Abstract
The mitotic spindle is a microtubule (MT)-based molecular machine that serves for equal segregation of chromosomes during cell division. The formation of the mitotic spindle requires the activity of MT motors, including members of the kinesin-14 family. Although evidence suggests that kinesins-14 act by driving the sliding of MT bundles in different areas of the spindle, such sliding activity had never been demonstrated directly. To test the hypothesis that kinesins-14 can induce MT sliding in living cells, we developed an in vivo assay, which involves overexpression of the kinesin-14 family member Drosophila Ncd in interphase mammalian fibroblasts. We found that green fluorescent protein (GFP)–Ncd colocalized with cytoplasmic MTs, whose distribution was determined by microinjection of Cy3 tubulin into GFP-transfected cells. Ncd overexpression resulted in the formation of MT bundles that exhibited dynamic “looping” behavior never observed in control cells. Photobleaching studies and fluorescence speckle microscopy analysis demonstrated that neighboring MTs in bundles could slide against each other with velocities of 0.1 μm/s, corresponding to the velocities of movement of the recombinant Ncd in in vitro motility assays. Our data, for the first time, demonstrate generation of sliding forces between adjacent MTs by Ncd, and they confirm the proposed roles of kinesins-14 in the mitotic spindle morphogenesis.
INTRODUCTION
The mitotic spindle is a molecular machine that serves for equal segregation of chromosomes during mitosis. The principal structural components of the mitotic spindle are cytoplasmic microtubules (MTs) organized into two polarized (minus ends at the center) radial arrays, located at a distance from each other. MT organization in the mitotic spindle is achieved by MT motors, kinesins and dyneins, which generate force for movement toward the plus or minus ends of MTs and that participate in the transport of chromosomes to the spindle poles, spindle elongation during anaphase, regulation of the MT turnover rates, and spindle morphogenesis (for review, see Sharp et al., 2000b; Karsenti and Vernos, 2001; McIntosh et al., 2002; Scholey et al., 2003; Gadde and Heald, 2004; Wadsworth and Khodjakov, 2004).
A special role in the formation and maintenance of the mitotic spindle belongs to the minus-end–directed members of the kinesin-14 subfamily. Unlike conventional kinesin, kinesin-14 family members have motor domains at the carboxy terminus (for review, see Ovechkina and Wordeman, 2003). The best studied kinesin-14 members are Drosophila Ncd (Komma et al., 1991; Matthies et al., 1996) and Saccharomyces cerevisiae Kar3 (Meluh and Rose, 1990). These MT motors, and their mammalian (Matuliene et al., 1999; Mountain et al., 1999; Zhu et al., 2005) and plant (Vanstraelen et al., 2006) homologues, play essential roles in mitosis and meiosis. Their activities are important for the formation of the mitotic spindle poles (Goshima and Vale, 2003; Goshima et al., 2005; Morales-Mulia and Scholey, 2005; Zhu et al., 2005) and for regulation of the distance between the poles in the mitotic spindle (Saunders and Hoyt, 1992; Saunders et al., 1997; Sharp et al., 1999, 2000a; Troxell et al., 2001). Loss of a kinesin-14 function may cause different mitotic and meiotic spindle defects even within the same organism. For example, in Drosophila S2 cells and female meiosis, loss of Ncd by RNA interference (RNAi) or null mutants causes disorganization of spindle poles (Theurkauf and Hawley, 1992; Goshima et al., 2005; Morales-Mulia and Scholey, 2005), whereas in embryos, it causes changes in spindle length and a decrease in the persistence of steady-state structures (Sharp et al., 1999; Brust-Mascher and Scholey, 2002).
Evidence suggests that kinesins-14 act by driving the sliding of parallel or antiparallel MT bundles in different areas of the spindle (for review, see Sharp et al., 2000b,c; McIntosh et al., 2002; Gadde and Heald, 2004). These motors have two MT binding sites, an ATP-dependent site in the motor domain and an ATP-independent site in the tail, and they have been shown to induce MT bundling (McDonald et al., 1990; Chandra et al., 1993; Karabay and Walker, 1999; Matuliene et al., 1999). Recent small interfering RNA (siRNA) studies demonstrate that knockdown of the kinesin-14 family member Ncd results in the formation of splayed mitotic spindle poles, suggesting that the activity of this motor is required for focusing of the MTs at the pole (Goshima and Vale, 2003; Goshima et al., 2005; Morales-Mulia and Scholey, 2005). It has been found that loss of kinesin-14 function in S. cerevisiae and Drosophila leads to an increase in the spindle length and that it rescues spindle pole separation defects seen in cells lacking members of the kinesin-5 family (Saunders and Hoyt, 1992; Saunders et al., 1997; Sharp et al., 1999; Sharp et al., 2000a) and that these two motors oppose each other in in vitro motility assays (Tao et al., 2006), suggesting that the balance of their activities is required for maintaining the correct distance between the spindle poles. It is hypothesized that kinesins-14 achieve this balance by mediating antiparallel sliding of MTs emanating from the opposite poles (Sharp et al., 2000b,c; McIntosh et al., 2002; Gadde and Heald, 2004). However, despite the abundance of indirect evidence and computational models that suggest an essential role for kinesins-14 in MT sliding (Mogilner et al., 2006), such sliding activity of kinesins-14 had never been demonstrated directly before.
In this study, we developed an in vivo assay to test the hypothesis that kinesin-14 family member Ncd can induce sliding of MTs against each other in living cells. We found that green fluorescent protein (GFP)–Ncd overexpressed in cultured human fibroblasts colocalized with cytoplasmic MTs, whose localization and behavior were monitored by microinjection of Cy3 tubulin into GFP-transfected cells. Ncd overexpression resulted in the formation of MT bundles that exhibited dynamic “looping” behavior never observed in control cells. Photobleaching studies demonstrated that neighboring MTs in bundles could slide against each other with the velocities of 0.1 μm/s, corresponding to the velocities of movement of the recombinant Ncd in in vitro motility assays. Our data confirm the hypothesis that kinesin-14 family members generate sliding forces between adjacent MTs, and they constitute the first demonstration of such sliding activity of MT motors in vivo.
MATERIALS AND METHODS
Plasmids for Expression of DNA in Human Fibroblasts
For expression in human fibroblasts, full-size Ncd (cDNA was a gift from Dr. Steven Rogers, University of North Carolina), an N-terminal tail domain of Ncd (amino acid residues 1–204), or a C-terminal motor domain (amino acid residues 327–700) was amplified using polymerase chain reaction (PCR) and subcloned into pEGFPC2 mammalian expression vector (BD Biosciences Clontech, Palo Alto, CA) to add a reporter GFP tag to the N termini of expressing proteins. Previous work showed that GFP-tagged Ncd fully rescued spindle defects caused by RNAi of endogenous protein and therefore remained fully functional (Goshima and Vale, 2005). For the amplification of the full-size molecule or tail domain, we used the same forward primer 5′-CTC GTC GAC ATG GAA TCC CGG CTA CCG AAA CCG-3′ but different reverse primers, 5′-CTC CTC GAG TTT ATC GAA ACT GCC GCT GTT GTT-3′ and 5′-CTC CTC GAG TTT ATC GAA ACT GCC GCT GTT GTT-3′, respectively. The gel-purified fragment was then digested with EcoR1/Sal1 and ligated into EcoR1/Sal1-digested pEGFPC2. Forward and reverse primers 5′-GAG ACC TGC AAA GAA TTC CTC TTC CAG TCG-3′ and 5′-GGT GAT TTG TCG ACA GAA CCG-3′ were used for amplification of the C-terminal motor domain. This fragment was then cloned into pEGFPC2 vector after restriction digestion with BamH1 and EcoR1.
An Ncd rigor-type mutant was made by introducing a point mutation T441N at the ATP-binding consensus motif. A similar kinesin mutation in the heavy chain of conventional kinesin (T93N) caused tight irreversible ATP-insensitive binding of kinesin molecules to MTs in vivo and in vitro (Nakata and Hirokawa, 1995). The forward and reverse primers used for the generation of Ncd rigor mutant were 5′CAG ACT GGT TCG GGA AAA AAT TAC ACT ATG GAC GGG GTG-3′ and 5′-CAC CCC GTC CAT AGT GTA ATT TTT TCC CGA ACC AGT CTG 3′, respectively. Mutant Ncd DNA was cloned into pEGFP2 expression vector similar to wild-type full-size Ncd DNA. Drosophila Kinesin-1 DNA (clone SD02406; Drosophila Genomics Resource Center, Indiana University, Bloomington, IN) was provided by Dr. William Saxton. For fusion with GFP, Kinesin-1 DNA was amplified using PCR (forward and reverse primers were 5′-GGA ATT CAT GTC CGC GGA ACG AGA G-3′ and 5′-GGA TCC ACG AGT TGA CAG GAT TAA CCT G-3′, respectively). The gel-purified fragment was then digested with EcoR1/BamH1 and ligated into EcoR1/BamH1-digested pEGFPN1 vector (Clontech, Mountain View, CA). GFP-tau plasmid (GFP-tau23; Samsonov et al., 2004) was a gift from Dr. Sergei Popov (University of Illinois at Chicago).
Cell Culture and Transfection
Human 356 fibroblasts were cultured in F-10 medium supplemented with 10% fetal bovine serum, and antibiotics, at 37°C. Cells were transfected with GFP-tagged full-size or truncated Ncd DNAs by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and they were incubated for 12–16 h at 37°C to allow for the protein expression.
Microinjection and Live Cell Imaging
For fluorescent labeling of MTs, cells were pressure microinjected with Cy3-tagged bovine brain tubulin prepared by labeling with Cy3-reactive dye as described previously (Vorobjev et al., 2001) and used at a needle concentration of 8–10 mg/ml. For fluorescence speckle microscopy (Waterman-Storer et al., 1998), the needle concentration of tubulin was reduced to 0.3–0.5 mg/ml. Injected cells were incubated for at least 1 h at 37°C in a CO2 incubator to allow for incorporation of labeled tubulin into MTs.
Fluorescence images of cells were acquired using a Nikon Diaphot 300 inverted microscope equipped with a Plan ×100 1.25 numerical aperture objective. Images were collected with a slow-scan back-illuminated cooled charge-coupled device camera (CH350; Roper Scientific, Trenton, NJ) driven by MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA). In some experiments, fluorescence images were acquired using a spinning disk confocal microscope (PerkinElmer Life and Analytical Sciences, Boston, MA). To reduce photobleaching and photodamage, before image acquisition cells were treated with the oxygen-depleting agent Oxyrase (Oxyrase, Mansfield, OH). During image acquisition the temperature was maintained at 37°C using the Delta T controlled culture dish system (Biopthechs, Butler, PA).
Fluorescence Redistribution after Photobleaching Assay
For photobleaching experiments, cells with Cy3-labeled MTs were locally irradiated with a laser beam focused through the objective lens. Our past work indicated that illumination leaves MTs intact but that it results in the bleaching of fluorochrome in the illuminated zone (Rodionov et al., 1994). For photobleaching, the beam of a 1-W argon ion laser (Laser Physics, West Jordan, UT) was channeled into the epi-illumination system of a Nikon Diaphot 300 inverted microscope (Nikon, Tokyo, Japan). A cylindrical lens was positioned to focus the laser beam through a 100×, 1.25 aperture objective lens into an ∼3-μm-wide bar in the specimen plane. The laser was operated at 514 nm and 100 mW. Some photobleaching experiments involved illuminating MTs with an argon ion laser beam by using a custom-made illumination system attached to PerkinElmer spinning disk confocal microscope, which produced 0.5–1 μm wide bar-shaped zones on MTs. Images of bleached MTs were captured with 16- or 12-bit digital cameras. Quantification of the photobleaching data was performed using MetaMorph Image acquisition and analysis software. The bleached regions were delineated with rectangles, and average fluorescence intensity values of the pixels in delineated areas were computed. After background subtraction, the data were expressed as a percentage of the average fluorescence intensity recovered after photobleaching. The 100% value was taken as the fluorescence intensity in the same region immediately before the bleach. Data were plotted an analyzed using Origin scientific graphing and analysis software (OriginLab, Northampton, MA). Quantification of the maximum rate of MT movement involved approximation of the slopes of fluorescence recovery curves using Origin software.
Immunostaining of MTs
For immunostaining with tubulin antibody, cells were fixed with methanol for 5 min at −20°C, rehydrated in phosphate-buffered saline (PBS) containing 1% bovine serum albumin, and successively incubated with rabbit affinity-purified antibody against tyrosilated tubulin (a gift from Dr. Gregg Gundersen, Columbia University) and goat anti-rabbit antibody conjugated with tetramethylrhodamine B isothiocyanate (Kirkegaard and Perry Laboratories, Gaithersburg, MD). Images of immunostained MTs were acquired using a charge-coupled device camera as described above.
RESULTS
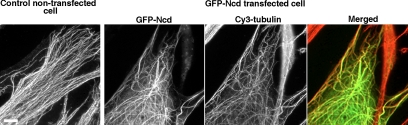
To examine the role of Ncd in MT organization, we overexpressed Drosophila Ncd containing an N-terminal GFP tag in human 356 fibroblasts, and we examined the distribution and behavior of microtubules in the transfected cells by microinjection of Cy3-labeled tubulin. In the control cells, MTs formed a dense radial network typical for fibroblast-type cells (Figure 1, left). In contrast, in cells overexpressing GFP–Ncd, MTs formed thick, relatively sparse cables, and they had a curled appearance (Figure 1, middle). Analysis of the cytoplasmic distribution of GFP indicated that GFP–Ncd in such cells colocalized with the MTs (Figure 1, right). We therefore conclude that GFP–Ncd overexpressed in interphase mammalian fibroblasts colocalized with cytoplasmic MTs, whose shape and density seemed to change as a result of GFP–Ncd overexpression.
Figure 1.
Overexpressed GFP–Ncd colocalizes with cytoplasmic MTs and changes their intracellular distribution. Left, MT distribution visualized by Cy3 fluorescence in a control untransfected cell injected with Cy3–tubulin. Middle, GFP (middle left) and Cy3 (middle right) fluorescence in a GFP–Ncd-expressing cell injected with Cy3–tubulin. Right, merge of the two middle images (green, GFP; red, Cy3). Unlike control untransfected cells MTs in GFP–Ncd-transfected cells form thick, sparse cables that are often curved. As seen from the merged image on the right, GFP–Ncd constructs colocalizes with MTs. Bar, 10 μm.
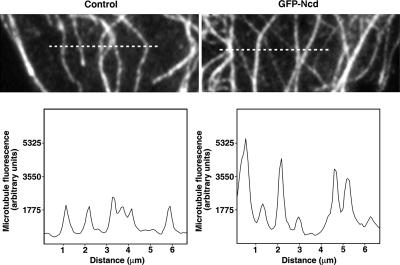
To examine the possibility that the observed Ncd-dependent reduction of MT density and apparent increase in their thickness was caused by MT cross-linking, we compared the distribution of MT fluorescence along a line drawn parallel to the cell edge (across the fiber axes) at the periphery of control and GFP–Ncd-expressing cells. We found that in control cells, the intensity of tubulin fluorescence in all the fibers was similar—an indication that these filaments likely corresponded to single MTs (Figure 2, left). In contrast, line scans across the fluorescent filaments in GFP–Ncd-transfected cells revealed a nonuniform pattern of labeling. Many of the filaments that incorporated fluorescently tagged tubulin were significantly brighter than others, as would be expected if they were composed of several MTs (Figure 2, right). Therefore, the labeling pattern of MTs in Ncd-expressing cells is consistent with the results of in vitro experiments (McDonald et al., 1990; Chandra et al., 1993) that indicate that Ncd causes the formation of MT bundles.
Figure 2.
MTs in the presence of Ncd form cytoplasmic bundles. Images of MTs (top), and profiles of MT fluorescence (bottom) along the line drawn across the MT axes in the lamella of control (left) and GFP-transfected cells (right). The lines used for measurement are shown as dashed lines on both images. The peak intensities of tubulin fluorescence in the control cell are uniform along the line, whereas in the GFP–Ncd-transfected cell the peaks are heterogeneous, indicating the formation of MT bundles.
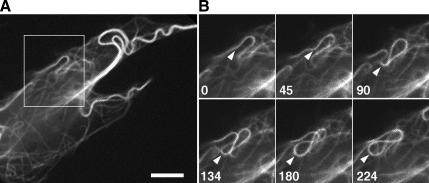
To test directly whether Ncd-dependent MT cross-linking results in movement of MTs against each other, we examined the behavior of MTs in cells overexpressing GFP–Ncd. Live imaging of MT behavior in Ncd-transfected cells showed that in such cells, the shapes of MT cables constantly changed. The most common pattern of movement involved the formation of MT loops whose size and curvature altered with time (Figure 3 and Supplemental Movie 2). Such MT behavior was never observed in control nontransfected cells. We conclude that overexpression of Ncd facilitates MT movement in the cytoplasm and that it produces a specific MT movement pattern, which involves the formation and expansion of the MT loops.
Figure 3.
MT looping in a cell overexpressing GFP–Ncd. (A) Low-magnification image of MTs. (B) Successive high-magnification images of MTs in the boxed area depicted in A. Numbers indicate time in seconds. Arrow points on a segment of a MT whose curvature changes with time, resulting in the formation of a loop. See also Supplemental Movie 2. Bar, 10 μm.
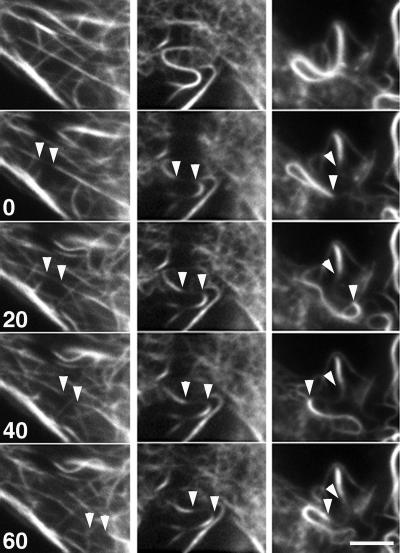
To further demonstrate MT movement in GFP-overexpressing cells, we used two independent experimental approaches to introduce fiduciary marks on MTs: fluorescent speckle microscopy (Waterman-Storer et al., 1998) and local photobleaching of tubulin fluorescence with a laser microbeam. To suppress MT dynamic instability and its contribution to MT movement, we conducted these observations in the presence of the MT-stabilizing drug Taxol under conditions that dramatically reduced the dynamic behavior of free MT ends as seen by time-lapse imaging (Supplemental Movie 1). Playback of the image sequences indicated that fluorescent speckles (Supplemental Movie 3) and bleached zones on MTs (Figure 4 and Supplemental Movie 4) moved in the cytoplasm in the presence of Ncd. Such movement has never been observed in control cells (data not shown). Because MT movement continued in the presence of Taxol, we conclude that MT movement cannot be explained by forces generated by MT dynamics. Instead, our data suggest that the observed MT movement was produced via sliding forces generated by MT-bound Ncd molecules.
Figure 4.
The movement of marks on Taxol-stabilized MTs generated by photobleaching in cells overexpressing GFP–Ncd. Three sets of successive images shown on the panel correspond to three independent photobleaching experiments. Numbers indicate time in seconds. Arrowheads show positions of the margins of the bleached zones. The bleached zones displaced in a linear manner (left and middle columns) or followed a curved path (right column). Also see Supplemental Movie 4. Bar, 10 μm.
To measure the rate of Ncd-dependent MT movement, we quantified the movement of the bleached zones on individual MTs by direct measurement of the displacement of the edges of the bleached zones over time (Figure 4). The individual rates of MT movements ranged from 0.09 to 0.165 μm/s (average 0.118 μm/s; n = 10). In an independent approach, we also measured the rates of MT movement by measuring the kinetics of recovery of average fluorescence intensity in the bleached area (Figure 5B). Because in the absence of MT dynamics fluorescence recovery reflects the movement of MTs into the bleached area, a linear increase in the average fluorescence provides a measure of the MT movement rate. The rate of MT movement calculated by dividing the width of the bleached zone by the time of the linear increase of the fluorescence in the bleached zone was 0.105 ± 0.001 μm/s. The numbers obtained by two independent experimental approaches were consistent with each other and with the velocity of Ncd-based MT movement in vitro determined using a MT gliding assay (0.06–0.16 μm/s (McDonald et al., 1990; Walker et al., 1990; deCastro et al., 1999; Tao et al., 2006). These results strongly suggest that MT movement seen in GFP–Ncd-overexpressing cells is indeed generated by the motor activity of Ncd.
Figure 5.
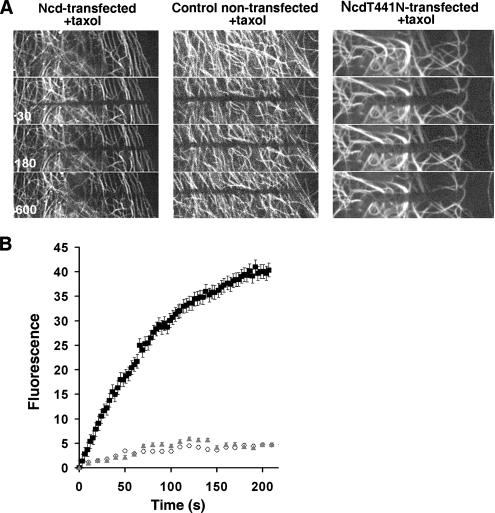
Kinetics of fluorescence redistribution after photobleaching in a control cell and in cells expressing wild-type GFP–Ncd or GFP–Ncd rigor mutant. (A) Images of MTs before (top) and 30, 180, and 600 s after bleaching in a cell overexpressing GFP–Ncd (left), GFP (middle), or GFP–NcdT441N (right). (B) Quantification of fluorescence recovery in the bleached zones; white diamonds, GFP-overexpressing cells; black squares, GFP–Ncd-overexpressing cells; gray triangles, GFP–NcdT441N-overexpressing cells. Error bars represent SEM for measurements in five different cells. Recovery of tubulin fluorescence is significantly faster in cells overexpressing GFP–Ncd, compared with GFP or GFP–Ncd rigor mutant-overexpressing cells. The linear region of the GFP–Ncd curve in B (corresponding to the time interval between 0 and 27 s) was used for the motility rate calculation described in Results.
To confirm that the observed MT gliding indeed requires the presence of enzymatically active Ncd, we generated a mutant form of Ncd by introducing a single amino-acid substitution (T441N) in the ATP-binding consensus motif. This substitution is known to cause a tight irreversible ATP-insensitive binding of kinesin family members to MTs in vivo and in vitro (Nakata and Hirokawa, 1995). Therefore, mutant Ncd molecules are expected to have MT-bundling activity but to lack the ability to support MT sliding against each other. Indeed, overexpression of GFP-NcdT441N in fibroblasts caused changes in MT organization similar to that seen in cells overexpressing wild-type GFP–Ncd (Figure 5). However, looping of microtubules has been never observed in the rigor mutant-transfected cells (Supplemental Movie 7), and photobleaching experiments indicated that in GFP–NcdT441N-overexpressing cells, MTs did not show movement above that observed in control untransfected cells (Supplemental Table 1 and Figure 5, A and B). These results suggest the NcdT441N mutant, unlike wild-type Ncd, was unable to induce MT sliding.
To confirm that MT sliding in cells overexpressing Ncd is explained by an inherent ability of this MT motor to concurrently bind two neighboring MTs via the binding sites located in the N-terminal tail and the C-terminal motor domains, and to rule out the possibility that sliding of MTs is a nonspecific consequence of their bundling, we performed several control experiments. We examined the organization and behavior of MTs in cells overexpressing GFP-tagged N-terminal tail domain (amino acids 1–204) or C-terminal motor domain (amino acids 327–700) of Ncd, or Drosophila kinesin-1 fused to GFP. Although the C terminus of Ncd and kinesin-1 do have motor activity, and can induce MT movement, none of these proteins has two MT binding sites; thus, they are not able to bundle MTs. We also overexpressed in cells GFP-tau fusion protein (GFP-tau23; Samsonov et al., 2004), which has a high MT-bundling activity, but lacks a motor domain and therefore cannot move MTs against each other. Fluorescence microscopy analysis of MTs in the transfected cells showed that, as expected, overexpression of tau-GFP, but not other proteins, induced the formation of MT bundles (Supplemental Movies 5 and 6 and Supplemental Figure 1). Importantly, playback of time sequences of Cy3-labeled MTs indicated that neither bundled MTs in GFP-tau–overexpressing cells, nor single MTs in cells overexpressing GFP–Ncd1–204, GFP–Ncd327–700, or GFP–kinesin-1 moved significantly, and that MT looping characteristic for cells transfected with full-size Ncd has never occurred (Supplemental Movies 5 and 6; data not shown). We conclude that MT movement seen in GFP–Ncd-overexpresssing cells is specific to Ncd; therefore, the ability to slide MTs against each other is a fundamental property of this MT motor.
DISCUSSION
Our results demonstrate that the kinesin-14 family of MT motors Drosophila Ncd induces sliding of MTs against each other in the cytoplasm of interphase mammalian fibroblasts. It has been previously shown that Ncd exhibits motile properties in vitro and that in MT gliding assays recombinant Ncd behaves as a nonprocessive minus end-directed MT motor (McDonald et al., 1990; Walker et al., 1990; Foster and Gilbert, 2000). Our study is the first demonstration of the motor activity of Ncd in living cells. In this work, we developed a new in vivo assay that provides the ability to examine the motility properties of MT motors in a cellular context.
It is thought that Ncd exerts its functions by bundling and sliding of MTs against each other (McDonald et al., 1990; Chandra et al., 1993; Sharp et al., 2000c). This MT bundling activity is explained by the simultaneous attachment to two neighboring MTs via its ATP-dependent motor domain and ATP-independent tail located on the opposite sides of the molecule (McDonald et al., 1990; Chandra et al., 1993; Karabay and Walker, 1999). In the presence of ATP such attachment would result in relative sliding of the neighboring MTs against each other. It has been previously demonstrated in in vitro assays that Ncd causes the formation of MT bundles; however, sliding of MTs in Ncd-induced bundles has never been seen in in vivo experiments. Our in vivo assay allowed us for the first time to observe the Ncd-dependent sliding of MTs against each other.
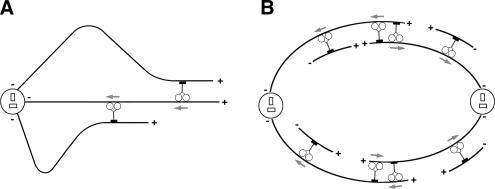
MT sliding seen in our experiments results in a remarkable pattern of MT behavior that involves the formation of dynamic MT loops with continuously changing curvature. Our results indicate that the formation and expansion of MT loops do not require MT dynamics, are specific for Ncd-overexpressing cells, depend on the Ncd motor activity, and involve both MT binding domains located on opposite ends of Ncd molecule. Based on these observations, we propose that the looping behavior of MTs in our assay is caused by the fact that in addition to being bundled by Ncd these MTs are also anchored on the centrosome, which restricts MT movement, resulting in the bending and buckling of the MTs during Ncd-dependent MT-MT sliding (Figure 6, left). In dividing cells, MT sliding activity generated by Ncd and other members of the kinesin-14 family is thought to be involved in two aspects of mitotic and meiotic spindle morphogenesis. For one aspect, kinesin-14 family members control mitotic spindle length by generating pulling forces on the overlapping antiparallel MTs in the spindle midzone that oppose the pushing forces produced by members of the kinesin-5 family (Saunders and Hoyt, 1992; Sharp et al., 1999, 2000a,b; Kapitein et al., 2005; Tao et al., 2006) (Figure 6, right). In addition, kinesins-14 are involved in the formation of the spindle poles themselves, presumably via minus-end–directed transport of MTs nucleated by noncentrosomal mechanisms in the cytoplasm (Goshima et al., 2005; Morales-Mulia and Scholey, 2005) (Figure 6, right). In our study, we demonstrate for the first time that kinesins-14 are indeed involved in MT–MT sliding in vivo; this result supports the hypothesis about the mitotic roles of kinesin-14 members and it provides experimental evidence for both aspects of their functioning in mitosis. In vitro reconstitution experiments are underway to confirm the sliding activity of kinesin-14 family members and to reproduce the key aspects of the mitotic spindle assembly in a purified system.
Figure 6.
Generation of MT sliding forces by kinesin-14 family MT motors in interphase and mitotic cells. In interphase cells (A), where minus ends of MTs are attached to the centrosome, sliding of MTs against each other by kinesin-14 MT motors causes the formation of MT loops. In mitotic cells (B), sliding forces applied on MT by kinesin-14 members play dual role in the mitotic spindle morphogenesis. Cross-linking and sliding of antiparallel MTs against each other in the midzone helps to maintain the distance between the mitotic spindle poles. Minus-end–directed transport of MTs that emerge by noncentrosomal nucleation maintains the focused organization of the mitotic spindle poles. Gray arrows indicate the direction of movement of individual Ncd motors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Anna Kashina for critical reading and editing the manuscript, Dr. Boris Slepchenko for advice about the quantification of MT movement, and Drs. William Saxton and Sergei Popov for generous gifts of plasmids. This work was supported by National Institutes of Health grant GM-062290 (to V.I.R.) and National Institutes of Health predoctoral fellowship 1F31GM075369 (to A.O.).
Abbreviations used:
- MT
microtubule.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1085) on June 27, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Brust-Mascher I., Scholey J. M. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol. Biol. Cell. 2002;13:3967–3975. doi: 10.1091/mbc.02-05-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R., Salmon E. D., Erickson H. P., Lockhart A., Endow S. A. Structural and functional domains of the Drosophila ncd microtubule motor protein. J. Biol. Chem. 1993;268:9005–9013. [PubMed] [Google Scholar]
- deCastro M. J., Ho C. H., Stewart R. J. Motility of dimeric ncd on a metal-chelating surfactant: evidence that ncd is not processive. Biochemistry. 1999;38:5076–5081. doi: 10.1021/bi9829175. [DOI] [PubMed] [Google Scholar]
- Foster K. A., Gilbert S. P. Kinetic studies of dimeric Ncd: evidence that Ncd is not processive. Biochemistry. 2000;39:1784–1791. doi: 10.1021/bi991500b. [DOI] [PubMed] [Google Scholar]
- Gadde S., Heald R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Goshima G., Nedelec F., Vale R. D. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L. C., Peterman E. J., Kwok B. H., Kim J. H., Kapoor T. M., Schmidt C. F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Karabay A., Walker R. A. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 1999;38:1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Komma D. J., Horne A. S., Endow S. A. Separation of meiotic and mitotic effects of claret non-disjunctional on chromosome segregation in Drosophila. EMBO J. 1991;10:419–424. doi: 10.1002/j.1460-2075.1991.tb07963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H. J., McDonald H. B., Goldstein L. S., Theurkauf W. E. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuliene J., Essner R., Ryu J., Hamaguchi Y., Baas P. W., Haraguchi T., Hiraoka Y., Kuriyama R. Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J Cell Sci. 1999;112:4041–4050. doi: 10.1242/jcs.112.22.4041. [DOI] [PubMed] [Google Scholar]
- McDonald H. B., Stewart R. J., Goldstein L. S. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., West R. R. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Rose M. D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Mogilner A., Wollman R., Civelekoglu-Scholey G., Scholey J. Modeling mitosis. Trends Cell Biol. 2006;16:88–96. doi: 10.1016/j.tcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Morales-Mulia S., Scholey J. M. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., Compton D. A. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T., Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina Y., Wordeman L. Unconventional motoring: an overview of the Kin C and Kin I kinesins. Traffic. 2003;4:367–375. doi: 10.1034/j.1600-0854.2003.00099.x. [DOI] [PubMed] [Google Scholar]
- Rodionov V. I., Lim S. S., Gelfand V. I., Borisy G. G. Microtubule dynamics in fish melanophores. J. Cell Biol. 1994;126:1455–1464. doi: 10.1083/jcb.126.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonov A., Yu J. Z., Rasenick M., Popov S. V. Tau interaction with microtubules in vivo. J. Cell Sci. 2004;117:6129–6141. doi: 10.1242/jcs.01531. [DOI] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M. A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. S., Hoyt M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Scholey J. M., Brust-Mascher I., Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Brown H. M., Kwon M., Rogers G. C., Holland G., Scholey J. M. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 2000a;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Microtubule motors in mitosis. Nature. 2000b;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Roles of motor proteins in building microtubule-based structures: a basic principle of cellular design. Biochim. Biophys. Acta. 2000c;1496:128–141. doi: 10.1016/s0167-4889(00)00014-8. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Yu K. R., Sisson J. C., Sullivan W., Scholey J. M. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Tao L., Mogilner A., Civelekoglu-Scholey G., Wollman R., Evans J., Stahlberg H., Scholey J. M. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr. Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E., Hawley R. S. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell C. L., Sweezy M. A., West R. R., Reed K. D., Carson B. D., Pidoux A. L., Cande W. Z., McIntosh J. R. pkl1(+)and klp2(+): two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol. Biol. Cell. 2001;12:3476–3488. doi: 10.1091/mbc.12.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M., Inze D., Geelen D. Mitosis-specific kinesins in Arabidopsis. Trends Plant Sci. 2006;11:167–175. doi: 10.1016/j.tplants.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Vorobjev I., Malikov V., Rodionov V. Self-organization of a radial microtubule array by dynein-dependent nucleation of microtubules. Proc. Natl. Acad. Sci. USA. 2001;98:10160–10165. doi: 10.1073/pnas.181354198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P., Khodjakov A. E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol. 2004;14:413–419. doi: 10.1016/j.tcb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Walker R. A., Salmon E. D., Endow S. A. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C. M., Desai A., Bulinski J. C., Salmon E. D. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.