Abstract
The ecological niche that a species can occupy is determined by its resource requirements and the physical conditions necessary for survival. The niche to which an organism is most highly adapted is the realized niche, whereas the complete range of habitats that an organism can occupy represents the fundamental niche. The growth and development of Cryptococcus neoformans and Cryptococcus gattii on pigeon guano were examined to determine whether these two species occupy the same or different ecological niches. C. neoformans is a cosmopolitan pathogenic yeast that infects predominantly immunocompromised individuals, exists in two varieties (grubii [serotype A] and neoformans [serotype D]), and is commonly isolated from pigeon guano worldwide. By contrast, C. gattii often infects immunocompetent individuals and is associated with geographically restricted environments, most notably, eucalyptus trees. Pigeon guano supported the growth of both species, and a brown pigment related to melanin, a key virulence factor, was produced. C. neoformans exhibited prolific mating on pigeon guano, whereas C. gattii did not. The observations that C. neoformans completes the life cycle on pigeon guano but that C. gattii does not indicates that pigeon guano could represent the realized ecological niche for C. neoformans. Because C. gattii grows on pigeon guano but cannot sexually reproduce, pigeon guano represents a fundamental but not a realized niche for C. gattii. Based on these studies, we hypothesize that an ancestral Cryptococcus strain gained the ability to sexually reproduce in pigeon guano and then swept the globe.
Emerging infectious diseases are those that have newly appeared in a population or geographic range (60). One route to understand and ultimately prevent these diseases is to define the specific factors precipitating their emergence. Responsible factors can include ecological changes, human demographic alterations, travel and commerce, technology, microbial adaptation, and breakdown of public health measures (35, 37, 81). Expansion of the pathogen's geographic range can also result in disease outbreaks, such as the ongoing outbreak of the fungal pathogen Cryptococcus gattii on Vancouver Island (VI) in British Columbia, Canada (28).
Most infections are caused by pathogens already present in the environment that gain a selective advantage by changing conditions or have an opportunity to infect new hosts (61). For example, Legionnaires' disease is caused by the intracellular bacterium Legionella pneumophila, which colonizes amoebae and, when present in cooling towers, is exposed to and infects humans (23, 77). Cooling towers thus provide a “man-made” reservoir for L. pneumophila growth that simulates the ecological niche of L. pneumophila—ponds where amoebae are readily available to sustain the organism's reproduction and survival (2, 3). Thus, when studying human infections acquired from environmental sources, knowledge of not only the infecting reservoir but also the natural ecology and life cycle of the microorganism is important.
Certain combinations of environmental conditions are necessary for species to tolerate the physical environment, obtain energy and nutrients, and evade predators. The total requirements of a species for resources and physical conditions determine its abundance and distribution in nature. In ecology, these requirements govern the niche for a species or population in an ecosystem. More formally, the niche includes how the population responds to available resources and competitors and establishes the organism's life history, habitat, and place in the food chain. However, according to the competitive exclusion principle, no two species can occupy the same niche in the same environment for a prolonged period, which has resulted in a distinction between fundamental and realized niches (32). The full range of environmental conditions (biological and physical) under which an organism can exist defines its fundamental niche. However, as a result of pressure from interactions with other organisms, as well as changes in the environment, species are usually forced to occupy a niche that is narrower than the fundamental niche. This is termed the realized niche and represents the environment to which a species becomes most highly adapted.
Cryptococcus neoformans and the closely related species Cryptococcus gattii are human fungal pathogens. Humans are thought to be exposed by inhalation of basidiospores, which are small enough to lodge in the alveoli of the lung (78). The organism can then spread from the lungs to the central nervous system to cause meningoencephalitis (11, 33, 47). C. neoformans occurs in two varieties—grubii (serotype A) and neoformans (serotype D)—and diverged from C. gattii ∼40 million years ago (11, 90). The grubii and neoformans varieties have different disease epidemiologies, with var. grubii causing the vast majority of cryptococcosis worldwide (11, 83). While the C. neoformans varieties are cosmopolitan and cause disease predominantly in immunocompromised individuals, C. gattii is found predominantly in tropical regions and frequently causes disease in individuals with no known immune deficiency (33). An outbreak of C. gattii is currently ongoing in British Columbia (28).
A complete life cycle, including a sexual cycle, has been described for both Cryptococcus species (21, 38, 43, 44, 50). The sexual cycle was first described for var. neoformans, and early studies examining genetic virulence determinants were conducted with this variety (43, 45). The sexual cycle for the more commonly pathogenic variety, grubii, has recently been characterized and applied to define virulence characteristics (38, 57, 62, 63, 65). While C. gattii mating had been identified 30 years ago, evidence of recombination has only recently been shown in the Australian Northern Territory, which may have played a role in the cryptococcosis outbreak on VI (9, 10, 20, 21, 44).
Over the past 2 decades, C. neoformans infections have increased in prevalence as the population of immunocompromised individuals expanded due to the AIDS pandemic, aggressive cancer therapy, and organ transplantation. The sporadic nature of human cryptococcosis and rarity of documented human-to-human transmission indicate that infection is acquired from the environment (11). Pigeon guano is a common source for infectious propagules of C. neoformans and is postulated to play a central role in transmission from the environment to humans (11, 15, 22, 27, 29, 39, 40, 52, 70, 72, 76, 79, 80, 91, 92). C. neoformans can readily be isolated from pigeon guano and has been shown to grow and mate on medium containing pigeon guano (31, 73-75, 85). The closely related species C. gattii is not isolated from pigeon guano and is instead associated with various tree species (13, 14, 71). The different environmental sources of these two species has led to the hypothesis that C. neoformans is ubiquitous in the environment due to dissemination by pigeons following migratory and trade routes and that C. gattii is restricted to tropical/subtropical regions because it is not associated with pigeons. If this is the case, then how has the outbreak of C. gattii developed in the Pacific Northwest? Two highly related strains have been identified on VI, both of which are associated with soil and various tree species (41). Furthermore, the major genotype has been found only in the Pacific Northwest, leading to the hypothesis that this new strain has gained the ability to proliferate in a temperate environment and/or is highly virulent.
This study characterizes the growth of C. neoformans and C. gattii strains on pigeon guano so that we may understand factors influencing species survival. We show that both species are capable of growth on pigeon guano. Moreover, C. neoformans undergoes robust sexual reproduction on pigeon guano, whereas C. gattii does not. These results provide evidence that pigeon guano could be the realized niche for C. neoformans and highlight why it is not a preferred ecological niche for C. gattii. These results also illuminate a possible explanation for why C. neoformans is cosmopolitan and C. gattii is geographically restricted. The implications of these studies for the emergence of global infectious diseases are considered.
MATERIALS AND METHODS
Strains and media.
Strains used in this study are listed in Table 1. Strains were grown on yeast peptone dextrose (YPD) prior to transfer to other medium types. Yeast nitrogen base (YNB) minimal medium, V8 (pH 5.0 and pH 7.0) medium, and Murashige and Skoog (MS) medium were as described previously (63).
TABLE 1.
Strains used in this study
| Species | Strain | Genotype | Reference |
|---|---|---|---|
| C. neoformans var. grubii | H99 | MATα | 84 |
| KN99a | MATa | 63 | |
| KN99α | MATα | 63 | |
| YSB119 | MATα NAT | 4 | |
| YSB121 | MATaNEO | 4 | |
| F99 | MATα ura5 | 87 | |
| MO49 | MATα ade2 | 84 | |
| ST303B12 | MATα arg3 | 34 | |
| ST225B9 | MATα pro2 | 34 | |
| ST232C4 | MATα arg8 | 34 | |
| MDC16 | MATα lac1 | 68 | |
| RCP26 | MATα lac2 | 68 | |
| RCP29 | MATα lac1 lac2 | 68 | |
| KN119/21-1 | MATα/MATaNAT NEO | This paper | |
| KN119/21-2 | MATα/MATaNAT NEO | This paper | |
| KN119/21-3 | MATα/MATaNAT NEO | This paper | |
| KN119/21-4 | MATα/MATaNAT NEO | This paper | |
| KN119/21-5 | MATα/MATaNAT NEO | This paper | |
| KN119/21-6 | MATα/MATaNAT NEO | This paper | |
| C. neoformans var. neoformans | JEC21 | MATα | 45 |
| JEC20 | MATa | 45 | |
| XL405 | MATα hxk2::NEO | This paper | |
| XL465 | MATagsy1::NAT | 46 | |
| JEC31 | MATα lys1 | 59 | |
| JEC33 | MATα lys2 | 59 | |
| JEC38 | MATα met1 | 59 | |
| JEC43 | MATα ura5 | 59 | |
| JEC47 | MATα arg1 | 59 | |
| JEC50 | MATα ade2 | 59 | |
| KN405/465-1 | MATα/MATaNAT NEO | This paper | |
| KN405/465-2 | MATα/MATaNAT NEO | This paper | |
| KN405/465-3 | MATα/MATaNAT NEO | This paper | |
| KN405/465-4 | MATα/MATaNAT NEO | This paper | |
| KN405/465-5 | MATα/MATaNAT NEO | This paper | |
| KN405/465-6 | MATα/MATaNAT NEO | This paper | |
| C. gattii | NIH312 | MATα | 21 |
| B4546 | MATa | 21 | |
| R265 | MATα | 21 | |
| JF65 | MATα NAT | 21 | |
| JF66 | MATaNEO ura5 | 21 | |
| KN65/66-1 | MATα/MATaNAT NEO | This paper | |
| KN65/66-2 | MATα/MATaNAT NEO | This paper | |
| KN65/66-3 | MATα/MATaNAT NEO | This paper | |
| KN65/66-4 | MATα/MATaNAT NEO | This paper | |
| KN65/66-5 | MATα/MATaNAT NEO | This paper | |
| KN65/66-6 | MATα/MATaNAT NEO | This paper | |
| Candida albicans | SC5314 | MTLα/MTLa | 19 |
Guano media.
Pigeon guano was collected in Durham, NC, at the intersection of state road NC147 and interstate highway 40 (35°53′51.37′′N, 78°52′34.34"W), where pigeons were observed continuously roosting for 3 years. “Contaminated” soil was taken from the same area as the pure pigeon guano after the guano had been removed. “Uncontaminated” soil was collected 10 m from the pure pigeon guano samples, where no guano was observed. Desert bat, dry-bar cave bat, fossilized seabird, and original seabird guanos were purchased from Guano Co. International, Inc. Guano or soil was ground to a fine powder using a coffee grinder. Guano (2.5%, 12%, 25% [wt/vol]) or soil was added to boiling distilled water, incubated for 5 min with occasional stirring, and then filtered through a coffee press (style 1028; Bonjour, Inc.). Agar (4% [wt/vol]) was added to the mixture and autoclaved for 50 min. For combination media, 12.5% (wt/vol) of each guano was combined to give a final guano concentration of 25%. For UV irradiation, plates were exposed to 240 mJ of UV light in a Stratalinker apparatus (Stratagene, La Jolla, CA).
Medium with and without l-DOPA.
A minimal medium containing 15.0 mM glucose, 10.0 mM MgSO4, 29.4 mM KH2PO4, 13.0 mM glycine, 3.0 μM thiamine, and 2% (wt/vol) agar with a pH of 5.5 was prepared. For positive 3,4-dihydroxy-l-phenylalanine (l-DOPA) plates, 1 mM l-DOPA (Sigma Chemical Co., St. Louis, MO) was also added.
Environmental-isolation medium.
Minimal medium for environmental isolation contained 50 mM glucose, 100 mM glycine, 200 mM KH2PO4, 20 mM MgSO4·7H2O, 5 μM CuSO4, 2 mg/ml l-DOPA, 10 μg/ml thiamine-HCl, 0.5 μg/ml biotin, 0.5 mg/ml ampicillin, 40 μg/ml rose bengal, and 2% (wt/vol) agar (pH 5.6).
Growth and pigmentation comparisons.
YPD broth overnight cultures were inoculated with the desired yeast strain and incubated with shaking at 30°C overnight. The overnight culture was centrifuged at 4,000 rpm for 5 min to pellet cells and then resuspended in phosphate-buffered saline (PBS). Serial dilutions from 1:1 to 1:106 were prepared in PBS. Two microliters of each dilution was plated, allowed to dry completely, and then incubated in the dark with Parafilm and/or Ziploc bags as protection against contamination. For studies including membranes, a sheet of sterilized 3.5- or 14-kDa-cutoff dialysis membrane (Spectrum Laboratories) was placed on the plate prior to inoculation with 0.5 μl of culture. Membranes were placed on moist filter paper for analysis. To quantify growth, colonies grown on membranes were suspended in 1 ml PBS and then CFU were enumerated from serial dilutions onto YPD medium.
Mating and monokaryotic fruiting assays.
Strains were suspended in PBS, and 5-μl droplets were plated onto all medium types, allowed to dry completely, and then incubated at 25°C in the dark. Alternatively, the strains were mated as described previously (63). In crosses with genetically marked strains on 25% pigeon guano medium, spores were microdissected and progeny analyzed for genetic-recombination events.
Fusion and filamentation assays.
To assess the level of cell fusion on various medium types, cell fusion assays were performed as described previously (4). Variety grubii parental strains were YSB119α (nourseothricin resistant [NATr]) and YSB121a (neomycin resistant [NEOr]); var. neoformans parental strains were XL465α (NEOr) and XL405a (NATr); and C. gattii parental strains were JF65 (NATr) and JF66 (NEOr). Briefly, 108 cells of each parental mating type were mixed in equal volumes, and 5 μl was spotted onto the various medium types. For C. gattii, the V8 and PG media were supplemented with uracil to allow growth of JF66. After incubation for 24 h in the dark, the cells were scraped from the plate and resuspended in 1 ml water. Serial dilutions were prepared and spread onto YPD plates containing both NAT and NEO. Plates were incubated at 37°C for 72 h to induce diploid formation. Strains that were thermally dimorphic (budding at 37°C and filamentous at 30°C) and contained both sex-determining genes, SXI1α and SXI2a, were isolated. Six strains each were isolated for var. grubii (named KN119/21-1 to KN119/21-6), var. neoformans (KN405/465-1 to KN405/465-6), and C. gattii (KN65/66-1 to KN65/66-6) to verify that measurements were representative for multiple strains. To examine filament length, 108 cells of the diploid strains for each variety/species were spotted as 5-μl drops onto the various medium types. Filament length was measured after incubation for 7 days in the dark at 30°C.
Environmental isolation.
One gram of guano or soil was suspended in 5 ml PBS, and then 50 μl was spread onto culture medium containing l-DOPA and rose bengal. Pigmented colonies were isolated and DNA was obtained as described previously (49). PCR with STE20 primers was used to identify C. neoformans strains as well as their serotypes and mating types (5).
Elemental analysis.
Guanos were analyzed for percent carbon, hydrogen, nitrogen, oxygen, and sulfur (Galbraith Laboratories, Knoxville, TN). For glucose concentration determination, powdered guano was added to boiling distilled water, incubated for 5 min, filtered through a French press, and autoclaved for 50 min. The medium was allowed to cool and solids to settle out of the solution. The glucose concentration in the resulting supernatant was determined using the QuantiChrom glucose assay kit (BioAssay Systems, Hayward, CA).
RESULTS
Pigeon guano supports the growth of C. neoformans and C. gattii.
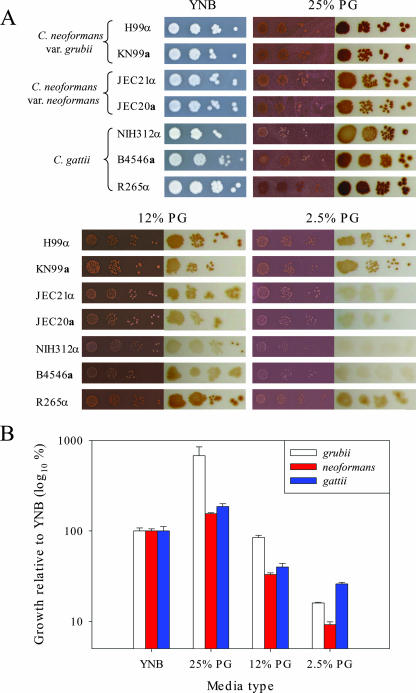
C. neoformans is readily isolated from pigeon guano, and var. neoformans strains have been cultured and mated on medium containing pigeon guano extracts (31, 73-75, 85), but C. gattii is not typically isolated from avian excreta (13, 14, 71), suggesting that C. gattii might not grow well in pigeon guano (11). This hypothesis was tested by examining the growth of C. neoformans (var. grubii and neoformans) and C. gattii on pigeon guano medium in which sterilized pigeon guano serves as the sole nutrient source. As shown in Fig. 1, both varieties of C. neoformans as well as C. gattii exhibited robust growth on pigeon guano medium. All strains tested had higher growth on medium containing 25% (wt/vol) pigeon guano than on YNB minimal medium (six times higher for var. grubii). The growth was reduced when pigeon guano in the medium was reduced (25% to 12% or 2.5%) (Fig. 1). No significant difference in growth was observed for the VI outbreak major strain, R265. Additional global C. neoformans and C. gattii strains were screened on pigeon guano medium, and no significant differences in growth were observed (data not shown).
FIG. 1.
Growth and pigmentation of Cryptococcus species on medium containing pigeon guano. C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii strains were grown overnight at 30°C in YPD medium, washed with PBS, and 10-fold serially diluted (103 to 106 dilutions). (A) Two microliters of each diluted cell suspension was spotted directly onto YNB or pigeon guano medium containing 25%, 12%, or 2.5% pigeon guano and incubated at 25°C for 7 days. To examine pigmentation, a sterilized 14-kDa-cutoff dialysis membrane was placed on the medium surface and then 0.5 μl of each diluted cell suspension was spotted onto the membrane. After 7 days of incubation at 25°C, membranes were removed from the medium and placed on moist filter paper to examine colony pigmentation. (B) To quantify growth, the 103 dilutions of JEC21α (neoformans), KN99α (grubii), and B4546α (gattii) grown on membranes were removed after 7 days and placed in PBS, and CFU were counted by serial dilution on YPD. Numbers of CFU resulting from growth on the various media are expressed as percentages of the number for the strain grown on YNB.
We next tested commercially available excreta for their ability to support Cryptococcus growth (data not shown). Medium containing desert bat guano supported the growth of all strains, but at a lower level than YNB. The growth on seabird guano, fossilized seabird guano, and dry-bar cave bat media was extremely poor. Two possibilities could explain the inability of Cryptococcus to grow on these media: the media could contain compounds that inhibit growth, or the medium types could lack required nutrients. To test this, growth was examined on media containing 12.5% seabird guano and 12.5% pigeon guano. Supplementation of seabird medium with pigeon guano restored growth (data not shown). Similar results were seen with other medium types tested, suggesting that other guanos lack an essential nutrient. Supplementation of seabird medium with components of YNB, including amino acids, nitrogen, and glucose, revealed growth only with glucose addition (data not shown). Elemental analysis revealed similar levels of total carbon in pigeon, bat, and seabird guanos; however, an analysis of glucose concentration revealed decreased levels in bat and seabird guanos compared to that in pigeon guano (Table 2). These data suggest that seabird medium lacks a sufficient utilizable carbon source.
TABLE 2.
Elemental analysis of guanosa
| Element | % in:
|
||
|---|---|---|---|
| Pigeon guano | Bat guano | Seabird guano | |
| Total carbon | 14.38 | 13.73 | 16.70 |
| Hydrogen | 2.12 | 2.09 | 3.92 |
| Nitrogen | 1.51 | 3.89 | 14.41 |
| Oxygen | 14.69 | 8.93 | 29.40 |
| Sulfur | 0.21 | 0.46 | 1.92 |
The glucose concentrations for pigeon, bat, and seabird guanos were 1,000, 24, and 12 ppm, respectively.
The pigeon guano used for these studies was obtained from the environment. Because guano was collected from the ground, we sought to verify that growth on pigeon guano medium was due to pigeon guano and not contamination with other nutrients from the collection site. Soil isolated from the same area resulted in very poor growth of Cryptococcus (data not shown). As with results with other guanos, growth on soil medium could be achieved by supplementation with pigeon guano, suggesting that soil medium is nutrient limited (data not shown). There was no increase in the growth of Cryptococcus on soil contaminated with pigeon guano compared to growth on an uncontaminated sample, suggesting that growth factors in pigeon guano do not readily diffuse into the surrounding soil or that these factors are metabolized by resident soil microbes. Cryptococcus was isolated from pigeon guano but not from soil at the collection site, confirming pigeon guano as the source of Cryptococcus from this environment (data not shown).
Cryptococcus produces pigment during growth on pigeon guano medium.
C. neoformans and C. gattii grown on pigeon guano medium produced brown pigmentation, which we hypothesized could be melanin. However, we found that this pigmentation was only partially generated via the well-characterized melanin biosynthesis pathway and that black particles resulting from another pigment production pathway could also be isolated from cells grown on pigeon guano.
Pigmentation consistently increased as the concentration of pigeon guano in the medium increased (Fig. 1). Treatment of the guano with activated charcoal significantly decreased pigmentation, suggesting that the components in pigeon guano stimulating pigmentation can be absorbed to a carbonaceous surface. When a dialysis membrane (cutoff, either 3.5 kDa or 14 kDa) was used to separate cryptococcal cells from the medium, pigmentation was still observed, suggesting that the component(s) involved in pigment formation is less than 3.5 kDa in size and can readily diffuse through the membrane (Fig. 1).
While the formation of a brownish pigment when Cryptococcus is grown on medium containing pigeon guano has been observed previously (74), pigment formation has not been characterized. The well-defined laccase pathway produces the brown/black pigment melanin (33). Recent studies have identified two laccase genes (LAC1 and LAC2) in Cryptococcus (56, 68, 93). Mutation of the LAC1 gene blocks melanin production, while mutation of the LAC2 gene has no discernible effect on melanin production on medium containing l-DOPA (68, 86). Previous studies have also identified melanized cells present in pigeon guano (66). Thus, we hypothesized that the brown pigment produced on pigeon guano medium is melanin.
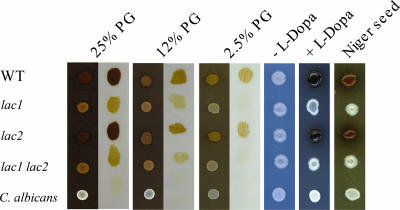
The wild-type strain, laccase mutant strains, and Candida albicans, which does not produce melanin, were tested for pigment production on pigeon guano medium (Fig. 2). The lac1 and lac1 lac2 mutant strains, while less pigmented than the isogenic wild-type strain, still produced pigment on pigeon guano medium but were unpigmented on minimal medium containing l-DOPA. In contrast, pigmentation of the lac2 mutant was unaffected on either pigeon guano medium or l-DOPA medium. The absolute amount of pigmentation observed with the laccase mutants was somewhat varied, with some batches of pigeon guano medium producing a slighter difference in pigmentation between the wild-type and laccase mutant strains (data not shown). Melanin ghosts were recovered from wild-type and lac2 strains grown on pigeon guano medium but not from the lac1 or lac1 lac2 mutants. Small black particles were observed in these mutants instead of cell-sized melanin ghosts (J. D. Nosanchuk, K. Nielsen, and J. Heitman, unpublished data). These results suggest that only some of the brown pigment observed on pigeon guano medium is produced via the classical laccase-dependent melanin pathway and that another, as-yet-uncharacterized pigment is also generated on pigeon guano medium.
FIG. 2.
Laccase mutant strains retain partial pigmentation on medium containing pigeon guano but not on medium containing l-DOPA. C. neoformans var. grubii H99 (wild type [WT]), lac1 and lac2 single mutants, a lac1 lac2 double mutant, and a Candida albicans strain were washed with PBS, and 2 μl of each cell suspension was spotted directly onto minimal medium (medium lacking l-DOPA [−l-DOPA]), Niger seed medium, medium containing l-DOPA to induce melanin production, or onto medium containing 25%, 12%, or 2.5% pigeon guano (PG). To examine the pigmentation of the strains grown on the pigeon guano medium, a sterilized 14-kDa-cutoff dialysis membrane was placed on the medium surface and then 0.5 μl of each diluted cell suspension was spotted onto the membrane. After 7 days of incubation at 25°C, membranes were removed from the medium, placed on moist filter paper, and photographed.
C. neoformans but not C. gattii strains mate on pigeon guano medium.
No difference in the ability of C. neoformans and C. gattii to grow on pigeon guano medium was observed even though C. gattii is not typically isolated from pigeon guano. We next determined whether both Cryptococcus species can complete their life cycle by undergoing sexual reproduction on pigeon guano, a hallmark of realized ecological niches, and found that only C. neoformans strains were able to robustly mate on pigeon guano medium; C. gattii strains did not.
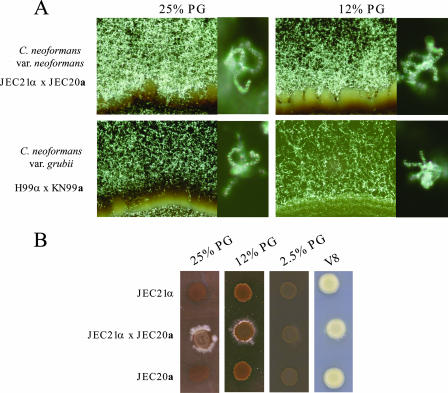
Crosses were performed between a and α strains of both C. neoformans (var. grubii and neoformans) (Fig. 3) and C. gattii (Fig. 4). The C. neoformans var. neoformans and var. grubii strains exhibited prolific mating on pigeon guano medium. The robustness of mating increased with the concentration of pigeon guano in the medium and exceeded that on V8 mating medium (Fig. 3). Spores produced from matings with genetically marked strains were microdissected and germinated, and based on marker analysis, they exhibited classical Mendelian segregation consistent with sexual reproduction (Table 3). C. neoformans strains were also able to mate at higher temperatures on pigeon guano medium than on V8 medium (data not shown). Mating was observed at 37°C on pigeon guano medium but not on V8 medium. In contrast, C. gattii mating was significantly reduced on pigeon guano medium compared to that on V8 or MS mating medium, and the inhibition of mating increased as the concentration of pigeon guano in the medium increased (Fig. 4). The mating of the C. gattii VI outbreak strain R265 was also significantly reduced (data not shown).
FIG. 3.
Cryptococcus neoformans var. grubii and var. neoformans mate robustly on pigeon guano. Opposite-mating-type strains of C. neoformans var. grubii and var. neoformans were washed with PBS, and equal volumes of each mating type were combined. The mixture was placed as a 10-μl drop onto V8 medium (pH 5 for var. grubii and pH 7 for var. neoformans) or medium containing 25%, 12%, or 2.5% pigeon guano (PG). Plates were incubated in the dark at 25°C for 7 days. (A) Filamentation (×20) and sporulation (×400) of var. neoformans and var. grubii strains on media containing 25% and 12% pigeon guano. (B) Comparison of C. neoformans var. neoformans mating colonies on V8 (pH 7) medium or pigeon guano medium containing increasing levels of pigeon guano.
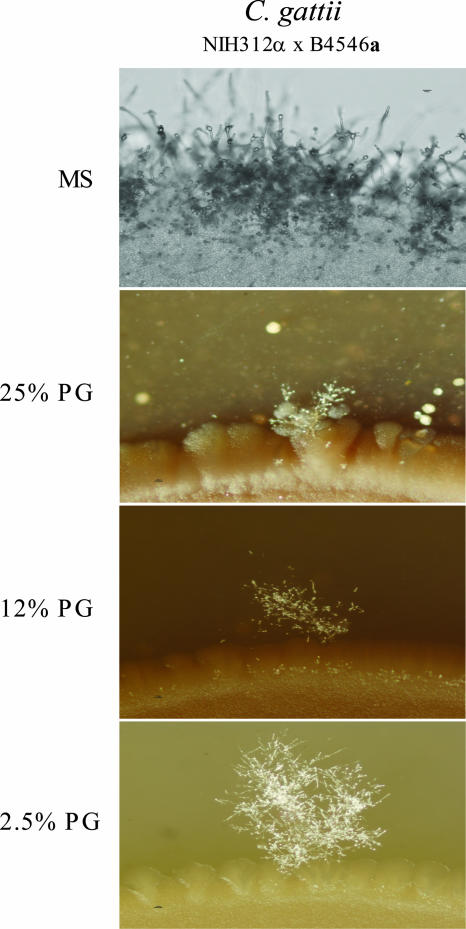
FIG. 4.
Cryptococcus gattii mating is inhibited on pigeon guano medium (PG). C. gattii strains of opposite mating types were washed with PBS, and equal volumes of each mating type were combined. The mixture was placed as a 10-μl drop onto MS medium or medium containing 25%, 12%, or 2.5% pigeon guano. Plates were incubated in the dark at 25°C for 3 days and then photographed at a ×20 magnification. The top panel shows a random patch at the edge of the mating colony on MS medium. The lower three panels show patches of the mating colonies where filamentation was observed on media containing 2.5%, 12%, and 25% pigeon guano, respectively.
TABLE 3.
Recombinational analysis of mating of C. neoformans KN99α NAT with KN99a NEO on medium containing pigeon guanoa
| Marker combination | No. of strains (%) |
|---|---|
| α NAT | 5 (11) (parental) |
| a NEO | 7 (16) (parental) |
| α | 5 (11) |
| a | 3 (7) |
| α NEO | 7 (16) |
| a NAT | 6 (13) |
| α NAT NEO | 9 (20) |
| a NAT NEO | 3 (7) |
| Total | 45 (100) |
Opposite-mating-type strains of C. neoformans var. grubii genetically marked with a NAT or NEO resistance gene were washed with PBS, and equal volumes of each mating type were combined. The mixture was placed as a 10-μl drop onto medium containing 25% pigeon guano. Plates were incubated in the dark at 25°C for 7 days. Spores were microdissected, and the resulting colonies were screened for mating type and the presence of each marker.
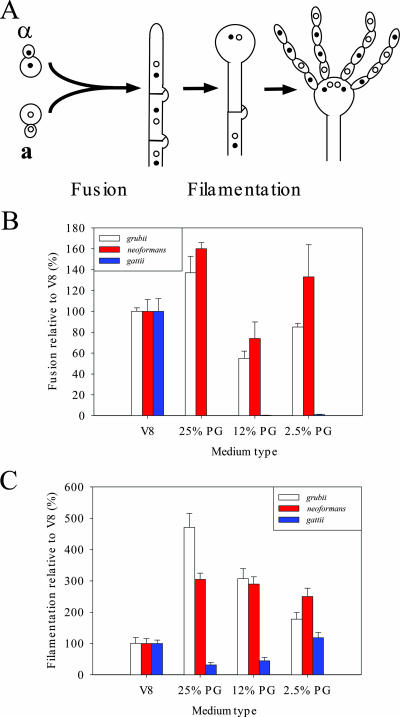
Mating is generally thought to occur in response to nutrient limitation. The two cell types produce peptide pheromones that trigger conjugation tube formation in α cells and uniform cell expansion of a cells, leading to cell fusion. Nuclear fusion is delayed, and the resulting heterokaryon adopts a filamentous state. The filaments ultimately produce basidia, where nuclear fusion and meiosis occur, and long chains of recombinant basidiospores are produced (33, 55). Thus, enhanced mating of C. neoformans on pigeon guano medium could occur at two stages of mating: cell fusion and filamentation (Fig. 5A).
FIG. 5.
Regulation of C. neoformans and C. gattii mating on V8 medium compared to that on medium containing pigeon guano (PG). (A) Schematic diagram of Cryptococcus mating. (B) Cells (108) of opposite mating types, genetically marked with a NAT or NEO resistance marker, were mixed, and 5-μl spots were placed onto V8 medium (pH 5 for C. neoformans var. grubii and pH 7 for C. neoformans var. neoformans and C. gattii) or medium containing 25%, 12%, or 2.5% pigeon guano. After 24 h of incubation, the resulting cells were screened for fusion products containing both markers. The numbers of fusion events on the various media are expressed as percentages of the number of fusion events occurring on V8 medium for each mating pair. (C) Six diploid strains for each variety/species were washed with PBS, inoculated onto V8 medium (pH 5 for C. neoformans var. grubii and pH 7 for C. neoformans var. neoformans and C. gattii) or pigeon guano medium contain differing levels of pigeon guano, and incubated in the dark for 7 days. Filament length is based on the average distance from the edge of the colony to the outer edge of filamentation and is expressed as a percentage of the average filament length on V8 medium.
To examine cell fusion of the C. neoformans varieties on pigeon guano medium, strains were neutrally marked with dominant NAT and NEO markers and cultured on pigeon guano or V8 medium and the levels of cell-cell fusion were compared. An increase in cell fusion was observed on 25% pigeon guano medium compared to cell fusion on V8 medium for both C. neoformans var. grubii and C. neoformans var. neoformans strains (Fig. 5B). Thus, components in pigeon guano stimulate C. neoformans a-α cell fusion during mating.
Next, the impact of pigeon guano medium on filamentation was analyzed using diploid a/α strains to allow an examination of filament length irrespective of fusion. Both var. neoformans and var. grubii diploid strains exhibited significantly increased filament lengths on pigeon guano medium (3-fold and 4.5-fold, respectively) compared to lengths on V8 medium. Even 2.5% pigeon guano medium increased filament length twofold in both C. neoformans varieties, indicating that pigeon guano stimulates filamentation, ultimately leading to the production of basidia and infectious basidiospores (Fig. 5C).
In contrast to C. neoformans strains, C. gattii strains showed significantly reduced mating on pigeon guano medium. Instead of the prolific mating observed on V8 or MS medium surrounding the entire colony periphery (Fig. 4A), only a few regions of mating were observed on pigeon guano medium, and the size and number of these regions decreased as the concentration of pigeon guano increased (Fig. 4). To determine whether C. gattii cell fusion is reduced on pigeon guano medium, the levels of fusion of NATr and NEOr strains were compared on pigeon guano and V8 media and found to be dramatically reduced (20-fold) on pigeon guano medium (Fig. 5B). Furthermore, the filament length of C. gattii a/α diploid strains was decreased by 80% on pigeon guano medium compared to that on V8 medium (Fig. 5C). Interestingly, as the concentration of pigeon guano in the medium decreased, filament length increased and was the same on 2.5% pigeon guano and V8 media. These results indicate that pigeon guano inhibits both the cell-cell fusion and the filamentation of C. gattii during mating.
DISCUSSION
Our results indicate that pigeon guano has the properties expected of the realized ecological niche for C. neoformans. The fact that C. neoformans var. grubii and neoformans produced higher numbers of CFU on pigeon guano than on YNB suggests that the nutritional composition of pigeon guano provides a highly favorable environment for the growth of Cryptococcus.
Increased growth with increasing pigeon guano concentration correlates with glucose levels in the medium. The fact that seabird guano did not support the growth of Cryptococcus unless it was supplemented with glucose suggests that guanos differ in the amounts of utilizable carbon sources that they contain. Bat guano also supported the growth of Cryptococcus, but to a lesser degree than pigeon guano, and Cryptococcus has occasionally been isolated from bat guano and caves (25, 58). The growth of Cryptococcus on chicken guano was previously found to be inhibited due to high alkalinity and the presence of a low-molecular-weight substance (85). The guanos tested here did not differ in alkalinity (data not shown). The fact that Cryptococcus was able to grow on all of the media tested when supplemented with pigeon guano or glucose indicates that nutrients are limiting in these guanos rather than that an inhibitory agent is present.
Many studies have examined the presence of Cryptococcus in association with birds and their guanos, including in aviaries where multiple species are housed in close proximity (6, 7, 12, 24, 26, 42, 51, 54, 67, 75). These studies have identified specific avian species as carriers for Cryptococcus, but no reason for this specificity has been determined. Our data comparing the levels of growth of Cryptococcus on multiple guanos suggests that the preference of Cryptococcus for certain avian species is likely due to the nutrient composition of the corresponding guano. This finding may help to more clearly define the role of birds and bird excreta, particularly pigeons, in the transmission of C. neoformans. The growth of these organisms on pigeon guano provides a potential mechanism for explaining how pigeons might play a role in harboring C. neoformans, either internally or on external parts of their anatomy that come into contact with guano, such as their feathers or feet (1, 8, 15, 30, 36, 39, 48, 54, 67, 80).
Pigmentation is observed when Cryptococcus is grown on pigeon guano and increases as the concentration of guano in the medium increases. Pigmentation was also observed in other media with carbon source supplementation. The pigment melanin is a virulence factor of Cryptococcus. Interestingly, melanin production is suppressed in standard laboratory media by carbon supplementation (88), yet a utilizable carbon source does not appear to be limiting in pigeon guano medium. The pigment observed on pigeon guano is in part produced by the classical laccase pathway involved in melanin production because laccase mutants exhibit a reduction in pigmentation. However, the laccase pathway is not solely responsible for the pigmentation, and lac1 and lac1 lac2 mutants still produce pigment on pigeon guano. It is unclear whether the remaining pigment is actively produced by cryptococcal cells or results from the transport of pigmented compounds from the medium into the cells. If the latter, it is specific to Cryptococcus, as a C. albicans strain did not accumulate pigment and produced white colonies on pigeon guano medium.
That C. neoformans var. grubii and var. neoformans strains grow and mate on pigeon guano medium and therefore complete their entire life cycle supports the hypothesis that pigeon guano is a realized ecological niche for C. neoformans. Finding the true realized ecological niche for an organism is challenging. While all evidence to date suggests that pigeon guano is an ecological niche for C. neoformans, we cannot exclude the possibility that there is another, as-yet-uncharacterized niche to which this organism is even better adapted.
While C. neoformans is well adapted to survive and sexually reproduce on pigeon guano, C. gattii is not well suited for long-term survival in this environment. C. gattii growth on pigeon guano is equivalent to that of C. neoformans, showing that pigeon guano is a fundamental niche for C. gattii and can sustain its growth. However, C. gattii mates poorly on pigeon guano. The inability to reproduce efficiently on pigeon guano shows that pigeon guano is not a suitable substrate overall for the species survival of C. gattii and therefore is not a realized ecological niche for this organism. These findings and our conclusions about the differing realized ecological niches for C. neoformans and C. gattii correlate well with environmental-isolation studies that show that C. neoformans is readily isolated from pigeon guano but that C. gattii is not. While the mating results highlight the importance of mating in pathogenic fungi, they also raise a paradox. In both C. neoformans and C. gattii, sexual reproduction is limited by a nearly unisexual population in which sexual reproduction might be uncommon (reviewed in reference 64). If sexual reproduction is a significant component of species survival, as these findings suggest, why is the population largely unisexual?
A monokaryotic fruiting cycle that produces spores has been identified in C. neoformans var. neoformans (89) and has recently been shown to produce sexual recombinant progeny (47). While this cycle has not been characterized yet in the laboratory for C. neoformans var. grubii or C. gattii, recent evidence suggests that monokaryotic fruiting may occur in nature (20, 69). C. neoformans var. neoformans strains were able to undergo filamentation on pigeon guano medium, but no spore production was observed (K. Nielsen, X. Lin, and J. Heitman, unpublished results). That filamentation could be induced suggests that monokaryotic fruiting might occur on pigeon guano under appropriate environmental conditions. If so, then both same-sex and a-α sexual reproduction may contribute to species survival.
Pigeon guano as a realized ecological niche for C. neoformans provides a plausible explanation for the cosmopolitan nature of this organism. Because of the intimate interaction between C. neoformans and pigeons (and possibly other avian species), the organism can disseminate worldwide along bird migratory routes and, due to the domestication of the pigeon, along trade routes. In contrast, C. gattii is associated with sedentary trees and thus has a more restricted global movement thought to be associated with tree export and planting. These observations also suggest that mating and sexual reproduction are required for the long-term survival of C. gattii, and thus the spread of the organism is limited. The C. gattii VI major outbreak strain exhibited no increase in mating on pigeon guano, suggesting that its introduction into the Pacific Northwest was not due to mating on pigeon guano. Instead recent studies suggest that the emergence of C. gattii in temperate environments is likely due to the expansion or alteration of the ecological niche by a subset of the population that allows for environmental proliferation predominantly in soil instead of in association with tree species (41, 53). Based on these studies, we hypothesize that at least two distinct events significantly altered the Cryptococcus ecology. First, an ancestral Cryptococcus strain gained the ability to sexually reproduce in pigeon guano and then swept the globe, likely as a result of the seafaring migration of humans and associated birds. Second, and perhaps more recently, another ecological-niche change has resulted in the survival of C. gattii in a temperate environment to allow further spread of a subset of this species.
The distribution of most primary fungal pathogens, including the dimorphic species Coccidioides immitis and Coccidioides posadasii, Histoplasma capsulatum, Penicillium marneffei, Paracoccidioides brasiliensis, and Blastomyces dermatitidis, is geographically restricted, likely due to an inability to reproduce or survive outside of their realized environmental niches. In many of these organisms, sexual reproduction and the requirements for reproduction are not clearly defined. However, with Cryptococcus neoformans and now C. gattii as examples, expansion of the environmental niches for the dimorphic primary pathogens could result in pandemic disease. The spread of C. immitis from North America to South America concomitant with Amerindian colonization exemplifies the ability of pathogenic fungi to adapt to environmental change (16). C. immitis outbreaks in regions of endemicity occur due to climactic changes rather than due to the emergence of pathogenic strains (17). However, the population exhibits high levels of genetic exchange, and thus the emergence of a new strain with the expansion of an ecological niche is conceivable and may have contributed to the migration of the population from North to South America (18). By studying differences between C. neoformans and C. gattii, we may be able to identify key events punctuating environmental-niche expansion that might apply to the emergence of or increased risk for other environmental fungal pathogens.
The findings presented here on emerging fungal pathogens are also applicable to other microbial pathogens. Both ecological changes and microbial evolution are significant determinants for bacterial- and viral-disease emergence. For example, all pandemic and epidemic influenza A virus outbreaks arise by genetic drift or reassortment to generate new viruses with differing pathogeneses (81, 82). This illustrates the pressing need to understand not only the driving force behind genetic alterations but also how these genetic changes affect the ecology of the organism and thereby impact disease prevalence.
Acknowledgments
We thank Josh Nosanchuk and Arturo Casadevall for melanin ghost analysis; Andrew Alspaugh, Yong-Sun Bahn, James A. Fraser, Alexander Idnurm, and Xiaorong Lin for strains; Diane Inglis for discussions and suggestions; and Ana Litvintseva and Xiaorong Lin for helpful comments on the manuscript.
This work was supported by NIAID R01 grant AI50113 to Joseph Heitman. Kirsten Nielsen was supported by NIAID grant T32 and Ruth L. Kirschstein NRSA in Molecular Mycology and Pathogenesis grant AI052080.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Abou-Gabal, M., and M. Atia. 1978. Study of the role of pigeons in the dissemination of Cryptococcus neoformans in nature. Sabouraudia 16:63-68. [DOI] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., L. Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y., C. Venkataraman, O. S. Harb, and L. Y. Gao. 1998. Signal transduction in the protozoan host Hartmannella vermiformis upon attachment and invasion by Legionella micdadei. Appl. Environ. Microbiol. 64:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahn, Y. S., J. K. Hicks, S. S. Giles, G. M. Cox, and J. Heitman. 2004. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot. Cell 3:1476-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreto de Oliveira, M. T., T. Boekhout, B. Theelen, F. Hagen, F. A. Baroni, M. S. Lazera, K. B. Lengeler, J. Heitman, I. N. Rivera, and C. R. Paula. 2004. Cryptococcus neoformans shows a remarkable genotypic diversity in Brazil. J. Clin. Microbiol. 42:1356-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauwens, L., D. Swinne, C. De Vroey, and W. De Meurichy. 1986. Isolation of Cryptococcus neoformans var. neoformans in the aviaries of the Antwerp Zoological Gardens. Mykosen 29:291-294. [PubMed] [Google Scholar]
- 7.Bauwens, L., F. Vercammen, C. Wuytack, K. Van Looveren, and D. Swinne. 2004. Isolation of Cryptococcus neoformans in Antwerp Zoo's nocturnal house. Mycoses 47:292-296. [DOI] [PubMed] [Google Scholar]
- 8.Bohm, K. H., G. Trautwein, E. Weiland, and I. S. Abdallah. 1974. Experimental infection of pigeons and chickens with Cryptococcus neoformans and further epidemiologic studies. Mycopathol. Mycol. Appl. 54:317-328. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, L. T., B. J. Currie, M. Krockenberger, R. Malik, W. Meyer, J. Heitman, and D. A. Carter. 2005. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot. Cell 4:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, L. T., J. A. Fraser, C. B. Nichols, F. S. Dietrich, D. A. Carter, and J. Heitman. 2005. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryot. Cell 4:1410-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, DC.
- 12.Criseo, G., M. S. Bolignano, F. De Leo, and F. Staib. 1995. Evidence of canary droppings as an important reservoir of Cryptococcus neoformans. Zentbl. Bakteriol. 282:244-254. [PubMed] [Google Scholar]
- 13.Ellis, D. H., and T. J. Pfeiffer. 1990. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336:923-925. [DOI] [PubMed] [Google Scholar]
- 14.Ellis, D. H., and T. J. Pfeiffer. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ensley, P. K., C. E. Davis, M. P. Anderson, and K. C. Fletcher. 1979. Cryptococcosis in a male Beccari's crowned pigeon. J. Am. Vet. Med. Assoc. 175:992-994. [PubMed] [Google Scholar]
- 16.Fisher, M. C., G. L. Koenig, T. J. White, G. San-Blas, R. Negroni, I. G. Alvarez, B. Wanke, and J. W. Taylor. 2001. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc. Natl. Acad. Sci. USA 98:4558-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher, M. C., G. L. Koenig, T. J. White, and J. W. Taylor. 2000. Pathogenic clones versus environmentally driven population increase: analysis of an epidemic of the human fungal pathogen Coccidioides immitis. J. Clin. Microbiol. 38:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher, M. C., B. Rannala, V. Chaturvedi, and J. W. Taylor. 2002. Disease surveillance in recombining pathogens: multilocus genotypes identify sources of human Coccidioides infections. Proc. Natl. Acad. Sci. USA 99:9067-9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser, J. A., S. S. Giles, E. C. Wenink, S. G. Geunes-Boyer, J. R. Wright, S. Diezmann, A. Allen, J. E. Stajich, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360-1364. [DOI] [PubMed] [Google Scholar]
- 21.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallo, M. G., P. Cabeli, and V. Vidotto. 1989. Presence of pathogenic yeasts in the feces of the semi-domesticated pigeon (Columba livia, Gmelin 1789, urban type) from the city of Turin. Parasitologia 31:207-212. [PubMed] [Google Scholar]
- 23.Garrett, L. 1994. The coming plague: newly emerging diseases in a world out of balance. Farrar, Straus, and Giroux, New York, NY.
- 24.Gokulshankar, S., S. Ranganathan, M. S. Ranjith, and A. J. Ranjithsingh. 2004. Prevalence, serotypes and mating patterns of Cryptococcus neoformans in the pellets of different avifauna in Madras, India. Mycoses 47:310-314. [DOI] [PubMed] [Google Scholar]
- 25.Grose, E., C. J. Marinkelle, and C. Streigel. 1968. The use of tissue cultures in the identification of Cryptococcus neoformans isolated from Colombian bats. Sabouraudia 6:127-132. [PubMed] [Google Scholar]
- 26.Gugnani, H. C., N. P. Gupta, and J. B. Shrivastav. 1972. Prevalence of Cryptococcus neoformans in Delhi Zoological Park and its recovery from the sputum of an employee. Indian J. Med. Res. 60:182-185. [PubMed] [Google Scholar]
- 27.Haag-Wackernagel, D., and H. Moch. 2004. Health hazards posed by feral pigeons. J. Infect. 48:307-313. [DOI] [PubMed] [Google Scholar]
- 28.Hoang, L. M., J. A. Maguire, P. Doyle, M. Fyfe, and D. L. Roscoe. 2004. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997-2002): epidemiology, microbiology and histopathology. J. Med. Microbiol. 53:935-940. [DOI] [PubMed] [Google Scholar]
- 29.Hotzel, H., P. Kielstein, R. Blaschke-Hellmessen, J. Wendisch, and W. Bar. 1998. Phenotypic and genotypic differentiation of several human and avian isolates of Cryptococcus neoformans. Mycoses 41:389-396. [DOI] [PubMed] [Google Scholar]
- 30.Hubalek, Z. 1975. Distribution of Cryptococcus neoformans in a pigeon habitat. Folia Parasitol. (Praha) 22:73-79. [PubMed] [Google Scholar]
- 31.Hubalek, Z., and Z. Prikazsky. 1975. Growth of Cryptococcus neoformans in UV-irradiated excreta of pigeons. Folia Microbiol. (Praha) 20:231-235. [DOI] [PubMed] [Google Scholar]
- 32.Hutchinson, G. E. 1958. Concluding remarks. Cold Spring Harbor Symp. Quant. Biol. 22:415-427. [Google Scholar]
- 33.Idnurm, A., Y. S. Bahn, K. Nielsen, X. Lin, J. A. Fraser, and J. Heitman. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753-764. [DOI] [PubMed] [Google Scholar]
- 34.Idnurm, A., J. L. Reedy, J. C. Nussbaum, and J. Heitman. 2004. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell 3:420-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Institute of Medicine. 1992. Emerging infections: microbial threats to health in the United States. National Academy Press, Washington, DC. [PubMed]
- 36.Kao, C. J., and J. Schwarz. 1957. The isolation of Cryptococcus neoformans from pigeon nests; with remarks on the identification of virulent cryptococci. Am. J. Clin. Pathol. 27:652-663. [DOI] [PubMed] [Google Scholar]
- 37.Keele, B. F., F. Van Heuverswyn, Y. Li, E. Bailes, J. Takehisa, M. L. Santiago, F. Bibollet-Ruche, Y. Chen, L. V. Wain, F. Liegeois, S. Loul, E. M. Ngole, Y. Bienvenue, E. Delaporte, J. F. Brookfield, P. M. Sharp, G. M. Shaw, M. Peeters, and B. H. Hahn. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller, S. M., M. A. Viviani, M. C. Esposto, M. Cogliati, and B. L. Wickes. 2003. Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate. Microbiology 149:131-142. [DOI] [PubMed] [Google Scholar]
- 39.Khan, Z. U., M. Pal, H. S. Randhawa, and R. S. Sandhu. 1978. Carriage of Cryptococcus neoformans in the crops of pigeons. J. Med. Microbiol. 11:215-218. [DOI] [PubMed] [Google Scholar]
- 40.Khosravi, A. R. 1997. Isolation of Cryptococcus neoformans from pigeon (Columbia livia) droppings in northern Iran. Mycopathologia 139:93-95. [DOI] [PubMed] [Google Scholar]
- 41.Kidd, S. E., Y. Chow, S. Mak, P. J. Bach, H. Chen, A. O. Hingston, J. W. Kronstad, and K. H. Bartlett. 2007. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and Pacific Northwest of the United States. Appl. Environ. Microbiol. 73:1433-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kielstein, P., H. Hotzel, A. Schmalreck, D. Khaschabi, and W. Glawischnig. 2000. Occurrence of Cryptococcus spp. in excreta of pigeons and pet birds. Mycoses 43:7-15. [DOI] [PubMed] [Google Scholar]
- 43.Kwon-Chung, K. J. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197-1200. [PubMed] [Google Scholar]
- 44.Kwon-Chung, K. J. 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68:943-946. [PubMed] [Google Scholar]
- 45.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, X., and J. Heitman. 2005. Chlamydospore formation during hyphal growth in Cryptococcus neoformans. Eukaryot. Cell 4:1746-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin, X., C. M. Hull, and J. Heitman. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017-1021. [DOI] [PubMed] [Google Scholar]
- 48.Littman, M. L., and R. Borok. 1968. Relation of the pigeon to cryptococcosis: natural carrier state, heat resistance and survival of Cryptococcus neoformans. Mycopathol. Mycol. Appl. 35:329-345. [DOI] [PubMed] [Google Scholar]
- 49.Litvintseva, A. P., L. Kestenbaum, R. Vilgalys, and T. G. Mitchell. 2005. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J. Clin. Microbiol. 43:556-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Litvintseva, A. P., R. E. Marra, K. Nielsen, J. Heitman, R. Vilgalys, and T. G. Mitchell. 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell 2:1162-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Martinez, R., and L. R. Castanon-Olivares. 1995. Isolation of Cryptococcus neoformans var. neoformans from bird droppings, fruits and vegetables in Mexico City. Mycopathologia 129:25-28. [DOI] [PubMed] [Google Scholar]
- 52.Lopez-Martinez, R., J. L. Soto-Hernandez, L. Ostrosky-Zeichner, L. R. Castanon-Olivares, V. Angeles-Morales, and J. Sotelo. 1996. Cryptococcus neoformans var. gattii among patients with cryptococcal meningitis in Mexico. First observations. Mycopathologia 134:61-64. [DOI] [PubMed] [Google Scholar]
- 53.MacDougall, L., S. E. Kidd, E. Galanis, S. Mak, M. J. Leslie, P. R. Cieslak, J. W. Kronstad, M. G. Morshed, and K. H. Bartlett. 2007. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg. Infect. Dis. 13:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malik, R., M. B. Krockenberger, G. Cross, R. Doneley, D. N. Madill, D. Black, P. McWhirter, A. Rozenwax, K. Rose, M. Alley, D. Forshaw, I. Russell-Brown, A. C. Johnstone, P. Martin, C. R. O'Brien, and D. N. Love. 2003. Avian cryptococcosis. Med. Mycol. 41:115-124. [DOI] [PubMed] [Google Scholar]
- 55.McClelland, C. M., Y. C. Chang, A. Varma, and K. J. Kwon-Chung. 2004. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 12:208-212. [DOI] [PubMed] [Google Scholar]
- 56.Missall, T. A., J. M. Moran, J. A. Corbett, and J. K. Lodge. 2005. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot. Cell 4:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mogensen, E. G., G. Janbon, J. Chaloupka, C. Steegborn, M. S. Fu, F. Moyrand, T. Klengel, D. S. Pearson, M. A. Geeves, J. Buck, L. R. Levin, and F. A. Muhlschlegel. 2006. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryot. Cell 5:103-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montagna, M. T., M. P. Santacroce, G. Caggiano, D. Tato, and L. Ajello. 2003. Cavernicolous habitats harbouring Cryptococcus neoformans: results of a speleological survey in Apulia, Italy, 1999-2000. Med. Mycol. 41:451-455. [DOI] [PubMed] [Google Scholar]
- 59.Moore, T. D., and J. C. Edman. 1993. The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morse, S. S. 2004. Factors and determinants of disease emergence. Rev. Sci. Tech. 23:443-451. [DOI] [PubMed] [Google Scholar]
- 61.Morse, S. S. 1995. Factors in the emergence of infectious diseases. Emerg. Infect. Dis. 1:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen, K., G. M. Cox, A. P. Litvintseva, E. Mylonakis, S. D. Malliaris, D. K. Benjamin, Jr., S. S. Giles, T. G. Mitchell, A. Casadevall, J. R. Perfect, and J. Heitman. 2005. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 73:4922-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen, K., and J. Heitman. 2007. Sex and virulence of human pathogenic fungi. Adv. Genet. 57:143-174. [DOI] [PubMed] [Google Scholar]
- 65.Nielsen, K., R. E. Marra, F. Hagen, T. Boekhout, T. G. Mitchell, G. M. Cox, and J. Heitman. 2005. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics 171:975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nosanchuk, J. D., J. Rudolph, A. L. Rosas, and A. Casadevall. 1999. Evidence that Cryptococcus neoformans is melanized in pigeon excreta: implications for pathogenesis. Infect. Immun. 67:5477-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nosanchuk, J. D., S. Shoham, B. C. Fries, D. S. Shapiro, S. M. Levitz, and A. Casadevall. 2000. Evidence of zoonotic transmission of Cryptococcus neoformans from a pet cockatoo to an immunocompromised patient. Ann. Intern. Med. 132:205-208. [DOI] [PubMed] [Google Scholar]
- 68.Pukkila-Worley, R., Q. D. Gerrald, P. R. Kraus, M. J. Boily, M. J. Davis, S. S. Giles, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ren, P., D. J. Springer, M. J. Behr, W. A. Samsonoff, S. Chaturvedi, and V. Chaturvedi. 2006. Transcription factor STE12alpha has distinct roles in morphogenesis, virulence, and ecological fitness of the primary pathogenic yeast Cryptococcus gattii. Eukaryot. Cell 5:1065-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schonheyder, H., and A. Stenderup. 1982. Isolation of Cryptococcus neoformans from pigeon manure on two media inducing pigment formation. Sabouraudia 20:193-197. [PubMed] [Google Scholar]
- 71.Sorrell, T. C., A. G. Brownlee, P. Ruma, R. Malik, T. J. Pfeiffer, and D. H. Ellis. 1996. Natural environmental sources of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 34:1261-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sriburee, P., S. Khayhan, C. Khamwan, S. Panjaisee, and P. Tharavichitkul. 2004. Serotype and PCR-fingerprints of clinical and environmental isolates of Cryptococcus neoformans in Chiang Mai, Thailand. Mycopathologia 158:25-31. [DOI] [PubMed] [Google Scholar]
- 73.Staib, F. 1981. The perfect state of Cryptococcus neoformans, Filobasidiella neoformans, on pigeon manure filtrate agar. Zentbl. Bakteriol. A 248:575-578. [PubMed] [Google Scholar]
- 74.Staib, F., and A. Blisse. 1982. Bird manure filtrate agar for the formation of the perfect state of Cryptococcus neoformans, Filobasidiella neoformans. A comparative study of the agars prepared from pigeon and canary manure. Zentbl. Bakteriol. Mikrobiol. Hyg. Ser. A 251:554-562. [PubMed] [Google Scholar]
- 75.Staib, F., and J. Schulz-Dieterich. 1984. Cryptococcus neoformans in fecal matter of birds kept in cages—control of C. neoformans habitats. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. B 179:179-186. [PubMed] [Google Scholar]
- 76.Stenderup, J., K. Flensted, C. Jorgensen, A. H. Sorensen, N. C. Hansen, and H. C. Siersted. 1989. Occurrence of the yeast, Cryptococcus (Cr) neoformans, in pigeon droppings. Ugeskr. Laeger 151:2974-2975. [PubMed] [Google Scholar]
- 77.Stout, J. E., and V. L. Yu. 1997. Legionellosis. N. Engl. J. Med. 337:682-687. [DOI] [PubMed] [Google Scholar]
- 78.Sukroongreung, S., K. Kitiniyom, C. Nilakul, and S. Tantimavanich. 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med. Mycol. 36:419-424. [PubMed] [Google Scholar]
- 79.Swinne, D., M. Deppner, R. Laroche, J. J. Floch, and P. Kadende. 1989. Isolation of Cryptococcus neoformans from houses of AIDS-associated cryptococcosis patients in Bujumbura (Burundi). AIDS 3:389-390. [DOI] [PubMed] [Google Scholar]
- 80.Swinne-Desgain, D. 1976. Cryptococcus neoformans in the crops of pigeons following its experimental administration. Sabouraudia 14:313-317. [DOI] [PubMed] [Google Scholar]
- 81.Taubenberger, J. K., and D. M. Morens. 2006. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 12:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taubenberger, J. K., and D. M. Morens. 2006. Influenza revisited. Emerg. Infect. Dis. 12:1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tintelnot, K., K. Lemmer, H. Losert, G. Schar, and A. Polak. 2004. Follow-up of epidemiological data of cryptococcosis in Austria, Germany and Switzerland with special focus on the characterization of clinical isolates. Mycoses 47:455-464. [DOI] [PubMed] [Google Scholar]
- 84.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walter, J. E., and R. B. Yee. 1968. Factors that determine the growth of Cryptococcus neoformans in avian excreta. Am. J. Epidemiol. 88:445-450. [DOI] [PubMed] [Google Scholar]
- 86.Walton, F. J., A. Idnurm, and J. Heitman. 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 57:1381-1396. [DOI] [PubMed] [Google Scholar]
- 87.Wang, P., M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO Rep. 2:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang, P., J. Cutler, J. King, and D. Palmer. 2004. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot. Cell 3:1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimor-phism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu, J., R. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto, Y., S. Kohno, T. Noda, H. Kakeya, K. Yanagihara, H. Ohno, K. Ogawa, S. Kawamura, T. Ohtsubo, K. Tomono, et al. 1995. Isolation of Cryptococcus neoformans from environments (pigeon excreta) in Nagasaki. Kansenshogaku Zasshi 69:642-645. [DOI] [PubMed] [Google Scholar]
- 92.Yilmaz, A., G. Goral, S. Helvaci, K. Kilicturgay, F. Gokirmak, O. Tore, and S. Gedikoglu. 1989. Distribution of Cryptococcus neoformans in pigeon feces. Mikrobiyol. Bul. 23:121-126. [PubMed] [Google Scholar]
- 93.Zhu, X., and P. R. Williamson. 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 5:1-10. [DOI] [PubMed] [Google Scholar]