Abstract
The RimM protein has been implicated in the maturation of the 30S ribosomal subunit. It binds to ribosomal protein S19, located in the head domain of the 30S subunit. Multiple sequence alignments predicted that RimM possesses two domains in its N- and C-terminal regions. In the present study, we have produced Thermus thermophilus RimM in both the full-length form (162 residues) and its N-terminal fragment, spanning residues 1 to 85, as soluble proteins in Escherichia coli and have performed structural analyses by nuclear magnetic resonance spectroscopy. Residues 1 to 80 of the RimM protein fold into a single structural domain adopting a six-stranded β-barrel fold. On the other hand, the C-terminal region of RimM (residues 81 to 162) is partly folded in solution. Analyses of 1H-15N heteronuclear single quantum correlation spectra revealed that a wide range of residues in the C-terminal region, as well as the residues in the vicinity of a hydrophobic patch in the N-terminal domain, were dramatically affected upon complex formation with ribosomal protein S19.
Ribosomes are large ribonucleoprotein particles consisting of two individual subunits: small (30S) and large (50S) subunits in prokaryotes. The 30S subunit is composed of 16S rRNA and more than 20 ribosomal proteins (r-proteins) and plays a crucial role in both decoding of mRNA and translation fidelity. Recent crystallographic studies revealed the complicated architectures of ribonucleoprotein complexes (32). However, the assembly process of these particles remains to be elucidated. For example, in vitro reconstitution experiments showed that the self-assembly of rRNA and proteins requires a high temperature, high ionic strength, and a long incubation time compared to the physiological conditions (30, 37). This implies that ribosome maturation in vivo is mediated by additional nonribosomal factors. Actually, several nonribosomal factors which are responsible for the assembly of ribosomal particles have been characterized for bacteria (1, 8). Their specific and transient interactions with nascent pre-rRNA and ribosomal proteins are necessary for the assembly of the ribosomal particles. Therefore, these ribosome assembly factors in bacteria could be novel antibacterial drug targets (8).
The RimM protein was first identified as one of the components of the trmD operon of Escherichia coli (6). It is widely conserved among bacteria, and RimM-related proteins have also been found in at least four eukaryotic species: the malaria parasites Plasmodium falciparum and Plasmodium yoelii, the malaria mosquito Anopheles gambiae, and the chloroplast of the plant Arabidopsis thaliana (26). The RimM protein reportedly associated only with the free 30S subunit and not with the 30S subunit incorporated in the 70S ribosome (4). An unprocessed precursor of 16S rRNA, 17S rRNA, was accumulated in an E. coli rimM disruptant (5). These results suggest that the RimM protein plays an important role in the maturation of the 30S ribosomal particle.
Previous genetic approaches suggested that the RimM protein is involved in the maturation of a specific region, composed of helices 31 and 33b of 16S rRNA, as well as r-proteins S13 and S19, in the head domain of the 30S subunit (26). The results of a glutathione S-transferase pull-down assay also revealed that the RimM protein binds to r-protein S19 (26). Furthermore, on the basis of the “assembly order map” and “kinetics order map” for the maturation of the 30S particles (13, 17, 28, 33), in which r-proteins are classified as early, middle, middle-late, and late binders, r-proteins S2, S13, and S19 are included among the late binders for the assembly of the head of the 30S subunit (16). The binding of r-protein S19 with helix 33b of 16S rRNA causes conformational changes in the 3′ major domain of 16S rRNA (20). Therefore, the RimM protein seems to be involved in the binding of r-protein S19 to 16S rRNA and to play an important role in the maturation of the head domain of the 30S subunit (26).
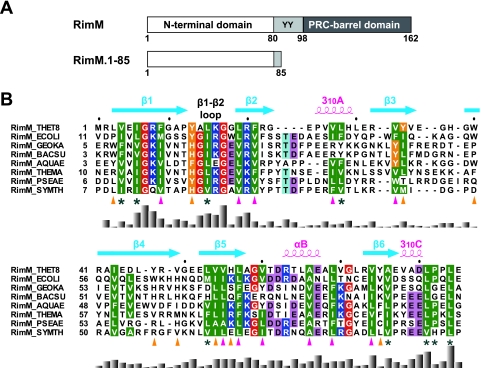
Recently the RimM protein was predicted to possess two domains, based on multiple sequence alignments (Fig. 1A): the RimM N-terminal domain (Pfam accession number PF01782) and the PRC-barrel domain in the C-terminal region (3). In many cases, the PRC-barrel domain, which is named for the H subunit of the photosynthetic reaction center, has been suggested to mediate specific interactions through multiple surfaces in the assembly of complexes (3). Meanwhile, the RimM N-terminal domain is unique among the PRC-barrel-containing proteins, and the N-terminal domain of RimM was predicted to include a β-strand-rich structure (3). Recently the crystal structure of P. aeruginosa RimM was deposited in the Protein Data Bank (PDB) (identifier, 2F1L), which confirmed that the N-terminal domain is composed of β-strands. However, it is still not clear which domain is responsible for the binding to r-protein S19. Thus, more structural information has to be accumulated for the RimM protein.
FIG. 1.
(A) Predicted domain structure of the bacterial RimM proteins (top) and the RimM fragment used for our structural and biochemical studies (bottom). Residue numbers are for the RimM protein from T. thermophilus. The two conserved tyrosine residues are indicated by the one-letter amino acid code. (B) Amino acid sequence alignment of residues 1 to 85 in T. thermophilus RimM from different bacterial species. The positions of secondary structure elements, as observed in RimM.1-85 from T. thermophilus, are shown above the sequence. The side chains of the conserved hydrophobic residues involved in the core and on the side surfaces of the β-barrel are marked below with magenta or orange triangles, respectively. Conserved but solvent-exposed hydrophobic residues are marked with asterisks. Conserved glycine, hydrophobic, and aromatic residues are shown in red, green, or orange, respectively. Positively and negatively charged conserved residues are highlighted in blue or pink, respectively. The multiple sequence alignment was performed using ClustalX (7) and was adjusted manually to align the structurally significant residues. The histogram below the sequence indicates degrees of similarity. Species abbreviations: THET8, Thermus thermophilus HB8; SYMTH, Symbiobacterium thermophilum; AQUAE, Aquifex aeolicus; THEMA, Thermotoga maritima; GEOKA, Geobacillus kaustophilus; BACSU, Bacillus subtilis; ECOLI, Escherichia coli; PSEAE, Pseudomonas aeruginosa.
Here we report the structural features of the RimM N-terminal domain (residues 1 to 80) from Thermus thermophilus and full-length RimM in solution. The tertiary structures within the N-terminal domain of T. thermophilus RimM were similar to those of P. aeruginosa RimM. We also analyzed the interactions between the RimM protein and r-protein S19, based on a structural characterization of the RimM protein by 1H-15N heteronuclear single quantum correlation (HSQC) spectra.
MATERIALS AND METHODS
Expression and purification.
The cDNA fragments encoding full-length RimM (TTHA1033) and truncated RimM.1-85 (residues 1 to 85; hereafter referred to as RimM.1-85) were amplified by PCR from the Thermus thermophilus HB8 genome and were ligated into the expression vector pET11b (Novagen). E. coli strain BL21(DE3) cells harboring the expression vector were grown at 37°C in M9 minimal medium containing 15NH4Cl and/or uniformly 13C-labeled glucose. The expression of RimM.1-85 and full-length RimM was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside when the culture attained an A600 value of about 0.7, and the cells were cultivated at 18°C for 23 h for RimM.1-85 and 20 h for full-length RimM. To purify the overexpressed RimM.1-85 and full-length RimM, the cells were lysed via sonication in 20 mM Tris-HCl buffer (pH 8.0) containing 1 M NaCl and 1 mM EDTA and were centrifuged at 15,000 × g for 15 min. The supernatants were heated at 70°C for 10 min to denature the E. coli proteins and were centrifuged at 15,000 × g for 15 min. After dialysis against 20 mM sodium phosphate buffer (pH 6.5), each solution was loaded onto a HiTrap Q HP column (GE Healthcare), which was washed with the same buffer. The proteins were eluted with a linear gradient of 1 M NaCl (0 to 100%). The fractions containing each protein were collected and subsequently purified by a series of column chromatography steps, including RESOURCE S and RESOURCE Q columns (GE Healthcare), using the same elution conditions as those with the HiTrap Q HP column. Finally, the fractions containing RimM.1-85 and full-length RimM were subjected to gel filtration column chromatography, using Superdex 75 HR10/30 (GE Healthcare), and the molecular weights of the purified proteins were estimated with a Low Molecular Weight Gel Filtration Calibration kit (GE Healthcare). The molecular weights were also confirmed by comparison to molecular weight standards (TEFCO) in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The fractions containing purified 13C/15N-labeled RimM.1-85 and 13C/15N-labeled full-length RimM were pooled, and the buffers were exchanged to 20 mM sodium phosphate buffer (pH 6.5).
The T. thermophilus HB8 rpsS gene (TTHA1688) was cloned into the expression vector pET11a by the RIKEN Structural Genomics/Proteomics Initiative (39), and the recombinant r-protein S19 (hereafter termed S19) was expressed in E. coli strain BL21(DE3). The cells were cultivated at 37°C. When the culture attained an A600 of 0.6, protein expression was induced with isopropyl-β-d-thiogalactopyranoside (0.5 mM). After 4 h of induction, the cells were harvested by centrifugation. The purification procedure for S19 was basically the same as that for full-length RimM, using HiTrap Q HP, RESOURCE S, and Superdex 75 HR10/30 columns (GE Healthcare).
For the nuclear magnetic resonance (NMR) studies of the interaction between 13C/15N-labeled full-length RimM and unlabeled S19, we copurified the two proteins. The cells producing each protein were mixed so that the amount of S19 would be greater than that of full-length RimM, and then they were lysed via sonication in 20 mM HEPES buffer (pH 7.0) containing 300 mM NaCl. The purification procedure of the complex was performed using HiTrap SP HP and RESOURCE S columns. Finally, the buffers of the complex (∼60 μM in total) and full-length RimM in the free state (∼0.2 mM) were exchanged into 20 mM HEPES buffer (pH 7.0) containing 300 mM NaCl and 90% H2O-10% 2H2O, respectively.
NMR spectroscopy and resonance assignments.
The RimM.1-85 protein was prepared in 20 mM sodium phosphate buffer (pH 6.5) in 90% H2O-10% 2H2O, and NMR data were collected for the resonance assignments at 25°C, using a Bruker AVANCE 600-MHz NMR spectrometer equipped with a cryogenic triple-resonance probe. Backbone and side chain resonance assignments were achieved by two-dimensional (2D) 1H-15N and 1H-13C HSQC and standard triple-resonance experiments, including HNCACB, CBCA(CO)NH, HNCO, H(CACO)CANH, HNCA, HN(CO)CA, C(CO)NH, HBHA(CO)NH, H(CCO)NH, HCCH correlation spectroscopy (COSY), and HCCH and CCH total correlation spectroscopy (TOCSY) experiments. Distance restraints were derived from three-dimensional (3D) 15N-separated and 13C-separated nuclear Overhauser effect spectroscopy (NOESY)-HSQC spectra on a Bruker AVANCE 800-MHz spectrometer, with a mixing time of 80 ms. All NMR data were processed using the NMRPipe software system (10) and were analyzed using the Olivia software (M. Yokochi, S. Sekiguchi, and F. Inagaki, Hokkaido University, Sapporo, Japan) and KUJIRA, a program suite for interactive NMR analysis (N. Kobayashi et al., unpublished program), working with NMRView (21).
In order to perform the structural analyses of full-length RimM, uniformly 13C/15N-labeled full-length RimM proteins were purified and subjected to NMR structure analyses at pH 6.5. The standard double- and triple-resonance NMR experiments for the resonance assignments and the NOESY experiments were performed at 45°C. The chemical shifts of 1H, 15N, and 13C were assigned. A 2D 1H-15N HSQC experiment was performed at 25°C for comparison with that of RimM.1-85.
For the NMR studies of full-length RimM with and without S19, NMR data were collected for 13C/15N-labeled full-length RimM, using Bruker AVANCE 600- and 800-MHz NMR spectrometers equipped with cryogenic triple-resonance probes at 45°C. Backbone resonance assignments of free full-length RimM were achieved by 2D 1H-15N HSQC, 3D HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HNCA, HN(CO)CA, and C(CO)NH experiments. For the complex of full-length RimM and S19, backbone resonance assignments were performed using 2D 1H-15N HSQC, 3D HNCA, HN(CO)CA, and 15N-separated NOESY-HSQC experiments.
In the case of the NMR studies of RimM.1-85 with and without S19, 0.2 mM 13C/15N-labeled RimM.1-85 and 1:1 and 1:2 molar mixtures with unlabeled S19 were exchanged into 100 mM phosphate buffer (pH 7.1) containing 90% H2O-10% 2H2O. For each sample, a 2D 1H-15N HSQC spectrum was acquired at a probe temperature of 45°C.
Structure calculations.
Structure calculations of RimM.1-85 were performed using the program CYANA 2.0.17 (14, 15, 18), by means of 40,000 torsion angle dynamics steps. In each structure calculation cycle, 100 randomized starting structures were generated, and the best 20 conformers were selected. Dihedral angles derived from the program TALOS (9) were also used for the φ and ψ torsion angle restraints. Stereospecific assignments of methylene protons, as well as χ1 and χ2 torsion angle restraints (classified as 60° ± 20°, 180° ± 20°, and −60° ± 20°) were obtained from the analyses based on the NOESY spectra and the HNHB and HN(CO)HB experiments. Hydrogen bond restraints within secondary structures were established from the nuclear Overhauser effect (NOE) patterns and the proximities of the donor and acceptor groups in the initial structures, calculated primarily with the NOE-derived restraints. Four distance restraints between H—O (1.7 Å to 2.2 Å), H—C (2.6 Å to 3.5 Å), N—O (2.6 Å to 3.3 Å), and N—C (3.6 Å to 4.6 Å) of the amide and carbonyl groups were used for the final calculation of the RimM.1-85 structure. The quality of each obtained structure was analyzed by using the MOLMOL program (23) and was assessed using the PROCHECK-NMR (25) software. Detailed experimental data and structural statistics are summarized in Table 1. Molecular images and ribbon diagrams were drawn with MOLSCRIPT and Raster3D (24, 27). The structural similarity search was performed using the DALI server (www.ebi.ac.uk/dali/index.html). The sequence alignments were generated with ClustalX (7).
TABLE 1.
Structural statistics for the 20 structures of RimM.1-85
| Parameter | Value |
|---|---|
| No. of NOE distance constraints | |
| Total exptl restraints | 1,550 |
| Intraresidue | 404 |
| Sequential (|i − j| = 1) | 385 |
| Medium range (1 < |i − j| < 5) | 189 |
| Long range (|i − j| ≥ 5) | 572 |
| No. of dihedral angle restraintsa | |
| φ angles | 48 |
| ψ angles | 52 |
| χ angles | 49 |
| No. of hydrogen bond restraints | 22 |
| RMSDs for residues 3-77, Å | |
| Backbone RMSD to mean | 0.18 ± 0.04 |
| Heavy atom RMSD to mean | 0.60 ± 0.04 |
| Avg of CYANA target function value (Å2) | 0.02 |
| No. of restraint violations | |
| Distance restraint (>0.10 Å) | 0 |
| van der Waals (>0.20 Å) | 0 |
| Dihedral angle restraint (>5.0°) | 0 |
| Ramachandran plotb | |
| Residues in most-favored regions (%) | 73.4 |
| Residues in additional allowed regions (%) | 26.6 |
| Residues in generously allowed regions (%) | 0.0 |
| Residues in disallowed regions (%) | 0.0 |
φ and ψ angles are derived from the program TALOS, based on the 13Cα, 13Cβ, 13CO, 1Hα, and 15N chemical shifts. The χ angles contain 34 χ1 and 15 χ2 angle restraints.
From PROCHECK-NMR for 20 models. All residues in RimM.1-85 were used.
NMR dynamics.
For the NMR dynamics studies, 15N R1, 15N R2, and steady-state 1H-15N NOE measurements were recorded on a 600-MHz NMR spectrometer, using the standard method (11). The 15N R1 values were derived from eight 1H-15N spectra with different delays: 5, 65, 145, 246, 366, 527, 757, and 1,148 ms. Similarly, the 15N R2 values were derived from 1H-15N spectra with eight different delays: 32, 48, 64, 80, 96, 112, 128, and 144 ms. The 1H-15N NOE measurement was performed with and without 1H saturation during relaxation delays (1 s for RimM.1-85 and 3 s for full-length RimM) for the NOE and reference experiments, respectively. The R1 and R2 values were extracted by a curve-fitting subroutine included in the program SPARKY (T. D. Goddard and D. G. Kneeler, University of California, San Francisco). The 1H-15N NOE values were obtained by recording spectra with and without 1H saturation and by calculating the ratios of the peak intensities. The NOE errors were estimated using the root-mean-square value of the background noise (11).
Size exclusion chromatography.
Size exclusion chromatography was performed on a Superdex 75 10/30 column (GE Healthcare) at 0.3 ml/min/fraction with 100 mM sodium phosphate buffer (pH 7.1). The elution profiles were recorded by determining the A280 values. The molecular weights of fractions 2 and 4 were estimated by comparison with the 25-kDa (chymotrypsinogen A) and 13.7-kDa (RNase A) markers in the Low Molecular Weight Gel Filtration Calibration kit (GE Healthcare).
Protein structure accession numbers.
The final coordinates of the 20 lowest-energy structures of RimM.1-85 have been deposited in the PDB under the accession number 2DOG. The chemical shift assignments of RimM.1-85 and full-length RimM in the free and S19-bound states have been deposited in the BioMagResBank (BMRB) under the accession numbers 10138, 10139, and 10140, respectively.
RESULTS AND DISCUSSION
The construct of the N-terminal domain of RimM.
The full-length RimM protein from T. thermophilus is composed of 162 amino acid residues and includes the PRC-barrel structure, predicted from our sequence comparison, in the C-terminal region (residues 98 to 162) (Fig. 1A) (3). Simultaneously, based on our sequence alignment of the N-terminal regions of the bacterial RimM proteins with the Pfam sequence PF01782, the region spanning residues 1 to 80 of T. thermophilus RimM is predicted to be the RimM N-terminal domain (Fig. 1).
For the structural study, we designed two constructs of T. thermophilus RimM: residues 1 to 83 and 1 to 85. As for the protein spanning residues 1 to 83, the sample was unstable and aggregated during the final step of sampling for NMR measurement. In contrast, the protein spanning residues 1 to 85 was produced as a soluble monomer, as confirmed by size exclusion chromatography (see Materials and Methods). Therefore, the construct comprising residues 1 to 85 (referred to as RimM.1-85) was selected for the NMR analysis.
Resonance assignments and structure determination of RimM.1-85.
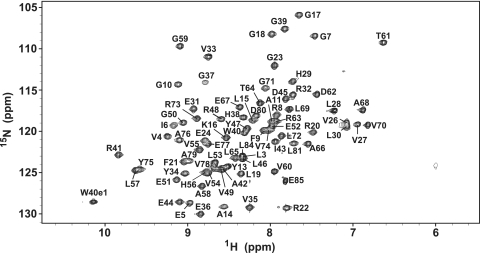
To elucidate the tertiary structure of RimM.1-85, standard 2 and 3D NMR experiments were performed, using approximately 1.2 mM of uniformly 13C/15N-labeled RimM.1-85. As indicated in Fig. 2, the 1H-15N HSQC spectrum showed sharp, well-dispersed NH signals for 98% of all nonproline residues. The chemical shifts of 1H, 15N, and 13C were assigned using standard heteronuclear NMR methods (35). The resonance assignments for the backbone amide 1H and 15N nuclei were completed, except for those of the two N-terminal residues (M1 and R2). For the side chains, more than 95% of the expected 1H and 13C chemical shifts were assigned.
FIG. 2.
1H-15N HSQC spectrum of RimM.1-85, recorded at 25°C on a 600-MHz spectrometer. Signals are labeled with the residue number and the one-letter amino acid code.
The structure calculation of RimM.1-85 was performed by using the program CYANA (14, 15, 18) based on NOE-derived distance restraints, dihedral angle constraints, and hydrogen bond information (Table 1). The backbone dihedral angle constraints were generated by the TALOS program (9) based on the 13Cα, 13Cβ, 13CO, 1Hα, and 15N chemical shifts. The χ angle constraints were obtained by analyzing the intensities of NOEs derived from NOESY spectra and by the HNHB, HN(CO)HB experiments. The strong dαN(i, i + 1) and weak dNN(i, i + 1) NOE connectivities were confirmed in β-strand conformations (β1, L3-P12; β2, L19-G23; β3, R32-V35; β4, G39-V49; β5, E52-L57; β6, R73-E77). The medium dNN(i, i + 1), dαN(i, i + 3), dαβ(i, i + 3), and/or dαN(i, i + 2) NOE connectivities were observed for most of the residues in helical conformations (310A, P25-H29; αB, R63-A68; 310C, V78-D80). Table 1 presents an overview of the statistics of the RimM.1-85 structure with the root-mean-square deviations (RMSD), indicating that the RimM.1-85 structure is well defined throughout the sequence.
Overall structure of the RimM N-terminal domain.
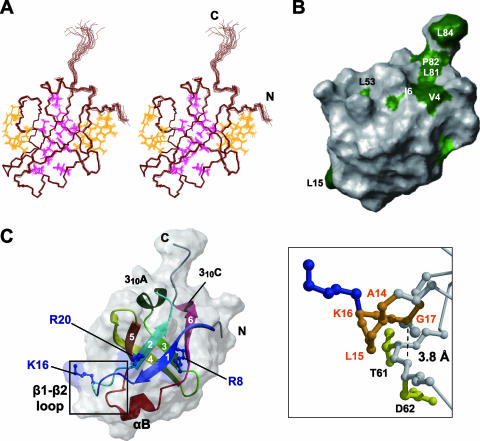
The overall structure of the predicted N-terminal domain (residues 1 to 80) within RimM.1-85 adopted a closed β-barrel fold, composed of six antiparallel β-strands in the order β1-β2-β5-β4-β3-β6-β1, with three short helices (Fig. 3). Residues 81 to 85 did not contain any secondary structure elements. The short 310A helix between β2-β3 and the short αB helix between β5-β6 were located on the top and the bottom of the β-barrel fold, respectively. As shown in Fig. 1B, the bacterial RimM proteins possess highly conserved, hydrophobic residues. In the tertiary structure of RimM.1-85, the side chains of most of the conserved, hydrophobic residues were buried in the molecule and participated in forming the hydrophobic core (Fig. 1B and Fig. 3A). Simultaneously, some conserved, hydrophobic residues were located on the side surfaces of the β-barrel, and they were intimately packed together (Fig. 1B and Fig. 3A). These interior and exterior hydrophobic residues stabilize the compact β-barrel structure of RimM.1-85. Other conserved, hydrophobic residues (e.g., V4, I6, L81, P82, and L84) resided on the surface of RimM.1-85, as shown in Fig. 3B. The side chains of these five residues were exposed to the solvent and formed a hydrophobic patch (Fig. 1B and 3B). Furthermore, we found another hydrophobic pocket in the space between the C terminus of β4 (D45-V49) and the 310A helix (P25-H29).
FIG. 3.
Tertiary structure of RimM.1-85. (A) Stereo view illustrating a trace of the backbone atoms for the ensemble of the 20 structures with the lowest CYANA target function of RimM.1-85. Interior and exterior side chains that stabilize the RimM.1-85 structure are shown in magenta and orange, respectively. The colors of these side chains correspond to those of the triangles in Fig. 1B. (B) Mapping of hydrophobic residues on the molecular surface of RimM.1-85, generated by the MOLMOL (23) program. This structure is rotated by 60° around the y axis, as in panel A. Conserved hydrophobic residues among bacterial RimM proteins are colored green, and conserved but exposed hydrophobic residues are dark green. A large number of conserved hydrophobic residues are invisible, suggesting that they are deeply buried in the core. (C) Ribbon representation of the RimM.1-85 structure (left) in the same orientation as in panel A. The strands in the β-sheet are indicated by arrows, and the secondary structure elements are labeled. The side chains of the three conserved basic residues (blue) are linearly arranged on the surface of RimM.1-85. Right, enlarged view of the framed area of the left panel. The highly conserved segment in the β1-β2 loop, which has been regarded as the GXXG motif, is represented as an orange ball-and-stick model. Some of the residues relevant to the discussion are labeled. The broken line represents the distance between the α-carbon atom of G17 and the nitrogen atom of D62.
Characteristic amino acid residues in RimM.1-85.
Sequence alignments of the regions corresponding to RimM.1-85 among various bacterial species revealed other conserved residues in its N-terminal portion in addition to the hydrophobic ones (Fig. 1B). Three basic residues (R8, K16, and R20 in T. thermophilus) and the highly conserved sequence from Y13 to G17 are shown in Fig. 3C. The side chains of the three basic residues were exposed to the solvent and were linearly arranged on the surface of RimM.1-85. It is tempting to speculate that these basic residues could contribute to interactions with targets, such as nucleotides. However, a previous mutational analysis of E. coli RimM indicated that alanine substitutions of two of these residues (corresponding to R8 and K16 in T. thermophilus) had no effect on the growth rate of the cells (26). The sequence from Y13 to G17 (β1-β2 loop) contains the highly conserved GXXG segment (corresponding to A14XXG17 in T. thermophilus), as shown in Fig. 1B. This conserved segment in the RimM proteins is suggested to be the “GXXG motif” (4), which is generally found in a well-known RNA binding motif, the KH domain. The GXXG segment in the β1-β2 loop adopted a β hairpin structure in RimM.1-85 (Fig. 3C), although the GXXG motif forms the turn structure connecting two α-helices in both the type I and type II KH domains (12). In the case of E. coli RimM, the substitution of the second glycine in the GXXG segment (G27R of E. coli corresponds to G17 in T. thermophilus) with a bulky amino acid severely impaired the stability and affected the function of the RimM protein (26). The present structure shows that G17 in the β1-β2 loop is very close to the residues T61 and D62, with a distance of approximately 3.8 Å between the α-carbon atom of G17 and the nitrogen atom of D62 (Fig. 3C). Therefore, the mutation would disrupt the tertiary structure of the RimM N-terminal domain because of its bulky side chain, abolishing the function of the RimM protein.
Structural comparison of RimM.1-85 with other proteins.
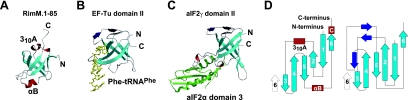
Searches for structurally related proteins in the PDB, using the program DALI (19), revealed that the structure of RimM.1-85 belongs to the reductase/isomerase/elongation factor fold (hereafter abbreviated as the RIEF fold) superfamily. As shown in Fig. 4, the RIEF fold contains six β-strands arranged to form a β-barrel and is involved in the recognition of its own target molecule (e.g., protein, RNA, or compound). There are two main interaction modes with the target molecules in their complex structures: one is via the side surface of their β-barrels (domain II of EF-Tu [31] and Gar1, which is one of the accessory proteins of Cbf5 [34]) and the other is via the bottom of the β-barrels (domain II of archaeal aIF2γ [38] and the fMet-tRNAfMet-binding domain of IF2 [2]) (Fig. 4). In the RimM.1-85 structure, however, the corresponding side chains on the side surface were packed too closely together to interact with another molecule (Fig. 3A), while the corresponding residues of domain II of EF-Tu (Fig. 4) and Gar1 are accessible for molecular interactions with tRNA and protein, respectively (31, 34).
FIG. 4.
Comparison of interfaces in RIEF fold proteins. The β-sheets in the RIEF folds are shown in cyan and are depicted in the same orientation as the RimM.1-85 structure shown in Fig. 3C. The target nucleotides and proteins are partially displayed and are colored yellow and green, respectively. The characteristic structural elements in translation proteins are colored navy. (A) RimM.1-85. (B) Domain II of T. aquaticus EF-Tu with Phe-tRNAPhe (PDB identifier 1TTT) (31). (C) Domain II of archaeal aIF2γ with domain 3 of aIF2α (PDB identifier 2AHO) (37). (D) Topology diagrams of RimM.1-85 (left) and translation proteins (right). The six common strands are cyan, and the dissimilar secondary structures, namely helices and strands, are red and navy, respectively.
Considering the interaction modes via the top and the bottom of the β-barrel, the characteristic structural features were found in RimM.1-85 compared to other translation proteins. In the other translation proteins, the bottom of the β-barrel is open for the molecular interaction, but the top is completely covered by characteristic structural elements (three β-strands or extended loops), as shown in Fig. 4 (2, 38). On the other hand, in the case of RimM.1-85, the areas at the top and the bottom of the β-barrel were not completely open due to the presence of helices (310A and αB), and only narrow spaces were found along these helices. A small but distinct hydrophobic patch existed at the top of the β-barrel of the RimM.1-85 structure (Fig. 3B). Thus, RimM.1-85 has the potential to interact with small molecules, such as peptides, at the top and/or the bottom of the β-barrel.
Structural features of the full-length RimM protein.
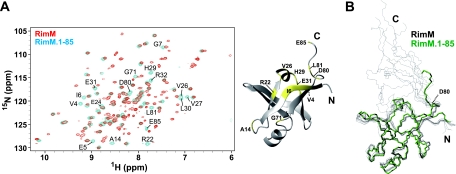
For the N-terminal domain of full-length RimM, the resonance assignments for the backbone and the side chains were completed, as for the RimM.1-85 construct (see Materials and Methods). In the 1H-15N HSQC spectra of full-length RimM and RimM.1-85, recorded at 25°C, most of the resonances originating from the N-terminal domain were observed at the same positions, whereas some resonances from the N-terminal domain of full-length RimM were slightly shifted compared to those for RimM.1-85 itself (Fig. 5A). Considering the spatial disposition of these residues in the calculated 20 conformers of full-length RimM (Fig. 5B), these chemical shift differences could be interpreted as resulting from the existence of a following flexible region (residues 81 to 162). Notably, the tertiary structure composed of residues 1 to 80 was maintained, even in full-length RimM (Fig. 5B). Thus, we conclude that the N-terminal domain of full-length RimM, spanning residues 1 to 80, is structurally independent of the rest of the protein, and it corresponds exactly to the RimM N-terminal domain predicted by multiple alignments.
FIG. 5.
Superposition of full-length RimM and RimM.1-85. (A) Left, 1H-15N HSQC spectra of full-length RimM (red) and RimM.1-85 (blue), recorded at 25°C on a 600-MHz spectrometer. Most of the resonances originating from the N-terminal domain were overlapped with each other. Only the shifted resonances for residues 1 to 85 of full-length RimM, compared to those for RimM.1-85 itself, are labeled with the residue number and the one-letter amino acid code: V4, E5, I6, G7, A14, R22, E24, V26, V27, H29, L30, E31, R32, G71, D80, L81, and E85. Right, mapping of these residues (yellow) on the RimM.1-85 structure. (B) Superposition of the backbones of the 10 best structures of full-length RimM (black) and the best structure of RimM.1-85 (green). The 10 structures of full-length RimM were calculated with CYANA 1.0.8. The structure of the C-terminal region (residues 96 to 162) is not displayed, because it does not adopt a rigid tertiary structure.
Most of the resonances for the backbone and the side chains of the C-terminal region in full-length RimM (residues 81 to 162) were assigned, except for some 1H-15N resonances at pH 7.0 (Y90, Y91, F92, L113, G116, A117, Q118, D119, V120, E128, L140, Q141, and G150). Among the identified residues in the C-terminal region, several residues from two parts (96 to 124 and 144 to 155) exhibited some obvious long-range NOEs. Furthermore, the prediction of the secondary structure elements by TALOS (9) suggested that approximately two-thirds of these residues adopt β-strand conformations (data not shown). Actually, in the structure of full-length RimM in solution, the C-terminal region seemed to possess β-sheet structures within two parts (residues 96 to 124 and 144 to 155). On the other hand, the central part (residues 125 to 143) of the C-terminal region exhibited very few long- or medium-range NOEs and adopted an unfolded structure despite the fact that residues 136 to 138 were also predicted to form a short β-strand. These findings indicate that the 3D structure of the C-terminal region was partly folded in solution, in contrast to the compact and stable structure of the N-terminal domain.
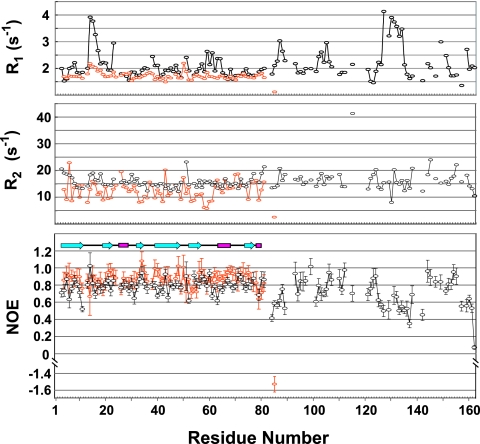
To confirm the result mentioned above, the measurements of the 15N R1, 15N R2, and steady-state 1H-15N NOE for full-length RimM and RimM.1-85 were performed at 45°C and 25°C, respectively (Fig. 6). Overlapped resonances and those with a very poor signal-to-noise ratio were excluded from relaxation analyses. The average NOE values observed for the N-terminal domain were ∼0.87 and ∼0.79 for RimM.1-85 and full-length RimM, respectively. These values indicate that the N-terminal domain is tightly packed in solution, independent of the C-terminal region (residues 81 to 162). By contrast, the NOE values observed for residues 84 to 89, 125 to 142, and 157 to 162 of full-length RimM were significantly lower than the average value of ∼0.75 for residues 1 to 162, indicating high mobility of the peptide backbone. Especially, the R1 value of L130 was higher than the average, but its R2 value was lower than the average (Fig. 6), which is characteristic of internal dynamics on the pico-to-nanosecond timescale (22). Therefore, residues 125 to 142 are involved in the central unfolded part in solution, as indicated above. The residues 84 to 89 and 157 to 162 seem to be involved in the linker between N- and C-terminal domains and the C-terminal tail, respectively. As a consequence, in the C-terminal region of full-length RimM, there are some β-strands in two parts (residues 96 to 124 and 144 to 155), and the other parts are unfolded in solution.
FIG. 6.
NMR dynamic studies for full-length RimM and RimM.1-85. 15N R1 (top) and 15N R2 (middle) relaxation rates and steady-state 1H-15N NOE data (bottom) are shown for full-length RimM (black) and RimM.1-85 (orange), respectively. β-Strands, helices, and loops are indicated schematically.
Before the completion of this work, the crystal structure of Pseudomonas aeruginosa RimM was deposited in the PDB (identifier 2F1L) by the Joint Center for Structural Genomics. A structure comparison using the DALI server revealed that the N-terminal domain of P. aeruginosa RimM possesses the same fold as that of T. thermophilus RimM (Z-score of 13.0 and an RMSD of 2.0 Å over 85 residues). The only notable difference was observed in the length and axial direction of the 310A helix, which apparently arose from the gap between the amino acid sequences of the two species (Fig. 1B). As for the C-terminal structure of P. aeruginosa RimM, it folded into a PRC-barrel structure. The residues in the central unfolded part (residues 125 to 142) of the C-terminal region in solution were composed of two loops and one short β-strand in the crystal structure (residues 138 to 154 in P. aeruginosa). In the crystal structure, several conserved hydrophobic residues are clustered around L149 on one surface of the PRC-barrel structure, and the cluster contacts the N-terminal domain of the symmetry-related molecule. By contrast, the corresponding residue, L137 in T. thermophilus, is located in the central unfolded part of the C-terminal region in solution, as indicated above. Therefore, this hydrophobic contact due to crystal packing may have stabilized the folding of the PRC-barrel in the crystal structure.
Interactions of ribosomal protein S19 with full-length RimM and RimM.1-85.
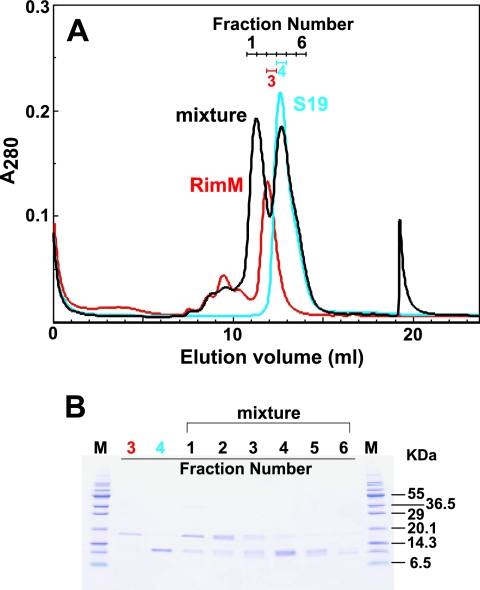
A previous glutathione S-transferase pull-down experiment indicated that the RimM protein associates with r-protein S19 (referred to as S19), as described above (26). Based on this report, we examined the ability of our purified full-length RimM to bind to S19 by size exclusion chromatography (Fig. 7). Prior to the chromatography, an excess of S19 was added to full-length RimM, to achieve a molar ratio of 2:1. This mixture was then loaded on a gel filtration column (Superdex 75 HR10/30). The retention times of full-length RimM (18.1 kDa), S19 (10.6 kDa), and the complex (28.7 kDa) were different from each other (Fig. 7), indicating that the RimM protein used in this study had S19 binding activity. When equimolar amounts of the proteins were mixed, the peak of the complex was broad but was observed at the same retention time (data not shown). Subsequently we attempted to investigate the association modes of full-length RimM with S19, using NMR analyses. Unfortunately, the gradual or equimolar addition of S19 to full-length RimM resulted in turbidity within a few days, and the analysis by SDS-polyacrylamide gel electrophoresis revealed that S19 and full-length RimM had precipitated. Therefore, we tried copurification of S19 and full-length RimM and succeeded in obtaining a complex suitable for NMR studies, as described in Materials and Methods. Due to its low solubility (∼60 μM in total), the complex could not be adequately concentrated for NMR data collection to solve the complex structure. However, this complex provided homogeneous 1H-15N HSQC spectra and was stable at 45°C for 1 month. The 1H-15N HSQC spectra of full-length RimM in the presence or absence of S19 are shown in the left panel of Fig. 8A. The resonance assignments of full-length RimM complexed with S19 were performed for the backbone atoms. Although some 1H-15N resonances were not assigned at pH 7.0 (M1, R2, I6, G37, G50, E51, D114, L121, L130, V138, E149, and I157), more residues in the C-terminal region could be assigned than was the case for those in the free state.
FIG. 7.
Complex formation between full-length RimM and S19. (A) Size exclusion chromatography, performed using a Superdex 75 10/30 column. The mixture of full-length RimM and S19 in the molar ratio of 1:2 is shown by the black line. Each chromatography step with full-length RimM (20 nmol; red) and S19 (40 nmol; cyan) was performed under the same conditions. Fraction numbers are indicated in the panel with the corresponding colors. (B) SDS-polyacrylamide gel electrophoresis of fractions collected from size exclusion chromatography in panel A. Fraction numbers are indicated at the top: fractions 3 (red number), 4 (cyan), and 1 to 6 (black) correspond to full-length RimM, S19, and their mixture, respectively. Molecular weight standards are displayed as “M.”
FIG. 8.
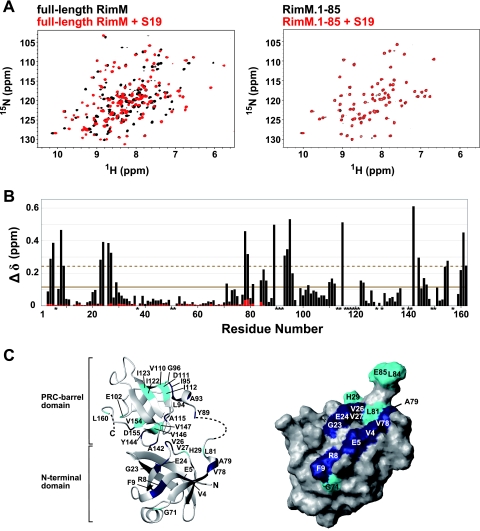
NMR studies of full-length RimM and RimM.1-85 with S19. (A) 1H-15N HSQC spectra of free full-length RimM (black) and the complex with S19 (red) obtained at 45°C (left) and spectra of free RimM.1-85 (black) and its 1:1 molar mixture with S19 (red) monitored at 45°C (right). (B) Chemical shift changes of full-length RimM (black bars) and RimM.1-85 (red bars) observed by the addition of S19. The chemical shift change, Δδ, was determined as follows: Δδ = [(ΔδHN)2 + (ΔδN/6.5)2]1/2 (29), where ΔδHN and ΔδN are the chemical shift differences for HN and 15N, respectively. The mean value is shown by a continuous line; the mean value plus 1 standard deviation is shown by a dashed line. Asterisks indicate residues with 1H-15N resonances that were not assigned in the S19-free or S19-bound form of full-length RimM. (C) Mapping of perturbed residues on the homology-modeled structure of full-length RimM (left) and the surface of RimM.1-85, which is presented in the same orientation as in Fig. 3B (right). The 3D model of full-length RimM from T. thermophilus was obtained using the homology modeling approach with the SWISS-MODEL Protein Modeling Server (http://swissmodel.expasy.org/) (36) on the basis of the crystal structure of P. aeruginosa RimM. Disordered regions in the crystal structure are depicted by a dashed line. Residues with chemical shift changes above average are cyan, and residues with chemical shift changes above the mean value plus 1 standard deviation are navy.
A comparison of the spectra revealed dramatic chemical shift changes (Δδ) for the resonances originating from residues 4 to 9, 23 to 27, and 78 to 162 (Fig. 8B). There are well-conserved, consecutive aromatic residues, corresponding to Y89Y90 in T. thermophilus (3, 4). Alanine substitutions (YY to AA) revealed that these residues are crucial for binding to S19 in the 30S subunit (26). Actually, the 1H-15N resonance of Y89 was dramatically shifted upon S19 binding (Fig. 8B), although we could not assign that of Y90. We then mapped the chemical shift changes of full-length RimM with and without S19 (Fig. 8C, left panel) on a homology-modeled structure generated from SWISS-MODEL (36). In the C-terminal region, the affected residues upon complex formation were spatially dispersed. Conceivably, the chemical shift differences for the residues in the C-terminal region (residues 81 to 162) reflected not only the direct association with S19 but also the internal conformational change upon S19 binding. In the N-terminal domain, however, some resonances originating from residues 4 to 9 and 23 to 27 within full-length RimM were clearly affected by the addition of S19 (Fig. 8B). Mapping of these residues on the surface of RimM.1-85 revealed that they were localized within the N-terminal domain (Fig. 8C, right panel). Thus, for the N-terminal domain, the shifts of the resonances indicate the interaction between these residues and S19. Interestingly, these residues are located in the vicinity of the hydrophobic patch described above (Fig. 3B).
To confirm whether the RimM N-terminal domain alone associates with S19, 1H-15N HSQC spectra for RimM.1-85 were obtained in the absence and presence of S19 with 1:1 (Fig. 8A, right panel) and with 1:2 molar mixtures. The calculated Δδ values represented negligible chemical shift changes (Fig. 8B): the average Δδ value of the 1:1 molar mixture was 0.005 (between 0.0007 and 0.04 of A79), and that of the 1:2 molar mixture was 0.007 (between 0.0001 and 0.05 of A79). This indicates that the RimM N-terminal domain alone could not tightly interact with S19.
Thus, we concluded that the residues in the vicinity of the N-terminal hydrophobic patch are involved in the interaction with S19 in the presence of the following C-terminal region. Considering that the RimM protein would participate in multiple interactions during the maturation of 30S ribosomal particles, it could be a target for new antibiotics. Therefore, it is important to clarify the functional role of the RimM protein in ribosome maturation more precisely. The present structural information will assist in promoting further investigations of the RimM protein.
Acknowledgments
This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analysis, Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank Naohiro Kobayashi for providing the KUJIRA software. We thank Takashi Nagata for technical assistance. We thank Roland Schmucki for critical reading of the manuscript and suggestions.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Alix, J. H. 1993. Extrinsic factors in ribosome assembly, p. 173-184. In K. H. Nierhaus, F. Franceschi, and A. R. Subramanian (ed.), The translational apparatus: structure, function, regulation, evolution. Plenum, New York, NY.
- 2.Allen, G. S., A. Zavialov, R. Gursky, M. Ehrenberg, and J. Frank. 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121:703-712. [DOI] [PubMed] [Google Scholar]
- 3.Anantharaman, V., and L. Aravind. 2002. The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism. Genome Biol. 3:RESEARCH0061.1-RESEARCH0061.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bylund, G. O., B. C. Persson, L. A. Lundberg, and P. M. Wikström. 1997. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J. Bacteriol. 179:4567-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bylund, G. O., L. C. Wipemo, L. A. Lundberg, and P. M. Wikström. 1998. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 180:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byström, A. S., K. J. Hjalmarsson, P. M. Wikström, and G. R. Björk. 1983. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 2:899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comartin, D. J., and E. D. Brown. 2006. Non-ribosomal factors in ribosome subunit assembly are emerging targets for new antibacterial drugs. Curr. Opin. Pharmacol. 6:453-458. [DOI] [PubMed] [Google Scholar]
- 9.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289-302. [DOI] [PubMed] [Google Scholar]
- 10.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 11.Farrow, N. A., R. Muhandiram, A. U. Singer, S. M. Pascal, C. M. Kay, G. Gish, S. E. Shoelson, T. Pawson, J. D. Forman-Kay, and L. E. Kay. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984-6003. [DOI] [PubMed] [Google Scholar]
- 12.Grishin, N. V. 2001. KH domain: one motif, two folds. Nucleic Acids Res. 29:638-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grondek, J. F., and G. M. Culver. 2004. Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch. RNA 10:1861-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Güntert, P. 2004. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278:353-378. [DOI] [PubMed] [Google Scholar]
- 15.Güntert, P., C. Mumenthaler, and K. Wüthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283-298. [DOI] [PubMed] [Google Scholar]
- 16.Hamacher, K., J. Trylska, and J. A. McCammon. 2006. Dependency map of proteins in the small ribosomal subunit. PLoS Comput. Biol. 2:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Held, W. A., B. Ballou, S. Mizushima, and M. Nomura. 1974. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 249:3103-3111. [PubMed] [Google Scholar]
- 18.Herrmann, T., P. Güntert, and K. Wüthrich. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319:209-227. [DOI] [PubMed] [Google Scholar]
- 19.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, K. L., and G. M. Culver. 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat. Struct. Mol. Biol. 11:179-186. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, B. A. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278:313-352. [DOI] [PubMed] [Google Scholar]
- 22.Kay, L. E., D. A. Torchia, and A. Bax. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28:8972-8979. [DOI] [PubMed] [Google Scholar]
- 23.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51-55. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 25.Laskowski, R. A., J. A. Rullmann, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 26.Lövgren, J. M., G. O. Bylund, M. K. Srivastava, L. A. Lundberg, O. P. Persson, G. Wingsle, and P. M. Wikström. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA 10:1798-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merritt, E. A., and M. E. Murphy. 1994. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50:869-873. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima, S., and M. Nomura. 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226:1214. [DOI] [PubMed] [Google Scholar]
- 29.Mulder, F. A., D. Schipper, R. Bott, and R. Boelens. 1999. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292:111-123. [DOI] [PubMed] [Google Scholar]
- 30.Nierhaus, K. H. 1991. The assembly of prokaryotic ribosomes. Biochimie 73:739-755. [DOI] [PubMed] [Google Scholar]
- 31.Nissen, P., M. Kjeldgaard, S. Thirup, G. Polekhina, L. Reshetnikova, B. F. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 32.Noller, H. F. 2005. RNA structure: reading the ribosome. Science 309:1508-1514. [DOI] [PubMed] [Google Scholar]
- 33.Powers, T., G. Daubresse, and H. F. Noller. 1993. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 232:362-374. [DOI] [PubMed] [Google Scholar]
- 34.Rashid, R., B. Liang, D. L. Baker, O. A. Youssef, Y. He, K. Phipps, R. M. Terns, M. P. Terns, and H. Li. 2006. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell 21:249-260. [DOI] [PubMed] [Google Scholar]
- 35.Sattler, M., J. Schleucher, and C. Griesinger. 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magnet. Reson. Spectrosc. 34:93-158. [Google Scholar]
- 36.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traub, P., and M. Nomura. 1968. Structure and function of Escherichia coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. USA 59:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yatime, L., Y. Mechulam, S. Blanquet, and E. Schmitt. 2006. Structural switch of the γ subunit in an archaeal aIF2αγ heterodimer. Structure 14:119-128. [DOI] [PubMed] [Google Scholar]
- 39.Yokoyama, S., H. Hirota, T. Kigawa, T. Yabuki, M. Shirouzu, T. Terada, Y. Ito, Y. Matsuo, Y. Kuroda, Y. Nishimura, Y. Kyogoku, K. Miki, R. Masui, and S. Kuramitsu. 2000. Structural genomics projects in Japan. Nat. Struct. Biol. 7:943-945. [DOI] [PubMed] [Google Scholar]