Abstract
The pyruvate dehydrogenase (PDH) multienzyme complex plays a key role in the metabolic interconnection between glycolysis and the citric acid cycle. Transcription of the Escherichia coli genes for all three components of the PDH complex in the pdhR-aceEF-lpdA operon is repressed by the pyruvate-sensing PdhR, a GntR family transcription regulator, and derepressed by pyruvate. After a systematic search for the regulation targets of PdhR using genomic systematic evolution of ligands by exponential enrichment (SELEX), we have identified two novel targets, ndh, encoding NADH dehydrogenase II, and cyoABCDE, encoding the cytochrome bo-type oxidase, both together forming the pathway of respiratory electron transport downstream from the PDH cycle. PDH generates NADH, while Ndh and CyoABCDE together transport electrons from NADH to oxygen. Using gel shift and DNase I footprinting assays, the PdhR-binding site (PdhR box) was defined, which includes a palindromic consensus sequence, ATTGGTNNNACCAAT. The binding in vitro of PdhR to the PdhR box decreased in the presence of pyruvate. Promoter assays in vivo using a two-fluorescent-protein vector also indicated that the newly identified operons are repressed by PdhR and derepressed by the addition of pyruvate. Taken together, we propose that PdhR is a master regulator for controlling the formation of not only the PDH complex but also the respiratory electron transport system.
The pyruvate dehydrogenase (PDH) complex of Escherichia coli contains three components, pyruvate dehydrogenase (E1p), dehydrolipoate acyltransferase (E2p), and dihydrolipoate dehydrogenase (E3), and catalyzes the NAD-linked oxidative decarboxylation of pyruvate and the concomitant formation of acetyl coenzyme A (acetyl-CoA) (25, 34), which then reacts with oxalacetate to produce citrate in the first reaction of the citric acid (tricarboxylic acid [TCA]) cycle. The E. coli PDH complex is composed of 24 units of E1p (AceE [pyruvate dehydrogenase]), 24 units of E2p (AceF [dehydrolipamide acyltranferase]), and 12 units of E3 (LpdA [dihydrolipoamide dehydrogenase]). The genes (aceE, aceF, and lpdA) encoding these three enzymes form a single operon, together with the pdhR gene, encoding a self-regulator of this operon, in the order pdhR-aceE-aceF-lpdA.
Transcription of the pdh operon is controlled by two major promoters: the upstream promoter Ppdh generates a pdhR-lpdA readthrough transcript, and the internal promoter Plpd generates independent lpdA transcript (26). The primary pdh promoter Ppdh is negatively autoregulated by PdhR, the product of the first gene in this pdh operon (26). PdhR is a member of the Gnt family of transcription factors and shares sequence similarity in its N-terminal DNA-binding domain with other members (14). PdhR senses the intracellular pyruvate pool, and its activity is controlled by pyruvate. Since the PdhR-pyruvate complex is unable to bind target DNA, the pdhR-aceEF-lpdA operon is derepressed when pyruvate is available (26). The secondary promoter Plpd is under the control of ArcA, which is the global repressor of TCA cycle genes under anaerobic conditions (8). Besides the pdhR operon, the only regulation target of PdhR identified so far is the yfiD gene, encoding a putative formate acetyltransferase (36), that is induced at low pH or by pyruvate (3).
Since the PDH multienzyme complex is a key enzyme for the metabolic interconnection between glycolysis and the TCA cycle, PdhR could be an important regulator for the steady-state maintenance of the central metabolism for energy production in response to changes in external environmental conditions. In order to identify the whole set of target genes under the control of PdhR, we performed a systematic search in vitro for PdhR-binding sequences in the E. coli genome using the newly developed genomic systematic evolution of ligands by exponential enrichment (SELEX) method (30). In this study, we identified two targets, the ndh gene, encoding NADH dehydrogenase II, and the cyoABCDE operon, encoding the cytochrome bo-type oxidase (4, 5, 17), both together forming the terminal respiratory electron transport system from NADH to oxygen, which is located downstream of the PDH system in the metabolic pathway of energy production. In E. coli, NADH dehydrogenase II is a primary dehydrogenase that plays a major role in aerobic and nitrate respiration. Under anaerobic conditions, the expression of ndh is repressed by FNR (regulator of fumarate and nitrate reduction) (13, 20, 21), while the expression of cyoABCDE is repressed by FNR under anaerobic growth conditions (6). Except for the involvement of FNR, the overall regulation of these two operons for the terminal electron transport system remained unsolved.
In this study, we performed a detailed analysis of the regulation of ndh and cyoABCDE promoters. Results described herein demonstrate that the ndh and cyoABCDE operons are under the direct control of PdhR. We then propose that PdhR is a master regulator of the genes involved in the main pathway of energy production, starting with the PDH system for the transfer of pyruvate, the final product of glycolysis, into the TCA cycle, followed by the Ndh-Cyo system for terminal electron transport from NADH to oxygen.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli KP7600 (W3110 lacIq lacZΔM15) (30) and JD20368 (pdhR deletion derivative of KP7600), used for the analysis of the regulatory roles of PdhR, were gifts from T. Miki (Fukuoka Dental College). The pdhR disruptant JD20368 was constructed after transposon insertion and was kanamycin resistant. E. coli KP74201 (KP7600 λcyoA::lacZ) and KP74202 (JD20368 λcyoA::lacZ), used for the single-copy assay of the cyoA promoter, were constructed according to a method described previously by Simons et al. (32). In brief, pRScyoA-Cm was recombined with phage λRS45, and the recombinant phages were lysogenized into KP7600 or JD20368 to isolate KP74201 or KP74202, respectively, on an LB agar plate with chloramphenicol. Plasmid pRScyoA-Cm was constructed from plasmid pRScyoA carrying the cyoA promoter between EcoRI and BamHI sites on pRS551, a lacZ reporter assay vector, after the replacement of its Km-resistant gene with a chloramphenicol (Cm)-resistant gene.
E. coli BL21(DE3) [F− ompT hsdSB(rB− mB−) dcm gal (DE3)] was used for the overproduction of PdhR. Cells were grown in LB medium at 37°C with shaking. When necessary, ampicillin was added at a final concentration of 100 μg/ml.
Purification of His-tagged PdhR protein.
His-tagged PdhR was overexpressed in E. coli BL21(DE3) using expression vector pPdhR and purified by affinity chromatography as described previously (37, 38).
SELEX search for PdhR-binding sequences.
The genetic SELEX method was performed as described previously (24, 30). A collection of 200- to 300-bp DNA fragments of the E. coli genome was prepared by PCR amplification using the E. coli DNA library plasmids as templates and a set of primers, primer-EcoRV-F (5′-CTTGGTTATGCCGGTACTGC-3′) and primer-EcoRV-R (5′-GCGATGCTGTCGGAATGGAC-3′), which hybridize with vector pBR322 at EcoRV junctions. PCR products thus generated were purified by 6% polyacrylamide gel electrophoresis (PAGE).
For the genomic SELEX screening of PdhR-binding sequences, 5 pmol DNA fragments and 20 pmol His-tagged PdhR were mixed in a binding buffer (10 mM Tris-HCl [pH 7.8 at 4°C], 3 mM Mg acetate, 150 mM NaCl, 1.25 μg/ml bovine serum albumin [BSA]) and incubated for 30 min at 37°C. The mixture was applied onto a Ni-nitrilotriacetic acid column, and after washing unbound DNA with the binding buffer containing 10 mM imidazole, DNA-PdhR complexes were eluted with an elution buffer containing 200 mM imidazole. If necessary, this SELEX cycle was repeated several times. For sequencing of PdhR-bound DNA fragments, DNA fragments were dissociated, isolated from DNA-PdhR complexes by PAGE, and PCR amplified. PCR products were cloned into the pT7 Blue-T vector (Novagen) using a blunt-end cloning kit (Takara) and transformed into E. coli DH5α. Fluorescently labeled DNA was prepared using 2T7P-primer (5′-TAATACGACTCACTATAGGG-3′), and sequencing was performed with an ABI DNA sequencer.
Plasmid construction.
The promoter assay vector pGRP carries two types of fluorescent protein genes, one for red fluorescent protein (RFP) under the control of reference promoter lacUV5 and the other for green fluorescent protein (GFP) under the control of a test promoter (19, 31). The ndh promoter sequence upstream from the initiation codon was amplified by PCR using the genomic DNA from E. coli KP7600 as a template and a pair of primers, K701S (5′-ACATCTGAAGAGATCTATCATTATTACGAG-3′) and K701T (5′-TCAATGGCGTATGCATCGTGACCCCCTTAA-3′), while the cyoA promoter sequence was amplified by PCR using a pair of primers, N073S (5′-CGATGCATCATTTAACGACCTCAATTCCACGG-3′) and N073T (5′-GAAGATCTCGTGCTTGGTGGTTTGCTGG-3′). These primers contain EcoT22I, BglII, or BamHI sites suitable for cloning into vector pGRP at the initiation codon of the GFP coding frame. The PCR products were digested with restriction enzymes and then ligated into the EcoT22I and BglII sites of pGRP. The sequences of the inserted promoter and junction with the GFP coding frame were confirmed by sequencing. The plasmids thus constructed were named pGRK701 and pGRN073, respectively.
For the construction of plasmid pPdhR for PdhR expression, a DNA fragment corresponding to the PdhR coding region was amplified by PCR using the E. coli KP7600 genome DNA as a template and a pair of primers, PDHRF (5′ to 3′ direction) and PDHRR (3′ to 5′ direction), which were designed to express the full-length PdhR from the initiation codon to the termination codon. Primer PDHRF contained the BamHI recognition sequence and the His tag sequence in this order, while primer PDHRR contained the NotI recognition sequence. After digestion with BamHI and NotI, the PCR product was cloned into pET21a(+) (Novagen) at the corresponding sites.
Measurement of the promoter activity.
Promoter strength was determined as described previously (19, 30). In brief, RFP was expressed under the control of test promoter lacUV5, while GFP was under the control of a test promoter. For measurement of the fluorescence intensity of RFP or GRP expressed in transformed E. coli, cells were grown in LB medium at 37°C for various lengths of time and harvested by centrifugation. Cells were resuspended in phosphate-buffered saline and diluted with phosphate-buffered saline to obtain approximately the same cell density (optical density at 600 nm [OD600] of 0.6) for all samples. For measurements of bulk fluorescence, aliquots of a 0.2-ml cell suspension were added to 96 0.4-ml flat-bottom wells, and the fluorescence was measured with a Wallace 1420 ARVOsx apparatus (Perkin-Elmer Life Sciences). GFP was measured using 485-nm excitation and 535-nm emission wavelengths, while RFP was measured using 544-nm excitation and 590-nm emission wavelengths, respectively. The fluorescence intensity of GFP by test promoter was normalized using the equation [X/Y]/[A/B], in which X and Y indicate the fluorescence intensities of GFP (test promoter) and RFP (lacUV5 promoter), respectively, while A and B represent the fluorescence intensities of GFP (lacUV5 promoter) and RFP (lacUV5 promoter), respectively. Promoter assays were repeated at least twice for confirmation.
Gel shift assay.
Probes were generated by PCR amplification of the ndh, cyoA, yfiD, and pdhR promoter regions (approximately 500 bp from the respective initiation codon) using a pair of primers, 5′ fluorescein isothiocyanate (FITC)-labeled T7pro-primer (5′-TAATACGACTCACTATAGGG-3′) and T7-R primer (5′-GGTTTTCCCAGTCACACGACG-3′), the genomic SELEX plasmids containing the respective promoters as templates, and Ex Taq DNA polymerase. PCR products with FITC at their termini were purified by PAGE. For gel shift assays, 0.5 pmol each of the FITC-labeled probes was incubated at 37°C for 30 min with various amounts of PdhR in 12.5 μl of gel shift buffer consisting of 10 mM Tris-HCl (pH 7.8), 150 mM NaCl, 3 mM Mg acetate, and 25 mg/ml BSA. After the addition of the DNA dye solution (40% glycerol, 0.025% bromophenol blue, 0.025% xylene cyanol), the mixture was directly subjected to 6% polyacrylamide gel electrophoresis.
DNase I footprinting assay.
A DNase I footprinting assay was carried out using the FITC-labeled DNA probes used for the gel shift assay. A total of 1 pmol each of the FITC-labeled probes was incubated at 37°C for 30 min with various amounts of PdhR in DNase I footprinting buffer consisting of 25 μl of 10 mM Tris-HCl (pH 7.8), 150 mM NaCl, 3 mM magnesium acetate, 5 mM CaCl2, and 25 μg/ml BSA. After incubation for 30 min, DNA digestion was initiated by the addition of 5 ng of DNase I (Takara). After digestion for 30 s at 25°C, the reaction was terminated by the addition of 45 μl of DNase I stop solution (20 mM EDTA, 200 mM NaCl, 1% sodium dodecyl sulfate, 250 μg/ml yeast tRNA) to the mixture. Digested products were precipitated with ethanol, dissolved in formamide dye solution, and analyzed by electrophoresis on a 6% polyacrylamide gel containing 8 M urea.
RESULTS
Isolation of PdhR-binding sequences by genomic SELEX.
For the identification of DNA sequences that are recognized by E. coli PdhR, we used the genomic SELEX method (24, 30), which uses a complete library of E. coli genome DNA fragments, instead of synthetic oligonucleotides, with all possible sequences used with the original SELEX method (33). First, we constructed the plasmid library, each carrying a piece of the size-fractionated DNA fragment (100 to 300 bp in length) from a pool of sonicated E. coli W3110 genome DNA. In each SELEX experiment, the fragment mixture was regenerated after amplification of the inserted DNA fragments by PCR. From mixtures of these DNA fragments and a fourfold molar excess of the purified His-tagged PdhR protein, the PdhR-bound DNA complexes were affinity purified. In the early period of SELEX cycles, the PdhR-bound DNA fragments gave smear bands on PAGE, as did the original genome fragment mixture. After four SELEX cycles, however, the width of the gel band decreased, indicating enrichment of specific DNA fragments with PdhR-binding activity. The SELEX DNA fragments were recovered from the gel and cloned into the pT7 Blue plasmid (Novagen) for sequencing (see Materials and Methods).
A total of 73 independent clones were isolated, sequenced, and classified into two different groups. Group A included 67 independent clones, which had unique sequences from four different spacer regions between two neighboring genes in the E. coli genome, i.e., 48 clones from the aroP-phdR spacer, 14 clones from the cyoA-ampG spacer, 3 clones from the yfiD-ung spacer, and 2 clones from the ycfD-ndh spacer (Table 1). On the other hand, group B included six clones, which carried parts of five protein-coding sequences, i.e., wzc, aas, cdaR, ybiT, and yhjE (Table 1). Since these sequences in the group B clones are widely separated from transcription initiation sites, it is unlikely that the group B sequences play some regulatory roles in transcription initiation, but possible influence of group B sequence-bound PdhR on transcription elongation cannot be ruled out. For the identification of the target genes under the control of PdhR, a detailed analysis has been focused on the group A sequences.
TABLE 1.
PdhR-bound DNA fragmentsa
| Group and no. of clones | Upstream gene | SELEX fragment | Downstream gene |
|---|---|---|---|
| A (SELEX fragments within spacer regions) | |||
| 48 | aroP← | (0122011) S (0122303) | →pdhR |
| 14 | cyoA← | (0450896) S (0451132) | ←ampG |
| 3 | yfiD← | (2714930) S (2715311) | →ung |
| 2 | ycfD→ | (1167498) S (1167753) | →ndh |
| B (SELEX fragments on open reading frames) | |||
| 2 | wcaA← | (2135986) S (wzc) (2136196) | ←wzb |
| 1 | ygeD← | (2974235) S (aas) (2974451) | →galR |
| 1 | degP→ | (0182534) S (cdaR) (0182770) | ←yaeH |
| 1 | ybiS← | (0857353) S (ybiT) (0857554) | ←ybiU |
| 1 | yhiD→ | (3964400) S (yhjE) (3964580) | ←yhjG |
Using the genomic SELEX method, a total of 73 DNA fragments have been isolated as complexes with the purified PdhR protein, which were all cloned into the sequencing vector. Group A clones contained the sequences from spacer regions between the indicated neighboring genes, while group B clones carried the sequences within the indicated coding frames. The numbers on both sides of each SELEX (S) fragment indicate the boundaries in the E. coli genome map (27). The arrows indicate the direction of transcription of the neighboring genes. The genes shown in boldface type indicate the PdhR regulation targets.
Judging from the orientation of the genes included in the group A clones, we predicted that the promoters for the aroP, pdhR, cyoA, yfiD, ung, and/or ndh gene could be the regulation targets of PdhR (Table 1). A total of 48 independent SELEX clones carried the sequences, which are located within the narrow 293-bp-long region between E. coli genome positions 122011 and 122303 (the location is based on the revised E. coli K-12 genome sequence) (26), upstream of aroP and pdhR (aroP and pdhR are transcribed divergently towards opposite directions). The target of the PdhR box within this region has previously been found to be the pdhR-aceEF-lpdA operon (15, 26).
Three SELEX fragments included the sequences from 382-bp-long region of the genome at positions 2714930 to 2715311, upstream of yfiD (putative formate acetyltransferase) and ung (uracil-DNA glycosylase) (Table 1). YfiD is one of the acid-inducible proteins with unidentified function. Between the two regulation targets yfiD and ung, PdhR has been indicated to be an anaerobic repressor of yfiD (36). The PdhR-binding sequence (cAaTGGTtttACCAATT, 12/14 match with the consensus [nonconsensus bases are in lowercase type]) within this region is similar to the consensus PdhR box.
A total of 14 group A clones carried segments within the 237-bp-long sequence upstream of the cyoA (cytochrome bo-type oxidase) gene, where PdhR binding has never been shown, although the presence of the PdhR box had been predicted (25), and two other clones included segments from the 256-bp-long sequence upstream of the ndh (NADH dehydrogenase II) gene (Table 1), where the binding of PdhR has never been detected. These results indicate that PdhR plays a key regulatory role in not only the formation of the PDH complex for the metabolic interconnection between glycolysis and the TCA cycle but also the synthesis of two key enzymes (Ndh and CyoA) in the terminal electron transport system.
Identification of PdhR-binding activity for SELEX DNA fragments.
The group A SELEX clones contain promoter regions for the pdhR-aceE-aceF operon, the cyoABCDE operon, the ndh gene, and the yfiD gene (Fig. 1). To confirm the presence of a PdhR-binding site in these four promoter regions, we performed a gel shift assay. Under standard conditions, we detected the PdhR-dependent gel shift for all the promoter fragments (Fig. 2A).
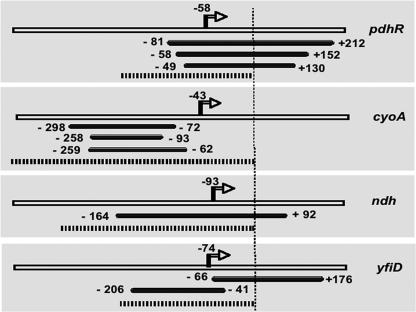
FIG. 1.
PdhR-bound DNA fragments. Using genomic SELEX, PdhR-bound DNA fragments were isolated, the indicated four groups of which contained the promoter regions from the pdhR, cyoA, ndh, and yfiD genes. Open bars indicate the genomic DNA sequence, while thick bars indicate the regions of fragments isolated by SELEX. The dotted lines show the DNA fragments used for gel shift and DNase I footprinting assays. The vertical line shows the translation initiation site. Numbers on each line represent the distance (bp) from the respective initiation codon. Transcription initiation sites, shown by arrows, are from pdhR (25), cyoA (22), ndh (13), and yfiD (12).
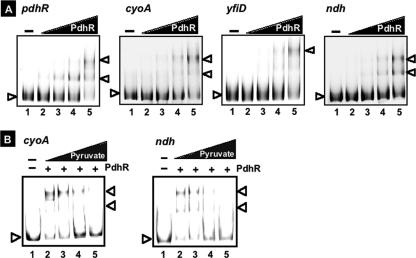
FIG. 2.
Gel shift assay. (A) Fluorescently labeled DNA probes, each containing a segment of the pdhR, cyoA, yfiD, or ndh promoter (for the region of each probe, see Fig. 1), were incubated at 37°C for 30 min with the indicated amounts (0, 0.625, 1.25, 2.5, and 5 pmol) of PdhR (lanes 1 to 5) and directly subjected to PAGE. (B) Fluorescently labeled DNA probes of cyoA or ndh were incubated at 37°C for 30 min in the absence (lane 1) or presence (lanes 2 to 5) of 5 pmol of PdhR and in the absence (lane 2) or presence (lane 3, 100 mM; lane 4, 200 mM; lane 5, 500 mM) of increasing concentrations of pyruvate. Samples were subjected directly to PAGE.
Previous studies on the regulation of the pdhR-aceE-aceF operon indicated that pyruvate prevents PdhR binding to the promoter of this operon, as detected by gel retardation assays (26). The same was found in the gel shift assays with the ndh and cyoABCDE promoter fragments, where the formation of PdhR-DNA complexes was reduced concomitantly with an increase in the pyruvate addition (Fig. 2B).
Identification of PdhR-binding sequences on the ndh and cyoABCDE promoters.
The PdhR-binding sites on the newly identified cyoABCDE and ndh promoter fragments were then examined by DNase I footprinting. PdhR was found to bind to a 26-bp sequence between positions −18 and −43 upstream of the ndh transcriptional start position (Fig. 3) and to a 28-bp sequence between positions −158 and −185 upstream of the cyoABCDE transcriptional start position (Fig. 4A and B). In both PdhR-binding sequences, there is a common 15-bp ATTGGTNNNACCAAT sequence, which includes an inverted ATTGGT repeat, referred to as the PdhR box (Fig. 5). The gel shift assay indicated the formation of two kinds of PdhR complexes with these promoter fragments (Fig. 2). One possible explanation is that two molecules of PdhR bind to each half of the inverted repeat of the PdhR box. Previously, Quail et al. (26) reported that on the pdhR-aceEF operon promoter, PdhR binds to the sequence between positions +8 and +28 on the coding strand and positions +10 and +33 on the noncoding strand downstream from transcriptional start site (+1). This sequence also includes the inverted repeat sequence AATTGGTNNNACCAATT of the PdhR box.
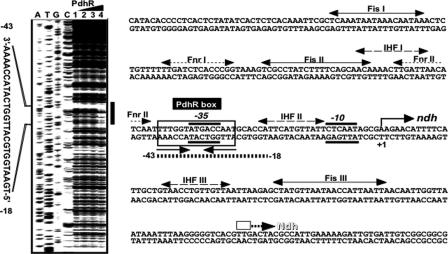
FIG. 3.
DNase I footprinting of the ndh promoter. (Left) The fluorescently labeled ndh promoter (Fig. 2) was incubated with increasing amounts of purified PdhR (lane 1, 0 pmol; lane 2, 2.5 pmol; lane 3, 5 pmol; lane 4, 10 pmol) and subjected to DNase I footprinting assays. Lanes A, T, G, and C represent the sequence ladders. The black bar on the right indicates the PdhR-binding region. (Right) The PdhR-binding site on the ndh promoter, determined as described above, is indicated by a thick bar between positions −43 and −18. The transcription initiation site according to Green and Guest (13) is marked by an arrow. Recognition sequences for Fis, FNR, and IHF (boxed) were described previously by Jackson et al. (18), Meng et al. (21), and Green et al. (11), respectively.
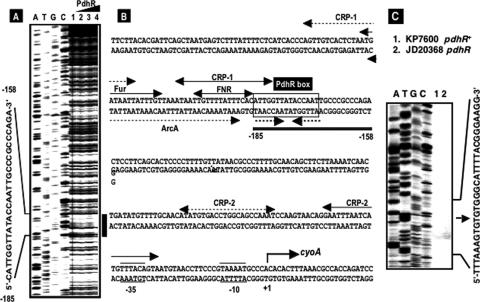
FIG. 4.
DNase I footprinting of the cyoA promoter. (A) The fluorescently labeled cyoA promoter was incubated with increasing amounts of purified PdhR (lane 1, 0 pmol; lane 2, 2.5 pmol; lane 3, 5 pmol; lane 4, 10 pmol) and subjected to DNase I footprinting assays. Lanes A, T, G, and C represent sequence ladders. (B) The black bar (positions −185 to −158) on the right indicates the PdhR-binding region located upstream of the cyoA promoter. The nucleotide numbers represent the distance from the transcription initiation site, which was proposed previously by Minagawa et al. (22) and confirmed in this study. Recognition sequences for Fur, ArcA, and Fnr were predicted based on sequence analyses reported previously by Stojiljkovic et al. (35), Shalel-Levanon et al. (29), and Salmon et al. (28), respectively. Two CRP-binding sequences, CRP1 and CRP2, were proposed previously, but at different positions, by Minagawa et al. (22) (dotted lines) and Zheng et al. (40) (solid line). (C) Primer extension assay of the transcription initiation site of the cyoA operon in both wild-type KP7600 and its pdhR disruptant, JD20368, was performed using a fluorescently labeled probe. Data for the site identified thus agreed with those reported previously by Minagawa et al. (22).
FIG. 5.

Consensus sequence for the PdhR box. The PdhR-binding sites are derived from the ndh and cyoA promoters determined in this study, the previously reported pdhR promoter (25), and the predicted yfiD promoter (36). After searching the PdhR-box-like sequence along the entire E. coli genome, three additional targets have been identified: ybaJ, with the sequence (480189)ATTGGTgacACtAAT(480203); hemL or clcA, with the sequence (175079)ATTNGTNNNACCAAT(175093); and lldP, with the sequence (3863230)ATTGGNNNNACCAAT(3863216).
The site of PdhR binding on the ndh promoter overlaps with its −35 region of the promoter in concert with the repression mode of PdhR. On the other hand, the PdhR-binding site on the cyoA promoter is far from the previously reported transcription initiation site (22). We then examined the transcription initiation site of the cyoA operon in the strains used. Primer extension assays indicated the presence of only a single initiation site at the same position with that previously identified (Fig. 4C).
PdhR-dependent repression of cyoABCDE and ndh transcription in vivo.
The pdhR-aceEF operon is repressed by PdhR and derepressed by pyruvate, the substrate of the PDH complex (26). To examine the mode of transcription regulation of ndh and cyoABCDE by PdhR in vivo, we employed the two-fluorescent-protein (TFP) promoter assay system (19, 31). The cyoABCDE and ndh promoters were inserted into the TFP vector so as to adjust the cyoA or ndh initiation codon to that of GFP (see Materials and Methods), while the RFP gene on the same plasmid was expressed under the control of the reference promoter lacUV5. The promoter activity can be accurately determined by measuring the GFP/RFP ratio.
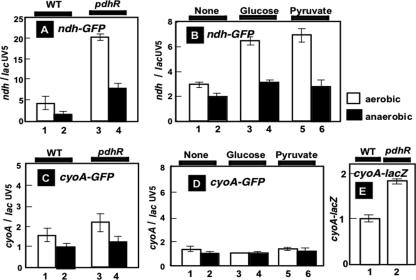
The promoter assays were carried out with both the wild type and a mutant lacking PdhR under both aerobic and anaerobic conditions. As expected, the activities of both ndh and cyoA promoters in the wild-type strain decreased under anaerobic growth conditions (Fig. 6A and C, lanes 1 and 2). In the pdhR deletion strain, the activities of both promoters increased under both anaerobic and aerobic conditions (Fig. 6A and C, lanes 3 and 4), supporting the repression model of these operons by PdhR. In particular, ndh promoter activity increased more than fourfold in the pdhR mutant (Fig. 6A, lanes 3 and 4). In the wild-type strain, the addition of pyruvate markedly increased the activity of the ndh promoter due to the inactivation of PdhR (Fig. 6B, lane 5). This pyruvate-induced derepression of ndh was significant under aerobic conditions (Fig. 6B, lane 5), where the respiratory electron transport system operates. The derepression of ndh was also observed with the addition of glucose (Fig. 6B, lanes 3 and 4). Under anaerobic conditions, the pyruvate- and glucose-induced derepressions were not so significant (Fig. 6B, lanes 2, 4, and 6). This is in good agreement with the previous observation that in E. coli, the expression of ndh is repressed under anaerobic growth conditions (13, 20). In contrast, the activity of the cyoABCDE promoter stayed unaffected even in the presence and absence of pyruvate and glucose (Fig. 6D). Under the culture conditions herein employed, i.e., shaking culture in LB medium, the activity of cyoABCDE must be activated even in the absence of a glucose or pyruvate addition.
FIG. 6.
Effect of PdhR on the ndh and cyoABCD promoters. (A and B) The promoter assay vectors containing the ndh or cyoABCD promoters were transformed into wild-type (KP7600) and pdhR deletion mutant (JD20368) cells and grown in LB medium under aerobic (open bars) and anaerobic (closed bars) conditions. Promoter activities were determined at a cell density of an OD600 of 0.6 and are represented as GFP/RFP ratios. (C and D) Wild-type cells and pdhR mutant transformants were grown in LB in the absence (lanes 1 and 2) or presence of 10 mM glucose (lanes 3 and 4) or 40 mM pyruvate (lanes 5 and 6) and under the aerobic (open bars) and anaerobic (closed bars) conditions. Promoter activities were determined at a cell density of an OD600 of 0.2. (E) The activity of a single-copy cyoA promoter-lacZ fusion in wild-type (KP74201) (lane 1) and pdhR mutant (KP74202) (lane 2) cells was determined by measuring β-galactosidase activity at a cell density of an OD600 of 0.6 under aerobic conditions.
The promoter assay was also carried out using a single-copy fusion on the chromosome. The cyoA promoter-lacZ fusion was inserted into both wild-type KP7600 and its pdhR mutant, JD20368, to generate KP74201 and KP74202, respectively, as described in Materials and Methods. In agreement with the results obtained using the multicopy promoter assay, cyoA promoter activity increased in the pdhR disruption mutant, as measured by β-galactosidase activity (Fig. 6E).
DISCUSSION
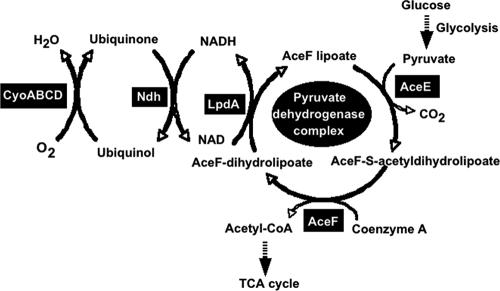
In the energy production pathway of E. coli under aerobic conditions, pyruvate, the terminal product of glycolysis, is oxidatively decarboxylated by the PDH multienzyme complex to produce CO2 and acetyl-CoA, with the concomitant reduction of NAD+ (9). The electron flow into respiration follows alternative routes to oxygen via an NADH dehydrogenase coupled or uncoupled to quinone (1, 9, 16). E. coli is capable of synthesizing two quinones, ubiquinone-8 and menaquinone-8 (17). NADH dehydrogenase II transfers electrons to ubiquinone-8 to form ubiquinol-8 (Fig. 7). The cytochrome bo-type oxidase (cyoABCDE gene) transfers electrons from ubiquinol-8 to oxygen, exporting two protons per electron (16, 17).
FIG. 7.
Role of PdhR as a master regulator of the metabolic pathways including both NAD-linked oxidative decarboxylation of pyruvate to acetyl-CoA and respiratory electron transport of NADH to oxygen.
The expression of the genes for metabolic energy production is influenced by the nature of the available terminal electron acceptor. For instance, the expression of cyoA is repressed in the absence of oxygen as the terminal electron acceptor under anaerobic conditions. The anaerobic repression of cyoA is known to be mediated, in part, by Fnr, as evidenced by a derepression of cyoA′-′lacZ expression in an fnr deletion strain (6). Using the genomic SELEX system followed by in vitro and in vivo transcription studies, we found that PdhR, the regulator of the genes for the PDH complex, regulates both the ndh gene and the cyoABCDE operon (Fig. 7). The ndh gene product (NADH dehydrogenase II) and the cyoABCDE gene products (cytochrome bo-type oxidase) are involved in the pathway of energy production downstream of the PDH pathway. Thus, we propose that PdhR is a master regulator in the entire pathway of oxidative energy production.
In this study, we identified the PdhR-binding site (PdhR box) on the cyoABCDE operon (between positions −158 and −185 upstream of transcriptional start position) near the upstream cyclic AMP receptor protein (CRP)-binding site (Fig. 4) (two different CRP1 sites have been predicted) (22, 40). The same site was detected previously in database searches for PdhR boxes by Quail and Guest (25), who suggested that the cyoABCDE operon might be regulated by PdhR. cyoABCDE promoter activity increased in a pdhR deletion strain under aerobic conditions (Fig. 4C and 6B). In general, the repression of transcription initiation by transcription factors takes place when the repressors bind at or downstream of the target promoters and interfere with either the binding of the RNA polymerase or its escape from the promoter complexes. In some instances, however, the upstream-bound repressors interact with the promoter-bound RNA polymerase and prevent its migration from promoters or even induce its dissociation from promoter complexes (37, 38). However, it should be noted that in addition to Fnr and CRP, ArcA was also proposed to associate near the PdhR site on the cyoA promoter (29) (Fig. 4); the mode of transcription regulation of the cyoA promoter must be complex. For instance, the apparent repression by PdhR could be due to an interference of activation by another factor such as CRP. The repression effect by PdhR was, however, not so strong under anaerobic conditions (Fig. 6B), implying that Fnr plays a major role in the repression of the cyoA gene under anaerobic conditions. However, it remains unsolved whether Fnr plays a direct role in the transcription regulation of the cyoABCDE operon (28).
On the other hand, NADH dehydrogenase II is a primary dehydrogenase used in E. coli during both aerobic and nitrate respiration. Under anaerobic conditions, the transcription factor Fnr represses ndh expression by binding at two sites centered at positions −94.5 and −50.5 (13, 21) (Fig. 3). The histone-like protein Fis binds to the three sites (centered at positions −123 [Fis I], −72 [Fis II], and +51 [Fis III]) in the ndh promoter (10, 11). Using ndh::lacZ promoter fusions carrying 5′ deletions or replacement mutations, it was shown that Fis that binds to site III functions as a repressor, while Fis that binds to sites I and II plays a role as an activator (18). Deletion of the C-terminal domain of the RNA polymerase α subunit abolished Fis-mediated activation of ndh expression, suggesting that ndh has a class I Fis-activated promoter (18). In accordance with the established pattern of Fis synthesis (2), ndh transcription was greatest during exponential growth (18). In the absence of Fnr, ndh expression is activated by the amino acid response regulator (Arr) during anaerobic growth in rich medium (11). Integration host factor (IHF) was shown to bind at three sites centered at positions +26 (IHF I), −17 (IHF II), and −58 (IHF III) in the ndh promoter (11). In this study, we identified the PdhR-binding site on the ndh promoter (between positions −18 and −43 upstream of the transcriptional start position), which overlaps with the −35 region of the promoter (Fig. 3), and ndh promoter activity increased in the pdhR deletion strain under both aerobic and anaerobic conditions (Fig. 6A). The promoter −35 and −10 regions are recognized by RNA polymerase-associated σ70 and isomerized into transcription-competent open promoter complexes. The marked repression of ndh gene expression by PdhR might be due to the competitive inhibition of RNA polymerase binding to the promoter.
Recent genetic and biochemical analyses with an F1-ATPase-defective mutant (enhanced glucose metabolism) indicated that the upregulation of the PDH complex by pyruvate takes place in both transcription and enzyme activities (23). Interestingly, in the same mutant, the expression level of ndh was significantly elevated, and cyoA expression also showed a tendency toward upregulation, even though the effect was not statistically significant because of the low level of activation. The F1-ATPase-defective mutant was constructed by the transduction of a defective gene for the α subunit of F1-ATPase into an E. coli strain, a lipoic acid-requiring pyruvate producer (39). The pyruvate production in this F1-ATPase-defective strain was found to be improved markedly compared with that of the wild-type strain. Taken together with our observations reported here, these results suggest that the increased level of pyruvate leads to the inactivation of the repressor PdhR, leading to the derepression of the ndh and cyoA promoters in the F1-ATPase-defective strain.
Acknowledgments
We thank Takenori Miki (Fukuoka Dental College) for the pdhR disruptant and Ayako Kori (Hosei University) for technical assistance.
This work was supported by grants-in-aid (Scientific Research Priority Area 17076016; Scientific Research 18310133) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Alexeeva, S., B. de Kort, G. Sawers, K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2000. Effects of limited aeration and of the arcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182:4934-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam, A. T., A. Iwata, A. Nishimura, A. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in the protein composition of Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calhoun, M. W., and R. B. Gennis. 1993. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J. Bacteriol. 175:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calhoun, M. W., K. L. Oden, R. B. Gennis, M. J. T. Mattos, and O. M. Neijssel. 1993. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 175:3020-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, P. A., S. Darie, and R. P. Gunsalus. 1992. The effect of iron limitation on expression of the aerobic and anaerobic electron transport pathway genes in Escherichia coli. FEMS Microbiol. Lett. 100:227-232. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, L., D. Georgellis, J. Green, and J. R. Guest. 1998. Co-regulation of lipoamide dehydrogenase and 2-oxoglutarate dehydrogenase synthesis in Escherichia coli: characterization of an ArcA binding site in the lpd promoter. FEMS Microbiol. Lett. 169:403-408. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, J., and U. Henning. 1970. Regulation of pyruvate dehydrogenase complex synthesis in Escherichia coli K12. Eur. J. Biochem. 14:258-269. [DOI] [PubMed] [Google Scholar]
- 10.Green, J., M. F. Anjum, and J. R. Guest. 1996. The ndh-binding protein (Nbp) regulates the ndh gene of Escherichia coli in response to growth phase and is identical to Fis. Mol. Microbiol. 20:1043-1055. [DOI] [PubMed] [Google Scholar]
- 11.Green, J., M. F. Anjum, and J. R. Guest. 1997. Regulation of the ndh gene of Escherichia coli by integration host factor and a novel regulator Arr. Microbiology 143:2865-2875. [DOI] [PubMed] [Google Scholar]
- 12.Green, J., M. J. Baldwin, and J. Richardson. 1998. Downregulation of Escherichia coli yfiD expression by FNR occupying a site of −93.5 involves the AR1-containing face of FNR. Mol. Microbiol. 29:1112-1123. [DOI] [PubMed] [Google Scholar]
- 13.Green, J., and J. R. Guest. 1994. Regulation of transcription at the ndh promoter of Escherichia coli by FNR and novel factors. Mol. Microbiol. 12:433-444. [DOI] [PubMed] [Google Scholar]
- 14.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63:291-295. [DOI] [PubMed] [Google Scholar]
- 15.Haydon, D. J., M. A. Quail, and J. R. Guest. 1993. A mutation causing constitutive synthesis of the pyruvate dehydrogenase complex in Escherichia coli is located within the pdhR gene. FEBS Lett. 336:43-47. [DOI] [PubMed] [Google Scholar]
- 16.Helling, R. B. 2002. Speed versus efficiency in microbial growth and the role of parallel pathways. J. Bacteriol. 184:1041-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingledew, W. J., and R. K. Poole. 1984. The respiratory chains of Escherichia coli. Microbiol. Rev. 48:222-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, L., T. Blake, and J. Green. 2004. Regulation of ndh expression in Escherichia coli by Fis. Microbiology 150:407-413. [DOI] [PubMed] [Google Scholar]
- 19.Makinoshima, H., A. Nishimura, and A. Ishihama. 2002. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 43:269-279. [DOI] [PubMed] [Google Scholar]
- 20.Marshall, F. A., S. L. Messenger, N. R. Wybom, J. R. Guest, H. Wing, S. J. W. Busby, and J. Green. 2001. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol. Microbiol. 39:747-753. [DOI] [PubMed] [Google Scholar]
- 21.Meng, W., J. Green, and J. R. Guest. 1997. FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites. Microbiology 143:1521-1532. [DOI] [PubMed] [Google Scholar]
- 22.Minagawa, J., H. Nakamura, I. Yamato, T. Mogi, and Y. Anraku. 1990. Transcriptional regulation of the cytochrome b562-o complex in Escherichia coli. Gene expression and molecular characterization of the promoter. J. Biol. Chem. 265:11198-11203. [PubMed] [Google Scholar]
- 23.Noda, S., Y. Takezawa, T. Mizutani, T. Asakura, E. Nishiumi, K. Onoe, M. Wada, F. Tomita, K. Matsushita, and A. Yokota. 2006. Alterations of cellular physiology in Escherichia coli in response to oxidative phosphorylation impaired by defective F1-ATPase. J. Bacteriol. 188:6869-6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogasawara, H., A. Hasegawa, E. Kanda, T. Miki, K. Yamamoto, and A. Ishihama. 2007. Genomic SELEX search for target promoters under the control of PhoQP-RstBA cascade. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 25.Quail, M. A., and J. R. Guest. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 15:519-529. [DOI] [PubMed] [Google Scholar]
- 26.Quail, M. A., D. J. Haydon, and J. R. Guest. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 12:95-104. [DOI] [PubMed] [Google Scholar]
- 27.Riley, M., T. Abe, M. B. Arnaud, M. K. B. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett III, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 34:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salmon, K., S. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12: the effects of oxygen availability and Fnr. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 29.Shalel-Levanon, S., K. Y. San, and G. N. Bennett. 2005. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metab. Eng. 7:364-374. [DOI] [PubMed] [Google Scholar]
- 30.Shimada, T., N. Fujita, M. Maeda, and A. Ishihama. 2005. Systematic search for the Cra-binding promoters using genomic SELEX system. Genes Cells 10:907-918. [DOI] [PubMed] [Google Scholar]
- 31.Shimada, T., H. Makinoshima, Y. Ogawa, T. Miki, M. Maeda, and A. Ishihama. 2004. Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J. Bacteriol. 186:7112-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simons, R. W., F. Howman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusion. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 33.Singer, B. S., T. Shtatland, D. Brown, and L. Gold. 1997. Libraries for genomic SELEX. Nucleic Acids Res. 25:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens, P. E., M. G. Darlison, H. M. Lewis, and J. R. Guest. 1983. The pyruvate dehydrogenase complex of Escherichia coli K12. Eur. J. Biochem. 133:155-162. [DOI] [PubMed] [Google Scholar]
- 35.Stojiljkovic, I., A. J. Baeumler, and K. Hantke. 1994. Fur regulation in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 36.Wyborn, N. R., S. L. Messenger, R. A. Henderson, G. Sawers, R. E. Roberts, M. M. Attwood, and J. Green. 2002. Expression of the Escherichia coli yfiD gene responds to intracellular pH and reduces the accumulation of acidic metabolic end products. Microbiology 148:1015-1026. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto, K., and A. Ishihama. 2003. Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mol. Microbiol. 47:183-194. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto, K., H. Ogasawara, N. Fujita, R. Utsumi, and A. Ishihama. 2002. Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of two-component system sensing external magnesium availability. Mol. Microbiol. 45:423-438. [DOI] [PubMed] [Google Scholar]
- 39.Yokota, A., Y. Terasawa, N. Takaoka, H. Shimizu, and F. Tomita. 1994. Pyruvic acid production by an F1-ATPase-defective mutant of Escherichia coli W1485lip2. Biosci. Biotechnol. Biochem. 58:2164-2167. [DOI] [PubMed] [Google Scholar]
- 40.Zheng, D., C. Constantinidou, J. L. Hobman, and S. D. Minchin. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]