Abstract
Human immunodeficiency virus type 1 (HIV-1) protease has been continuously evolving and developing resistance to all of the protease inhibitors. This requires the development of new inhibitors that bind to the protease in a novel fashion. Most of the inhibitors that are on the market are peptidomimetics, where a conserved water molecule mediates hydrogen bonding interactions between the inhibitors and the flaps of the protease. Recently a new class of inhibitors, lysine sulfonamides, was developed to combat the resistant variants of HIV protease. Here we report the crystal structure of a lysine sulfonamide. This inhibitor binds to the active site of HIV-1 protease in a novel manner, displacing the conserved water and making extensive hydrogen bonds with every region of the active site.
Human immunodeficiency virus type 1 (HIV-1) protease plays an essential role in the viral life cycle by cleaving Gag and Gag-Pol polyproteins into structural and functional proteins necessary for viral assembly and maturation (3). Therefore, HIV-1 protease is a prime target of drugs developed to control HIV/AIDS, with nine protease inhibitor drugs approved for clinical use since 1995 by the U.S. Food and Drug Administration. The nine protease inhibitors are saquinavir, indinavir (IDV), ritonavir, nelfinavir (NFV), amprenavir (APV), lopinavir (LPV), atazanavir, tipranavir (TPV), and darunavir (DRV/TMC114). All of these drugs are competitive inhibitors that bind in the active site of HIV-1 protease, and all of these inhibitors, except for TPV, are peptidomimetics, i.e., they have a common hydroxyethylene or hydroxyethylamine core element instead of a peptide bond (22). These core elements act as noncleavable peptide isosteres to mimic the transition state formed by the HIV-1 protease substrates during cleavage, thereby effectively inhibiting the enzyme. HIV-1 protease inhibitors were the first drugs to successfully use structure-based drug design. Complexes between peptidomimetic inhibitors and HIV-1 protease are characterized by a noticeable structural feature, a conserved water molecule that mediates contacts between the P2/P1′ carbonyl oxygen atoms of the inhibitors and the amide groups of Ile50/Ile50′ of the enzyme (30). Replacing this conserved water was proposed as a way of making highly specific protease inhibitors (28). This approach was used to design nonpeptidic compounds with seven-membered cyclic urea and sulfamide rings as starting pharmacophores (11, 12). The crystal structures of HIV-1 protease complexes of these two cyclic compounds showed that oxygen atoms on urea and sulfamide groups replace the role of conserved water (1). One of the cyclic urea inhibitors, DMP-450, was shown to have excellent inhibitory properties, was highly potent against the virus in cell cultures, and was orally bioavailable in humans. DMP-450 showed promising results until phase I/II trials, when its development was discontinued due to safety concerns (25). TPV is another protease inhibitor in which the conserved water is replaced by the lactone oxygen atom of the inhibitor's dihydropyrone ring (29). TPV was the first nonpeptidic compound among the currently marketed protease inhibitors.
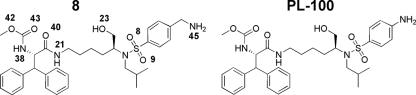
The development of protease inhibitors has improved the life of AIDS patients and contributed to the success of highly active antiretroviral therapy. However, the rapid emergence of resistance to these protease inhibitors has become a major issue. This problem has generated a pressing need to improve current drugs in terms of greater antiretroviral potency, bioavailability, toxicity, and higher activity towards drug-resistant mutant viruses. These goals are being targeted by the development of many second-generation protease inhibitors. One way of developing new drugs is to modify the substituents of existing protease inhibitors or to design totally new molecular cores. Recently lysine sulfonamides were developed as novel HIV-1 protease inhibitors (27). One of these lysine sulfonamides, PL-100, is highly potent against drug-resistant proteases and exhibits a favorable cross-resistance profile against the marketed protease inhibitors (31) (Fig. 1). PL-100 is in phase I human clinical trials with promising results thus far. In this study, we present the synthesis, characterization, and crystal structure of a related lysine sulfonamide-8 (Fig. 1 and Fig. 2) in complex with HIV-1 protease and show that it binds to the active site of protease in a novel mode by displacing the conserved water molecule.
FIG. 1.
The chemical structures of lysine sulfonamide-8 and PL-100. Atoms of lysine sulfonamide-8 that are involved in hydrogen bonding are labeled according to the numbering in the crystal structure.
FIG. 2.
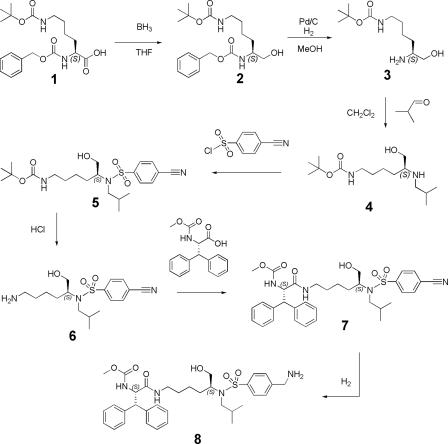
Synthesis of the lysine sulfonamide-8 inhibitor.
MATERIALS AND METHODS
Synthesis of lysine sulfonamide-8 [(S)-(S)-(1-{5-[(4-aminomethyl-benzenesulfonyl)-isobutyl-amino]-6-hydroxy-hexylcarbamoyl}-2,2-diphenyl-ethyl)- carbamic acid methyl ester].
Lysine sulfonamine-8 was synthesized from (S)-(5-benzyloxycarbonylamino-6-hydroxy-hexyl)-carbamic acid tert-butyl ester in a seven-step synthesis as shown in Fig. 2.
Synthesis of (S)-(5-benzyloxycarbonylamino-6-hydroxy-hexyl)-carbamic acid tert-butyl ester (compound 2).
Commercially available (S)-2-benzyloxycarbonylamino-6-tert-butoxycarbonylamino-hexanoic acid (compound 1) (14.89 g) was dissolved in 120 ml dry tetrahydrofuran. This solution was cooled to −10°C. BH3 (80 ml; 1 M in THF) was slowly added, and the resulting solution was stirred for 1 h below −5°C and was allowed to warm to room temperature overnight. The reaction was quenched with MeOH, evaporated to dryness, used as such in the next reaction.
Synthesis of (S)-(5-amino-6-hydroxy-hexyl)-carbamic acid tert-butyl ester (compound 3).
The residue from the first reaction was dissolved in MeOH (150 ml), and Pd/C (3 g) was added. The mixture was placed under an H2 atmosphere and hydrogenated overnight at room temperature (RT). The mixture was filtrated over a pad of Dicalite, evaporated to dryness. The crude compound was purified by column chromatography using ethyl acetate-methanol (MeOH)(NH3) 97-3 as the eluent. After evaporation, an overall yield (over two steps) of 75% was obtained.
Synthesis of (S)-(6-hydroxy-5-isobutylamino-hexyl)-carbamic acid tert-butyl ester (compound 4).
(S)-(5-amino-6-hydroxy-hexyl)-carbamic acid tert-butyl ester (compound 3) (6.85g) was dissolved in 200 ml CH2Cl2. Isobutyraldehyde (2.67 ml) was added, and this solution was stirred for 2 h. Sodium triacetoxyborohydride (1.1 eq) was added, and the solution was stirred for 2 h at RT. The solution was washed with saturated NaHCO3. The milky organic phase was separated and evaporated to dryness. The residue was purified by column chromatography using 99:1 EtOAc (100%) to EtOAc-MeOH(NH3) 99-1 as the eluent. Fractions containing the product were evaporated, yielding 2.60 g (31%) of the title compound.
Synthesis of (S)-{5-[(4-cyano-benzenesulfonyl)-isobutyl-amino]-6-hydroxy-hexyl}-carbamic acid tert-butyl ester (compound 5).
(S)-(6-hydroxy-5-isobutylamino-hexyl)-carbamic acid tert-butyl ester (compound 4) (1.85 g) was dissolved in 50 ml CH2Cl2. Triethylamine (1.05 eq) and 4-cyano benzene sulfonylchloride (1 eq) were added, and the mixture was stirred overnight. Saturated NaHCO3 (50 ml) was added, and the mixture was well shaken. The organic layer was separated, dried on MgSO4, filtrated, and evaporated. The crude compound was purified by column chromatography using EtOAc-heptane (1-3) as the eluent. Fractions containing the product were evaporated, yielding 1.06 g (36%) of solid.
Synthesis of (S)-N-(5-amino-1-hydroxymethyl-pentyl)-4-cyano-N-isobutyl-benzenesulfonamide (compound 6).
(S)-{5-[(4-cyano-benzenesulfonyl)-isobutyl-amino]-6-hydroxy-hexyl}-carbamic acid tert-butyl ester (compound 5) (1.06 g) was dissolved in 50 ml MeOH. HCl (5 to 6 N in isopropanol; 30 ml) was added, and the mixture was stirred overnight at RT. The mixture was evaporated to dryness and used as such in the next reaction.
Synthesis of (S)-(S)-(1-{5-[(4-cyano-benzenesulfonyl)-isobutyl-amino]-6-hydroxy-hexylcarbamoyl}-2,2-diphenyl-ethyl)-carbamic acid methyl ester (compound 7).
(S)-N-(5-amino-1-hydroxymethyl-pentyl)-4-cyano-N-isobutyl-benzenesulfonamide (compound 6) obtained in the previous reaction was dissolved in 50 ml CH2Cl2. BOP (1.2 eq) and triethylamine (5 eq) were added. After 10 min, (S)-2-methoxycarbonylamino-3,3-diphenyl-propionic acid (1 eq) was added and the mixture was stirred at RT for 3 h. Liquid chromatography-mass spectrometry indicated a complete reaction. The reaction mixture was washed with saturated NaHCO3. The aqueous-phase material was washed again with 100 ml CH2Cl2. The combined organic layers were dried with MgSO4, filtered over Dicalite, and evaporated to dryness. The residue was purified by column chromatography with CH2Cl2-MeOH(NH3) (97:3). The appropriate fraction was evaporated to dryness (1.31 g, 84% yield over two steps).
Synthesis of (S)-(S)-(1-{5-[(4-aminomethyl-benzenesulfonyl)-isobutyl-amino]-6-hydroxy-hexylcarbamoyl}-2,2-diphenyl-ethyl)-carbamic acid methyl ester (compound 8).
(S)-(S)-(1-{5-[(4-cyano-benzenesulfonyl)-isobutyl-amino]-6-hydroxy-hexylcarbamoyl{-2,2-diphenyl-ethyl)-carbamic acid methyl ester (compound 7) (1.31 g) was suspended in MeOH containing NH3 (7 N). An aqueous suspension of Ra-Ni was added. The mixture was placed under an atmosphere of H2 and stirred at RT. When the reaction was complete, the mixture was evaporated to dryness and the residue was purified by preparative high-performance liquid chromatography-mass spectrometry. After evaporation, 0.435 g (34%) of white solid, the final lysine sulfonamide (compound 8), was obtained. Light chromatography-mass spectrometry: m/z = 639. 1H nuclear magnetic resonance (CDCl3): 7.78 ppm (d, J = 8.01, 2H); 7.45 ppm (d, J = 7.56, 2H); 7.35 to 7.15 ppm (m, 10H); 6.61 ppm (br s, 1H, NH); 5.58 ppm (d, J = 8.65, 1H, NH); 4.86 ppm (dd, J = 9.74, J = 9.66, 1H); 4.42 ppm (d, J = 10.65, 1H); 4.04 ppm (d, J = 15.42, 1H); 4.02 ppm (d, J = 15.58, 1H); 3.58 ppm (s, 3H); 3.57 to 3.52 (m, 4H); 3.11 to 2.8 (m, 3H); 2.45 (m, 1H); 1.89 (sept, J = 7.11, 1H); 1.34 ppm (m, 2H); 1.1 (m, 1H); 0.99 to 0.96 ppm (m, 6H); 0.95 to 0.75 (m, 2H).
Cell-based anti-HIV and toxicity assay.
The antiviral activity was determined with a cell-based replication assay. This assay directly measures the ongoing replication of virus in MT4 cells via the specific interaction of HIV Tat with long-terminal-repeat sequences coupled to green fluorescent protein. In the toxicity assay, a reduced expression of the green fluorescent protein reporter protein serves as a marker for the cellular toxicity of a compound. Briefly, various concentrations of the test compound are brought into a 384-well microtiter plate. Subsequently, MT4 cells and HIV-1/LAI (wild type) are added to the plate at a concentration of 150,000 cells/ml or 200 cell culture 50% infectious doses, respectively. To determine the toxicity of the test compound, mock-infected cell cultures containing an identical compound concentration range are incubated for 3 days (37°C, 5% CO2) in parallel with the HIV-infected cell cultures. On the basis of the calculated percent inhibition for each compound concentration, dose-response curves are plotted and 50% effective (EC50) and cytotoxic (CC50) concentrations are calculated.
Protein expression and purification.
The wild-type protease was expressed from a synthetic gene optimized for Escherichia coli codon usage with Q7K to prevent autoproteolysis (23). The protease was expressed and purified as previously described (7). The protein was refolded by rapid dilution in a 10-fold volume of 0.05 M sodium acetate buffer at pH 5.5, containing 10% glycerol, 5% ethylene glycol, and 5 mM dithiothreitol (refolding buffer). To reduce the volume, the protease was concentrated and dialyzed to remove any residual acetic acid. Protease used for crystallization was further purified with a Pharmacia Superdex 75 fast-performance liquid chromatography column equilibrated with refolding buffer.
Crystallization and data collection.
Crystals were set up with a threefold molar excess of inhibitor to protease, which ensures ubiquitous binding. The concentration of the protein was 1.6 mg/ml in refolding buffer. The hanging drop method was used for crystallization as previously described (20). The reservoir solution consisted of 126 mM phosphate buffer at pH 6.2, 63 mM sodium citrate, and 26% ammonium sulfate.
Intensity data were collected on an in-house Rigaku X-ray generator equipped with an R-axis IV image plate system. Data were collected at −80°C. Approximately 180 3-min frames were collected with 1-degree oscillations and no overlap between frames. The frames were integrated and scaled using the programs DENZO (XDISPLAYF; W. Minor, Purdue University, West Lafayette, IN, 1999) and ScalePack (17), respectively. The data collection statistics are listed in Table 1.
TABLE 1.
Crystallographic statistics for the lysine sulfonamide (compound 8)-protease complex
| Parameter | Value or identifier |
|---|---|
| Data collection | |
| Space group | P212121 |
| Z | 4 |
| a (Å) | 51.11 |
| b (Å) | 58.10 |
| c (Å) | 61.60 |
| Resolution (Å) | 2.0 |
| Total no. of reflections | 81058 |
| No. of unique reflections | 12879 |
| Rmerge (%) | 7.2 |
| Completeness (%) | 99.2 |
| I/σI | 13.1 |
| Refinement statistics | |
| Rwork value (%) | 18.7 |
| Rfree value (%) | 23.2 |
| RMSDa | |
| Bond length (Å) | 0.007 |
| Bond angles (°) | 1.260 |
| No. of waters | 110 |
| Protein Data Bank code | 2Q3K |
Root mean square deviation.
Structure solution and crystallographic refinement.
The CCP4i interface to the CCP4 suite (2) was used to refine the structure. The structure was solved with the molecular replacement package AMoRe (16), with 1F7A (19) as the starting model. A radius of integration of 25 Å and X-ray data within 8.0 to 3.0 Å were used for the structure solution. The molecular replacement phases were further improved by using ARP/wARP (14) to build solvent molecules into the unaccounted regions of electron density. Difference Fourier maps were computed and inspected with the interactive graphic program O (6), and major structural changes were incorporated in the model, such as inclusion of ligand and solvent molecules. Conjugate gradient refinement using Refmac5 (15) was performed by incorporating the Schomaker and Trueblood tensor formulation of translation, libration, and screw rotation parameters (10, 26; I. J. Tickle and D. S. Moss, presented at the IUCr99 Computing School, London, United Kingdom, 1999). The working R value (Rfactor) and its cross-validation (Rfree) were monitored throughout the refinement. The refinement statistics are also shown in Table 1. All of the illustrations were made using the program PyMOL (4).
RESULTS
Binding affinity.
The binding affinity of the lysine sulfonamide-8 inhibitor was determined in vivo. In viral culture, lysine sulfonamide-8 had a 50% effective concentration of 0.030 μM against a laboratory HIV-1/LAI wild-type strain. Cytotoxicity measurements of the same culture showed that this inhibitor has a cytotoxicity level higher than 30 μM, resulting in a selective index of greater than 1,000.
Overall structure.
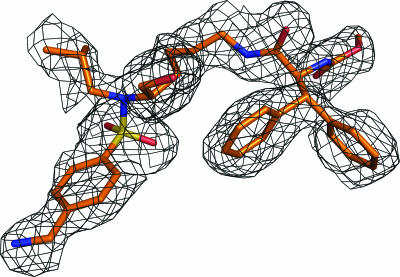
We solved and refined to 2.0 Å the crystal structure of the complex of wild-type protease with the lysine sulfonamide-8 inhibitor, which crystallized in the P212121 space group with one protease dimer per asymmetric unit. The inhibitor was modeled in one orientation, with continuous electron density for the entire molecule except for the isopropyl group at the P1′ position (Fig. 3). The final Rfactor value is 18.7% (Rfree = 23.2%). The crystallographic statistics are listed in Table 1.
FIG. 3.
Fo-Fc inhibitor omit electron density map contoured at 1.8 σ for lysine sulfonamide-8, showing the inhibitor is in single orientation.
Inhibitor protease hydrogen bonds.
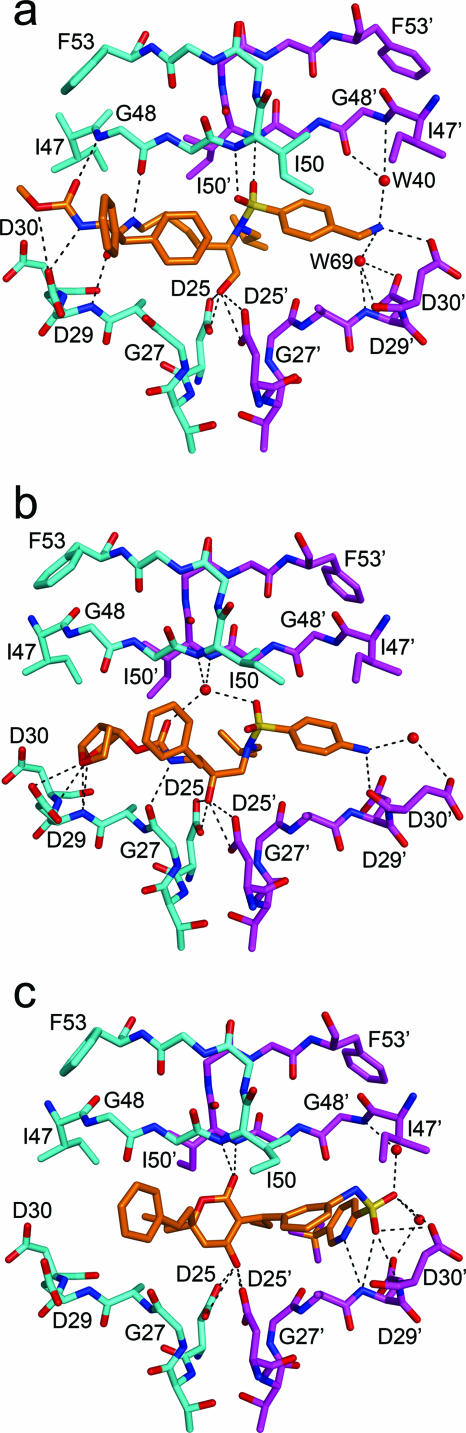
The lysine sulfonamide-8 inhibitor displays a novel hydrogen bonding pattern compared to other protease inhibitors. Table 2 shows the hydrogen bonding distances between the protease and the inhibitor. Figure 4a shows hydrogen bonding between atoms in the protease active site and atoms in the inhibitor, including those mediated by water. The catalytic aspartic acids of the enzyme, D25 and D25′, are within hydrogen bonding distance of the hydroxyl group of this lysine sulfonamide. The inhibitor makes 12 hydrogen bonds with the protease. Except for one interaction, all hydrogen bonds are between the inhibitor and either protease main-chain atoms or side-chain atoms of conserved residues (Asp25 and Asp29). The two nitrogen atoms of Ile50 and Ile50′ at the tips of the flaps form hydrogen bonds with the two oxygen atoms of the inhibitor's sulfonyl group. This hydrogen bonding is a novel structural feature; only the cyclic sulfamide inhibitors (which have not yet been clinically viable) have exhibited a similar interaction. Generally most protease-inhibitor and all protease-substrate complexes have a water molecule tetrahedrally coordinating the nitrogen atoms of Ile50 and Ile50′ with the inhibitor atoms.
TABLE 2.
Hydrogen bonding distances between protease and inhibitor atoms in the crystal structure of the lysine sulfonamide (compound 8)
| Protease atom | Inhibitor atom | Distance (Å) |
|---|---|---|
| Asp30′ OD2 | N45 | 2.8 |
| Water 40 | N45 | 2.9 |
| Water 69 | N45 | 3.1 |
| Ile50 N | O8 | 3.1 |
| Ile50′ N | O9 | 2.9 |
| Asp25 OD1 | O23 | 3.1 |
| Asp25 OD2 | O23 | 2.7 |
| Asp25′ OD1 | O23 | 2.9 |
| Asp25′ OD2 | O23 | 2.6 |
| Gly48 O | N21 | 2.8 |
| Asp29 OD2 | N38 | 2.9 |
| Asp29 N | O40 | 2.9 |
| Asp29 OD2 | O42 | 3.3 |
| Water 37 | O42 | 2.8 |
| Gly48 N | O43 | 3.1 |
FIG. 4.
(a) Hydrogen bonding interactions between the HIV-1 protease and lysine sulfonamide-8. The inhibitor makes hydrogen bonds directly to the flaps, unlike the case for other peptidomimetic inhibitors. (b) Hydrogen bonding interactions between the HIV-1 protease and the FDA-approved peptidomimetic inhibitor DRV. A water molecule mediates hydrogen bonds between the flaps of the protease and the inhibitor, which is typical for all of the peptidomimetic inhibitors. (c) Hydrogen bonding between the HIV-1 protease and the nonpeptidomimetic inhibitor TPV. Notice the absence of the water-mediated hydrogen bonds with the flaps. The inhibitor directly forms hydrogen bonds with the flaps of the protease.
The amide and carbamate groups of lysine sulfonamide-8 form hydrogen bonds with residue G48 in the flap and with residue D29 in the floor of the active site. At the other end of the inhibitor, the amine group forms a hydrogen bond with the carboxyl group of D30 of the protease. This amine group also forms water-mediated hydrogen bonds with the residues G48 in the flap and D29 at the bottom of the active site. The important feature of the lysine sulfonamide-8 inhibitor-protease hydrogen bonds is their involvement with residues in both the flap (G48 and I50) and the active site (D25, D29, and D30). This feature is distinct from the inhibitor-protease hydrogen bonds formed by peptidomimetic inhibitors (IDV, NFV, DRV, APV, and LPV), which do not form hydrogen bonds with the flap residues (18). The substrates (and acetyl-pepstatin), in contrast, form hydrogen bonds with the flap residue, G48 (9), and in certain cases even with M46 (21). Thus, in some ways lysine sulfonamide-8 better mimics the substrate hydrogen bonds.
vdW contacts.
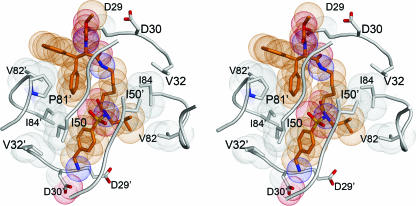
The lysine sulfonamide-8 inhibitor packs in an extended conformation in the active site by forming 134 van der Waals (vdW) contacts to the protease, with an interatomic distance of <4.2 Å (Fig. 5). The isopropyl group of the P1′ site is surrounded by residues L23, D25, I84, G27′, and I50′ of the protease. The benzyl amine group of P2′ forms vdW contacts with I50, G48′, and G49′ of the flap as well as with D30′ and V32′. Because of the lysine-like side chain in the center of the inhibitor, the inhibitor fills P1 and P2 in a novel manner. This aliphatic nonpolar chain is within vdW distance of the nonpolar residues A28, I47, G48, I50′, and I84 of the protease. An aromatic diphenyl methyl group of the lysine sulfonamide occupies the P3 position. One of the phenyl rings of this group points in the direction of the P1 site. This phenyl ring is within vdW distance of many protease residues (L23′, D25′, G27, G48, G49, I50, P81′, and I84′), whereas the other phenyl ring forms vdW contacts with only three residues (R8′, P81′, and V82′). Thus, the lysine sulfonamide inhibitor packs within the active site in a novel manner, making several vdW contacts.
FIG. 5.
Stereo view of the packing of the HIV-1 protease around the inhibitor lysine sulfonamide-8 (in orange).
DISCUSSION
By solving the structure of a complex between HIV-1 protease and a lysine sulfonamide inhibitor, we have shown that this inhibitor binds in a novel way to the active site compared to other protease inhibitors in clinical use. Lysine sulfonamide-8 is a nonpeptidic, competitive inhibitor similar to PL-100, which is in phase I clinical trials. The hydrogen bonding pattern of the PL-100-protease complex is likely to be similar to that of the protease complex of the lysine sulfonamide-8 inhibitor, except for bonds made by the amine group at P2′, where the benzyl amine is replaced by a phenyl amine in PL-100. The lysine sulfonamide-8 inhibitor forms hydrogen bonds mostly to main-chain atoms or conserved side-chain atoms of the protease. This hydrogen bonding pattern is a favorable situation for the inhibitor, since it is less likely to be affected by drug resistance mutations in protease residues of the active site. The only hydrogen-bonding interaction of lysine sulfonamide-8 with a nonconserved residue side chain is from the P2′ amine group to D30′. This interaction may not be present in PL-100, since it has a P2′ phenyl amine group, one methylene group less than the benzyl amine group of the lysine sulfonamide-8 presented here. Otherwise, PL-100 likely also forms water-mediated hydrogen bonds similar to what was observed (Fig. 4a). Overall the hydrogen bonding pattern between the two lysine sulfonamides is likely to be very similar.
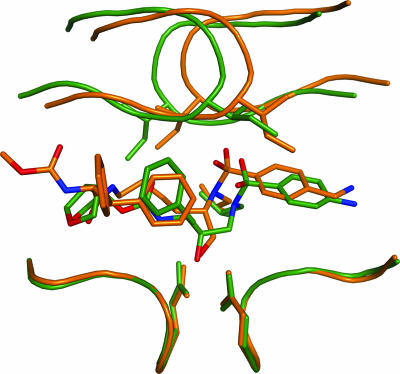
The lysine sulfonamide-8 inhibitor, and hence PL-100, has properties that contribute to its binding to HIV-1 protease differently from other protease inhibitors. It has a primary OH group that interacts with the two catalytic aspartic acids, D25 and D25′, whereas all other peptidomimetic protease inhibitors have a secondary OH group that interacts with D25 and D25′. The hydrogen bonding between this primary OH group in lysine sulfonamide-8 and D25 and D25′ is facilitated by the methylene group that connects the hydroxyl group and the inhibitor core; the methylene group pushes the entire inhibitor towards the protease flaps. This displacement in turn brings the two oxygen atoms of the inhibitor's sulfone group within hydrogen-bonding distance of I50 and I50′. All other peptidomimetic protease inhibitors lack the methylene group, likely accounting for the different binding of lysine sulfonamide-8. For instance, in DRV, a peptidomimetic inhibitor whose P1′ and P2′ are similar to those of this lysine sulfonamide, the secondary hydroxyl group forms hydrogen bonds with D25 and D25′ (Fig. 4b) (8). However, DRV is closer than lysine sulfonamide-8 to the bottom of the active-site cavity and further from the flap region of the protease. Compared to lysine sulfonamide-8, DRV has an additional carbon atom between the sulfonamide nitrogen and the carbon attached to the hydroxyl group. This intervening carbon atom displaces the sulfonyl group laterally and also towards the flaps of the protease. This displacement of the sulfonyl group may make direct hydrogen bonding to the flaps difficult, but it facilitates water-mediated hydrogen bonding with the flaps (Fig. 6). The presence of a primary hydroxyl group instead of a secondary hydroxyl group might be a primary reason for the novel binding of lysine sulfonamide-8 to the protease, resulting in the absence of the conserved water molecule in the crystal structure.
FIG. 6.
Superimposition of the crystal structure of the DRV complex (in green) on the lysine sulfonamide-8 complex (in orange).
TPV and the lysine sulfonamides are all nonpeptidomimetic protease inhibitors with a binding mode different from that of other protease inhibitors (Fig. 4c). The lysine sulfonamides and TPV have two features that distinguish them from other protease inhibitors. First, these inhibitors do not form hydrogen bonds with G27 in the floor of the protease active site. This interaction with G27 is conserved in all protease-substrate and protease-inhibitor complexes. Second, these inhibitors form hydrogen bonds directly with the protease flaps, without the mediation of a water molecule, although each inhibitor binds differently. In TPV, the lactone oxygen atom of the dihydropyrone ring forms hydrogen bonds with the amide nitrogen atoms of the flap residues, analogous to how water molecules in peptidomimetic inhibitors interact with flap residues. In the lysine sulfonamide-8 inhibitor, two oxygen atoms from the sulfone group form two hydrogen bonds with the flap residues. This kind of interaction distinguishes the binding of lysine sulfonamide-8 not only from that of TPV but also from that of all known crystal structures of protease-inhibitor complexes that are in clinical use.
Insights can be gained into PL-100's pattern of selecting for protease mutations in vitro from the crystal structure of the related lysine sulfonamide-8. When PL-100 was subjected to an in vitro test of its ability to select for protease mutants conferring resistance (31), a novel selection pattern of four mutations (K45R, M46I, T80I, and P81S) was found. T80 and P81 are previously invariant (conserved) residues in the patient population, while the K45R and M46I mutations are observed commonly with the other inhibitor therapy as compensatory mutations. The observed mutation pattern may be explained from the crystal structure of lysine sulfonamide-8, which forms extensive vdW contacts with the residue P81′ as shown in Fig. 5. These vdW contacts will likely be lost when P81 gets mutated to serine. The interactions of P81 are important for the binding of the lysine sulfonamide-8 inhibitor/PL-100 and hence the protease may be developing resistance to PL-100 by mutating the residue P81. Although T80, K45, and M46 do not make direct contacts with the inhibitor, they mutate to compensate for other changes. This may account for the fact that single, double, or triple viral mutants did not show resistance to PL-100 and only mild resistance was observed with the quadruple viral mutant. PL-100 does not show extensive cross-resistance to the clinical protease inhibitors APV, LPV, atazanavir, saquinavir, IDV, and NFV. All of these inhibitors select for signature protease mutations in the active site, which do not affect the hydrogen-bonding pattern of PL-100. Thus, the experimental observations for PL-100 are consistent with predictions of its structure from the similar crystal structure of the lysine sulfonamide-8 in complex with HIV-1 protease.
HIV-1 resists efforts to find a cure that will eradicate the virus from infected individuals or to develop a vaccine. Until a safe and effective vaccine against HIV-1 is developed, new drugs need to be developed that can reach high levels in plasma and possibly overcome the cross-resistance among various inhibitors. One approach to overcoming cross-resistance could be to design new inhibitors that not only bind tightly to mutant proteases but also bind in a mode different from that of existing inhibitors. Support for this approach comes from studies with TPV, which has a different binding mode and has been shown to bind to drug-resistant mutants (13, 24). The crystal structure of the lysine sulfonamide inhibitor-protease complex presented here provides a new direction for designing protease inhibitors. In fact, PL-100, which is similar to lysine sulfonamide-8, is currently in phase I clinical trials and showing promising results. Now, however, with the structure of lysine sulfonamide-8 in complex with HIV-1 protease, it is possible to design other inhibitors that fill the active-site cavity in a novel manner.
Acknowledgments
We acknowledge the assistance of Rajintha Bandaranayake, Anna Buabbud, Shivender Shandilya, and Claire Baldwin in the preparation of the manuscript.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Backbro, K., S. Lowgren, K. Osterlund, J. Atepo, T. Unge, J. Hulten, N. M. Bonham, W. Schaal, A. Karlen, and A. Hallberg. 1997. Unexpected binding mode of a cyclic sulfamide HIV-1 protease inhibitor. J. Med. Chem. 40:898-902. [DOI] [PubMed] [Google Scholar]
- 2.Collaborative-Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 3.Debouck, C. 1992. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retrovir. 8:153-164. [DOI] [PubMed] [Google Scholar]
- 4.DeLano, W. L. 2002. The PyMOL user's manual. DeLano Scientific, San Carlos, CA.
- 5.Fitzgerald, P. M. D., B. M. McKeever, J. F. VanMiddlesworth, J. P. Springer, J. C. Heimbach, C. T. Leu, W. K. Herber, R. A. F. Dixon, and P. L. Darke. 1990. Crystallographic analysis of a complex between human immunodeficiency virus type 1 protease and acetyl-pepstatin at 2.0-A resolution. J. Biol. Chem. 265:14209-14219. [PubMed] [Google Scholar]
- 6.Jones, T. A., M. Bergdoll, and M. Kjeldgaard. 1990. O: a macromolecular modeling environment, p. 189-195. In C. Bugg and S. Ealick (ed.), Crystallographic and modeling methods in molecular design. Springer-Verlag Press, Berlin, Germany.
- 7.King, N. M., L. Melnick, M. Prabu-Jeyabalan, E. A. Nalivaika, S. S. Yang, Y. Gao, X. Nie, C. Zepp, D. L. Heefner, and C. A. Schiffer. 2002. Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease. Protein Sci. 11:418-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, P. Wigerinck, M. P. de Bethune, and C. A. Schiffer. 2004. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 78:12012-12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar, M., V. Prashar, S. Mahale, and M. V. Hosur. 2005. Observation of a tetrahedral reaction intermediate in the HIV-1 protease-substrate complex. Biochem. J. 389:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuriyan, J., and W. I. Weis. 1991. Rigid protein motion as a model for crystallographic temperature factors. Proc. Natl. Acad. Sci. USA 88:2773-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam, P. Y. S., P. K. Jadhav, C. J. Eyermann, C. N. Hodge, G. V. DeLucca, and J. D. Rodgers. 1 September 1994. Patent WO9419329-A.
- 12.Lam, P. Y. S., P. K. Jadhav, C. J. Eyermann, C. N. Hodge, Y. Ru, L. T. Bacheler, J. L. Meek, M. J. Otto, M. M. Rayner, Y. N. Wong, et al. 1994. Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science 263:380-384. [DOI] [PubMed] [Google Scholar]
- 13.Larder, B. A., K. Hertogs, S. Bloor, C. H. van den Eynde, W. DeCian, Y. Wang, W. W. Freimuth, and G. Tarpley. 2000. Tipranavir inhibits broadly protease inhibitor-resistant HIV-1 clinical samples. AIDS 14:1943-1948. [DOI] [PubMed] [Google Scholar]
- 14.Morris, R. J., A. Perrakis, and V. S. Lamzin. 2002. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr. D 58:968-975. [DOI] [PubMed] [Google Scholar]
- 15.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 16.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. A 50:157-163. [Google Scholar]
- 17.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 18.Prabu-Jeyabalan, M., N. M. King, E. A. Nalivaika, G. Heilek-Snyder, N. Cammack, and C. A. Schiffer. 2006. Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease. Antimicrob. Agents Chemother. 50:1518-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prabu-Jeyabalan, M., E. Nalivaika, and C. A. Schiffer. 2000. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J. Mol. Biol. 301:1207-1220. [DOI] [PubMed] [Google Scholar]
- 20.Prabu-Jeyabalan, M., E. A. Nalivaika, N. M. King, and C. A. Schiffer. 2003. Viability of a drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy. J. Virol. 77:1306-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabu-Jeyabalan, M., E. A. Nalivaika, and C. A. Schiffer. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369-381. [DOI] [PubMed] [Google Scholar]
- 22.Randolph, J. T., and D. A. DeGoey. 2004. Peptidomimetic inhibitors of HIV protease. Curr. Top. Med. Chem. 4:1079-1095. [DOI] [PubMed] [Google Scholar]
- 23.Rose, J. R., R. Salto, and C. S. Craik. 1993. Regulation of HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J. Biol. Chem. 268:11939-11945. [PubMed] [Google Scholar]
- 24.Rusconi, S., S. La Seta Catamancio, P. Citterio, S. Kurtagic, M. Violin, C. Balotta, M. Moroni, M. Galli, and A. d'Arminio-Monforte. 2000. Susceptibility to PNU-140690 (Tipranavir) of human immunodeficiency virus type 1 isolates derived from patients with multidrug resistance to other protease inhibitors. Antimicrob Agents Chemother. 44:1328-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rusconi, S., and O. Vigano. 2006. New HIV protease inhibitors for drug-resistant viruses. Therapy 3:79-88. [Google Scholar]
- 26.Schomaker, V., and K. N. Trueblood. 1968. Acta Crystallogr. B 24:63-76. [Google Scholar]
- 27.Stranix, B. R., G. Sauve, A. Bouzide, A. Cote, G. Sevigny, and J. Yelle. 2003. Lysine sulfonamides as novel HIV-protease inhibitors: optimization of the Nepsilon-acyl-phenyl spacer. Bioorg Med. Chem. Lett. 13:4289-4292. [DOI] [PubMed] [Google Scholar]
- 28.Swain, A. L., M. M. Miller, J. Green, D. H. Rich, J. Schneider, S. B. Kent, and A. Wlodawer. 1990. X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor. Proc. Natl. Acad. Sci. USA 87:8805-8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner, S. R., J. W. Strohbach, R. A. Tommasi, P. A. Aristoff, P. D. Johnson, H. I. Skulnick, L. A. Dolak, E. P. Seest, P. K. Tomich, M. J. Bohanon, M. M. Horng, J. C. Lynn, K. T. Chong, R. R. Hinshaw, K. D. Watenpaugh, M. N. Janakiraman, and S. Thaisrivongs. 1998. Tipranavir (PNU-140690): a potent, orally bioavailable nonpeptidic HIV protease inhibitor of the 5,6-dihydro-4-hydroxy-2-pyrone sulfonamide class. J. Med. Chem. 41:3467-3476. [DOI] [PubMed] [Google Scholar]
- 30.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:543-585. [DOI] [PubMed] [Google Scholar]
- 31.Wu, J. J., S. Dandachel, B. Stranix, C. Panchal, and M. A. Wainberg. 2006. The HIV-1 protease inhibitor PL-100 has a high genetic barrier and selects a novel pattern of mutations, abstr. 136. Antivir. Ther. 11:S152.. [Google Scholar]