sequences (2, 11, 19). Here, we have shown that coupled translation also occurs in the pneumovirus PVM and the metapneumovirus APV. In both cases, the downstream (M2-2 gene) ORF has two potential AUG translation initiation codons. Mutational analysis showed that both of these codons can be used with maximal expression when both are present. When both start codons were removed, a very low level of expression was detected (Fig. 2C and 3C). The reason for the low but detectable level of expression in the absence of a functional initiation codon is not known. Mutation of the natural stop codon of the upstream (M2-1) ORF so that translation termination is relocated up to 138 and 57 nucleotides downstream of its usual position in PVM and APV, respectively, ablates translation of the second ORF in a distance-dependent manner. This would not be the case if the translation was independent of the first ORF, indicating that coupled translation is utilized by RSV, PVM, and APV to access the M2-2 ORF. Indeed, one of the mutations in the second translation initiation sequence of the PVM M2-2 ORF increased the similarity to the optimal consensus Kozak sequence and would be expected to enhance translation initiated by a conventional scanning process.
Using an ELISA system to accurately measure the levels of protein produced from the second ORF, we have shown that the levels of expression from the second ORFs of the pneumovirus M2 mRNA differ markedly. In the case of the RSV M2 transcript, the levels of coupled translation are approximately 6-fold and 67-fold higher than those seen in the PVM and APV M2 transcripts, respectively. Initial attempts to detect the APV M2-2 protein using monospecific antiserum were unsuccessful, suggesting that the protein may be present in only small amounts (1). Similar attempts to detect hMPV M2-2 protein using monospecific sera were also unsuccessful, also suggesting low levels of expression in vivo (6). The analysis described above showed that the APV M2-2 ORF was indeed accessed by coupled translation but that this was approximately 67-fold less efficient than is seen with RSV. This difference between the pneumoviruses and an MPV may be explained, in part, by consideration of the role of the M2-2 protein and the difference in genome organization of viruses in the two genera. The RSV M2-2 protein has been shown to inhibit virus mRNA synthesis in a concentration-dependent manner, and it has been suggested that it may be responsible for the switch between virus replication and packaging prior to virus particle production (1, 5). A similar activity has been detected for the APV M2-2 protein (Edworthy and Easton, unpublished observations). While the M2-2 ORF can be deleted from the RSV, APV, and hMPV genomes without significant detrimental effect on virus replication in tissue culture (6, 21; R. Ling and A. Easton, unpublished observations), expression of high levels of the M2-2 gene almost completely suppresses RNA synthesis (5). In contrast, the M2-1 protein of pneumoviruses enhances transcription of mRNA (9). Consequently, the balance between the levels of M2-1 and M2-2 proteins in infected cells is critical in determining the outcome of infection and overexpression of the M2-2 gene would be detrimental for the virus. The process of transcription in single-stranded negative-sense RNA viruses such as pneumoviruses occurs in a regulated fashion in which the transcription complex accesses the template at the 3′ end of the genome RNA and genes are transcribed in a linear fashion with progressively less mRNA from genes located further from the 3′ end (8). In this way, a “gradient” of transcription is established. The M2 genes of RSV and PVM are the ninth of the 10 virus genes, and so there will be relatively low levels of M2 mRNA in infected cells with correspondingly low levels of M2-2 protein. However, the genome organization of MPVs such as APV differs from that of the pneumoviruses in having only eight transcription units, which are organized in a slightly different order (8). The result is that the APV M2 gene is the fifth from the 3′ end. Consequently, the level of M2 mRNA relative to those of the other virus mRNAs will be much higher for APV and, if coupled translation were as efficient in this system as seen for RSV, the level of M2-2 protein produced would be likely to limit virus replication. Thus, the reduced efficiency of coupled translation is able to limit M2-2 protein production to acceptable levels. Recently, it was reported that the expression of the hMPV M2-2 protein could be detected even when the upstream ORF had been “silenced” by introduction of translational stop codons, and it was suggested that this indicated that coupled translation did not occur in hMPV (6). However, the authors did not discount the possibility that in the absence of a functional M2-1 ORF that ribosome scanning was used to initiate translation at one of two possible internal AUG codons in the M2-1 reading frame, so allowing coupling to occur, or that scanning ribosomes initiated M2-2 translation directly on the mutant mRNA.
Chimeric RSV and APV M2-1 ORFs were used to dissect further the functional segments in RSV that enable the substantial difference in the levels of coupled translation to be seen (Fig. 5). The data supported the previous findings that a region approximately 150 nucleotides upstream of the M2-1/M2-2 gene overlap region and which has been shown to contain a strong secondary structure feature is a key element for coupled translation. In the chimeric genes, the presence of this region segregated with the ability to couple translation. This showed that the sequences in the RSV M2-1 ORF that are required for efficient coupled translation can also function in conjunction with the overlap region from a different virus. Identification of the important regions in the RSV M2-1 ORF showed that while the region with secondary structure was important, other regions closer to the 5′ end of the mRNA also played a role. Comparison of the results for chimeras pR1A1C and pR2A2C with pR3A3C (Fig. 5B) shows that these additional upstream sequences are necessary for maximal coupled translation but are not sufficient alone to direct the process. The data previously presented using truncated RSV M2-1 constructs support these findings (11). Based on the low levels of coupled translation seen with the APV constructs, it was surprising to detect levels of coupled translation equivalent to those seen with the RSV M2 mRNA in pR3A3C that contains the APV overlap region. This indicates that the low levels of coupled translation seen in APV M2 transcript are due to the effect of the sequences upstream of the overlap and not to a malfunction within the APV overlap region itself. Analysis of the APV M2 mRNA using toeprinting (11) showed that there were significant levels of secondary structure in the mRNA (data not shown). The data from the PVM and APV systems demonstrate that coupled translation can be regulated by upstream sequences and that the precise nature of the structure engendered by these sequences is critical in determining the level of coupled translation that occurs approximately 150 nucleotides downstream. Currently, we are investigating why these structures cannot efficiently direct coupled expression in APV.
In conclusion, we have shown that the process of coupled translation is not restricted to human RSV but is utilized by another pneumovirus, PVM, and an MPV, APV. The levels of expression from the second ORFs directed by the coupling process differ in these viruses, particularly in the MPV, and this may be necessary to ensure correct levels of the M2-2 regulatory protein are produced. The lower level of coupling in the APV system is most likely to be due to the lack of stimulatory sequences in the M2-1 ORF region of the mRNA. The demonstration of coupled translation in an avian virus indicates that the process is also used in nonmammalian systems and is likely to be a general phenomenon available to most eukaryotic systems.
Acknowledgments
We thank Cathy Parry for technical assistance and Tony Marriott and Helen Stokes for the chimeric M2 gene plasmids, Lindsay Thorpe for plasmid pM2-1PVM, and Nicole Edworthy for plasmid pAPVM2-6.9.00.
This work was funded by Biotechnology and Biological Sciences Research Committee grant no. 88/P16683.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Ahmadian, G., P. Chambers, and A. J. Easton. 1999. Detection and characterization of proteins encoded by the second ORF of the M2 gene of pneumoviruses. J. Gen. Virol. 80:2011-2016. [DOI] [PubMed] [Google Scholar]
- 2.Ahmadian, G., J. S. Randhawa, and A. J. Easton. 2000. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 19:2681-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baird, S. D., M. Turcotte, R. G. Korneluk, and M. Holcik. 2006. Searching for IRES. RNA 12:1755-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J., P. Chambers, P. Harriott, C. R. Pringle, and A. J. Easton. 1994. Sequence of the phosphoprotein gene of pneumonia virus of mice: expression of multiple proteins from two overlapping reading frames. J. Virol. 68:5330-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bermingham, A., and P. L. Collins. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. USA 96:11259-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz, U. J., S. Biacchesi, Q. N. Pham, K. C. Tran, L. Yang, C. L. Luongo, M. H. Skiadopoulos, B. R. Murphy, and P. L. Collins. 2005. Deletion of M2 gene open reading frames 1 and 2 of human metapneumovirus: effects on RNA synthesis, attenuation, and immunogenicity. J. Virol. 79:6588-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran, J., and D. Kolakofsky. 1990. Sendai virus P gene produces multiple proteins from overlapping open reading frames. Enzyme 44:244-249. [DOI] [PubMed] [Google Scholar]
- 8.Easton, A. J., J. B. Domachowske, and H. F. Rosenberg. 2004. Animal pneumoviruses: molecular genetics and pathogenesis. Clin. Microbiol. Rev. 17:390-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gould, P. S., and A. J. Easton. 2005. Coupled translation of the respiratory syncytial virus M2 open reading frames requires upstream sequences. J. Biol. Chem. 280:21972-21980. [DOI] [PubMed] [Google Scholar]
- 12.Hemmings-Mieszczak, M., and T. Hohn. 1999. A stable hairpin preceded by a short open reading frame promotes nonlinear ribosome migration on a synthetic mRNA leader. RNA 5:1149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang, S. K. 2006. Internal initiation: IRES elements of picornaviruses and hepatitis c virus. Virus Res. 119:2-15. [DOI] [PubMed] [Google Scholar]
- 14.Jang, S. K., H.-G. Kräusslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapp, L. D., and J. R. Lorsch. 2004. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73:657-704. [DOI] [PubMed] [Google Scholar]
- 16.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latorre, P., D. Kolakofsky, and J. Curran. 1998. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 18:5021-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luttermann, C., and G. Meyers. 2007. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 282:7056-7065. [DOI] [PubMed] [Google Scholar]
- 19.Meyers, G. 2003. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 278:34051-34060. [DOI] [PubMed] [Google Scholar]
- 20.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 21.Teng, M. N., S. S. Whitehead, A. Bermingham, M. St. Claire, W. R. Elkins, B. R. Murphy, and P. L. Collins. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74:9317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yueh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557-1567. [DOI] [PubMed] [Google Scholar]