Abstract
Coordinated regulation of molecular chaperones is an important feature of the bacterial stress response. The small molecular chaperone gene acr2 of Mycobacterium tuberculosis is activated by exposure to several stresses, including heat and the detergent sodium dodecyl sulfate (SDS). In this study, we show that acr2 is directly regulated by the MprAB two-component system, and that MprAB has both positive and negative effects on acr2 expression. mRNA analyses showed that acr2 expression levels were lower under SDS stress and control conditions but higher under heat shock in an mprAB deletion mutant than they were in the parental strain. Parental expression patterns were restored in an mprAB-complemented strain. Western blotting using an anti-Acr2 antibody showed that Acr2 protein synthesis correlated with mRNA levels. Primer extension identified one transcriptional start point (TSP) for acr2 in all three strains under control and stress conditions. Electrophoresis mobility shift assays revealed multiple MprA binding sites in the acr2 promoter, including one downstream and three upstream of the acr2 TSP, with one overlapping the binding sites predicted for SigE, SigH, and HspR. DNA footprinting confirmed that MprA protected large sections of the acr2 promoter region. Expression of several housekeeping genes under SDS stress also was evaluated, revealing the upregulation of large molecular chaperone genes and, unexpectedly, sigA, with slightly lower sigA mRNA levels detected in the mprAB deletion mutant than in the wild type. In contrast to Acr2, SigA protein synthesis did not correlate with mRNA expression. Overall, the data indicated that MprA has complex interactions with the acr2 promoter and indirect effects on major housekeeping genes.
Mycobacterial species inhabit diverse environments, ranging from aquifers to macrophages (8, 17, 41), and survival in these often harsh habitats must involve the coordinated regulation of genes involved in the stress response. For the intracellular pathogen Mycobacterium tuberculosis, there has been considerable interest in the role of heat shock proteins (molecular chaperones) in stress survival (42, 58), in their impact on the immune response (19, 32, 43, 55), and in the factors that regulate their expression (10, 44).
The M. tuberculosis genome encodes two small heat shock proteins belonging to the α-crystallin family, Acr1 and Acr2 (7, 44, 51). The encoding genes, acr1 and acr2, are expressed following infection of macrophages and mice (39, 42, 55), and both proteins have chaperone activity in vitro (6, 47, 58). acr2 (hsp, hrpA, Rv0251c) is also the most highly induced gene in M. tuberculosis following heat shock at 45°C (44). The 18-kDa Acr2 protein has been detected in the ribosomal fractions of heat-stressed mycobacteria and is, therefore, also referred to as HrpA, for heat-stress-induced ribosome binding protein A (28). It has been proposed that Acr2 may stabilize the 30S subunit of the ribosome at high temperatures and, thereby, assist in translation initiation (28). acr2 also is induced by exposure to the detergent sodium dodecyl sulfate (SDS), starvation conditions, and oxidative stress produced by exposure to diamide or hydrogen peroxide (23, 39).
The available evidence indicates that the regulation of acr2 is multifactorial. It is downregulated by the heat shock repressor protein HspR, and a potential HspR binding site has been identified in the acr2 promoter (44). acr2 also is repressed, directly or indirectly, by the PhoPR two-component system (TCS) during exponential growth in broth culture (54). SigH is the major regulator of the responses to heat shock and oxidative stress and may directly regulate acr2 following exposure to these stresses (9, 22, 33). Under SDS stress, activation of acr2 is SigE dependent (23), and as SigE also is upregulated by heat shock and oxidative stress (21, 22, 33, 56), it may contribute to acr2 regulation under these conditions.
We (30) and others (13) have shown that, under SDS stress, sigE is regulated by the MprAB TCS, which consists of the response regulator MprA and the histidine kinase MprB. Although the mprAB operon is itself regulated by SigE (23), many genes activated by SDS exposure are downregulated in the absence of mprA, including sigE and acr2 (13, 30). MprA binds directly to the sigE promoter, and MprA binding sites have been detected in the promoters of sigB, pepD, and mprA (13, 14, 30), all members of the SigE regulon (23). As part of our ongoing studies on the role of MprAB in the regulation of stress-associated genes, we have determined that MprA directly regulates acr2 and that the interactions of MprA with the acr2 promoter are complex. Moreover, depending on the stress condition, MprAB can have either positive or negative effects on acr2 expression. Unexpectedly, during the course of these studies we found that the major housekeeping sigma factor gene, sigA, is activated by SDS stress and that MprAB may contribute to this activation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis strain Rv-D981 is an mprAB deletion mutant of the laboratory strain H37Rv, and it lacks a 1.1-kbp region encoding the predicted DNA binding domain of MprA and the N terminus of MprB, including a portion of the kinase domain (30). The mprAB-complemented strain, Rv-D981C, was generated by inserting mprAB under the control of the mprA promoter into the genome of Rv-D981 (30). M. tuberculosis mutants and parental strain H37Rv were grown at 37°C under normal atmospheric conditions in either Middlebrook 7H9 broth containing 0.05% Tween 80 or Middlebrook 7H10 agar (Difco), both enriched with 10% oleic acid-albumin-dextrose-catalase (Difco). Broth cultures were incubated with gentle shaking. Escherichia coli Novablue and Rosetta(DE3)pLysS (Novagen) were used as host strains for general cloning and gene expression, respectively. E. coli strains were grown on L agar or in L broth. Antibiotics were added to growth media as required.
Expression and purification of MprA.
The 690-bp predicted coding region of mprA was cloned into pBEn-SBP-SET1a (Stratagene), a Variflex expression vector containing an N-terminal streptavidin binding peptide tag and a solubility enhancement tag. The resulting plasmid, pSTH20, was used to transform E. coli Rosetta(DE3)pLysS (Novagen). Expression of mprA was induced by the addition of isopropyl-β-d-thiogalactopyranoside, and then bacteria were collected by centrifugation and resuspended in streptavidin binding buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl). Following sonication, lysates were centrifuged, and the tagged MprA was recovered from the supernatant using streptavidin agarose (Novagen) according to the manufacturer's instructions. Peptide tags were removed using the Thrombin CleanCleave kit (Sigma), and removal was verified by gel electrophoresis of the MprA protein before and after cleavage.
Electrophoresis mobility shift assays (EMSAs).
To analyze binding of MprA to the acr2 promoter, DNA regions were amplified using the following primer pairs: Acr2GST-F1/Acr2GST-R1, Acr2GST-F2/Acr2-R2, Acr2GST-F2/Acr2GST-R1, Acr2GST-F1/Acr2GST-R2, Acr2GST-F3/Acr2GST-R1, Acr2GST-F4/Acr2GST-R1, Acr2GST-F5/Acr2GST-R1, Acr2GST-F6/Acr2GST-R1, Acr2GST-F3/Acr2GST-R1, Acr2GST-F3/Acr2GST-R3, Acr2GST-F3/Acr2GST-R4, and Acr2GST-F3/Acr2GST-R5 (see the table in the supplemental material). PCR products were gel purified, end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega), and then separated from free isotope by Quick Spin column (Roche) filtration according to the manufacturer's instructions. Labeled DNA fragments were incubated with MprA using procedures described previously (37). For competition assays, a 100-, 200-, or 400-fold excess (in nanograms) of unlabeled competitor DNA was included. Reaction mixtures were loaded onto 5 to 6% nondenaturing polyacrylamide gels and were electrophoresed for 2 to 3 h at 140 V at 4°C in 0.5× Tris-Borate-EDTA buffer. A 278-bp fragment that lies upstream of the mprA promoter and that does not bind MprA was used as a negative control in competition assays.
Stress treatment and RNA isolation.
Cultures were grown to log phase (optical density at 600 nm of 0.3 to 0.4) and then were exposed to 0.05% SDS, 0.1% deoxycholate, 10 mM hydrogen peroxide, and 5 mM diamide or were left unexposed. RNA was extracted from control and stress-treated samples after 90 min of incubation and shaking at 37°C. For heat stress, cultures were divided into two aliquots, which were incubated for 1 h in either a 37°C water bath (control) or a 45°C water bath without shaking. Total RNA was isolated using TRIzol LS reagent (Invitrogen) according to the manufacturer's instructions, except that lysing matrix B and a FastPrep FP120 shaker (BIO 101) were used to disrupt the mycobacteria. Chromosomal DNA was removed with the DNA-free DNase treatment and removal kit (Ambion) according to the manufacturer's instructions.
Reverse transcription and relative quantification of mRNA by real-time PCR.
Total RNA (3 μg) was annealed with 2 μg of random hexamer primers (pdN6; Amersham Pharmacia Biotech, Inc.) and 20 U of RNasin RNase inhibitor (Promega). Following incubation at 65°C for 5 min, reverse transcription was carried out at 37°C for 60 min using 200 U of Moloney monkey leukemia virus reverse transcriptase (Invitrogen), 20 μm deoxynucleoside triphosphates (Roche), and first-strand buffer (Invitrogen). Primers and probes for real-time PCR were designed with PRIMER EXPRESS software (Applied Biosystems), and probes were labeled with 5′-fluorescein phosphoramidite and 3′-6-carboxytetramethylrhodamine. Assays were performed using the ABI Prism 7700 thermal cycler with 25-μl reaction volumes containing 1× TaqMan universal PCR master mix (ABI), 0.3 mM each primer, 0.2 mM probe, and 2.5 μl cDNA or genomic DNA as the template, and the following thermal cycles were carried out: 2 min at 50°C and 10 min at 95°C, followed by 40 repeats of 15 s at 95°C and 1 min at 60°C. Relative quantities of cDNA were determined from standard curves generated by amplification of serial 10-fold dilutions of H37Rv genomic DNA, using the appropriate probe and primers, and were normalized for amounts of 16S rRNA. The following primer/probe sets were used: for acr2, Rv0251QF, Rv0251QR, and Rv0251QP; for 16S rRNA, W16SF, W16SR, and W16SP; and for sigA, sigAQF2, sigAQR2, and sigAQP2 (see the table in the supplemental material).
Primer extension analysis.
Primers Rv0251cPE-1 and Rv0251cPE-2 were used to analyze acr2 transcripts. Primers were labeled with [γ-32P]ATP by T4 polynucleotide kinase as indicated above. For the annealing step, 7 μg of M. tuberculosis RNA was denatured at 90°C for 5 min, snap-cooled on ice, and then incubated for 20 min with 2 pmol of labeled primer in 1× reverse transcription buffer (50 mM Tris-HCl [pH 8.3], 50 mM KCl, 10 mM MgCl, 10 mM dithiothreitol, 1 mM each deoxynucleoside triphosphate, and 0.5 mM spermidine). Annealing temperatures were adjusted for each primer. Primer extension was performed using the primer extension system-avian myeloblastosis virus reverse transcriptase kit (Promega) as directed by the manufacturer, and reaction products were separated using an 8% polyacrylamide sequencing gel. Sequencing reactions performed with the same primer used in primer extensions were used to determine the start sites of the transcripts.
DNase I protection assay (DNA footprinting).
A 345-bp region upstream of acr2 was amplified by PCR using two different primer pairs; amplicons generated using Rv0251cFP-F/Rv0251cFP-R and Rv0251cFPAS-F/Rv0251cFPAS-R were used for analysis of the sense and antisense strand, respectively. Following cloning into pGEM-T-Easy (Promega), inserts were released by BamHI/HpaI digestion and were gel purified, and then the BamHI ends were labeled in an end-filling reaction with Klenow enzyme, [α-32P]dGTP, and other nucleotides. Approximately 40,000 cpm of labeled DNA was incubated with 10 μg of recombinant MprA protein in a reaction buffer containing 20 mM Tris-HCl (pH 7.5), 60 mM KCl, 2 mM EDTA, 4% Ficoll 400, and 0.2 μg/μl of poly(dI-dC). Samples were incubated for 15 min at room temperature, and then 1 U of RQ1 DNase I and DNase I buffer (Promega) was added. After 2 min of incubation at room temperature, reactions were processed essentially as described previously (14). Samples then were loaded onto an 8% denaturing polyacrylamide gel alongside a DNA sequencing ladder prepared with M13mp18 DNA and a size marker.
SDS-PAGE and Western blotting.
Total protein was extracted from broth cultures of M. tuberculosis, using the B-PER kit (Pierce) according to the manufacturer's instructions, except for the additional step of disrupting the mycobacteria with a FastPrep FP120 shaker and lysing matrix B. Protein concentrations were determined using the bicinchoninic acid method (Pierce). Using 10 μg of each protein extract, SDS-polyacrylamide gel electrophoresis (PAGE) was performed with a 12% polyacrylamide gel under reducing conditions essentially as described previously (20). The gel then was electroblotted in Tris-glycine buffer containing 20% methanol onto a nitrocellulose membrane (Trans-blot; Bio-Rad, Hercules, CA). The membrane then was processed as described previously (38) and was incubated with a 1:1,000 dilution of anti-Acr2 antibody (42), kindly provided by G. Stewart (University of Surrey, United Kingdom). After being washed with Tris-buffered saline-Tween, the membrane was incubated with goat anti-rabbit antibody conjugated with horseradish peroxidase (Bio-Rad) at a 1:10,000 dilution and rewashed, and then bands were detected by chemiluminescence using the ECL Western blotting kit (Amersham Pharmacia Biotech). For detection of SigA, the membrane was stripped and reprobed as described previously (38), using a 1:5,000 dilution of monoclonal antibody 2G10 (45) (Neoclone Biotechnology) and then a 1:5,000 dilution of horseradish peroxidase-conjugated goat anti-mouse antibody (Santa Cruz Biotechnology) as the secondary antibody.
RESULTS
Expression patterns of acr2 in an mprAB deletion mutant.
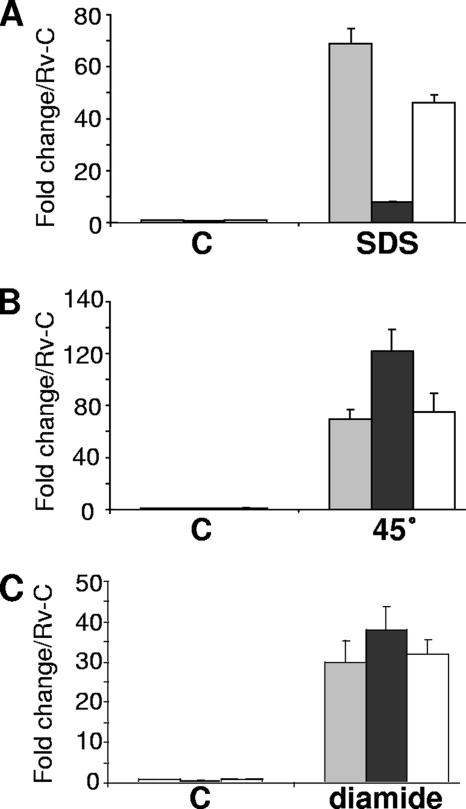
We previously generated the M. tuberculosis mprAB deletion mutant, Rv-D981, from the laboratory strain H37Rv, and, using DNA microarray analysis, we showed that acr2 expression was more highly induced in H37Rv than in Rv-D981 following exposure to the detergent SDS (30). To evaluate more fully the role of MprAB in acr2 expression, we used real-time PCR analyses to compare mRNA levels in M. tuberculosis strains exposed to various stresses. We verified that acr2 was highly upregulated in H37Rv by SDS, but we found that the level of upregulation was nearly 10-fold lower in Rv-D981 than in the wild type (Fig. 1A). In the presence of another detergent, deoxycholate, acr2 was induced approximately fivefold in H37Rv but showed less than twofold induction in Rv-D981 (data not shown). In the mprAB-complemented strain, Rv-D981C (30), acr2 expression patterns were similar to those of H37Rv following exposure to detergent (Fig. 1A and data not shown), indicating that the loss of mprAB was responsible for the observed differences in Rv-D981.
FIG. 1.
Effects of mprAB deletion on acr2 expression under various stresses. Expression of acr2 was determined by real-time PCR using RNA extracted from cultures exposed for 90 min to 0.05% SDS (A), 45°C (B), or 5 mM diamide (C). The C on the horizontal axis in each panel indicates control samples. Results were normalized for 16S RNA content and are shown as the fold change above the level of the H37Rv control (Rv-C), which was given a value of 1. Data are the means ± standard errors of the means from three separate experiments. H37Rv, gray bar; Rv-D981, black bar; Rv-D981C, white bar.
As acr2 is one of the most highly induced genes under heat stress (44), we examined expression at 45°C. acr2 was dramatically upregulated by heat shock in all three strains (Fig. 1B). However, in contrast to detergent stress, acr2 was upregulated more highly in Rv-D981 than in the other two strains, suggesting that when intact, MprAB partially represses acr2 under this condition. acr2 also was induced by exposure to hydrogen peroxide and diamide, by approximately 7-fold and 34-fold, respectively, above control levels, but similar levels of induction were detected in H37Rv and Rv-D981 (Fig. 1C and data not shown), suggesting that MprAB does not impact acr2 expression under these conditions. Interestingly, over multiple stress experiments, the average expression level of acr2 under control conditions was approximately 40% lower in Rv-D981 than in H37Rv (Fig. 1 and data not shown), suggesting that MprAB also contributes to basal expression levels of acr2.
Characterization of the acr2 TSP.
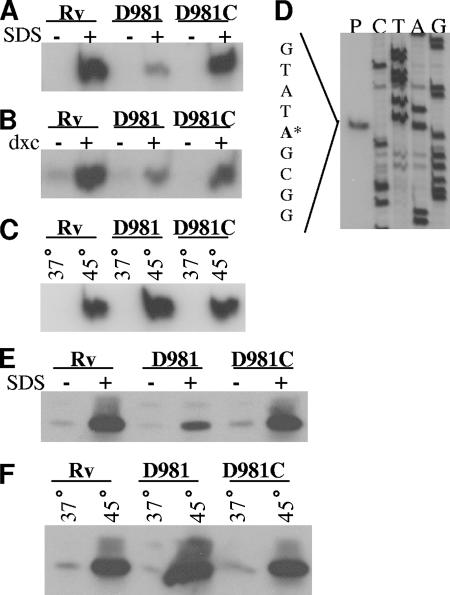
The above data suggested that, under certain stresses, MprA affects transcription of acr2. To confirm these data and to identify the acr2 transcriptional start point (TSP), we performed primer extension analysis using RNA from strains exposed to SDS (Fig. 2A), deoxycholate (Fig. 2B), or heat stress (Fig. 2C). The results again indicated that, following detergent exposure, transcript levels were higher in H37Rv and Rv-D981C than in Rv-D981, but that under heat stress Rv-D981 showed the highest expression levels of acr2.
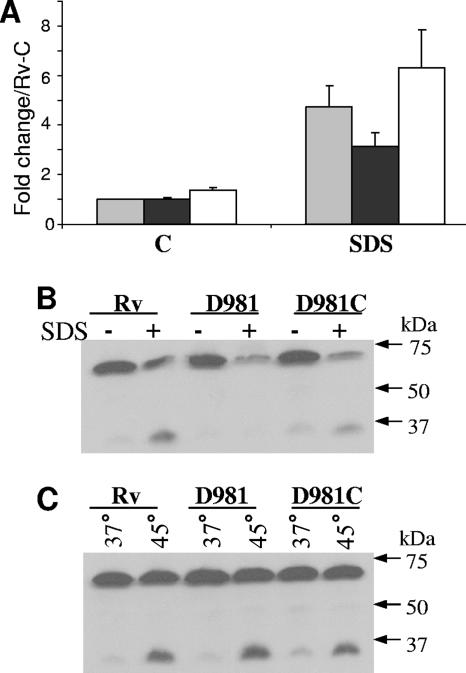
FIG. 2.
Analyses of the acr2 TSP and Acr2 synthesis under stress. (A to D) RNA was extracted from M. tuberculosis cultures, and the 5′ ends of the acr2 transcripts were mapped by primer extension using primer Rv0251cPE-2. Strains were incubated under control conditions or were exposed to 0.05% SDS (A), 0.1% deoxycholate (B), or 45°C (C). Because of the greater band intensity with SDS and heat shock exposures, smaller amounts of RNA were loaded for panels A and C than for panel B. Control bands were visible in panels A and C upon longer exposures. (D) Mapping of the acr2 TSP. The primer extension reaction (lane P) was performed with H37Rv RNA and was electrophoresed alongside a sequence ladder generated by the same primer. The TSP is the adenosine indicated by the asterisk in the sequence to the left of the panel. (E and F) Western blot analyses of Acr2. Cell extracts were prepared from cultures incubated under control conditions or exposed to 0.05% SDS (E) or 45°C (F). Samples of 10 μg protein were loaded. Electrophoresis was performed under reducing conditions. Western blots were prepared and probed with an anti-Acr2 antibody, followed by probing with goat anti-rabbit antibody conjugated with horseradish peroxidase. Rv, H37Rv; D981, Rv-D981; D981C, Rv-D981C. Control samples are indicated by a minus sign or 37°C. Stress samples are indicated by a plus sign or 45°C.
The TSP for acr2 was identified as an adenosine located 58 nucleotides upstream of the predicted start codon (Fig. 2D; also see Fig. 4A). This TSP was detected under all conditions tested and in all three strains (Fig. 2A to D and data not shown), indicating that the site of transcript initiation is not altered by stress and that it is not MprA dependent.
FIG. 4.
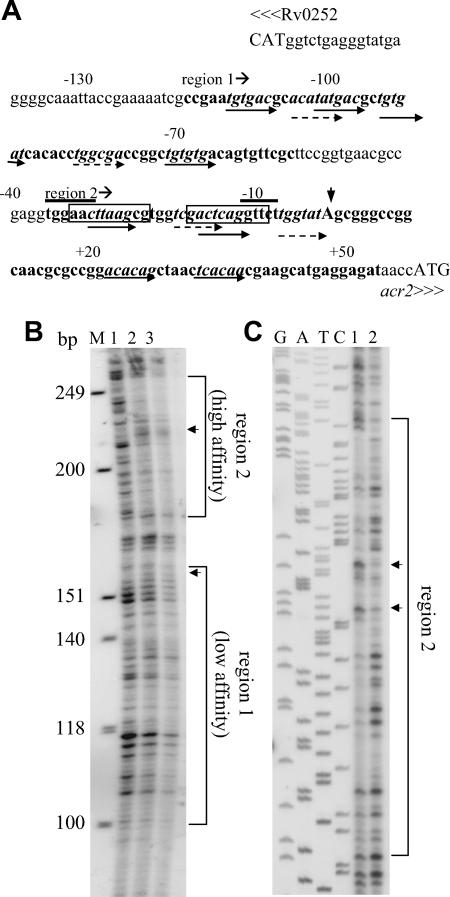
Association of MprA with the acr2 promoter region. (A) Features of the region upstream of acr2 (Rv0251c). Potential MprA binding sites are indicated by italicized sequences and paired arrows. Dashed and solid arrows indicate motif subunits that either share two or share three or more bases, respectively, with the MprA binding consensus TCTCAG. Numbers indicate the positions relative to the acr2 TSP. The overlined sequences are the overlapping −35 and −10 promoter consensus sequences for SigH and SigE (22, 23, 33). The boxed sequences are the predicted HspR binding sites (44). Capital letters indicate start codons. The vertical arrowhead indicates the acr2 TSP. Sequences in boldface are regions protected by MprA in DNA footprinting assays. The start of each region is indicated. (B and C) DNA footprinting analyses of the acr2 promoter region. Protected regions are bracketed. Arrowheads mark areas of DNase I hypersensitivity in the presence of MprA. (B) Analysis of the sense strand showing protected regions 1 and 2. Lane 1, no MprA and 1 U DNase I; lane 2, 2 μg MprA and 1 U of DNase I; lane 3, 8 μg MprA and 1 U of DNase I. M, molecular size markers (HinfI-digested φX174). The positions of the protected regions were verified using a sequence ladder included on the same gel (not shown). (C) Analysis of the antisense strand. Samples were run beside a DNA sequence ladder generated with M13mp18. Both regions were analyzed, but only the high-affinity region (region 2) is shown. Lane 1, 4 μg MprA and 1 U of DNase I; lane 2, no MprA and 1 U of DNase I. Note that the sample loadings in panels B and C are in reverse order.
Expression of Acr2 protein in Rv-D981.
To determine whether the altered expression pattern of acr2 in Rv-D981 was maintained at the protein level, we performed Western blotting. Crude cell lysates were obtained from M. tuberculosis cultures grown under control or stress conditions, and then protein blots were prepared and hybridized with an anti-Acr2 antibody (42). Compared to the levels in H37Rv and Rv-D981C, Acr2 protein levels were reduced in Rv-D981 under control conditions and under SDS stress (Fig. 2E). Under heat stress, Acr2 levels were markedly higher in Rv-D981 than in H37Rv and Rv-D981C (Fig. 2F). These data showed a direct correlation between acr2 mRNA (Fig. 1A and B and 2A and C) and protein levels (Fig. 2E and F).
MprA binds the acr2 promoter.
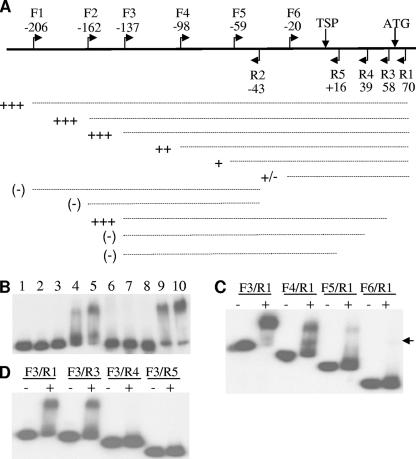
We next performed EMSAs to determine whether MprA had a direct role in regulating acr2. MprA bound a 276-bp fragment from the acr2 promoter, generated using primers F1 and R1 (Fig. 3A and data not shown). A nested series of fragments then was designed to localize the MprA binding site in the acr2 promoter. Removal of 70 bp from the 5′ end did not affect binding, as indicated by shifting of the F3/R1 fragment by MprA (Fig. 3A and B). However, further deletions from the 5′ end gradually reduced binding by MprA (Fig. 3A and C), with the smallest fragment (F6/R1) showing only very weak binding. These results indicated that a region from −137 to −20 bp upstream of the acr2 TSP is involved in MprA binding. However, no shift was observed with the F1/R2 fragment, which overlapped most of this region, indicating that this region alone was not sufficient for MprA binding (Fig. 3A and data not shown).
FIG. 3.
Localization of MprA binding sites in the acr2 promoter region by EMSA. (A) Schematic diagram of the regions flanking the acr2 TSP. Flags mark the location of primers used to generate nested PCR fragments. The numbers indicate the location of forward (F) and reverse (R) primers relative to the TSP. Vertical arrows mark the TSP and start codon. Dashed lines represent PCR probes used in EMSAs with MprA. The binding strength of MprA to each probe is indicated by the number of plus signs beside each dashed line, with +++ indicating the highest level of binding. A minus sign indicates that no binding was detected. (B to D) EMSAs with acr2 promoter probes and MprA. (B) A fixed amount of labeled DNA probe amplified by primers F3/R1 was incubated in reaction mixtures containing no MprA (lane 1); 0.12, 0.24, 0.48, and 0.72 μg MprA (lanes 2, 3, 4, and 5, respectively); 0.72 μg MprA and a 140-, 280-, or 420-fold excess of unlabeled F3/R1 (lanes 6, 7, and 8, respectively); 0.72 μg MprA and a 100- or 200-fold excess of unlabeled control fragment (lanes 9 and 10, respectively). Probes representing 5′ nested deletions (C) and 3′ nested deletions (D) were generated using the indicated primer pairs and were incubated with (+) or without (−) 0.72 μg MprA. The arrowhead in panel C marks the position of a weak band in the last lane.
A second set of deletions was used to evaluate sequences at the 3′ end of the F3/R1 fragment. The F3/R3 fragment, which ends 58 bp downstream of the TSP, bound MprA, but further deletions at the 3′ end abrogated binding (Fig. 3A and D). These deletions indicated that a section downstream of the acr2 TSP, between positions +39 and +58, also was involved in MprA binding. However, this section alone produced only weak binding, as indicated by the faint shift of the overlapping F6/R1 fragment (Fig. 3A and C). Together, the analyses indicated that more than one region of the acr2 promoter was involved in MprA binding and suggested that there may be cooperative binding between MprA bound to different sites. We performed primer extension analyses to determine whether there were other TSPs downstream of the +39 to +58 region, but none were detected (data not shown).
The acr2 promoter region has multiple MprA binding sites.
As the EMSA results suggested that MprA might bind to the acr2 promoter at several locations, we searched the entire region for potential MprA binding motifs. The motif (MprA box) originally was identified as tandem repeats of a conserved octamer with a 3-bp spacer (14). Subsequent analysis of several target promoters suggested that the conserved section was shorter and consisted of tandem repeats of the consensus hexameric sequence TCTCAG, separated by a 5-bp spacer (30), so the regions were examined for this sequence. Several possible MprA binding sites were identified (Fig. 4A), and although no matches identical to the conserved hexamer are present, each of the indicated hexamers contains at least two conserved bases and is separated from its paired hexamer by 5 bp. Furthermore, the locations of the putative MprA binding sites are consistent with the results of the EMSAs. Specifically, the progressive loss of MprA binding resulting from 5′ deletions to positions −98, −59, and −20 (Fig. 3A and C) correlates with the removal of complete or partial MprA binding motifs (Fig. 4A). Similarly, the 3′-end deletion to position +39, which abrogated MprA binding (Fig. 3A and D), ends immediately adjacent to one of the motifs (Fig. 4A) and presumably disrupts binding at this location.
We found two pairs of potential tandem binding sites for MprA in the region between the TSP and position −31 (Fig. 4A). Each of these sites partially overlaps the putative promoter sites for SigE (23) and SigH (22) previously identified in the acr2 promoter, and at least one of the predicted MprA binding sites is functional, as deletion of the −59 to −20 region reduced MprA binding to the acr2 promoter (Fig. 3A and C). Examination of the predicted −35 and −10 promoter sequences indicated that these also overlap the predicted binding site for HspR, which was previously identified on the basis of its similarity to the HAIR (HspR-associated inverted repeat) element (44).
To confirm the location of MprA binding sites in the acr2 promoter, we performed DNA footprinting and discovered that two large regions were protected by MprA (Fig. 4). Region 1 extends from approximately position −120 to −55 upstream of the TSP (Fig. 4A and B and data not shown). Region 2 is about 90 bp long and ends more than 50 bp downstream of the TSP (Fig. 4). Other studies have shown that when a single MprA box is present, MprA protects an overlapping region of only 30 to 40 bp, whereas larger protected regions are indicative of additional binding sites (13, 14). Therefore, the DNA footprinting analyses supported the EMSA results, which had suggested the presence of multiple MprA binding sites in the acr2 promoter region. Both regions had areas that were rendered hypersensitive to DNase I in the presence of MprA (Fig. 4B and C), a feature that has been observed for other promoters and that may result from MprA-induced conformational changes in the DNA structure (13, 14).
Band intensities were lower in region 2 than in region 1 (Fig. 4B), indicating that, compared to that of region 1, it may have higher affinity for MprA and, therefore, greater protection from DNase I cleavage. This is consistent with the EMSA results, which showed that the F3/R4 deletion (Fig. 3A and D), which removed the 3′ end of region 2 (Fig. 4A), had the most dramatic impact on MprA binding. Overall, these data indicate that multiple MprA binding sites are present in the acr2 promoter region, including one downstream of the TSP and another overlapping major regulatory sequences.
Expression of housekeeping genes in Rv-D981.
In view of the effects of MprAB on acr2 expression, we examined our array data (30) to determine whether expression patterns of the large housekeeping molecular chaperone genes dnaK, groEL1, and groEL2 also were altered in Rv-D981. Basal expression levels of groEL2 were significantly lower in Rv-D981 than in H37Rv (Table 1), but we did not detect any binding of MprA to the groEL2 promoter (data not shown), indicating that the effects of MprA are indirect. All three genes were induced by SDS stress, with higher levels of induction detected in Rv-D981, particularly for groEL2. However, final mRNA levels for groEL2 under SDS stress were similar between Rv-D981 and H37Rv, suggesting that the higher induction levels in the mutant under stress were required to compensate for the lower basal levels.
TABLE 1.
Expression pattern of large molecular chaperone genes in Rv-D981 by DNA microarray analyses
| Genea | Mean fold inductionb (±SEM) of first sample vs second sample of cohybridized pair
|
|||
|---|---|---|---|---|
| D981-ctrl/Rv-ctrl | Rv-SDS/Rv-ctrl | D981-SDS/D981-ctrl | D981-SDS/Rv-SDS | |
| dnaK (Rv0350) | −1.7 ± 0.1 | 12.7 ± 4.1* | 19.1 ± 2.8* | 0.58 ± 0.50 |
| groEL2 (Rv0440) | −3.2 ± 0.5* | (5.0 ± 2.2) | 28.1 ± 10.2* | 1.15 ± 0.46 |
| groEL1 (Rv3417c) | (−2.18 ± 0.33) | 4.2 ± 1.1* | 11.0 ± 1.3* | 1.05 ± 0.56 |
Data were obtained from DNA microarrays associated with a previously published work (30). For each cohybridized pair, results show the mean fold induction with three different RNA preparations and dye flips. Rv, H37Rv; D981, Rv-D981; ctrl, control sample; SDS, sample treated with 0.05% SDS for 90 min. Asterisks indicate a significant difference in expression in the first sample compared to that of the second sample of each pair, as determined by one-class significance analyses of microarrays (49). A mean change of at least twofold was considered significant (30). Values in parentheses were not significant by significance analyses of microarrays, as differences of twofold or greater were detected in fewer than five arrays.
While analyzing the array data, we noticed a minor, but consistent, reduction in the expression of the major housekeeping sigma factor gene, sigA, in Rv-D981. Although this difference was not significant, the observation prompted us to analyze sigA expression by real-time PCR. We used a primer/probe set that hybridized in a region at the 5′ end of sigA that had no similarity to the related sigma factor gene sigB. No differences were detected between Rv-D981 and H37Rv under control conditions. However, we observed an increase in sigA mRNA levels under SDS stress (Fig. 5A). In addition, in each separate experiment, the increases were consistently greater in H37Rv and Rv-D981C than in Rv-D981 (Fig. 5A and data not shown). These data were unexpected, as it had been reported that sigA is not induced by SDS stress (21). To confirm our data, we used a second primer/probe set that hybridized near the 3′ end of sigA transcripts. The results were similar to those described above, except that the fold increases under SDS stress were slightly lower (data not shown), indicating either a difference in probe sensitivity or reduced stability of the 3′ end of the transcript compared to that of the 5′ end. We also examined sigA expression under heat stress, but no induction was detected under this stress in any of the three strains (data not shown).
FIG. 5.
Expression pattern of sigA under stress. (A) RNA extracts were prepared and analyzed by real-time PCR, as described in the legend to Fig. 1, using a probe and primers that hybridized within the 5′ end of sigA. Data are the means ± standard errors of the means from three separate experiments. H37Rv, gray bar; Rv-D981, black bar; Rv-D981C, white bar; C, control cultures; SDS, SDS-treated cultures; Rv-C, H37Rv control. (B and C) Western blot analyses of SigA using extracts of samples exposed to 0.05% SDS (B) or 45°C (C). The blots used in experiments depicted in Fig. 2 were stripped and reprobed with an anti-SigA antibody. SigA migrates at ∼70 kDa under the conditions used and is visible as the strong upper band. The lower band is probably SigB, which is ∼36 kDa (2).
The effects of SDS exposure on SigA synthesis were examined using the monoclonal antibody 2G10 to σ70 (45), which we had previously used to analyze M. tuberculosis SigA (57). In sharp contrast to the mRNA profile, SigA levels were lower than control levels in all three strains after SDS exposure (Fig. 5B). Under heat stress, SigA levels were similar to control levels (Fig. 5C) and reflected the mRNA profile (data not shown). The anti-σ70 antibody cross-reacted with a protein of lower molecular size (Fig. 5B and C) that, based on its size and expression pattern, is probably SigB, which has high similarity to SigA (12). Moreover, sigB is induced by SDS and heat stress in H37Rv (21, 23) and has reduced expression in Rv-D981 under control conditions and SDS stress (30), consistent with the pattern observed for the smaller band. Overall, these analyses indicated that sigA can be induced by certain stresses, but that there is not always a direct correlation between sigA mRNA and protein levels. To determine whether MprA directly regulated sigA, we performed EMSA with the sigA promoter but did not detect binding (data not shown), indicating that, as with groEL2, the effects of MprA are indirect.
DISCUSSION
Our analyses indicate that the MprAB TCS directly regulates the small molecular chaperone gene acr2 and has indirect effects on other stress-associated genes. Depending on the stress condition, acr2 expression levels either were decreased or increased in the mprAB deletion mutant Rv-D981 relative to levels in H37Rv, suggesting that MprAB has both positive and negative regulatory effects on acr2. Further analyses indicated the presence of several MprA binding sites in the acr2 promoter region, whereas only one acr2 TSP was detected.
Several lines of evidence suggest that regulation of acr2 is complex. In M. tuberculosis and other actinomycetes, HAIR elements in the promoter of the large molecular chaperone gene hsp70 (dnaK) serve as binding sites for the repressor protein HspR (3, 4, 43, 44), and a HAIR-like sequence was discovered 71 bp upstream of the acr2 start codon (44). The expression pattern of acr2 is consistent with this functioning as a repressor site, as DNA microarray analyses showed that both acr2 and dnaK were derepressed at 37°C in an hspR mutant and had higher expression levels at 45°C (44). In the dnaK promoter, there are two separate HAIR elements, each located a few bases downstream of a TSP (43). In contrast, our analyses place the HAIR-like sequence about nine bases upstream of the acr2 TSP (Fig. 4A). Examination of the predicted −35 and −10 sequences for SigE (23) and SigH (22) in the acr2 promoter indicate that these overlap the HAIR-like sequence (Fig. 4A), so HspR may repress acr2 expression by blocking access to the promoter site.
One of the MprA binding sites overlaps the recognition motifs for SigE, SigH, and HspR (Fig. 4A), and it is likely that MprA and HspR compete for access in this region. Similar to HspR, binding of MprA to this region may repress transcription. However, it also is possible that MprA interacts with RNA polymerase to activate transcription from this site. MprA activates its own transcription (13, 14, 30), and the MprA binding site in the mprA promoter overlaps the −35 region for SigE (30). Our previous studies had suggested that MprA may have different mechanisms of transcriptional activation at different promoters, and in some cases MprA may act as a bridge between the promoter site and the transcriptional machinery (30), similar to the interactions of catabolite activator protein (CAP) at class 2 CAP-dependent promoters (5).
The predicted MprA binding sites at approximately −70 and −100 bp upstream of the TSP probably are activation sites. Their location is similar to the position of the MprA binding site in the sigB promoter, which is activated by MprA under control conditions and SDS stress (30). MprA may activate transcription from these sites through SigE, which also regulates acr2 following SDS exposure (23). The reduction in acr2 expression in Rv-D981 under SDS stress probably results both from the direct loss of MprA activity on the acr2 promoter and from the lower expression levels of sigE in the absence of mprA compared to those of H37Rv (13, 30).
We hypothesize that the MprA binding site at the 3′ end of region 2 is a repressor site, given its location more than 20 bp downstream of the TSP. Moreover, we were unable to detect a downstream TSP that would indicate that MprA activates transcription from this site. As acr2 expression was higher following heat shock in Rv-D981 than in H37Rv, we speculate that MprA may bind to this downstream site and prevent overexpression of acr2 under conditions in which HspR may be inactive. It is unclear, though, why removal of this site abrogated binding to the entire promoter region. This finding suggests that there is cooperative binding between MprA bound to this site and the other locations, similar to the situation detected at the two MprA binding sites in the sigE promoter (13). However, as we predict that MprA functions as an activator at the upstream sites in the acr2 promoter, in contrast to the downstream site, it may be that other transcription factors stabilize MprA at the weaker binding sites upstream. Additional studies are required, though, to clarify the role of each of these sites. It should be noted that none of the predicted MprA binding sites in the acr2 promoter region shows a strong match to the consensus sequence, but similar variation has been observed previously, particularly at the weaker MprA binding sites in the sigE and pepD promoters (13, 14).
The stress-associated signals that activate MprAB are unknown, but given the predicted association of the SigE regulon with envelope integrity (23, 39), we hypothesize that MprB senses changes in surface proteins or other structural elements. The E. coli σE regulon also is predicted to have a role in maintaining envelope integrity and appears to be activated by the interaction of unfolded outer membrane porins with transmembrane sensor proteins, leading to proteolysis of RseA, an anti-sigma factor of σE (1, 25, 53). MprB has two transmembrane domains with an intervening extracellular loop that is required for induction of sigE following detergent exposure (13). We predict that this loop either is activated by misfolded proteins or becomes altered itself under stress, triggering autophosphorylation and then transphosphorylation of MprA. In this case, the signal for induction of the SigE regulon would, therefore, be transmitted via a phosphorylation cascade rather than a proteolytic cascade. Interestingly, E. coli has a second pathway that also responds to misfolded proteins, and this is regulated by a TCS (36), although it is not directly associated with σE. Alternatively, MprB may sense damage to membrane lipids or cell wall components, as reported for other systems (15, 24). Whatever the activating signal may be, the ensuing transphosphorylation of MprA would result in upregulation of sigE, mprA itself, and the remainder of the SigE regulon. Although it is not required for DNA binding in EMSAs, phosphorylation of MprA is necessary for activation of some genes (14), and we presume that this is true for acr2 activation under detergent stress. However, it remains to be determined whether MprA must be phosphorylated to repress acr2 under heat stress.
It is noteworthy that the other α-crystallin gene, acr1 (hspX), also is directly regulated by a TCS. Following exposure to nitric oxide or hypoxic conditions, acr1 is activated by the response regulator DosR (DevR) (29, 31, 40, 52), which is part of the DosRS TCS (34). An 18- to 20-bp palindromic motif in the promoter of acr1 and other hypoxia-induced genes was identified as the DosR binding site (10, 31, 35).
In H37Rv, acr2 was highly induced following stress exposure, supporting data from DNA microarray analyses (23, 39, 44). Other bacterial α-crystallin genes have shown similarly high levels of stress-associated induction, in contrast with the very low levels of synthesis present under normal growth conditions (27). The main role of these proteins is to suppress aggregation of denatured proteins as part of the protein refolding pathway (16, 26, 27, 50), and although for the most part they are nonessential, disruption of α-crystallin genes can influence survival under stress (27). An acr2 deletion mutant of H37Rv was more sensitive than H37Rv to hydrogen peroxide (42) and heat stress (G. Stewart, personal communication). Mice infected with the acr2 mutant had a milder course of disease than mice infected with the parental strain (42), indicating that the gene is important in the disease process.
Although activation of the large molecular chaperone genes by SDS was not unexpected given their established role in the stress response, the upregulation of sigA was surprising. It had been reported that sigA transcripts remain at basal levels under a variety of stress conditions, including SDS exposure (21). A possible explanation for the discrepancy may be differences in the sensitivities of the analysis methods. We noted that the 3′ primer used for quantitative PCR in the original study (21) hybridizes more than 20 bp downstream of sigA, in the 3′-untranslated region of the transcript. We speculate that this region has reduced stability compared to that of the coding region of the transcript, which was reported to be quite stable (18), and therefore increases in mRNA levels have been underestimated. There also may be some differences in stability across the coding region, as we detected lower induction levels with a primer/probe set that hybridized near the 3′ end compared to results with the set that hybridized near the 5′ end of sigA. sigA also was upregulated in M. tuberculosis Beijing strains during infection of a human monocytic cell line (57) and in M. smegmatis during stationary phase (11). sigA transcript levels are sometimes used for normalization in quantitative PCR (13, 46), but these data indicate that caution should be used when choosing this gene as an internal reference. The slight reduction in sigA mRNA levels detected in Rv-D981 under SDS stress suggests that MprA may have indirect effects on regulation of sigA, possibly through SigE or SigB, which have reduced expression in Rv-D981 (30). In contrast to the mRNA levels, SigA protein levels decreased under SDS stress. A similar dichotomy was observed with M. smegmatis during stationary phase (11), and it was proposed that sigA mRNA accumulates so that translation can take place quickly when normal conditions return.
Previous evidence pointed to an important role for MprAB during in vitro stress (13, 30) and in host systems (30, 48, 59, 60), and our current findings reinforce the significance of MprAB in the stress response of M. tuberculosis. Further studies are required to reveal the entire gene cohort directly regulated by MprA, as well as the mechanisms by which MprAB itself is regulated.
Supplementary Material
Acknowledgments
We thank Graham Stewart for the anti-Acr2 antibody and for unpublished data on acr2. We also thank Peter F. Barnes for support of this work and for critical reading of the manuscript and Buka Samten, Shiping Wu, Galina Mukamolova, and Claudio Cortès for helpful discussions.
This research was supported by the Margaret E. Byers Cain Chair for Tuberculosis Research (to P.F.B.) and NIH grant R21 AI063229-01 (to S.T.H.).
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 2.Beggs, M. L., M. D. Cave, and K. D. Eisenach. 1996. Isolation and sequence of a Mycobacterium tuberculosis sigma factor. Gene 174:285-287. [DOI] [PubMed] [Google Scholar]
- 3.Bucca, G., G. Ferina, A. M. Puglia, and C. P. Smith. 1995. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol. Microbiol. 17:663-674. [DOI] [PubMed] [Google Scholar]
- 4.Bucca, G., Z. Hindle, and C. P. Smith. 1997. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J. Bacteriol. 179:5999-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Z., T. P. Primm, J. Jakana, I. H. Lee, I. Serysheva, W. Chiu, H. F. Gilbert, and F. A. Quiocho. 1996. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J. Biol. Chem. 271:7218-7223. [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Dannenberg, A. M., Jr., and G. A. W. Rook. 1994. Pathogenesis of pulmonary tuberculosis: an interplay of tissue-damaging and macrophage-activating immune responses—dual mechanisms that control bacillary multiplication, p. 459-483. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection, and control. American Society for Microbiology, Washington, DC.
- 9.Fernandes, N. D., Q. L. Wu, D. Kong, X. Puyang, S. Garg, and R. N. Husson. 1999. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181:4266-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez, M., L. Doukhan, G. Nair, and I. Smith. 1998. sigA is an essential gene in Mycobacterium smegmatis. Mol. Microbiol. 29:617-628. [DOI] [PubMed] [Google Scholar]
- 12.Gomez, M., and I. Smith. 2000. Determinants of mycobacterial gene expression, p. 111-129. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, DC.
- 13.He, H., R. Hovey, J. Kane, V. Singh, and T. C. Zahrt. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188:2134-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, H., and T. C. Zahrt. 2005. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J. Bacteriol. 187:202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, H. J., M. S. Paget, and M. J. Buttner. 2002. A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol. Microbiol. 44:1199-1211. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz, J. 1992. Alpha-crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA 89:10449-10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard, S. T., and T. F. Byrd. 2000. The rapidly growing mycobacteria: saprophytes and parasites. Microbes Infect. 2:1845-1853. [DOI] [PubMed] [Google Scholar]
- 18.Hu, Y., and A. R. Coates. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, Y., F. Movahedzadeh, N. G. Stoker, and A. R. Coates. 2006. Deletion of the Mycobacterium tuberculosis alpha-crystallin-like hspX gene causes increased bacterial growth in vivo. Infect. Immun. 74:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 22.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 23.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 24.Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 26.Merck, K. B., P. J. Groenen, C. E. Voorter, W. A. de Haard-Hoekman, J. Horwitz, H. Bloemendal, and W. W. de Jong. 1993. Structural and functional similarities of bovine alpha-crystallin and mouse small heat-shock protein. A family of chaperones. J. Biol. Chem. 268:1046-1052. [PubMed] [Google Scholar]
- 27.Narberhaus, F. 2002. Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 66:64-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohara, N., N. Ohara, M. Naito, C. Miyazaki, S. Matsumoto, Y. Tabira, and T. Yamada. 1997. HrpA, a new ribosome-associated protein which appears in heat-stressed Mycobacterium bovis bacillus Calmette-Guerin. J. Bacteriol. 179:6495-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Toole, R., M. J. Smeulders, M. C. Blokpoel, E. J. Kay, K. Lougheed, and H. D. Williams. 2003. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 185:1543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang, X., P. Vu, T. F. Byrd, S. Ghanny, P. Soteropoulos, G. V. Mukamolova, S. Wu, B. Samten, and S. T. Howard. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153:1229-1242. [DOI] [PubMed] [Google Scholar]
- 31.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qamra, R., S. C. Mande, A. R. Coates, and B. Henderson. 2005. The unusual chaperonins of Mycobacterium tuberculosis. Tuberculosis 85:385-394. [DOI] [PubMed] [Google Scholar]
- 33.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rison, S. C. G., S. L. Kendall, F. Movahedzadeh, and N. G. Stoker. 2005. The mycobacterial two-component regulatory systems, p. 29-69. In T. Parish (ed.), Mycobacterium molecular microbiology. Horizon Biosciences. Wydmondham, Norfolk, United Kingdom.
- 35.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz, N., and T. J. Silhavy. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 8:122-126. [DOI] [PubMed] [Google Scholar]
- 37.Samten, B., P. Ghosh, A. K. Yi, S. E. Weis, D. L. Lakey, R. Gonsky, U. Pendurthi, B. Wizel, Y. Zhang, M. Zhang, J. Gong, M. Fernandez, H. Safi, R. Vankayalapati, H. A. Young, and P. F. Barnes. 2002. Reduced expression of nuclear cyclic adenosine 5′-monophosphate response element-binding proteins and IFN-γ promoter function in disease due to an intracellular pathogen. J. Immunol. 168:3520-3526. [DOI] [PubMed] [Google Scholar]
- 38.Samten, B., B. Wizel, H. Shams, S. E. Weis, P. Klucar, S. Wu, R. Vankayalapati, E. K. Thomas, S. Okada, A. M. Krensky, and P. F. Barnes. 2003. CD40 ligand trimer enhances the response of CD8+ T cells to Mycobacterium tuberculosis. J. Immunol. 170:3180-3186. [DOI] [PubMed] [Google Scholar]
- 39.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solano-Serena, F., R. Marchal, S. Casaregola, C. Vasnier, J. M. Lebeault, and J. P. Vandecasteele. 2000. A Mycobacterium strain with extended capacities for degradation of gasoline hydrocarbons. Appl. Environ. Microbiol. 66:2392-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, G. R., S. M. Newton, K. A. Wilkinson, I. R. Humphreys, H. N. Murphy, B. D. Robertson, R. J. Wilkinson, and D. B. Young. 2005. The stress-responsive chaperone α-crystallin 2 is required for pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 55:1127-1137. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, G. R., V. A. Snewin, G. Walzl, T. Hussell, P. Tormay, P. O'Gaora, M. Goyal, J. Betts, I. N. Brown, and D. B. Young. 2001. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat. Med. 7:732-737. [DOI] [PubMed] [Google Scholar]
- 44.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 45.Strickland, M. S., N. E. Thompson, and R. R. Burgess. 1988. Structure and function of the σ70 subunit of Escherichia coli RNA polymerase. Monoclonal antibodies: localization of epitopes by peptide mapping and effects on transcription. Biochemistry 27:5755-5762. [DOI] [PubMed] [Google Scholar]
- 46.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 47.Tabira, Y., N. Ohara, and T. Yamada. 2000. Identification and characterization of the ribosome-associated protein, HrpA, of Bacillus Calmette-Guerin. Microb. Pathog. 29:213-222. [DOI] [PubMed] [Google Scholar]
- 48.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valdez, M. M., J. I. Clark, G. J. Wu, and P. J. Muchowski. 2002. Functional similarities between the small heat shock proteins Mycobacterium tuberculosis HSP 16.3 and human αB-crystallin. Eur. J. Biochem. 269:1806-1813. [DOI] [PubMed] [Google Scholar]
- 51.Verbon, A., R. A. Hartskeerl, A. Schuitema, A. H. Kolk, D. B. Young, and R. Lathigra. 1992. The 14,000-molecular-weight antigen of Mycobacterium tuberculosis is related to the α-crystallin family of low-molecular-weight heat shock proteins. J. Bacteriol. 174:1352-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 54.Walters, S. B., E. Dubnau, I. Kolesnikova, F. Laval, M. Daffe, and I. Smith. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60:312-330. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson, K. A., G. R. Stewart, S. M. Newton, H. M. Vordermeier, J. R. Wain, H. N. Murphy, K. Horner, D. B. Young, and R. J. Wilkinson. 2005. Infection biology of a novel α-crystallin of Mycobacterium tuberculosis: Acr2. J. Immunol. 174:4237-4243. [DOI] [PubMed] [Google Scholar]
- 56.Wu, Q. L., D. Kong, K. Lam, and R. N. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, S., S. T. Howard, D. L. Lakey, A. Kipnis, B. Samten, H. Safi, V. Gruppo, B. Wizel, H. Shams, R. J. Basaraba, I. M. Orme, and P. F. Barnes. 2004. The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis strains in vivo. Mol. Microbiol. 51:1551-1562. [DOI] [PubMed] [Google Scholar]
- 58.Yuan, Y., D. D. Crane, and C. E. Barry, 3rd. 1996. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J. Bacteriol. 178:4484-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zahrt, T. C., and V. Deretic. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. USA 98:12706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zahrt, T. C., C. Wozniak, D. Jones, and A. Trevett. 2003. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 71:6962-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.