Abstract
Using DNA microarrays, we identified 126 genes in Escherichia coli K-12 whose expression is increased at human body temperature (37°C) compared to growth at 23°C. Genes involved in the uptake and utilization of amino acids, carbohydrates, and iron dominated the list, supporting a model in which temperature serves as a host cue to increase expression of bacterial genes needed for growth. Using quantitative real-time PCR, we investigated the thermoregulatory response for representative genes in each of these three categories (hisJ, cysP, srlE, garP, fes, and cirA), along with the fimbrial gene papB. Increased expression at 37°C compared to 23°C was retained in both exponential and stationary phases for all of the genes and in most of the various media tested, supporting the relative importance of this cue in adapting to changing environments. Because iron acquisition is important for both growth and virulence, we analyzed the regulation of the iron utilization genes cirA and fes and found that growth in iron-depleted medium abrogated the thermoregulatory effect, with high-level expression at both temperatures, contrasting with papB thermoregulation, which was not greatly altered by limiting iron levels. A positive role for the environmental regulator H-NS was found for fes, cirA, hisJ, and srlE transcription, whereas it had a primarily negative effect on cysP and garP expression. Together, these studies indicate that temperature is a broadly used cue for regulating gene expression in E. coli and that H-NS regulates iron, carbohydrate, and amino acid utilization gene expression.
Bacteria have the ability to keenly sense a multitude of environmental stimuli, such as temperature, pH, osmolarity, oxygen levels, carbon sources, and concentrations of various ions and compounds, including iron (reviewed in references 11, 33, and 36). Elaborate sensory mechanisms have evolved in order for these microorganisms to detect environmental changes and thereby regulate cellular activities to adapt to their changing surroundings. This is particularly evident among bacterial pathogens, in which expression of virulence factors is highly regulated in response to the surrounding environment (reviewed in references 33 and 36). Pathogenic bacteria use environmental cues to distinguish between host and nonhost environments, and subsequent regulation of virulence gene expression can allow a more efficient utilization of resources. Environmental regulation of virulence gene expression is of particular importance in Escherichia coli, which can colonize the gut or translocate from that competitive environment to other niches, such as the sterile urinary tract.
Our laboratory has studied the effect of mammalian host temperature (37°C) in controlling pyelonephritis-associated pili, otherwise known as Pap or P fimbriae, that are expressed by uropathogenic strains of E. coli and are encoded by the pap operon (43, 44). P fimbriae are critical in the colonization of the host upper urinary tract, leading to acute upper urinary tract infections (43, 44). The thermoregulation of papBA transcription has been well characterized in the uropathogenic strain and at the molecular level as a transcriptional fusion in E. coli K-12 (4, 21-23, 70-74). Optimal expression occurs at 37°C, with a 52-fold reduction in papBA transcription at 23°C (70). The environmental cue of temperature is important, not just for pap, but in the control of a number of virulence genes in E. coli, including those encoding fimbriae and toxins (14, 17, 18, 20, 26, 34, 39, 50, 56, 74).
Two proteins, RimJ and H-NS, are known to be important in the regulation of papBA transcription in response to temperature and other environmental conditions. RimJ is the N-terminal acetyltransferase of the ribosomal protein S5 (12), and deletion of the rimJ gene leads to a loss of thermoregulation so that papBA transcription levels are equivalent at both 37°C and 23°C (71). The mechanism by which RimJ represses papBA transcription and how the modification of a ribosomal protein might be involved in this process are currently under investigation. H-NS is a histone-like nucleoid-structuring protein that binds to A-T-rich, bent regions of DNA and compacts them (1, 13, 66, 75). H-NS controls the expression of a number of environmentally controlled virulence genes in several gram-negative genera (1). In contrast to many other virulence genes in which H-NS plays only a repressive role, the protein plays both a positive and a negative role in papBA transcription (67, 70, 74). At 37°C, introduction of the hns651 mutation leads to a decrease in transcription, suggesting that H-NS is required for maximal transcription at this temperature. At 23°C, papBA transcription in the hns651 mutant strain is higher than in the wild-type strain and approaches the levels observed at 37°C for the mutant strain, indicating a loss of thermoregulation.
In this study, we investigated the genomewide effect of temperature (37°C) on transcription in E. coli K-12 to identify other genes that are regulated in a manner similar to that of pap and thus might be important in adaptation to and colonization of the host. Transcriptome analyses based on temperature have been conducted in group A Streptococcus (37° and 29°C) (60), Yersinia pestis (37°C and 26°C) (24, 38), and Borellia burgdorferi (37°C and 23°C) (7, 52), but to our knowledge, this is the first investigation and analysis of E. coli. Here, we present evidence that 126 genes are increased in expression at 37°C, with a significant number of carbohydrate, amino acid, and iron utilization genes represented within this list. We further characterize the thermoregulatory responses for representative genes within these three categories under a variety of growth conditions and demonstrate that, in the majority of cases, a temperature differential is maintained that favors increased expression of these genes at 37°C compared to growth at 23°C. Lastly, we demonstrate that H-NS controls the amino acid, carbohydrate, and iron utilization genes tested in this study, linking this global regulator to acquisition of these important substances required for growth.
MATERIALS AND METHODS
Strains and media.
The strains, plasmids, and bacteriophages used in this study are shown in Table 1. Luria-Bertani (LB), and M9 minimal media and antibiotics were prepared as described previously (37, 58).
TABLE 1.
Bacterial strains, bacteriophages, primers, and probes used in this study
| Strains and bacteriophages | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| MC4100 | F−araD139 Δ(lacIPOZYA-argF) U169 rpsL thi-1 | 9 |
| H1941 | MC4100 fur | K. Hantke |
| NH757 | B178 hns651 tyrTβ::Tn10 | 19 |
| DL1504 | MC4100 λ354 lysogen | 5 |
| DL1947 | DL1504 hns651 | 67 |
| CWZ388 | DL1504 ΔrimJ | 71 |
| CWZ479 | H1941 λ354 lysogen | This study |
| Bacteriophages | ||
| P1L4 | Virulent phage P1 | D. A. Low |
| λ354 | papBA-lacZYA fusion phage | 5 |
Construction of CWZ479.
Phage λ354 was retrieved from DL1504 by UV induction as described previously (58). The phage lysate was used to infect H1941 (MC4100 fur), and lysogens were selected on LB medium containing kanamycin to create CWZ479 (Table 1) as described previously (72).
Bacterial growth conditions.
Bacterial cultures were inoculated and grown as described previously (71, 74). For iron-replete growth conditions at 37°C and 23°C, the bacteria were cultured in M9 glycerol medium (M9 minimal liquid medium containing 2.45 μM ferric citrate, 30 μM thiamine, 100 μM calcium chloride, 1 mM magnesium sulfate, and 0.2% glycerol as a carbon source, pH 7) with aeration. For iron-depleted growth conditions, the bacteria were cultured in M9 glycerol medium containing 200 μM 2,2′-dipyridyl, an iron-chelating agent. An inoculum from a single colony grown at 37°C was used to initiate parallel, colony-matched cultures grown at 37°C and 23°C. The cells from these cultures were harvested at equivalent optical densities (OD) after approximately 9 to 11 generations of growth. With the exception of stationary-phase cultures, which were harvested at an OD at 600 nm (OD600) of 1.4 to 1.6, all cultures were harvested in early to mid-exponential phase (OD600 = 0.2 to 0.6). The cell pellets were subsequently frozen in liquid nitrogen and stored at −80°C for RNA isolation.
RNA isolation.
For microarray analyses, RNA was isolated by phenol-chloroform extraction as described previously (70) with the modifications that the RNA was subjected to a second DNase digestion in solution and cleaned using an RNeasy column according to the manufacturer's directions (QIAGEN). For quantitative real-time reverse transcription-PCR (qRT-PCR) experiments, RNA isolation was carried out as directed in the QIAGEN RNeasy Mini protocol with the modifications that cell pellets were resuspended in 100 μl Tris-EDTA containing lysozyme (400 μg/ml) and boiled for 90 seconds prior to application to the column, and the RNA samples were digested twice with DNase (first on column and then in solution). RNA concentrations and purity were determined by spectrophotometry. Isolated RNAs were stored at −80°C until they were used.
cDNA synthesis, labeling, and hybridization.
Synthesis, labeling of cDNA with Cy3/Cy5, and hybridization were performed using the 3DNA Array 350 RP Expression Array Detection Kit according to the manufacturer's protocol (Genisphere). cDNA for each condition was prepared from 2 μg total RNA. cDNAs from both temperatures (37°C and 23°C) were cohybridized to slides containing full-length PCR products from all 4,290 annotated open reading frames (ORFs) in E. coli MG1655. The slides were produced by the University of Wisconsin-Gene Expression Center (http://www.biotech.wisc.edu/GEC/) and obtained at a reduced cost through the Genome Consortium for Active Teaching (http://www.bio.davidson.edu/projects/gcat/gcat.html). The hybridization solution (10 μl of cDNA mixture, 1 μl of denatured rat Hybloc DNA [1 μg/μl], 20 μl of 2× sodium dodecyl sulfate [SDS] buffer, 2 μl of dT blocker, and 7 μl of nuclease-free water) was placed over a prewarmed microarray slide and covered with a Corning 22- by 40-mm coverslip. The slide was then sealed in a hybridization chamber and submerged for 3 days in a 60°C water bath. Microarray slides were washed for 10 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% SDS at 37°C, 10 min in 2× SSC at room temperature, and 10 min in 0.2× SSC at room temperature. The slides were dried by centrifugation at 10,000 rpm for 2 minutes. For the second hybridization to allow the 3DNA capture reagent to bind to the complementary Cy3- or Cy5-specific 3DNA capture sequences ligated to the cDNAs, each microarray received 42 μl of hybridization solution (2.5 μl of Cy3 capture reagent, 2.5 μl of Cy5 capture reagent, 21 μl of 2× SDS buffer, 0.5 μl antifade reagent, and 15.5 μl of nuclease-free water) and was hybridized as before via submersion for 3 to 4 h in a 61°C water bath. The slides were washed, dried, and kept in the dark until they were scanned and analyzed.
Microarray data analysis.
Five slides were used in the analysis with cDNAs, representing three independent growth experiments and two technical replicates. Hybridized slides were scanned using GenePix Pro 4.1 software (Axon Instruments, Inc.). Photomultiplier tube voltage was altered for the initial scanning to ensure proper channel balance and decrease background. Any gene feature that had a signal-to-background ratio of <2.0 was rejected and not further analyzed. The ratios of median values were used for further analysis. The five slides used for detailed studies were normalized by a Lowess fit using GPROCESSOR, developed by Zhong Guan (http://bioinformatics.med.yale.edu). Replicate spots on the arrays were merged using GEPAS (http://gepas.bioinfo.cipf.es/). Significance analysis was completed as a one-class response using significance analysis of microarrays (65) with a delta of 0.65 and a median false-discovery rate of 1%. An ORF was considered temperature regulated if it demonstrated a statistically significant change in expression greater than 1.7-fold.
qRT-PCR.
Primers and dual-labeled fluorogenic probes for qRT-PCR were created using Primer Express (v.1.5) software from Applied Biosystems. One-step reactions were prepared as directed by the manufacturer (QIAGEN QuantiTect kit or Invitrogen SuperScript III Paltinum SYBR Green kit) using 50 ng of total RNA per reaction. An Applied Biosystems 7700 instrument was used for thermocycling with the following conditions: 50°C for 30 min, 95°C for 15 min, 40 cycles of 94°C for 15 seconds, and 60°C for 1 min. All reactions were performed in triplicate, with no reverse transcriptase controls run for each RNA sample to detect DNA contamination.
All reactions were normalized by using the same amount of total RNA (50 ng) in each reaction. Relative levels of gene expression were calculated as previously described (32) using the following equation: ΔCT′ = CT gene at x°C, X genotype − CT gene at 37°C M9 glyc, wild type and then transformed to relative changes (n-fold) using 2−ΔCT′. CT is the cycle number at which the real-time amplification curve crosses the user-defined threshold, x°C is the temperature at which the RNA was isolated (37°C or 23°C), and X is the genotype of the strain (wild type or mutant). Differences in average CT values were determined to be statistically significant (P < 0.05) by two-way analysis of variance using STATA SE software (StataCorp). The data represent the average change (n-fold) determined from at least three independent experiments.
RESULTS
Microarray design to identify temperature-regulated genes in E. coli K-12 MC4100.
Because many virulence genes in E. coli and other pathogens are regulated by temperature, we wanted to determine on a genomewide scale which genes are modulated in adapting to both human host temperature (37°C) and ambient room temperature (23°C). Growth and medium conditions were chosen (M9 glycerol medium with aeration) to maximize papBA transcription at 37°C so we could use this operon as a temperature-regulated positive control in our microarrays. Parallel cultures grown at 37°C and 23°C were harvested in exponential phase at 9 to 11 generations of growth after inoculation. Thus, the results presented here reflect the adapted state and signify genes whose expression is differentially maintained over long-term growth at a given temperature.
Overall, 126 genes were found to be more highly expressed at 37°C (Table 2), whereas 297 genes were more highly expressed at 23°C (C. A. White-Ziegler, S. Um, N. M. Pérez, A. L. Berns, A. J. Malhowski, and S. Young, unpublished data). The lacZ, lacY, and lacA genes, contained on a chromosomally located papBA transcriptional fusion and driven by the pBA promoter, showed the greatest increase at 37°C (13.7-fold, 7.5-fold, and 5.4-fold, respectively), supporting the validity of the experimental design.
TABLE 2.
Genes demonstrating increased expression at 37°C
| Genea | Blattner no. | Productb | Change (n-fold) at 37°/23°Cc | |||
|---|---|---|---|---|---|---|
| Amino acid transport and metabolism | ||||||
| argD | b3359 | Acetylornithine delta-aminotransferase | 2.4 | |||
| aroF | b2601 | 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase (DAHP synthetase), tyrosine-repressible | 2.2 | |||
| aroG | b0754 | 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase (DAHP synthetase) | 2.0 | |||
| asnA | b3744 | Asparagine synthetase A | 2.4 | |||
| carB | b0033 | Carbamoyl phosphate synthase, large subunit | 2.8 | |||
| cysD | b2752 | ATP-sulfurylase, subunit 2 (ATP-sulfate adenylyltransferase) | 2.2 | |||
| cysH | b2762 | 3′-Phosphoadenosine 5′-phosphosulfate (PAPS) reductase | 2.6 | |||
| cysM | b2421 | Cysteine synthase B (O-acetylserine sulfhydrolase B) | 2.4 | |||
| gcvP | b2903 | Glycine cleavage complex protein P, glycine decarboxylase, PLP dependent | 1.9 | |||
| hisC | b2021 | Histidinol phosphate aminotransferase | 1.9 | |||
| hisJ | b2309 | Histidine transport protein (ABC superfamily, peri_bind) | 2.6 | |||
| hisQ | b2308 | Histidine and lysine/arginine/ornithine transport system (ABC superfamily, membrane) | 2.2 | |||
| idnD | b4267 | l-idonate 5-dehydrogenase, NAD binding | 1.8 | |||
| livG | b3455 | ATP-binding component of high-affinity branched-chain amino acid transport system | 1.9 | |||
| livK | b3458 | High-affinity branched-chain amino acid transport protein (ABC superfamily, peri_bind) | 2.4 | |||
| lysC | b4024 | Aspartokinase III, lysine sensitive | 2.6 | |||
| lysP | b2156 | Lysine-specific permease | 2.5 | |||
| nanA | b3225 | N-Acetylneuraminate lyase (aldolase) | 2.1 | |||
| tesA | b0494 | Acyl-coenzyme A thioesterase I; also functions as protease I | 2.0 | |||
| trpD | b1263 | Anthranilate synthase component II | 2.1 | |||
| ybiK | b0828 | l-asparaginase | 1.9 | |||
| yecC | b1917 | Putative ATP-binding component of a transport system | 2.5 | |||
| yecS | b1918 | Putative amino acid transport protein (ABC superfamily, membrane) | 2.1 | |||
| Carbohydrate transport and metabolism | ||||||
| araA | b0062 | l-arabinose isomerase | 1.7 | |||
| galK | b0757 | Galactokinase | 3.3 | |||
| garD | b3128 | (d)-galactarate dehydrogenase | 3.0 | |||
| garL | b3126 | Alpha-dehydro-beta-deoxy-d-glucarate aldolase | 5.4 | |||
| garP | b3127 | Putative transport protein | 6.3 | |||
| lamB | b4036 | Maltoporin, high-affinity receptor for maltose and maltoseoligosaccharides; phage lambda receptor | 2.3 | |||
| mglA | b2149 | ATP-binding component of methyl-galactoside transport and galactose taxis | 3.3 | |||
| mglB | b2150 | Galactose transport protein (ABC superfamily, peri_bind) | 3.1 | |||
| mglC | b2148 | Methyl-galactoside transport and galactose taxis | 4.4 | |||
| nagE | b0679 | PTS family enzyme IIC (N terminal); enzyme IIB (center); enzyme IIC (C terminal) | 2.4 | |||
| sgbH | b3581 | 3-Keto-l-gulonate 6-phosphate decarboxylase | 1.8 | |||
| srlB | b2704 | PTS family enzyme IIA, glucitol/sorbitol specific | 2.2 | |||
| uidA | b1617 | Beta-d-glucuronidase | 2.3 | |||
| uxuB | b4323 | d-mannonate oxidoreductase | 2.1 | |||
| yeiC | b2166 | Putative kinase | 2.4 | |||
| Cell motility and secretion | ||||||
| fliQ | b1949 | Flagellar biosynthesis | 1.9 | |||
| Cell wall/membrane biogenesis | ||||||
| nmpC | b0553 | DLP12 prophage; outer membrane porin, at locus of qsr prophage | 10.0 | |||
| ompT | b0565 | DLP12 prophage; protease VII, outer membrane protein 3b (a), putative porin | 3.3 | |||
| yciD | b1256 | Outer membrane protein W; colicin S4 receptor; putative transport protein | 3.0 | |||
| Coenzyme transport and metabolism | ||||||
| btuB | b3966 | Outer membrane porin: vitamin B12/cobalamin transport, receptor for E colicins | 2.1 | |||
| entC | b0593 | Isochorismate hydroxymutase 2, enterochelin biosynthesis | 2.0 | |||
| folE | b2153 | GTP cyclohydrolase I | 2.7 | |||
| ubiA | b4040 | 4-Hydroxybenzoate-octaprenyltransferase | 2.1 | |||
| Defense mechanisms | ||||||
| hsdR | b4350 | Endonuclease R, host restriction | 2.8 | |||
| Energy production and conversion | ||||||
| fumB | b4122 | Fumarase B (fumarate hydratase class I), anaerobic isozyme | 1.9 | |||
| galT | b0758 | Galactose-1-phosphate uridylyltransferase | 2.6 | |||
| hybA | b2996 | Hydrogenase-2 small subunit | 2.1 | |||
| napF | b2208 | Fe-S ferredoxin-type protein, electron transfer | 1.7 | |||
| nirB | b3365 | Nitrite reductase [NAD(P)H] subunit | 3.3 | |||
| sucD | b0729 | Succinyl-coenzyme A synthetase, alpha subunit | 1.9 | |||
| ybiC | b0801 | Putative dehydrogenase | 3.1 | |||
| yqhD | b3011 | Putative alcohol dehydrogenase | 3.3 | |||
| Inorganic-ion transport and metabolism | ||||||
| cirA | b2155 | Outer membrane pore protein, receptor for colicin I, requires TonB | 2.8 | |||
| cysA | b2422 | ATP-binding component of sulfate permease A protein; chromate resistance | 2.3 | |||
| cysC | b2750 | Adenosine 5′-phosphosulfate kinase | 2.7 | |||
| cysI | b2763 | Sulfite reductase, alpha subunit | 2.2 | |||
| cysJ | b2764 | Sulfite reductase (NADPH), flavoprotein beta subunit | 2.7 | |||
| cysN | b2751 | ATP-sulfurylase (ATP:sulfate adenylyltransferase), subunit 1, probably a GTPase | 2.1 | |||
| cysP | b2425 | Thiosulfate binding protein | 3.1 | |||
| fecA | b4291 | KpLE2 phage-like element; outer membrane porin, receptor for ferric citrate | 2.5 | |||
| fecR | b4292 | KpLE2 phage-like element; regulator in multicomponent regulatory system with FecI/FecA | 3.3 | |||
| fepA | b0584 | Outer membrane porin, receptor for ferric enterobactin and colicins B and D | 2.0 | |||
| fepB | b0592 | Ferric enterobactin (enterochelin) binding protein; periplasmic component | 1.9 | |||
| fepC | b0588 | ATP-binding component of ferric enterobactin transport | 2.0 | |||
| fepG | b0589 | Ferric enterobactin transport protein | 2.0 | |||
| fes | b0585 | Enterochelin esterase | 2.7 | |||
| ycdO | b1018 | Hypothetical protein | 1.8 | |||
| Lipid transport and metabolism | ||||||
| acs | b4069 | Acetyl-coenzyme A synthetase | 2.4 | |||
| fadE | b0221 | Medium-long-chain fatty acyl-coenzyme A dehydrogenase | 1.8 | |||
| garR | b3125 | Tartronate semialdehyde reductase (TSAR) | 1.8 | |||
| Nucleotide transport and metabolism | ||||||
| allB | b0512 | Allantoinase | 1.9 | |||
| rihA | b0651 | Pyrimidine-specific nucleoside hydrolase | 2.0 | |||
| rihC | b0030 | Nucleoside hydrolase | 3.3 | |||
| Posttranslational modification, protein turnover, chaperones | ||||||
| ccmC | b2199 | Heme export protein (ABC superfamily, membrane) | 2.3 | |||
| ccmG | b2195 | Heme lyase/disulfide oxidoreductase (thiol-disulfide interchange protein) | 1.8 | |||
| cysU | b2424 | Sulfate, thiosulfate transport system permease T protein | 2.7 | |||
| yeeD | b2012 | Hypothetical protein | 3.0 | |||
| Replication, recombination, and repair | ||||||
| nohB | b0560 | DLP12 prophage; bacteriophage DNA- packaging protein | 2.0 | |||
| sbcB | b2011 | Exonuclease I, 3′→ ′-specific; deoxyribophosphodiesterase | 2.0 | |||
| trs5_1 | b0259 | CP4-6 prophage; IS5 protein 1 | 2.0 | |||
| trs5_10 | b3218 | IS5 protein 10 | 2.3 | |||
| trs5_11 | b3505 | IS5 protein 11 | 2.2 | |||
| trs5_3 | b0656 | IS5 protein | 2.2 | |||
| trs5_4 | b1331 | IS5 protein | 2.1 | |||
| trs5_5 | b1370 | IS5Y transposase | 2.4 | |||
| trs5_6 | b1994 | IS5 protein | 2.4 | |||
| trs5_7 | b2030 | IS5 protein | 2.6 | |||
| trs5_8 | b2192 | IS5 protein | 2.5 | |||
| trs5_9 | b2982 | IS5 protein | 3.2 | |||
| Secondary-metabolite biosynthesis, transport and catabolism | ||||||
| entB | b0595 | 2,3-Dihydro-2,3-dihydroxybenzoate synthetase, isochroismatase | 2.1 | |||
| entE | b0594 | 2,3-Dihydroxybenzoate-AMP ligase | 2.2 | |||
| idnO | b4266 | 5-Keto-d-gluconate 5-reductase | 2.0 | |||
| srlD | b2705 | Glucitol (sorbitol)-6-phosphate dehydrogenase | 1.9 | |||
| yeiN | b2165 | Hypothetical protein | 3.1 | |||
| Signal transduction mechanisms | ||||||
| ycdT | b1025 | Putative transmembrane protein | 1.7 | |||
| Transcription | ||||||
| fecI | b4293 | KpLE2 phage-like element; sigma 19 factor of RNA polymerase | 2.2 | |||
| lrhA | b2289 | NADH dehydrogenase transcriptional regulator, LysR family; modulates SprE (RssB) activity | 4.4 | |||
| yijC | b3963 | Putative regulator (TetR/AcrR family) | 2.4 | |||
| General function prediction or function unknown | ||||||
| yadS | b0157 | Putative membrane protein | 2.2 | |||
| yddA | b1496 | Putative ABC transport system ATP-binding protein | 4.8 | |||
| yeeE | b2013 | Putative membrane component of transport system | 2.2 | |||
| yfiD | b2579 | Putative formate acetyltransferase | 4.4 | |||
| Not in COGS database | ||||||
| b1172 | b1172 | Hypothetical protein | 2.0 | |||
| b2191 | b2191 | Unknown CDS | 2.3 | |||
| cdaR | b0162 | Regulator of d-galactarate, d-glucarate, and d-glycerate metabolism | 1.9 | |||
| cysW | b2423 | Sulfate transport system permease W protein | 2.6 | |||
| malM | b4037 | Periplasmic protein of mal regulon | 2.4 | |||
| pppA | b2972 | Bifunctional prepilin peptidase | 2.7 | |||
| srlA | b2702 | PTS family enzyme IIC, glucitol/sorbitol specific | 2.0 | |||
| srlE | b2703 | PTS family enzyme IIBC, glucitol/sorbitol specific | 2.5 | |||
| ybcT | b0556 | DLP12 prophage; bacteriophage lambda endopeptidase homolog | 3.5 | |||
| ybcU | b0557 | DLP12 prophage; bacteriophage lambda Bor lipoprotein homolog, involved in serum resistance | 3.5 | |||
| yddB | b1495 | Hypothetical protein | 8.0 | |||
| yedO | b1919 | d-cysteine desulfhydrase, PLP-dependent enzyme | 2.4 | |||
| ygaW | b2670 | Putative membrane protein | 4.9 | |||
| yhcE | b3217 | Unknown CDS | 2.1 | |||
| yijD | b3964 | Putative membrane protein | 3.4 | |||
| yjiD | b4326 | Unknown CDS | 2.4 | |||
| ylaC | b0458 | Putative membrane protein | 2.1 | |||
| ymfL | b1147 | e14 prophage; putative negative regulator | 2.3 | |||
Genes involved in iron utilization and/or known to be Fe2+-Fur regulated are shown in boldface.
Product descriptions and functional categories are based upon the E. coli K-12 COGS categorization (63). Descriptions were shortened in some instances.
Change is indicated as the average ratio of medians (37°C/23°C). All genes included showed a statistically significant increase at 37°C as described in Materials and Methods and based on the results from five microarrays.
COGS categorization of genes.
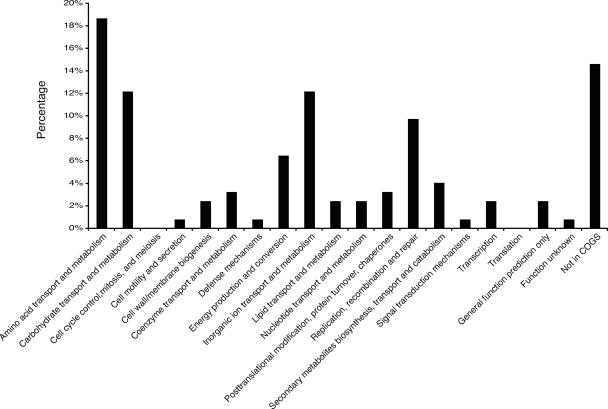
The 126 genes with increased expression at 37°C were categorized using the Clusters of Orthologous Groups of Proteins (COGS) database (63). At least one gene was found in each category, with the exception of cell cycle control (Table 2 and Fig. 1). Twelve genes within the category of replication, recombination, and repair were increased at 37°C; with the exception of sbcB, all encode proteins with phage/transposon-related functions (Table 2 and Fig. 1). Approximately 12% of the genes with increased transcription at 37°C have unknown functions (Table 2 and Fig. 1). The other three categories of genes that dominated the response were those associated with amino acid (19%), carbohydrate (12%), and inorganic-ion (12%) transport and metabolism.
FIG. 1.
Percentages of ORFs demonstrating increased expression at 37°C compared to 23°C in each of 20 functional categories. ORFS were placed in functional categories according to the E. coli K-12 COGS categorization (63).
Human body temperature increases carbohydrate and amino acid uptake and utilization gene expression.
Expression levels of 15 genes involved in carbon transport and utilization and 23 genes involved in synthesis and transport of amino acids were significantly increased at 37°C in comparison to 23°C (Table 2). These genes showed increased expression in the absence of the cognate sugar and, in the case of amino acid transporters, in the absence of the specific amino acid. The carbohydrate utilization genes that were increased at 37°C are involved in the catabolism of a diversity of sugars, including arabinose, maltose, galactose, d-galactarate, d-glucarate, d-gluconuride, glucitol, and sorbitol. A variety of amino acid transporters demonstrated increased expression at 37°C, whereas biosynthesis was focused on histidine and cysteine (Table 2). Genes required for the uptake and utilization of other important compounds for growth, including sulfur/thiosulfate (cysDHM) and cobalamin (btuB), were similarly increased at 37°C.
Human body temperature increases expression of genes for iron uptake and utilization.
Interestingly, 12 genes involved in iron utilization were identified, representing 10% of all genes with increased expression at 37°C (Table 2). They represent ferric enterobactin uptake genes (fepA, fepB, fepC, and fepG), enterobactin synthesis genes (entB, entC, and entE), and the enterobactin esterase gene fes. In addition, the genes fecA, fecI, fecR, and cirA, involved in utilization of other iron forms (ferric-dicitrate, ferrous iron, ferri-copragen/rhodotorulic acid, ferrioxamine B, and ferric-dihydroybenzoate), were increased in expression at 37°C compared to 23°C.
Based on other studies, it is known that transcription of all of these genes is responsive to the iron concentration, showing high expression under iron-depleted conditions but repressed by the transcriptional regulator Fur (ferric uptake regulator) when iron is present (reviewed in references 16 and 25). In our microarrays, the bacteria were grown in iron-replete medium, where Fur binds iron and act as a repressor. Thus, these results indicate that the increased expression of these genes is due to the effect of temperature and not to limiting iron conditions.
Several additional genes that are regulated by Fe2+-Fur demonstrated higher expression at 37°C than at 23°C (35). Due to their similarity to ABC transporters (YddA) and TonB-dependent outer membrane receptors (YddB), it has been hypothesized that these two proteins may form part of a new iron uptake system (35). Interestingly, the genes were also identified in a transposon mutagenesis screen and found to be required for optimal growth at 37°C in rich medium (57). They are two of the most highly expressed genes at 37°C compared to 23°C (4.8- and 8.0-fold, respectively). yciD (ompW) encodes a S4 colicin receptor and putative transport protein (48), similar to the iron acquisition genes cirA and fepA, which also serve as colicin receptors. garPLR, napF, nirB, and ycdO are also included in Fe2+-Fur-controlled operons (35) and demonstrated temperature-regulated expression in this study.
Thermoregulation of amino acid, carbohydrate, and iron utilization genes occurs in both exponential and stationary phases.
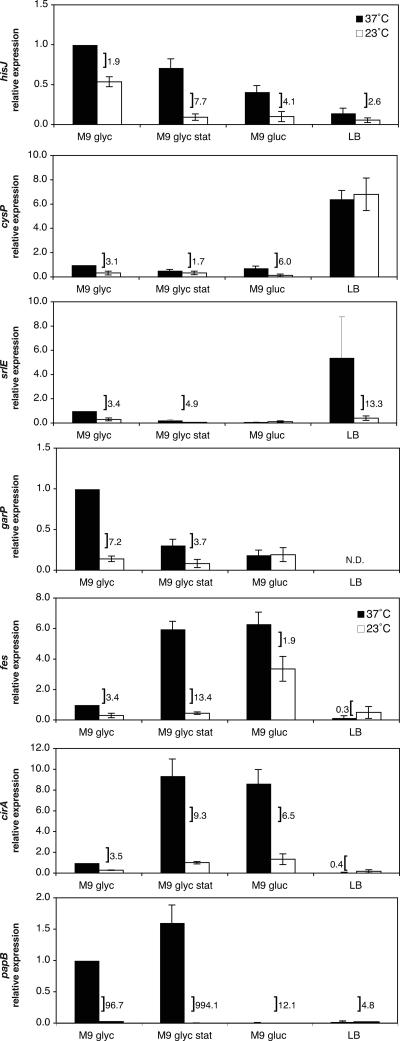
We analyzed the expression of six genes, two from each of the amino acid, carbohydrate, and iron utilization gene categories, at 37°C and 23°C under a variety of growth conditions. Both cysP and hisJ encode periplasmic binding proteins that are involved in thiosulfate (59) and histidine (31) transport, respectively. For carbohydrate utilization genes, we examined srlE, a subunit of the glucitol/sorbitol PTS permease (49), and garP, a putative glucarate transporter (46). For iron utilization genes, we studied fes and cirA. Fes is a cytoplasm protein that functions in releasing iron bound to the bacterial iron chelator enterobactin (6). CirA is an outer membrane receptor with broad specificity that has been postulated to transport iron complexes of enterobactin and is known to transport colicins I/V and catechol-substituted cephalosporins (42). We also analyzed the expression of papB, a known temperature-regulated virulence gene. papB is the first gene in the multicistronic papBA operon that encodes pyelonephritis-associated pili (Pap) (reviewed in reference 28).
In concordance with our microarray results, all six genes demonstated statistically significant thermoregulatory responses in exponentially growing cells in M9 glycerol medium (Fig. 2). The amino acid utilization genes hisJ and cysP demonstrated the smallest differential in expression based on temperature, with 1.9- and 3.1-fold-higher expression, respectively, at 37°C compared to 23°C. garP and srlE demonstrated higher differentials in expression due to temperature, with 7.2- and 3.4-fold increases, respectively, in their transcription at 37°C compared to 23°C. The relative expression levels of fes and cirA were 3.4-fold and 3.5-fold greater, respectively, at 37°C than at 23°C. papB demonstrated a 96.7-fold-greater level of expression at 37°C than at 23°C, similar to previous results using a β-galactosidase assay, where transcription from the papBA promoter was 52-fold higher at 37°C than at 23°C (70). Together, our data support a model in which temperature serves as an important environmental cue that modulates gene expression.
FIG. 2.
Effects of growth phase and medium on the thermoregulation of gene expression for amino acid, carbohydrate, and iron utilization genes. The bars indicate relative levels of expression measured in the wild-type strain DL1504 in M9 glycerol medium in exponential phase (M9 glyc), M9 glycerol medium in stationary phase (M9 glyc stat), M9 glucose medium in exponential phase (M9 gluc), and LB medium in exponential phase (LB). Relative levels of expression are expressed in comparison to the wild-type strain grown in M9 glycerol medium at 37°C. A bracket indicates a statistically significant difference in expression levels based on temperature under the given condition and is accompanied by the ratio of expression (37°C/23°C). Error is expressed as ±1 standard deviation from the mean.
All cultures grown to stationary phase in M9 glycerol medium still retained a thermoregulatory response, indicating that temperature serves to maintain higher levels of expression at 37°C independent of the growth phase (Fig. 2). For the iron utilization genes (fes and cirA) and papB, transcription at 37°C was increased significantly in stationary phase, leading to a higher differential in expression between 37°C and 23°C (13.4-, 9.3-, and 994-fold, respectively). For the carbohydrate utilization genes (garP and srlE) and hisJ, expression was decreased at both 37°C and 23°C in stationary phase, but a statistically significant thermoregulatory response was retained (3.7-, 4.9-, and 7.7-fold higher at 37°C, respectively). Growth in stationary phase decreased cysP expression at 37°C but did not alter transcription significantly at 23°C, leading to a smaller differential in gene expression based on temperature during this growth phase.
Glucose as a carbon source abrogates thermoregulation for the carbohydrate genes, but not the amino acid or iron utilization genes.
Growth in M9 medium utilizing glucose as a carbon source abrogated the thermoregulatory response for the carbohydrate utilization genes (garP and srlE) so that they had equivalent expression at both 37°C and 23°C (Fig. 2). All of the other genes retained a statistically significant differential in expression based on temperature when grown in M9 glucose medium (Fig. 2). Similar to stationary phase, growth on glucose increased expression of the iron utilization gene cirA at 37°C, resulting in a 6.5-fold differential in expression between the two temperatures, whereas glucose increased fes expression at both 37°C and 23°C so that the differential in their expression was only 1.9-fold. Growth in M9 glucose decreased the expression of hisJ and cysP at both temperatures in comparison to glycerol as a carbon source and resulted in a greater differential in expression based on temperature (4.1-fold and 6.0-fold, respectively). papB demonstrated greatly reduced levels of transcription at both temperatures so that the differential in expression between 37°C and 23°C was only 12.4-fold compared to the 97-fold observed in M9 glycerol.
Growth in LB medium had varied effects on the thermoregulatory response.
Growth in LB medium greatly increased expression of cysP so that equivalent expression levels of these genes were observed at both 37°C and 23°C, abrogating the thermoregulatory response (Fig. 2). For papB and hisJ, LB decreased expression at both temperatures, but statistically significant temperature differentials (4.8 and 2.6, respectively) were retained (Fig. 2). Interestingly, for the iron utilization genes cirA and fes, LB significantly decreased the expression of both genes at 37°C but had little effect on expression at 23°C, so that fes and cirA were more highly expressed at 23°C than at 37°C under these growth conditions (Fig. 2). For srlE, LB increased expression at 37°C, leading to a 13.3-fold differential between its expression at 37°C and at 23°C. garP expression increased at both 37°C and 23°C, but there was significant colony variability, so that an accurate relative expression could not be determined (data not shown).
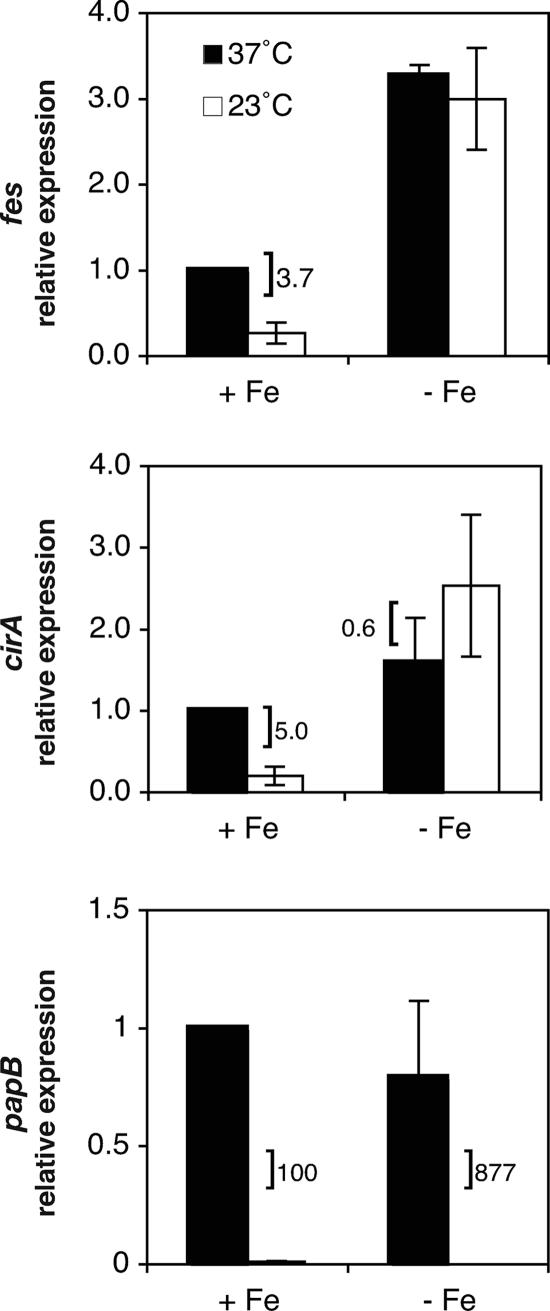
Low-iron conditions abrogate the thermoregulatory effect on cirA and fes, but not pap, transcription.
Because iron acquisition is essential to bacterial growth and impacts uropathogenic E. coli virulence (30, 54, 64), we further assessed the relative importance of iron availability and temperature as cues for regulating transcription of the iron utilization genes fes and cirA by measuring expression under iron-depleted conditions at both 37°C and 23°C in the wild-type strain (Fig. 3). Iron-depleted conditions were obtained by adding 200 μM 2,2′-dipyridyl, an iron-chelating agent, to M9 glycerol medium for growth. cirA and fes transcription levels were higher under iron-depleted conditions than under iron-replete conditions at both temperatures. fes demonstrated 3.3- and 3.0-fold increases at 37°C and 23°C, respectively, under iron-depleted conditions compared to iron-replete conditions at 37°C (Fig. 3). Similarly, cirA transcription demonstrated 1.6- and 2.5-fold increases at 37°C and 23°C, respectively, in iron-depleted medium compared to iron-replete medium at 37°C (Fig. 3). These results demonstrate that a lack of iron has a stimulatory effect on the transcription of these genes at both temperatures and that the thermoregulatory effect on cirA and fes transcription is abrogated when iron is limiting.
FIG. 3.
Effects of temperature and iron concentration on cirA, fes, and papBA transcription. The bars indicate relative levels of expression measured in the wild-type strain DL1504 under iron-replete (+ Fe) and iron-depeleted (− Fe) conditions. Relative levels of expression are expressed in comparison to the wild-type strain grown in iron-replete (M9 glycerol) medium at 37°C. A bracket indicates a statistically significant difference in expression levels based on temperature under the given condition and is accompanied by the ratio of expression (37°C/23°C). Error is expressed as ±1 standard deviation from the mean.
Because research has indicated that the urinary tract is an iron-limited environment (61) and that iron affects the expression of certain fimbrial operons (27), we investigated whether iron availability, in addition to temperature, might serve as a cue for controlling papBA gene expression. Our data demonstrate that expression levels for papBA are similar under iron-depleted conditions to those seen in iron-replete media at both temperatures and that thermoregulation of papBA transcription still occurs when iron is limiting (Fig. 3). Thus, while fes, cirA, and papBA exhibited thermoregulated expression under iron-replete conditions, they showed much different expression patterns under iron-limiting conditions, where fes and cirA are no longer affected by temperature but papBA remains thermoregulated.
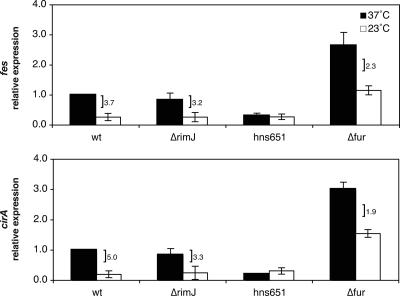
The thermoregulator H-NS, but not RimJ, alters cirA and fes transcription.
We wanted to assess whether temperature regulation of cirA and fes under iron-replete conditions was mediated by thermoregulators known to control papBA transcription, thereby suggesting a coordinate thermoregulation of fimbrial expression with iron acquisition. Thermoregulation of cirA and fes was investigated by analyzing the relative levels of gene expression at 37°C and 23°C in the hns651 strain. The hns651 mutation contains an insertion sequence that leads to the loss of H-NS expression (70). At 37°C, both fes and cirA expression levels were decreased an average of 3.3- and 4.9-fold, respectively, in the hns651 strain compared to the wild type at 37°C (Fig. 4). This decrease in expression in the mutant, in the absence of a temperature change, indicates that H-NS has a positive effect on cirA and fes transcription at 37°C. At 23°C, relative levels of cirA and fes in the hns651 mutant were similar to that measured in the wild-type strain at 23°C (Fig. 4). In the hns651 mutant, there was no longer a significant difference in the levels of cirA and fes transcription based on temperature, primarily due to decreased expression at 37° rather than an alleviation of a repressive effect of H-NS at 23°C.
FIG. 4.
Effects of the ΔrimJ, hns651, and fur mutations on cirA and fes transcription. The bars indicate relative levels of expression measured in the wild-type strain DL1504 (wt), in the ΔrimJ mutant strain CWZ388 (ΔrimJ), in the hns651 mutant strain DL1947 (hns651), and in the fur mutant strain CWZ479 (Δfur). Relative levels of expression are expressed in comparison to the wild-type strain grown in M9 glycerol medium at 37°C. A bracket indicates a statistically significant difference in expression levels based on temperature under the given condition and is accompanied by the ratio of expression (37°C/23°C). Error is expressed as ±1 standard deviation from the mean.
RimJ is an N-terminal acetyltransferase that represses papBA transcription (71-73), and thermoregulatory repression was relieved in the ΔrimJ mutant so that papBA transcription levels were equivalent at 37°C and 23°C (71). In a ΔrimJ strain, there was an average 3.2- and 3.3-fold difference in the levels of fes and cirA expression, respectively, at 37°C compared to 23°C (Fig. 4). These results are similar to those observed in the wild-type strain, indicating that cirA and fes are not under the thermoregulatory control of RimJ.
Thermoregulation of cirA and fes transcription occurs independently of Fur.
Although there was no previous research suggesting a thermoregulatory role for Fur, we investigated whether Fur might have a dual regulatory role in sensing both iron concentration and temperature to control fes and cirA transcription. As expected, introduction of the fur mutation caused an increase in transcription of fes and cirA at both 37°C and 23°C in the absence of this transcriptional repressor (Fig. 4). However, unlike the experiment in which iron was limiting and cirA and fes transcription was no longer thermoregulated (Fig. 3), analysis of fes and cirA transcription in the fur mutant strain showed a thermoregulatory trend similar to that seen in the wild-type strain (Fig. 4). fes and cirA showed 2.3- and 1.9-fold differences, respectively, in expression levels at 37°C compared to 23°C in the fur mutant (Fig. 4). Thus, our results indicate that thermoregulation of fes and cirA occurs in the absence of Fur.
H-NS controls the regulation of carbohydrate and amino acid utilization genes.
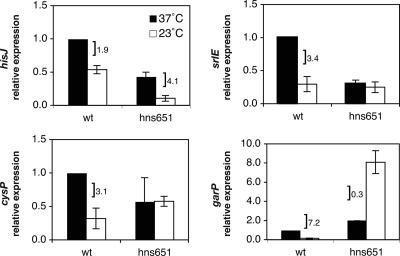
Because of the role of H-NS in controlling iron utilization gene transcription, we wanted to explore if H-NS played a similar regulatory role for the amino acid and carbohydrate utilization genes in this study. Similar to the iron utilization genes, a hns651 mutation led to a 3.4-fold decrease in srlE expression at 37°C compared to the wild-type strain at 37°C, thereby abrogating any differential in expression due to temperature and indicating a positive role for H-NS in the regulation of srlE (Fig. 5). For hisJ, there were statistically significant decreases in transcription at both 37°C and 23°C by the hns651 mutation (2.3- and 5.1-fold, respectively), indicating that H-NS has a positive role in controlling transcription of this gene at both temperatures (Fig. 5). H-NS plays a predominantly repressive role for transcription of cysP at low temperature, with the hns651 mutation leading to statistically significant increased transcription at 23°C and a subsequent loss of a thermoregulatory differential in gene expression. garP expression demonstrated a purely negative effect of H-NS on transcription, as an hns651 mutation led to increased expression of garP at both temperatures (Fig. 5). Together, our results indicate that H-NS may have a broad regulatory role in controlling gene expression that is responsive to temperature but that there are a variety of modes by which H-NS exerts this influence.
FIG. 5.
Effects of the hns651mutation on hisJ, cysP, srlE, and garP transcription. The bars indicate relative levels of expression measured in the wild-type strain DL1504 (wt) and in the hns651 mutant strain DL1947 (hns651). Relative levels of expression are expressed in comparison to the wild-type strain grown in M9 glycerol medium at 37°C. A bracket indicates a statistically significant difference in expression levels based on temperature under the given condition and is accompanied by the ratio of expression (37°C/23°C). Error is expressed as ±1 standard deviation from the mean.
DISCUSSION
Our microarray data indicate that mammalian host temperature (37°C) serves to increase and maintain iron, carbohydrate, and amino acid utilization genes at a higher steady-state level of expression than at 23°C. While temperature has been widely studied as a factor controlling virulence gene expression, our study and others indicate that temperature is used genomewide as a cue for adapting gene expression that may facilitate colonization of a mammalian host (7, 8, 24, 38, 45, 60). In particular, our study shows commonality with Y. pestis, in which the expression levels of a significant proportion of genes for amino acid biosynthesis and carbohydrate utilization were increased at 37°C compared to growth at 26°C (24, 38), and with group A Streptococcus, in which temperature was identified as an important cue in controlling the transcription of iron utilization genes (60).
We propose that temperature may serve as a sentinel cue to quickly allow adaptation of the organism to its environment. Upon entry into a human host, temperature would increase and maintain higher levels of expression of the genes needed for growth. This would allow the bacterium to preemptively prepare for and utilize a diverse set of carbohydrate, amino acid, and iron sources that it might encounter as it transits through different niches in the host, allowing it to more efficiently draw upon the available resources. Interestingly, a recent study demonstrated that a number of iron, carbohydrate, and amino acid utilization genes were increased in expression in bacteria obtained from patients colonized with the asymptomatic-bacteriuria E. coli strain 83972 (53). Together with preliminary experiments from our laboratory demonstrating that a shift from 23°C to 37°C causes a quick increase in iron and fimbrial gene transcription (data not shown), these data support the model that temperature may be an important in vivo cue for modulating bacterial gene expression.
The role of temperature in regulating iron utilization genes is particularly interesting because of evidence that these genes are increased in expression within a mammalian host and that they are required for virulence in vivo. As mentioned previously, bacteria collected from patients colonized by the asymptomatic-bacteriuria E. coli strain 83972 demonstrated increased expression of genes within the fep, ent, fhu, and iut iron systems (53). Similarly, bladder infection in mice from uropathogenic E. coli led to increased expression of a number of iron acquisition genes (61), while in a mouse model of peritonitis, transcriptional fusions to iron acquisition genes were isolated (51). Torres et al. assessed the effect of iron acquisition on virulence, demonstrating that uropathogenic E. coli compromised in the ability to acquire iron through multiple pathways (tonB mutants) were attenuated and those deficient in a single pathway were less successful at colonization in mixed competition in mice (64). Other studies demonstrated urovirulence for individual iron acquisition systems and the presence of additional iron utilization systems in the genomes of pathogenic E. coli (10, 15, 30, 47, 55, 62, 69). Together, these studies provide in vivo evidence for the importance of iron acquisition for E. coli virulence.
Our study delineates the increased expression of several genes in E. coli K-12 in the enterobactin system (ent, fep, and fes) and the ferric citrate (fec) uptake systems at human body temperature, supporting the broad use of this cue in regulating multiple iron uptake pathways. This is further supported by our results indicating that the Fhu system (fhuA), involved in the uptake of ferrichrome and other iron forms, is also temperature regulated (data not shown). Uropathogenic and enterohemorrhagic E. coli strains are known to contain additional systems for iron uptake beyond those found in E. coli K-12 (47, 69), and it would be interesting to investigate if temperature regulation is broadly applicable to these iron utilization systems as well.
Our assessment of temperature regulation demonstrated that the majority of genes retained a higher level of expression at 37°C than at 23°C under a variety of growth conditions, including medium, growth phase, and carbon source, supporting the importance of temperature in modulating gene expression. At the same time, it should be noted that these conditions often influenced gene expression so that the level of expression at 37°C and/or 23°C was altered, leading to an expansion or contraction of the temperature differential, depending on the input of additional environmental cues and reflecting the ability of E. coli to integrate multiple environmental cues and adapt accordingly. Some clear exceptions to this generality were observed in which a given environmental cue led to an abrogation of the thermoregulatory response.
Regulation of fes and cirA by iron and temperature provide an example in which multiple cues are used for adapting gene expression. One could imagine that it could be advantageous to the bacterium to use both cues to adapt to various conditions within the host. In our in vitro experiments, limiting iron is the overriding cue and has the ability to abrogate the thermoregulatory response, significantly increasing expression of cirA and fes regardless of temperature. This response would be what one would expect, given the requirement for iron for bacterial growth and the regulation of these genes by Fur. When might it be advantageous to have temperature regulation of iron acquisition genes? Either upon initial entry into the host or in niches where iron is available within the host, using the cue of host temperature to increase iron acquisition utilization gene expression would benefit the bacterium by allowing it to more efficiently capture and store iron that could subsequently be used when iron-limited niches are encountered.
Similar to the iron utilization genes in our study, others have noted the regulation of virulence gene expression by both temperature and iron availability. In Pseudomonas aeuroginosa, the tolRQ and tolA genes, involved in pyocin transport, are optimally expressed at 37°C under iron-limiting conditions; low temperature and high iron concentrations decrease the expression of these genes (29). Similarly, Shiga toxin synthesis in Shigella dysenteriae is increased by 37°C temperature and low-iron conditions (68). In group A Streptococcus, temperature is an important cue in controlling the transcription of iron utilization genes and hemolysins (60). This coordinate regulation suggests a model in which a temperature of 37°C and iron-limiting conditions cause high expression of a number of genes required for virulence, conditions that most closely replicate the host environment.
Based on these observations and the evidence suggesting that the mammalian host is a 37°C, iron-limited environment (53, 61), we hypothesized that the pap fimbrial genes might be more highly expressed under iron-limiting conditions. This, however, was not the case, as our results showed that papB was transcribed at similar levels at 37°C when grown under iron-depleted conditions and iron-replete conditions (Fig. 3). Previous experiments demonstrated that high iron concentrations do not affect papBA transcription (74), and our results presented here do not support a role for iron or the Fur protein in controlling papBA gene expression (data not shown). Experiments growing uropathogenic E. coli in vitro in human urine at 37°C induced pap transcription (61), and previous studies identified the roles of several individual environmental stimuli in controlling papBA transcription (74), indicating that while iron may not play a role in pap transcription, there are likely multiple overlapping cues, including temperature, that control pap expression in vivo. It should be noted that our experiments were conducted in an E. coli K-12 strain, and future studies will address whether papBA transcription is modulated in the same manner in a uropathogenic E. coli strain.
Our results indicate that H-NS regulates iron, carbohydrate, and amino acid utilization gene expression in E. coli, supporting a broad role for this regulator in the adaptation to temperature. For cirA and fes, the hns651 mutation causes decreased expression of these genes at 37°C, suggesting an activating role for H-NS in the transcription of the genes at host temperature. We also found this to be the case for hisJ and srlE, where H-NS has an activating role at one or both temperatures. This regulation is different from that of many H-NS controlled genes, where H-NS serves as a transcriptional silencer in response to a variety of environmental cues and deletion of H-NS leads to high-level expression under the normally repressive cue (reviewed in references 1 and 13). Since there is no evidence that H-NS can act as a transcriptional activator, it has been postulated that H-NS may play an indirect role (reviewed in reference 13). For the Erwinia chrysanthemi pectate lyase synthesis genes (40, 41) and E. coli flagellar synthesis genes (2, 3), it has been found that H-NS exerts it control, not on these promoters directly, but at the promoters of transcriptional repressors that act upon these target genes. Similarly, it may be that H-NS disrupts the regulation of cirA, fes, srlE, and hisJ through indirect effects. It is interesting that the hns651 mutation has a similar effect on the papBA operon, causing a 7.6-fold decrease in transcription at 37°C (67, 70, 74). Recent microarray results in our laboratory indicate that H-NS controls a majority of the temperature-regulated genes identified in this study (unpublished data), supporting a broad role of H-NS in adapting to host and ambient temperature conditions.
While temperature has been known to be a control for the expression of virulence genes, such as fimbriae, toxins, and adhesins (33), this study and others demonstrate that temperature may have a broader effect to fine tune and regulate a number of genes to presumably allow more efficient colonization of the host. Future studies to investigate and understand the pathways of this thermal regulation could yield valuable anti-infective targets for chemotherapeutic drugs that would decrease the ability of bacteria to compete and survive within the host.
Acknowledgments
We thank the Genome Consortium for Active Teaching and the University of Wisconsin Gene Expression Center for making microarrays available at a reduced cost for use by the two undergraduates, S.Y. and A.M., who completed the microarray experiments in this study. We are grateful to present and former Smith College students, staff, and faculty for their technical assistance and advice, including Michelle Ploutz, Scott Edmands, Eva Ladow, Adam Hall, Nick Horton, and Lori Sanders.
This work was supported by National Institutes of Health grant GM62792 to C.W.-Z., by the Albert F. Blakeslee Trust, and by Smith College. A.M. and S.Y. were supported by student grants through the Albert F. Blakeslee Trust and by the Howard Hughes Medical Institute grant awarded to Smith College.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Bertin, P., F. Hommais, E. Krin, O. Soutourina, C. Tendeng, S. Derzelle, and A. Danchin. 2001. H-NS and H-NS-like proteins in Gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie 83:235-241. [DOI] [PubMed] [Google Scholar]
- 3.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blyn, L. B., B. A. Braaten, C. A. White-Ziegler, D. H. Rolfson, and D. A. Low. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braaten, B. A., X. Nou, L. S. Kaltenbach, and D. A. Low. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577-588. [DOI] [PubMed] [Google Scholar]
- 6.Brickman, T. J., and M. A. McIntosh. 1992. Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem. 267:12350-12355. [PubMed] [Google Scholar]
- 7.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caimano, M. J., C. H. Eggers, K. R. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 72:6433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban, M. 1976. Transposition and fusion of the lac genes to selected promoters in E. coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 10.Chapman, T. A., X. Y. Wu, I. Barchia, K. A. Bettelheim, S. Driesen, D. Trott, M. Wilson, and J. J. Chin. 2006. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ Microbiol. 72:4782-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. A., and J. F. Miller. 1998. In vivo and ex vivo regulation of bacterial virulence gene expression. Curr. Opin. Microbiol. 1:17-26. [DOI] [PubMed] [Google Scholar]
- 12.Cumberlidge, A. G., and K. Isono. 1979. Ribosomal protein modification in Escherichia coli. I. A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J. Mol. Biol 131:169-189. [DOI] [PubMed] [Google Scholar]
- 13.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 14.Dorman, C. J., and N. N. Bhriain. 1992. Thermal regulation of fimA, the Escherichia coli gene coding for the type 1 fimbrial subunit protein. FEMS Microbiol. Lett. 78:125-130. [DOI] [PubMed] [Google Scholar]
- 15.Durant, L., A. Metais, C. Soulama-Mouze, J. M. Genevard, X. Nassif, and S. Escaich. 2006. Identification of candidates for a subunit vaccine against extraintestinal pathogenic Escherichia coli. Infect. Immun. 75:1916-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Earhart, C. F. 1996. Uptake and metabolism of iron and molybdenum, p. 1075-1090. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 17.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 18.Edwards, R. A., and D. M. Schifferli. 1997. Differential regulation of fasA and fasH expression of Escherichia coli 987P fimbriae by environmental cues. Mol. Microbiol. 25:797-809. [DOI] [PubMed] [Google Scholar]
- 19.Falconi, M., V. McGovern, C. Gualerzi, D. Hillyard, and N. P. Higgins. 1991. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 3:615-625. [PubMed] [Google Scholar]
- 20.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Göransson, M., K. Forsman, and B. E. Uhlin. 1989. Regulatory genes in the thermoregulation of Escherichia coli pili gene transcription. Genes Dev. 3:123-130. [DOI] [PubMed] [Google Scholar]
- 22.Göransson, M., B. Sonden, P. Nilsson, B. Dagberg, K. Forsman, K. Emanuelsson, and B. E. Uhlin. 1990. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature 344:682-685. [DOI] [PubMed] [Google Scholar]
- 23.Göransson, M., and B. E. Uhlin. 1984. Environmental temperature regulates transcription of a virulence pili operon in E. coli. EMBO J. 3:2885-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, Y., D. Zhou, X. Pang, Y. Song, L. Zhang, J. Bao, Z. Tong, J. Wang, Z. Guo, J. Zhai, Z. Du, X. Wang, X. Zhang, J. Wang, P. Huang, and R. Yang. 2004. Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol. Immunol. 48:791-805. [DOI] [PubMed] [Google Scholar]
- 25.Hantke, K., and V. Braun. 2000. The art of keeping low and high iron concentrations in balance, p. 275-288. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 26.Jordi, B. J., B. Dagberg, L. A. de Haan, A. M. Hamers, B. A. van der Zeijst, W. Gaastra, and B. E. Uhlin. 1992. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 11:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karjalainen, T. K., D. G. Evans, D. J. Evans, Jr., D. Y. Graham, and C. H. Lee. 1991. Iron represses the expression of CFA/I fimbriae of enterotoxigenic E. coli. Microb. Pathog. 11:317-323. [DOI] [PubMed] [Google Scholar]
- 28.Krabbe, M., N. Weyand, and D. Low. 2000. Environmental control of pilus gene expression, p. 305-322. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 29.Lafontaine, E. R., and P. A. Sokol. 1998. Effects of iron and temperature on expression of the Pseudomonas aeruginosa tolQRA genes: role of the ferric uptake regulator. J. Bacteriol. 180:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leveille, S., M. Caza, J. R. Johnson, C. Clabots, M. Sabri, and C. M. Dozois. 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 74:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, P. Q., and G. F. Ames. 1998. In vitro disassembly and reassembly of an ABC transporter, the histidine permease. Proc. Natl. Acad. Sci. USA 95:3495-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 33.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1996. Environmental regulation of virulence gene expression in Escherichia, Salmonella, and Shigella spp., p. 2803-2816. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 34.Martinez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153-166. [DOI] [PubMed] [Google Scholar]
- 35.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 36.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratories, Cold Spring Harbor, NY.
- 38.Motin, V. L., A. M. Georgescu, J. P. Fitch, P. P. Gu, D. O. Nelson, S. L. Mabery, J. B. Garnham, B. A. Sokhansanj, L. L. Ott, M. A. Coleman, J. M. Elliott, L. M. Kegelmeyer, A. J. Wyrobek, T. R. Slezak, R. R. Brubaker, and E. Garcia. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, L. K., T. Mackenzie, D. J. Pickard, and G. Dougan. 1986. Effects of immune colostrum on the expression of a K88 plasmid encoded determinant: Role of plasmid stability and influence of phenotypic expression of K88 fimbriae. J. Gen. Microbiol. 132:2497-2503. [DOI] [PubMed] [Google Scholar]
- 40.Nasser, W., M. Faelen, N. Hugouvieux-Cotte-Pattat, and S. Reverchon. 2001. Role of the nucleoid-associated protein H-NS in the synthesis of virulence factors in the phytopathogenic bacterium Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 14:10-20. [DOI] [PubMed] [Google Scholar]
- 41.Nasser, W., and S. Reverchon. 2002. H-NS-dependent activation of pectate lyases synthesis in the phytopathogenic bacterium Erwinia chrysanthemi is mediated by the PecT repressor. Mol. Microbiol. 43:733-748. [DOI] [PubMed] [Google Scholar]
- 42.Nikaido, H., and E. Y. Rosenberg. 1990. Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J. Bacteriol. 172:1361-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Normark, S., D. Lark, R. Hull, M. Norgren, M. Bœga, P. O'Hanley, G. Schoolnik, and S. Falkow. 1983. Genetics of digalactoside-binding adhesion from a uropathogenic Escherchia coli. Infect. Immun. 41:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Hanley, P., D. A. Low, I. Romero, D. Lark, K. Vosti, S. Falkow, and G. Schoolnik. 1985. Gal-Gal binding and hemolysin phenotypes and genotypes associated with uropathogenic Escherichia coli. N. Engl. J. Med. 313:414-420. [DOI] [PubMed] [Google Scholar]
- 45.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 48.Pilsl, H., D. Smajs, and V. Braun. 1999. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J. Bacteriol. 181:3578-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 51.Redford, P., P. L. Roesch, and R. A. Welch. 2003. DegS is necessary for virulence and is among extraintestinal Escherichia coli genes induced in murine peritonitis. Infect. Immun. 71:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roos, V., and P. Klemm. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect. Immun. 74:3565-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo, T. A., U. B. Carlino, A. Mong, and S. T. Jodush. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 67:5306-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, T. J. Barnard, and J. R. Johnson. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmoll, T., M. Ott, B. Oudega, and J. Hacker. 1990. Use of a wild-type gene fusion to determine the influence of environmental conditions on expression of the S fimbrial adhesin in an Escherichia coli pathogen. J. Bacteriol. 172:5103-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serina, S., F. Nozza, G. Nicastro, F. Faggioni, H. Mottl, G. Deho, and A. Polissi. 2004. Scanning the Escherichia coli chromosome by random transposon mutagenesis and multiple phenotypic screening. Res. Microbiol. 155:692-701. [DOI] [PubMed] [Google Scholar]
- 58.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratories, Cold Spring Harbor, NY.
- 59.Sirko, A., M. Zatyka, E. Sadowy, and D. Hulanicka. 1995. Sulfate and thiosulfate transport in Escherichia coli K-12: evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J. Bacteriol. 177:4134-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steele, M., K. Ziebell, Y. Zhang, A. Benson, P. Konczy, R. Johnson, and V. Gannon. 2006. Identification of Escherichia coli O157:H7 genomic regions conserved in genotypes associated with human infection. Appl. Environ. Microbiol. [DOI] [PMC free article] [PubMed]
- 63.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 64.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ussery, D. W., J. C. D. Hinton, B. J. A. M. Jordi, P. E. Granum, A. Seirafi, R. J. Stephen, A. E. Tupper, G. Berridge, J. M. Sidebotham, and C. F. Higgins. 1994. The chromatin-associated protein H-NS. Biochimie 76:968-980. [DOI] [PubMed] [Google Scholar]
- 67.van der Woude, M. W., L. S. Kaltenbach, and D. A. Low. 1995. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol. Microbiol. 17:303-312. [DOI] [PubMed] [Google Scholar]
- 68.Weinstein, D. L., R. K. Holmes, and A. D. O'Brien. 1988. Effects of iron and temperature on Shiga-like toxin I production by Escherichia coli. Infect. Immun. 56:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White-Ziegler, C. A., M. L. Angus Hill, B. A. Braaten, M. W. van der Woude, and D. A. Low. 1998. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 28:1121-1137. [DOI] [PubMed] [Google Scholar]
- 71.White-Ziegler, C. A., A. M. Black, S. H. Eliades, S. Young, and K. Porter. 2002. The N-acetyltransferase RimJ responds to environmental stimuli to repress pap fimbrial transcription in Escherichia coli. J. Bacteriol. 184:4334-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White-Ziegler, C. A., L. B. Blyn, B. A. Braaten, and D. A. Low. 1990. Identification of a genetic locus in Escherichia coli involved in the thermoregulation of the pap operon. J. Bacteriol. 172:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.White-Ziegler, C. A., and D. A. Low. 1992. Thermoregulation of the pap operon: evidence for the involvement of RimJ, the N-terminal acetylase of ribosomal protein S5. J. Bacteriol. 174:7003-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White-Ziegler, C. A., A. Villapakkam, K. Ronaszeki, and S. D. Young. 2000. H-NS controls pap and daa fimbrial transcription Escherichia coli in response to multiple environmental cues. J. Bacteriol. 182:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175-185. [DOI] [PubMed] [Google Scholar]