Abstract
We have previously shown that synthesis of poliovirus protein 3CD in uninfected HeLa cell extracts induces an increased association with membranes of the cellular Arf GTPases, which are key players in cellular membrane traffic. Arfs cycle between an inactive, cytoplasmic, GDP-bound form and an active, membrane-associated, GTP-bound form. 3CD promotes binding of Arf to membranes by initiating recruitment to membranes of guanine nucleotide exchange factors (GEFs), BIG1 and BIG2. GEFs activate Arf by replacing GDP with GTP. In poliovirus-infected cells, there is a dramatic redistribution of cellular Arf pools that coincides with the reorganization of membranes used to form viral RNA replication complexes. Here we demonstrate that Arf translocation in vitro can be induced by purified recombinant 3CD protein; thus, concurrent translation of viral RNA is not required. Coexpression of 3C and 3D proteins was not sufficient to target Arf to membranes. 3CD expressed in HeLa cells was retained after treatment of the cells with digitonin, indicating that it may interact with a membrane-bound host factor. A F441S mutant of 3CD was shown previously to have lost Arf translocation activity and was also defective in attracting the corresponding GEFs to membranes. A series of other mutations were introduced at 3CD residue F441. Mutations that retained Arf translocation activity of 3CD also supported efficient growth of virus, regardless of their effects on 3D polymerase elongation activity. Those that abrogated Arf activation by 3CD generated quasi-infectious RNAs that produced some plaques from which revertants that always restored the Arf activation property of 3CD were rescued.
Picornaviridae is a family of small RNA viruses, of which poliovirus is perhaps the best-studied member. Its genome is a single-stranded RNA of about 7,500 nucleotides in length that initiates translation into a single large polyprotein via an internal ribosome entry site mechanism. The polyprotein is cleaved in cis and in trans by virus-encoded proteases to produce about 10 final products as well as a number of cleavage intermediates. Often intermediate cleavage products perform specific independent functions in the viral replication cycle, distinct from those of the final proteins processed from complete polyprotein maturation, thereby effectively increasing the coding capacity of the relatively small genome. Protein 3CD is an uncleaved precursor composed of protease 3C and RNA-dependent RNA polymerase 3D sequences. 3C is responsible for the majority of proteolytic events that generate the viral nonstructural replication proteins, while 3D catalyzes reactions of initiation and elongation of both plus and minus RNA strands during poliovirus genome replication. 3CD manifests no detectable polymerase activity, but it possesses protease activity with substrate recognition properties distinct from those of 3C protein. The recently resolved atomic structure of 3CD shows that the protein retains most structural aspects of its component 3C and 3D domains, despite their linkage (23, 28, 36).
Numerous functions have been attributed to protein 3CD during the poliovirus replication cycle. Cleavage of the viral capsid proteins, located in the N-terminal portion of the poliovirus polyprotein, requires 3CD-catalyzed proteolysis (44). In addition, 3CD protease activity has been implicated in cleavage of several host proteins. An unidentified cellular factor that generated products incorporated into the viral replication complex was shown to be cleaved by 3CD (33). 3CD also may participate in cleavage of cellular polypyrimidine tract binding protein, which facilitates internal ribosome entry site-mediated translation of poliovirus RNA, and may contribute to the switch from RNA translation to replication (3). Sharma et al. demonstrated that 3CD enters the nuclei of infected cells, where it can initiate degradation of specific transcription factors, leading to shutoff of cellular transcription (34).
In addition to its proteolytic activities, 3CD also has been shown to mediate multiple steps in the pathway of viral RNA replication by virtue of its specific interactions with viral RNA structures and viral or cellular proteins. The protein binds to the cloverleaf structure at the 5′ terminus of poliovirus RNA, and this binding was shown to be important for virus replication (1, 20, 30). This interaction of 3CD with the 5′ nontranslated region of poliovirus RNA was proposed to divert RNA from directing translation to serving as a template for replication (21). 3CD also stimulates uridylylation of viral protein VPg, catalyzed by 3D, which is required for initiation of RNA replication, by binding to the template for uridylylation on the cis-acting replication element (31, 43). The cellular heterogeneous nuclear ribonucleoprotein (hnRNP) C1/C2, which forms a specific RNP complex with the poliovirus 3′ nontranslated region that is thought to be important for replication, was shown to interact with 3CD in vitro (9). Finally, addition of 3CD to an in vitro translation-replication system for poliovirus RNA was reported to significantly stimulate the yield of infectious particles, although the mechanism of this stimulation remains unclear (19, 37).
During recent work initiated in this laboratory to study the role of membranes in the poliovirus RNA replication reaction, another activity of viral protein 3CD was discovered. We demonstrated that 3CD (as well as another poliovirus protein, 3A), induces the recruitment to membranes of cellular proteins from the family of small GTPases, designated Arfs (ADP ribosylation factors) (7). The binding of Arfs to membranes appears to be essential for viral RNA replication, as the fungal metabolite brefeldin A, which inhibits Arf activation, also inhibits poliovirus replication in vivo and in vitro (13, 24). Arfs are important components of intracellular membrane trafficking pathways. They cycle between GTP- and GDP-bound states, with Arf-GTP, referred to as the activated form, bound to membrane and Arf-GDP resulting from cellular GTPase activity, being released to the cytoplasm. Exchange of the nucleoside diphosphate to regenerate Arf-GTP requires interaction with guanine nucleotide exchange factors (GEFs). Different GEFs are associated with different intracellular compartments and preferentially work with a limited subset of Arf family members, thus conferring specificity to the process of Arf activation. Membrane-associated Arf-GTP induces changes in membrane curvature and recruits other proteins that facilitate formation of transport vesicles from intracellular membrane organelles. They also regulate the activities of enzymes involved in lipid metabolism and cytoskeleton function (4, 29). Viral proteins 3CD and 3A exploit different pathways to induce targeting of Arf to membranes. While 3A induces such membrane binding through involvement of the GEF GBF1, 3CD-dependent Arf activation appears to rely on two other GEFs, BIG1 and BIG2 (5). 3A is a membrane-associated protein, and available data suggest that it can directly interact with GBF1 (reference 40 and our unpublished results). 3CD, on the other hand, does not have any intrinsic membrane binding property, and therefore the mechanism by which it interferes with the metabolism of membrane-associated proteins is not obvious. In this study we characterized Arf recruitment by 3CD and investigated the effect of this process on poliovirus replication.
MATERIALS AND METHODS
Plasmids.
pXpA-SH, containing full-length cDNA of poliovirus type 1 under control of the T7 promoter with the hammerhead ribozyme-coding sequence at the 5′end of the poliovirus sequence and additional unique restriction sites within the viral cDNA, and plasmids used for transcription of RNAs coding for individual poliovirus proteins have been previously described (7). The pXpA-RenR plasmid, encoding a poliovirus replicon with the Renilla luciferase gene, was described previously (5). A plasmid coding for the Sec61-green fluorescent protein (GFP) fusion protein was a gift from Nihal Altan-Bonnet, Rutgers University, Newark, NJ. Plasmid pMA-3CD for expression of 3CD under control of the cytomegalovirus promoter was produced by inserting the SalI-HpaI fragment of pXpA-3CD (7) into expression vector pM1-MT (Roche) cut with SalI-EcoRV.
Recombinant proteins.
Untagged purified recombinant 3CD, 3D, and 3AB proteins were gifts from Oliver Richards, University of Colorado.
Antibodies.
Antibodies against BIG1 and BIG2 were the generous gifts of M. Vaughan and J. Moss (NHLBI). Monoclonal anti-poliovirus 3D was a gift from K. Bienz and D. Egger, University of Basel, Switzerland. Anti-Arf and anti-Rab9 monoclonal antibodies were from Affinity Bioreagents. Anti-GGA3 antibodies were from Becton Dickinson. Secondary antibody conjugates used for immunofluorescence were from Molecular Probes; those used in Western blotting were from Amersham.
In vitro translation.
In vitro transcription and translation of viral RNAs and immunoblotting of the membrane fractions were performed essentially as described previously (7). Briefly, HeLa cell S10 extracts were programmed with poliovirus-specific RNAs and incubated for 3.5 h at 34°C, and the membrane-associated material was collected by centrifugation at 16,100 × g for 20 min at 4°C prior to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Effects of 3CD on production of infectious virus in vitro.
Virion RNA (500 ng) was translated in HeLa S10 extract at 34°C in the presence of unlabeled amino acids and nucleotides in a total reaction volume of 25 μl (13, 27). 3CD (wild type [wt]) or 3CD (mutant) mRNA (140 ng) was added at the start of the reaction as indicated. After incubation for 12 to 15 h, the samples were diluted with phosphate-buffered saline (PBS) and were added to HeLa cell monolayers. Virus titers were determined by plaque assay, as described previously (22).
Digitonin treatment.
Cells grown on coverslips were washed once with KHM buffer (110 mM K-acetate, 2 mM MgCl2, 20 mM HEPES-KOH, pH 7.4), treated for 1 min with 50 μM digitonin in KHM buffer, and washed with KHM buffer. Cells were then fixed for 20 min with 4% formaldehyde in PBS and processed for immunofluorescence microscopy.
Microscopy.
HeLa cells, grown on coverslips and fixed with 4% paraformaldehyde-PBS for 20 min, were permeabilized with 0.2% Triton X-100 in PBS for 5 min. The cells were then incubated in 3% nonfat dry milk solution for 1 h to block nonspecific binding sites. This solution also was used for dilution of primary and secondary antibodies with which cells were sequentially incubated for 1 h each. Images were taken with a Leica DMIRE microscope. Digital images were processed with Adobe Photoshop software.
Expression of 3CD for electron microscopy.
HeLa cells grown in 35-mm plates were simultaneously transfected with plasmid pXpA-3CD, encoding 3CD under T7 promoter control, and infected with vaccinia virus vTF7-3, expressing T7 polymerase, as described previously (35). After 16 h, cells were fixed and processed for electron microscopy.
Transfection.
DNA transfections were performed with Fugene 6 reagent (Roche), and RNA transfections utilized the Trans-It mRNA transfection kit (Mirus) according to the manufacturers’ instructions.
3D polymerase elongation assay.
Two microliters of in vitro translation reaction product as a source of 3D protein was added to a total of 30 μl of replication mix, containing 500 μM ATP, CTP, and GTP; 100 μM UTP; 3 μg poly(A); 16.7 μM oligo(U)12-15; and 0.2 μCi [α-32P]UTP (3,000 Ci/mmol) in replication buffer (50 mM HEPES-KOH [pH 7.5], 3 mM Mg-acetate, 600 μM ZnCl2, 0.01% NP-40). Reaction mixtures were assembled and incubated on ice for 10 min and then transferred to 30°C and incubated for 30 min. These conditions were determined to correspond to the linear phase of the elongation reaction. Reactions were stopped with 5 μl of 0.5 M EDTA solution; then 10 μl was spotted onto Whatman DE81 paper disks, air dried, washed three times with 5% Na2HPO4, and dried, and radioactivity was counted by scintillation spectroscopy.
Renilla luciferase replicon assay.
HeLa cells grown in 96-well plates were transfected in normal growth medium with poliovirus replicon RNA (0.8 ng/well) transcribed from pXpA-RenR. The transfection mix contained 60 μM EnduRen Renilla luciferase substrate (Promega). Light from Renilla luciferase activity was measured at hourly intervals with a Molecular Devices MV microplate reader.
RESULTS
Intact 3CD protein is necessary for Arf translocation.
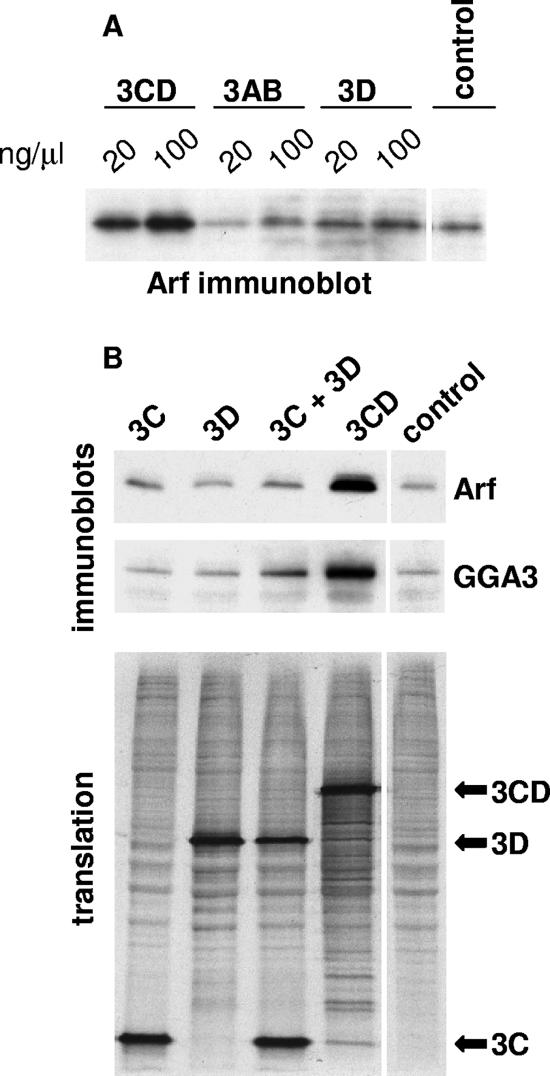
Our previous data showed that translation of poliovirus 3CD-encoding RNAs in HeLa S10 extracts resulted in increased binding of at least some Arf species to membranes. We decided to test whether Arf recruitment was coupled to translation of 3CD mRNA to generate the protein, perhaps at a membrane site, or whether addition of soluble 3CD protein itself induced Arf activation and membrane association. Purified recombinant 3CD protein, as well as control protein 3AB or 3D, was added directly to HeLa S10 extracts and incubated for 3.5 h at 34°C (the same conditions used for translation experiments). Membranous fractions were collected and analyzed by immunoblot assay with anti-Arf antibodies. Figure 1A shows that addition of 3CD protein, but not 3AB or 3D proteins, to cell extracts produced a strong increase in the association of Arf with membranes.
FIG. 1.
Translocation of Arf to membranes in vitro. (A) Recombinant proteins 3CD, 3AB, and 3D were added to HeLa S10 extracts and incubated for 3.5 h. The membranous fraction was collected by centrifugation and assessed by immunoblotting with anti-Arf antibodies. (B) RNAs coding for proteins 3C, 3D, and 3CD were translated in HeLa S10 extracts. After translation, the membranous fractions were collected by centrifugation and assessed by immunoblotting with anti-Arf antibodies (upper panel). The immunoblot membrane was stripped and probed with antibodies to GGA3, which bind the GTP-bound form of Arf (middle panel). The lower panel shows autoradiography of the gel showing [35S]methionine-labeled products of the translation reactions used in the Arf and GGA3 immunoblots.
Previous results demonstrated that induction of Arf translocation did not require that 3CD be an active proteinase or contain a functional 3D polymerase domain (7). In addition, translation of neither 3C- nor 3D-encoding RNA separately induced Arf translocation (7). To determine whether Arf translocation required intact 3CD protein or merely the presence of both 3C and 3D domains simultaneously, we translated 3C and 3D RNAs in HeLa S10 extracts in one reaction, collected the membrane fraction by centrifugation, and assessed the amount of Arf associated with membranes by immunoblot analysis with antibodies that recognize all human Arf species except Arf 4. [35S]methionine was added to an aliquot of each reaction mixture to monitor translation of the RNAs (Fig. 1B, lower panel). As shown in Fig. 1B (upper panel), cotranslation of 3C and 3D was not sufficient to induce Arf translocation to membranes, suggesting that this process is specific to the intact 3CD protein.
3CD-induced Arf recruited to membranes is in its active form.
Usually Arfs acquire the ability to stably associate with membranes when they are in their activated, GTP-bound form. Binding to GTP results in a conformational change of the molecule and exposure of the N-terminal amphipathic helix that tethers Arf to the lipid bilayer (2). Nevertheless Wessels et al. recently reported that expression in cells of protein 3A from coxsackievirus B3, a close relative of poliovirus, results in entrapment of Arf on membranes in its inactive, GDP-bound form (40). To determine the state of Arf accumulated on membranes after translation of 3CD in our assay, we took advantage of Arf-GTP's ability to specifically and strongly interact with adaptor proteins of the GGA family, which participate in trans-Golgi membrane transport. GGA proteins accumulate on membranes only in the presence of activated Arf-GTP (8). The membrane from the Arf translocation assay shown in the upper panel of Fig. 1B was stripped of anti-Arf antibodies and reprobed with anti-GGA3 antibodies. The middle panel of Fig. 1B shows a significant increase in the GGA3 signal in the sample that was positive for Arf after translation of 3CD. Thus, poliovirus protein 3CD induces the accumulation on membranes of the active, GTP-bound form of Arf.
F441S mutant of 3CD fails to induce association of GEFs with membranes.
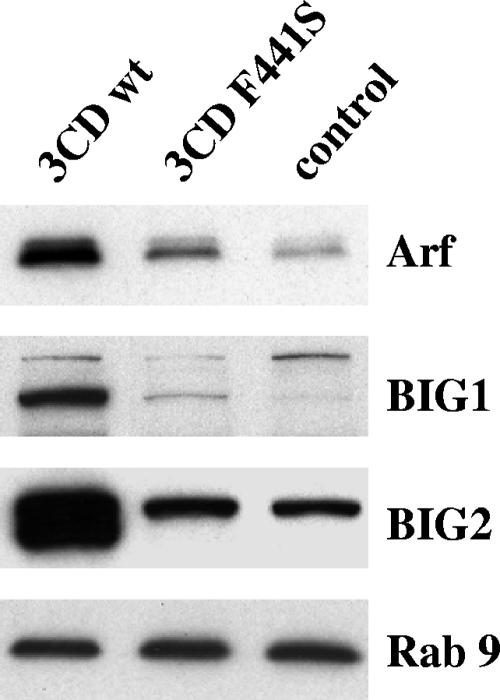
An F441S mutation in the 3D domain of 3CD that abolished its ability to stimulate Arf binding to membranes was described previously (7). Subsequent studies showed that 3CD-induced Arf translocation is mediated by cellular GEFs BIG1 and BIG2 (5), which normally participate in membrane traffic in the Golgi apparatus (11, 42). To determine what step in 3CD-induced Arf recruitment to membranes is affected by the 3CD F441S mutation, we compared recruitment of BIG1 and BIG2 by wt and F441S mutant 3CD proteins. RNAs coding for these proteins were translated in HeLa S10 extracts, and the membrane fractions were analyzed by immunoblotting with anti-BIG1 or anti-BIG2 antibodies. Equal translation efficiencies of the two RNAs were confirmed in portions of the reaction mixtures labeled with [35S]methionine (not shown). The blots were stained with antibodies to another cellular membrane-associated small GTPase, Rab9, as a loading control. Figure 2 shows that translation of the mutant 3CD RNA did not induce translocation of GEFs BIG1 and BIG2 to membranes, indicating that the failure to activate Arf by the F441S mutant of 3CD is not due to formation of defective complexes between this protein and cellular GEFs but must occur somewhere earlier in the cascade of events leading to the recruitment of cellular GEFs to membranes by 3CD.
FIG. 2.
Mutant 3CD F441S is defective in recruitment of Arf and specific GEFs to membranes. RNAs coding for wt 3CD or for the F441S 3CD mutant were translated in HeLa S10 extracts, and the membranous fraction was collected by centrifugation and assessed by immunoblotting with anti-Arf, -BIG1, -BIG2, and -Rab9 antibodies.
Interaction of 3CD with membranes.
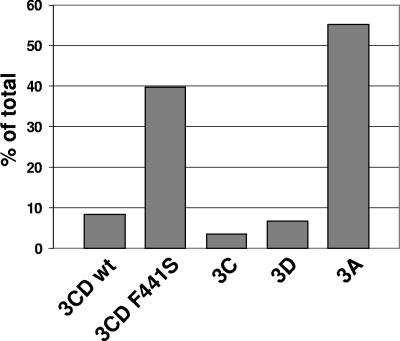
Poliovirus 3CD protein does not have obvious membrane binding sequences or membrane binding properties; therefore, it is puzzling how it may be responsible for altering the metabolism of membrane-targeted proteins. It is known that 3CD interacts with the poliovirus membrane-associated protein 3AB (26, 41), which could provide a mechanism for recruiting 3CD to membranes and thus for 3CD to induce alterations in other membrane protein metabolism. However, at least in a HeLa S10 translation-replication extract in vitro, 3CD alone induces increased binding of Arf and GEFs to membranes without any auxiliary role contributed by other viral proteins. This may be an aberrant property of the in vitro system, or it may suggest that 3CD is able to interact with some unidentified host membrane proteins. We therefore investigated the distribution of 3CD between the membranous and supernatant fractions of the HeLa S10 extracts. The 3CD RNA was translated in the presence of [35S]methionine, samples were collected before and after centrifugation, the proteins were resolved by SDS-PAGE, and the radioactivities in the 3CD bands from membrane fraction and total translation reaction mixture were quantified by phosphorimaging. The results shown in Fig. 3 demonstrate that although the majority of 3CD protein remained in the supernatant fraction after centrifugation, approximately 8% sedimented with the membranes. The amount of 3D recovered in the pellet fraction was slightly less than 7%, while only about 3% of 3C was associated with the membranous fraction. Note that more than 50% of protein 3A, with well-documented membrane binding properties, was recovered in the pellet fraction. Interestingly, the F441S mutation strongly increased the amount of 3CD in the pellet, up to 40% of total protein, suggesting either that it might form an unproductive complex with the membrane-associated host factor(s) needed for 3CD-induced translocation of GEFs and Arf to membranes or that this mutation may reduce solubility of the protein, causing it to pellet with membranes without specific association.
FIG. 3.
Association of poliovirus proteins with membranes in vitro. RNAs coding for wt 3CD, the 3CD F441S mutant, 3C, 3D, or 3A were translated in HeLa S10 extracts in the presence of [35S]methionine, and the membranous fraction was collected by centrifugation. Total material recovered after centrifugation and 1/10th of the material from unfractionated translation reactions were resolved by SDS-PAGE, and the distribution of proteins between fractions was determined using phosphorimaging.
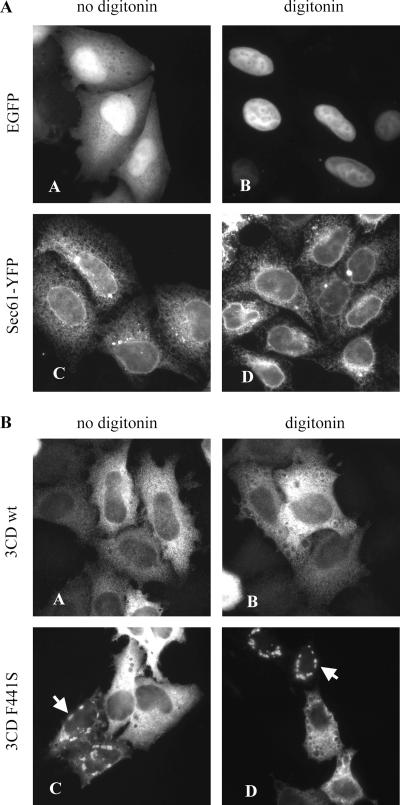
To determine whether 3CD localized to cellular membranes in vivo, we expressed the protein in HeLa cells and detected it by immunofluorescence with monoclonal anti-3D antibodies. Cells expressing 3CD were treated with digitonin prior to fixation to selectively perforate the cellular plasma membrane containing high concentrations of cholesterol, thus allowing soluble cytoplasmic components to be washed out from the cells (32). At the same time, intracellular membrane organelles with their associated proteins are left relatively intact. To validate the digitonin permeabilization of cells for our assay, we treated cells expressing free enhanced GFP (EGFP) and cells expressing the endoplasmic reticulum (ER) protein Sec61 fused to another fluorescent protein, yellow fluorescent protein (YFP) (a modified version of GFP). Cells transfected with the EGFP expression plasmid displayed diffuse cytoplasmic fluorescence as well as accumulation of EGFP in nuclei, while in cells expressing the Sec61-YFP fusion protein, fluorescence was confined to an ER-like complex network (Fig. 4A, panels A and C). Treatment of EGFP-expressing cells with digitonin resulted in complete loss of cytoplasmic fluorescence, whereas these cells retained nuclear EGFP fluorescence, confirming that nuclear membrane permeability was not damaged during this treatment (Fig. 4A, panel B). When digitonin was applied to the cells expressing Sec61-YFP, the ER network staining remained intact, indicating that the internal cellular membrane structures were not significantly affected (Fig. 4A, panel D).
FIG. 4.
Association of 3CD with cellular structures in HeLa cells. (A) HeLa cells were transfected with plasmids expressing either free EGFP or ER-targeted Sec61-YFP fusion protein. After 24 h, the cells were treated with digitonin, fixed, and examined by fluorescence microscopy. (B) HeLa cells were transfected with plasmids expressing either wt 3CD or the F441S 3CD mutant and treated with digitonin as for panel A. The cells were fixed, stained with anti-3D antibody, and examined by fluorescence microscopy.
3CD did not show a preferential distribution to specific cell compartments; however, the staining was not diffuse as might be expected from a soluble, cytoplasmic protein, but instead the protein was concentrated in numerous small foci, scattered throughout the cell (Fig. 4B, panel A). This punctate staining was clearly different from the diffuse pattern of fluorescence in cells expressing EGFP that underwent the same fixation procedure (Fig. 4A, panel A), suggesting that the concentrated 3CD staining in small foci may be due to interaction of the protein with some unidentified cellular structures. We did not observe any significant cytotoxicity of 3CD expression to the cells during 24 h of incubation after transfection. Nevertheless to exclude possible nonspecific effects due to 3CD's protease activity, we repeated the experiment with expression of a proteolytically inactive mutant of 3CD. The staining pattern and distribution of the mutant 3CD was indistinguishable from that of the wt molecule (not shown). When we expressed in cells the F441S mutant of 3CD, which is inactive in the Arf translocation assay, a significant proportion of cells developed large, round foci of concentrated fluorescence, usually located around the nucleus (Fig. 4B, panel C, arrowhead). This pattern was very rarely observed in cells transfected with the wt 3CD expression plasmid, suggesting that interaction of the F441S 3CD mutant with cellular structures is modified compared with that of the wt 3CD, consistent with the previously described in vitro data.
Digitonin permeabilization slightly reduced the overall intensity of 3CD staining, but the characteristic pattern was retained, both for the wt molecule and the F441S mutant, indicating that 3CD was somehow tethered on intracellular structures before fixation (Fig. 4B, panels B and D).
To see if expression of 3CD in cells resulted in apparent morphological reorganization of intracellular membranes, we examined HeLa cells expressing 3CD protein from a vaccinia virus-based expression system by electron microscopy. Cells were transfected with a plasmid encoding 3CD under control of the T7 RNA polymerase promoter and infected with a vaccinia virus expressing T7 RNA polymerase. No major distortion of any intracellular organelles was observed, although Golgi stacks appeared slightly enlarged and elongated in a significant fraction of cells expressing 3CD (not shown).
These results suggest that protein 3CD likely interacts with some cellular membrane-associated factors, which might account for its participation in reactions resulting in membrane alterations.
To search for possible cellular interaction partners of 3CD we utilized Myriad Genetics’ Pro-Net yeast two-hybrid screen service with cDNA libraries from human breast cancer, prostate cancer, and brain tissue. Three cellular proteins that interacted with 3CD were identified. Two of them, KHDRBS1 (Sam68) (25) and HNRPC (hnRNP C1/C2) (9), have been previously reported to bind to 3CD, which confirmed the validity of the screening; the third protein, UAP1L1 (UDP-N-acteyl-glucosamine pyrophosphorylase 1-like 1), was previously unknown to interact with poliovirus peptides. None of these potential host factors provide an obvious explanation for the proposed attachment of 3CD to cellular membranes. Thus, 3CD tethering to cellular structures may be complex and require multiple proteins, or the result could reflect a limitation of the two-hybrid system, which is based on activation of transcription in the yeast nucleus, in identifying membrane-targeted proteins.
Effect of mutations at position F441 of 3CD on virus.
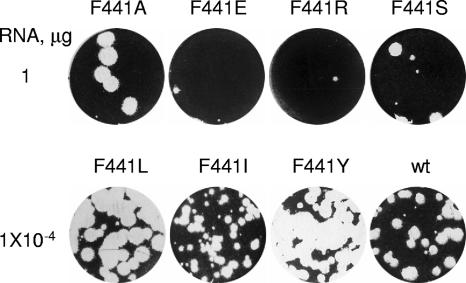
The F441S mutation in 3CD caused a loss of the protein's Arf-activating function and simultaneously caused a drastic loss of infectivity of the corresponding viral RNA containing this mutation (7). The mutation is located in the 3D domain of 3CD (residue 3CD-441 corresponds to position 258 in 3D). We generated a series of viral RNAs encoding other amino acids at position 441 to investigate their effects on Arf activation and virus viability. We substituted the original F441 in the 3CD sequence for either Y (most closely resembling the original F), a small neutral amino acid (A), amino acids with hydrophobic side groups (L and I), or charged amino acids (R and E). RNA transcripts bearing these mutations were tested for their infectivity after transfection into HeLa cells. RNAs with F441I, F441L, and F441Y substitutions showed wt specific infectivity of full-length transcripts, although plaques produced after transfection with F441I were smaller than those from wt RNA (Fig. 5). Other substitutions resulted in significant loss of infectivity, generating only a few plaques after transfection with very high RNA concentrations. This is a characteristic feature of a quasi-infectious phenotype and indicates that those plaques likely resulted from growth of revertant viruses (note that the single plaques shown in Fig. 5 for quasi-infectious mutants F441S, F441A, F441R, and F441E were obtained at RNA concentrations 10,000 times higher than that shown for RNAs with wt infectivity). Viruses from those plaques were collected and amplified once in HeLa cells, and regions containing the original mutations were sequenced. In all cases, reversions were found at the original mutation site. Reversions were either to wt F or to L and I (which were among the original mutations tested) or to V residues (Table 1). Contamination by F441L and F441I RNAs in the laboratory was ruled out, since we found a different codon for the same amino acid in at least one of the revertants. When a V codon was engineered in the full-length RNA, it also showed wt infectivity upon transfection into HeLa cell monolayers, although with a small-plaque phenotype (not shown), similar to the case for I. Thus, it seems that only amino acids with relatively large hydrophobic side chains are tolerated at position 441 of poliovirus 3CD protein. We noted that the revertant virus isolated from transfections with the F441A mutant RNA required changes in all three nucleotides of the alanine codon in order to restore the original wt residue; in this case, we cannot rule out the possibility that the “revertant” plaque might have been a contaminant from wt virus.
FIG. 5.
Viability of poliovirus RNAs with mutations in 3CD. HeLa cell monolayers were transfected with serial dilutions of full-length RNA transcripts, overlaid with agarose, incubated for ∼48 h, and stained for plaques. Plaques from appropriate dilutions/representative experiments are shown.
TABLE 1.
Infectivity of full-length RNA transcripts correlates with Arf activation by 3CD
| 3CD amino acid 441 (codon) | % of Arf activation by 3CDa | Full-length RNA specific infectivity | Position 441 reversion (codon) |
|---|---|---|---|
| F (TTC) (wt) | 100 | wt | NAb |
| Y (TAC) | 86 | wt | NA |
| I (ATT) | 71 | wt | NA |
| L (TTA) | 60 | wt | NA |
| S (TCC) | 9 | Quasi-infectious | F (TTC) |
| S (TCA) | NDc | Quasi-infectious | L (TTA) |
| A (GCA) | 35 | Quasi-infectious | F (TTC) |
| R (AGA) | 4 | Quasi-infectious | I (ATA) |
| E (GAA) | 1 | Quasi-infectious | V (GTA) |
The percentage of Arf activation was quantitated by analyzing the Arf immunoblot image with Total Lab 1D software.
NA, not applicable.
ND, not determined.
Revertants restored the Arf translocation property of 3CD.
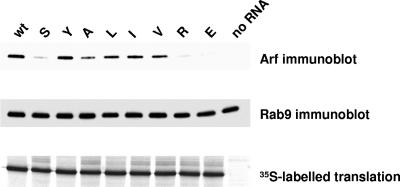
To see if the infectivity of the mutant RNAs correlated with the abilities of the corresponding 3CD proteins to induce Arf translocation, we generated RNAs coding for 3CDs with the original mutations as well as with the newly recovered F441V substitution. These RNAs were translated in HeLa S10 extracts, and the membrane fractions were tested for the amount of associated Arf. Mutations in position 441 of the 3CD-coding sequence did not affect the translational efficiencies of the corresponding RNAs (Fig. 6, bottom panel). The top panel of Fig. 6 shows that 3CD with Y in position 441 was as potent as the wt protein in inducing Arf translocation to membranes. Proteins with F441I, F441L, and F441V mutations were also active. All other substitutions resulted in significant loss of Arf recruitment activity. Thus, all mutations that gave high infectivity of the original full-length transcripts (Y, I, and L) as well as the V revertant were positive for 3CD-induced Arf translocation, while those that produced only quasi-infectious virus were either completely negative (E and R) or significantly impaired (A). The membrane used in the Arf immunoblot was also assessed with antibodies to another cellular small GTPase, Rab9, which is involved in membrane traffic, and demonstrated uniform distribution of this protein among the samples, confirming the specificity of Arf recruitment by 3CD proteins and serving as a loading control in this experiment (Fig. 6, middle panel). The result shows that the infectivity of viral RNA strongly correlates with the ability of 3CD to induce binding of Arf to membranes.
FIG. 6.
Translocation of Arf to membranes induced by mutant 3CD proteins. RNAs coding for wt 3CD or for 3CDs with the indicated mutations in position 441 were translated in HeLa S10 extracts. The membranous fraction was collected by centrifugation and assessed by immunoblotting with anti-Arf antibodies (upper panel). The same membrane used for the Arf immunoblot was stripped and probed with antibodies to Rab9, as a loading control (middle panel). The lower panel shows the [35S]methionine-labeled products of the translation reactions.
Stimulation of virus production in vitro by 3CD mutants does not correlate with RNA infectivity.
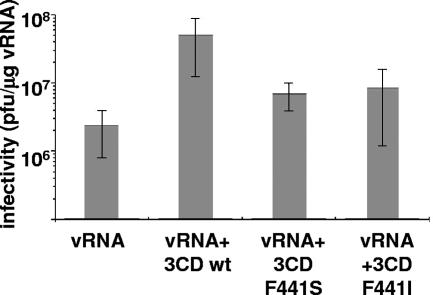
It has been reported that 3CD stimulates the production of infectious virus when synthesized or added to in vitro translation-replication reactions (19, 37), although the mechanism of 3CD's effect on virus yield was not elucidated. We examined whether this activity was affected by mutations at residue 441 and how it correlated with the viability of the virus. wt, F441S (nonviable), and F441I (viable with nearly wt phenotype) 3CD RNAs were cotranslated with the wt full-length poliovirus RNA in a HeLa cell S10 translation-replication system, and the yield of infectious virus was measured by plaque assay. wt 3CD stimulated production of virus 20-fold, consistent with previous findings, although the effect observed was not as marked as reported previously. Stimulation by either F441S or F441I was significantly lower than that by wt 3CD but showed no differences from each other (Fig. 7). Thus, this stimulatory activity of 3CD in vitro did not correlate with the virus growth phenotype in vivo.
FIG. 7.
Stimulation of poliovirus virion production in vitro by 3CD. Poliovirus RNA (vRNA) alone or in combination with RNA coding for wt 3CD or the F441S or F441I 3CD mutant were translated and replicated in HeLa S10 extracts for 12 to 15 h. Formation of infectious virus was determined by plaque assay on HeLa cells monolayers. Error bars indicate standard deviations.
3D polymerase activity does not determine viral growth rates.
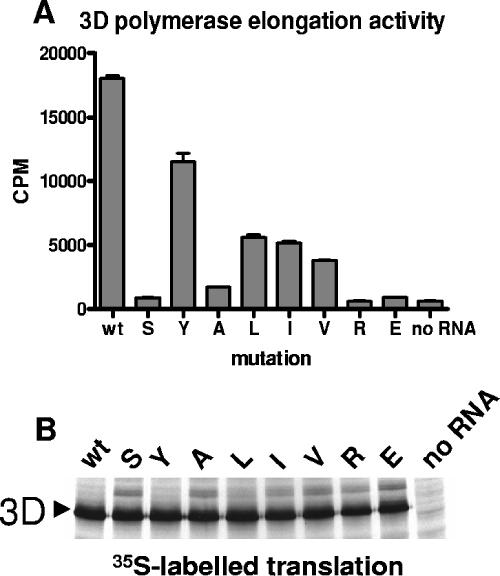
A common property of proteins encoded by small viral genomes with limited coding capacity is that multiple activities have evolved to reside in single polypeptide chains. This property often complicates genetic analysis of viral proteins, since mutations in a protein-coding sequence may affect multiple different functions of the protein or of other proteins containing that sequence. We observed that the F441S mutation in 3CD caused a subtle defect in capsid protein processing, which is catalyzed by 3CD; however, that did not explain the observed loss in infectivity, since the mutation severely inhibited viral RNA replication, which is not dependent on structural proteins (7). In addition to several known activities of 3CD, a potentially detrimental effect on virus growth could result from this mutation impairing one or more of the activities of 3D, the viral RNA-dependent RNA polymerase. We therefore assayed RNA chain elongation activity in the 3D proteins harboring each of the substitutions at position 441 of 3CD (amino acid residue 258 in 3D) to determine the effects on RNA polymerase activity of 3D. RNAs coding for each of the 3D mutants were translated in HeLa S10 extracts, and aliquots of the translation reaction mixtures were assayed for RNA-dependent RNA polymerase activity without further purification. Figure 8 shows that each substitution decreased the polymerase activity of mutant 3D proteins. The polymerase activity of 3D with F441Y was least affected; the F441L, F441I, and F441V mutations, which manifested wt levels of RNA infectivity in the full-length RNA context as well as high levels of Arf activation in 3CD, caused significantly lower activities than that of the wt polymerase, although higher activities than those of F441A, F441R and F441E mutants. It is surprising that viruses expressing 3D with <30% of wt 3D polymerase activity nevertheless grew almost as well as the wt, suggesting that the amount or activity of 3D polymerase is not rate limiting for virus replication.
FIG. 8.
RNA polymerase elongation activity of mutant 3D proteins. (A) RNAs coding for wt 3D or for 3Ds with the indicated mutations in position 258, corresponding to amino acid 441 of 3CD, were translated in HeLa S10 extracts, and 2 μl of translation mix was used as a source of 3D protein in an RNA chain elongation assay with poly(A) template and oligo(U) primer. (B) Autoradiogram of the gel showing [35S]methionine labeled products of translation reactions used in the elongation assay. Error bars indicate standard deviations.
A replicon bearing a 3D debilitating mutation shows wt replication.
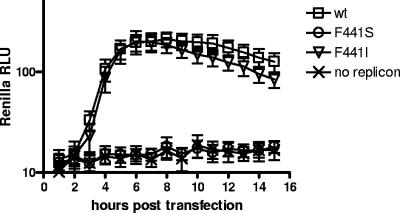
The surprising result that wt infectivity was observed from RNAs with mutations that significantly reduced RNA elongation activity of the corresponding 3D polymerase prompted us to investigate RNA replication in another model. We introduced the original F441S mutation, which generated quasi-infectious virus, and the F441I substitution, which demonstrated wt levels of infectivity although produced smaller plaques, in the Renilla luciferase poliovirus replicon, which was described earlier (5). Replicon RNAs were transfected into HeLa cells, and their replication was monitored by measurement of luciferase activity in vivo. As evidenced from Fig. 9, replication of the mutant with the F441I substitution in 3CD was virtually indistinguishable from that of its wt counterpart, while replication of the mutant with the F441S mutation was nondetectable. The small-plaque phenotype observed after transfection of HeLa cell monolayers with F441I mutant RNA therefore may have been due to the defect in capsid protein processing by mutated 3CD protease, similar to the one observed in the case of F441S (7). Thus, we confirmed that RNA replication of the mutants was not affected by the reduced level of 3D polymerase activity but strongly correlated with the ability of the corresponding 3CDs to induce Arf translocation to membranes.
FIG. 9.
Replication of poliovirus replicons harboring mutations that inhibit 3D elongation activity. Renilla luciferase poliovirus replicon RNA transcripts were transfected into HeLa cells growing in 96-well plates and incubated in the presence of cell-permeative Renilla luciferase substrate. Light readings were taken hourly with a Molecular Devices MV reader. RLU, relative light units. Error bars indicate standard deviations.
DISCUSSION
Modulation of cellular pathways by viral proteins is a signature of virus infection, which is required to divert cell metabolic machinery into production of viral progeny. Here we investigated the interaction of poliovirus protein 3CD with host factors involved in membrane traffic and the role that such interactions might play in viral infection. Protein 3CD performs multiple functions in the poliovirus replication cycle: it catalyzes proteolysis of capsid protein precursors and binds RNA in the processes of VPg uridylylation and initiation of RNA synthesis. We recently found that 3CD induces translocation of the small cellular Arf GTPases to membranes where poliovirus replication occurs (7). Here we show that this activity of 3CD appears to be very important for virus growth. After translation of 3CD mRNA in HeLa cell extracts, Arf accumulated on membranes in its active, GTP-bound form. This result likely explains our previous finding that Arf-GTP levels increase in poliovirus-infected cells (5).
3CD apparently initiates Arf activation by promoting the binding to membranes of cellular GEFs BIG1 and BIG2, which catalyze nucleotide exchange converting Arf-GDP to Arf-GTP, which in turn stabilizes the membrane binding of Arf. Mutant 3CD with a change of F in position 441 to S was unable to recruit cellular GEFs to membranes and thus failed to activate Arf. The role of Arf-GTP on membranes of poliovirus replication complexes remains unknown. There are six members of the Arf family, and five of them are expressed in human cells. Arfs 1, 3, 4, and 5 are implicated in intracellular membrane traffic, while Arf 6 is mostly believed to regulate plasma membrane metabolism. In uninfected cells, the association of Arf-GTP with membranes induces curvature of the lipid bilayer that is believed to facilitate formation of secretory vesicles. Arf-GTP also stimulates phospholipase D activity, which modifies membrane lipid composition. Finally, the membrane-bound, activated Arf interacts with and recruits to membranes a wide variety of effector proteins (4, 17, 29). Any or all of these properties of Arf may be used by poliovirus for remodeling cellular membranes into replication complexes and for recruitment of host factors that might be utilized during the viral RNA replication process.
3CD is not the only poliovirus protein involved in Arf metabolism. We reported previously that 3A also induces binding of Arf to membranes via recruitment of another cellular GEF, GBF1. GBF1-dependent pathways appear to be important in the virus life cycle, since overexpression of GBF1 partially rescued poliovirus RNA replication in the presence of brefeldin A, a potent inhibitor of nucleotide exchange by GEFs in the cellular secretory pathway (5) and also of poliovirus infection (15, 24). Thus, it seems that these two proteins, 3A and 3CD, could synergistically modify cellular membrane traffic pathways to develop viral replication complexes, in accordance with the recently proposed model (6). Interestingly, Wessels et al. (38-40) found that direct binding of the 3A proteins of poliovirus and the closely related coxsackievirus B3 with GBF1 is responsible for inhibition of cellular secretion observed in cells infected with poliovirus or coxsackieviruses or expressing their 3A proteins (10, 12, 14, 16). Virus defective in this function of 3A was less pathogenic in mice (40). These data indicate that manipulation of Arf-activating pathways in infected cells may be necessary for the virus not only to replicate its RNA but also to ensure survival in a multicellular animal host.
The mechanism by which 3CD induces the association of specific GEFs with membranes is not clear. The amino acid sequence of 3CD does not show identifiable membrane-targeting motifs. Nevertheless, when we performed fractionation of HeLa S10 extracts after translation of 3CD RNA, we found that a portion of the translated 3CD protein was associated with the membrane fraction. When we expressed 3CD in HeLa cells and treated them with digitonin, which selectively permeabilized the plasma membrane and allowed soluble components of the cytoplasm to escape from the cell, most of 3CD was retained, obviously bound to some unidentified intracellular structures. It seems plausible that 3CD can interact with some cellular membrane-associated protein(s) and that such interaction can initiate 3CD-dependent binding of GEFs to membranes. Although we did not determine the amount of 3CD protein that remained bound to intracellular structures after treatment of the cells with digitonin, it appeared greater than the amount of the protein bound to membranes in the in vitro translation study. This may reflect a deficiency of morphological organization of the membranes in vitro (18) or of the factor(s) needed for association of 3CD with membranes. For example, about half of the 3A protein synthesized in vitro was found in the supernatant fraction, despite its known membrane binding properties. This may indicate that the in vitro system generally lacks sufficient appropriate membrane structures to accommodate poliovirus proteins. Interestingly, the F441S mutation, which eliminated the Arf-translocating properties of 3CD, strongly increased recovery of the mutated protein in the pellet fraction after translation of the corresponding RNA in HeLa S10 extract and also resulted in an altered distribution of the protein when expressed in intact cells. These data suggest that this mutation may impair interaction of 3CD with host factors.
Our attempt to identify possible host factors that could promote association of 3CD with membranes was unsuccessful. A yeast two-hybrid screen yielded three interactions, but all of them seem to be with soluble cytosolic or nuclear proteins and they did not provide a clear clue to how 3CD could be bound to cellular structures. Protein Sam68 was reported previously to interact with polymerase 3D and was found on replication complexes, suggesting a possible role in RNA replication (25). Nuclear RNA binding protein hnRNP C1/C2 was found to coimmunoprecipitate with 3CD and bind to poliovirus RNA, enhancing replication (9). Currently there is no available information about the third 3CD-interacting protein identified in this screen, UDP-N-acteyl-glucosamine pyrophosphorylase 1-like 1, but sequence analysis does not show any potential membrane-interacting domains. This may suggest that interaction of 3CD with membranes may include multiprotein associations, or the failure to find membrane-targeted 3CD-interacting protein may be due to limitations of the yeast two-hybrid screen to identify membrane-bound proteins, since the system depends on nuclear activation of transcription.
Since the F441S mutation generated a 3CD protein defective in Arf translocation activity, we performed further mutagenesis of this F residue. We found that only amino acids with rather bulky hydrophobic side chains, i.e., I, L,V, and Y, were tolerated in this position and produced 3CDs with a high level of Arf translocation activity. Residue 441 is located on the outer surface of the finger region of the 3D polymerase domain of 3CD (23, 36). This location is consistent with the idea that this portion of the molecule may be involved in protein-protein interactions important for 3CD-mediated Arf activation.
Mutations that abolished the Arf activation property of 3CD manifested a quasi-infectious phenotype when incorporated into full-length poliovirus RNA. All the revertants obtained after transfection of HeLa cells with those RNAs had restored the Arf translocation property of 3CD by nucleotide changes encoding I, L, V, Y, or the wt F. Although all these mutations reduced 3D polymerase elongation activity, full-length RNAs harboring those mutations demonstrated wt infectivities and growth curves. The lack of correlation between reduced polymerase elongation activity and RNA replication level was confirmed in experiments with a poliovirus replicon engineered to carry the F441I substitution, which replicated just like the wt construct. We also saw no correlation between the virion assembly-stimulating activity of 3CD in vitro and the infectivity of our mutant RNAs.
Viruses with small genomes generally produce proteins that display multiple functions, which makes the task of affecting one activity without imposing any effect on another activity uncertain. Other known and yet-unidentified functions of 3D and/or 3CD might also be impaired by these mutations and therefore could have contributed to the observed loss of viability. Although this complicates the assessment and attribution of their individual inputs into a given virus phenotype, the data presented here show that Arf activation by 3CD strongly correlates with successful virus replication.
Acknowledgments
We thank Oliver Richards, Kim Green, and Gail Belliot for their help with the 3D polymerase assay.
This work was supported in part by the intramural research programs of the NIAID, NIH.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonny, B., S. Beraud-Dufour, P. Chardin, and M. Chabre. 1997. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36:4675-4684. [DOI] [PubMed] [Google Scholar]
- 3.Back, S. H., Y. K. Kim, W. J. Kim, S. Cho, H. R. Oh, J. E. Kim, and S. K. Jang. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro). J. Virol. 76:2529-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behnia, R., and S. Munro. 2005. Organelle identity and the signposts for membrane traffic. Nature 438:597-604. [DOI] [PubMed] [Google Scholar]
- 5.Belov, G. A., N. Altan-Bonnet, G. Kovtunovych, C. L. Jackson, J. Lippincott-Schwartz, and E. Ehrenfeld. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov, G. A., and E. Ehrenfeld. 2007. Involvement of cellular membrane traffic proteins in poliovirus replication. Cell Cycle 6:36-38. [DOI] [PubMed] [Google Scholar]
- 7.Belov, G. A., M. H. Fogg, and E. Ehrenfeld. 2005. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J. Virol. 79:7207-7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boman, A. L. 2001. GGA proteins: new players in the sorting game. J. Cell Sci. 114:3413-3418. [DOI] [PubMed] [Google Scholar]
- 9.Brunner, J. E., J. H. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe, S. S., D. A. Dodd, and K. Kirkegaard. 2005. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337:18-29. [DOI] [PubMed] [Google Scholar]
- 11.Claude, A., B. P. Zhao, C. E. Kuziemsky, S. Dahan, S. J. Berger, J. P. Yan, A. D. Armold, E. M. Sullivan, and P. Melancon. 1999. GBF1: a novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 146:71-84. [PMC free article] [PubMed] [Google Scholar]
- 12.Cornell, C. T., W. B. Kiosses, S. Harkins, and J. L. Whitton. 2006. Inhibition of protein trafficking by coxsackievirus B3: multiple viral proteins target a single organelle. J. Virol. 80:6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuconati, A., A. Molla, and E. Wimmer. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 72:6456-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doedens, J., L. A. Maynell, M. W. Klymkowsky, and K. Kirkegaard. 1994. Secretory pathway function, but not cytoskeletal integrity, is required in poliovirus infection. Arch. Virol. Suppl. 9:159-172. [DOI] [PubMed] [Google Scholar]
- 16.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza-Schorey, C., and P. Chavrier. 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell. Biol. 7:347-358. [DOI] [PubMed] [Google Scholar]
- 18.Fogg, M. H., N. L. Teterina, and E. Ehrenfeld. 2003. Membrane requirements for uridylylation of the poliovirus VPg protein and viral RNA synthesis in vitro. J. Virol. 77:11408-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franco, D., H. B. Pathak, C. E. Cameron, B. Rombaut, E. Wimmer, and A. V. Paul. 2005. Stimulation of poliovirus synthesis in a HeLa cell-free in vitro translation-RNA replication system by viral protein 3CDpro. J. Virol. 79:6358-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, H. H., C. F. Yang, A. D. Murdin, M. H. Klein, J. J. Harber, O. M. Kew, and E. Wimmer. 1994. Mouse neurovirulence determinants of poliovirus type 1 strain LS—a map to the coding regions of capsid protein VP1 and proteinase 2Apro. J. Virol. 68:7507-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcotte, L. L., A. B. Wass, D. W. Gohara, H. B. Pathak, J. J. Arnold, D. J. Filman, C. E. Cameron, and J. M. Hogle. 2007. Crystal structure of poliovirus 3CD: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J. Virol. 81:3583-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maynell, L. A., K. Kirkegaard, and M. W. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 93:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269:27015-27020. [PubMed] [Google Scholar]
- 27.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 28.Mosimann, S. C., M. M. Cherney, S. Sia, S. Plotch, and M. N. James. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273:1032-1047. [DOI] [PubMed] [Google Scholar]
- 29.Nie, Z., D. S. Hirsch, and P. A. Randazzo. 2003. Arf and its many interactors. Curr. Opin. Cell Biol. 15:396-404. [DOI] [PubMed] [Google Scholar]
- 30.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 31.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plutner, H., H. W. Davidson, J. Saraste, and W. E. Balch. 1992. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 119:1097-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roehl, H. H., T. B. Parsley, T. V. Ho, and B. L. Semler. 1997. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J. Virol. 71:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma, R., S. Raychaudhuri, and A. Dasgupta. 2004. Nuclear entry of poliovirus protease-polymerase precursor 3CD: implications for host cell transcription shut-off. Virology 320:195-205. [DOI] [PubMed] [Google Scholar]
- 35.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, A. A., and O. B. Peersen. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verlinden, Y., A. Cuconati, E. Wimmer, and B. Rombau. 2002. The viral protein 3CD induces an equilibrium between the viral protein and RNA synthesis in a cell-free system for poliovirus replication. Arch. Virol. 147:731-744. [DOI] [PubMed] [Google Scholar]
- 38.Wessels, E., D. Duijsings, K. H. Lanke, W. J. Melchers, C. L. Jackson, and F. J. van Kuppeveld. 2007. Molecular determinants of the interaction between the coxsackievirus protein 3A and the guanine nucleotide exchange factor GBF1. J. Virol. 81:5238-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wessels, E., D. Duijsings, K. H. Lanke, S. H. van Dooren, C. L. Jackson, W. J. Melchers, and F. J. van Kuppeveld. 2006. Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COP-I recruitment. J. Virol. 80:11852-11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wessels, E., D. Duijsings, T. K. Niu, S. Neumann, V. M. Oorschot, F. de Lange, K. H. Lanke, J. Klumperman, A. Henke, C. L. Jackson, W. J. Melchers, and F. J. van Kuppeveld. 2006. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell 11:191-201. [DOI] [PubMed] [Google Scholar]
- 41.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaji, R., R. Adamik, K. Takeda, A. Togawa, G. Pacheco-Rodriguez, V. J. Ferrans, J. Moss, and M. Vaughan. 2000. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc. Natl. Acad. Sci. USA 97:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, Y., R. Rijnbrand, S. Watowich, and S. M. Lemon. 2004. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J. Biol. Chem. 279:12659-12667. [DOI] [PubMed] [Google Scholar]
- 44.Ypma-Wong, M. F., P. G. Dewalt, V. H. Johnson, J. G. Lamb, and B. L. Semler. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265-270. [DOI] [PubMed] [Google Scholar]