Abstract
Here we report the rapid identification of the proteins of the spliceosomal U1 small nuclear ribonucleoprotein (snRNP) from the yeast Saccharomyces cerevisiae by searching mass spectrometric data in genomic sequence databases. The U1 snRNP, containing a histidine-tagged 70K protein, was isolated from cell extracts by anti m3G-cap immunoaffinity and subsequent nickel nitrilotriacetic acid chromatography. A U1 snRNP fraction containing 20 proteins was obtained. Further purification by glycerol gradient centrifugation identified nine U1 snRNP specific and six common proteins. The U1 snRNP proteins were partially sequenced by nanoelectrospray mass spectrometry, and their genes were identified in the data base via multiple peptide sequence tags. Apart from the already known common proteins D1, D3, F, and G, the D2 and E homologs were also identified. The same six common proteins were detected in core U2 snRNP, which was purified and analyzed separately. The biochemical association of these six proteins with yeast snRNPs is shown here for the first time. Intriguingly, the Sm B/B′ homolog was not detected. In addition to the well characterized yeast U1 specific proteins [U1-70K (Snp1p), U1-A (Mud1p), Prp39p, and Prp40p] the homolog of the U1-C protein was identified together with four additional novel U1 specific proteins, which are not found in mammalian U1. This is the first time that the components of a multiprotein complex from an organism with a sequenced genome have been characterized by mass spectrometry. The technique should be applicable to any protein complex that can be biochemically purified from an organism whose genome is known.
Many vital functions of the cell are carried out by large protein complexes. Although sophisticated biochemical purification methods allow the isolation of such complexes, the characterization of their components has been difficult and time consuming at best and often precluded altogether by the low amount of isolated protein available.
Mass spectrometric data can now be obtained from proteins at a high sensitivity. Recently, methods have been developed to screen such data against sequence databases (for review, see ref. 1) and a first large scale protein identification project has been completed (2). Partial peptide sequence information obtained by electrospray (3, 4) tandem mass spectrometry can be assembled easily into a “peptide sequence tag,” basically a short stretch of amino acids combined with the measured distance in mass to the N and C termini of the peptide (5). In this scheme, two to three amino acids are sufficient to identify a unique peptide in large sequence databases. Using nanoelectrospray mass spectrometry (6, 7) of unseparated peptide mixtures, proteins from silver-stained gels can be sequenced in the subpicomole range (8). The combination of these analytical tools with complete genome information in model organisms such as yeast now provides a ready route to any gene using the small amounts of proteins obtained by excision from gels.
A promising field for application of this new technique is the splicing machinery, especially in the yeast Saccharomyces cerevisiae. The U1, U2, U4, U5, and U6 small nuclear ribonucleoprotein particles (snRNPs) are essential components of the spliceosome, the complex that catalyzes the excision of introns from the primary gene transcripts (refs. 9 and 10 and references therein). Mammalian snRNPs consist of one RNA molecule and two sets of proteins. Eight common proteins, B/B′, D1, D2, D3, E, F, and G (also called Sm proteins) are associated with all of the snRNPs except U6. In addition, every snRNP contains several particle specific proteins (11).
Most research on the splicing process is performed in human (HeLa) cells and the yeast S. cerevisiae. The high copy number of snRNPs in HeLa cells allows biochemical experiments, while genetics cannot be performed. In yeast, in contrast, genes involved in splicing were generally identified by genetics or by searching for homologs of already characterized human proteins (9).
Recently, the isolation of yeast snRNPs was successfully performed by anti-m3G-cap affinity chromatography on whole cell extracts (12). Characterization of the protein components by SDS/PAGE indicated the existence of common and particle specific snRNP proteins in yeast, as in the mammalian case. The identification of individual yeast snRNP proteins was not possible, due to insufficient material for conventional microsequencing and impurity of the fractions obtained. Moreover, efficient methods of protein determination, similar to the one described here, had not yet been developed.
U1 snRNP is the first snRNP contacting the pre-mRNA by base pairing of the 5′ end of its RNA with the 5′ splice site of the pre-mRNA (9, 10). The yeast U1 snRNP is larger (18S particle) and contains more proteins (at least 15) than its human counterpart (12S particle, 11 proteins) (12). The different size and protein composition of human and yeast U1 snRNPs might well reflect differences in the early steps of the splicing process between the two species (12).
Several yeast U1 snRNP proteins have already been identified by genetic means or homology searches in the yeast genome sequence. The yeast U1 specific proteins Snp1p (homolog of the human U1-70K protein), Mud1p (homolog of the human U1-A), Prp39p, and Prp40p, which have no known human counterpart, are well studied (13–16). Some of the homologs of human Sm proteins have been identified also: D1, D3, F, and G (17–19). These four proteins have been cloned, and an association with yeast snRNAs was demonstrated by immunoprecipitation assays (17–19). Yeast homologs of the Sm proteins B/B′, D2, and E, as well as the U1 snRNP specific protein C, however, have not been described yet.
Here we report the preparative isolation of U1 snRNPs from yeast using a strain that expresses the U1 specific protein Snp1p with a histidine tag on its N terminus. Highly purified U1 snRNP particles were isolated, their proteins were partially sequenced by nanoelectrospray mass spectrometry, and the corresponding genes were identified in the database. The D1, D2, D3, E, F, and G proteins were all identified unequivocally. The association of D2 and E with yeast snRNPs is shown here for the first time. In a separate analysis, U2 snRNP was shown to contain the same six common proteins as U1 snRNP. Surprisingly, no Sm B/B′ homologs appear to be present in the yeast snRNPs analyzed so far. Among the specific proteins of the yeast U1 snRNP, we clearly detect U1-70K (Snp1p), U1-A (Mud1p), Prp39p, and Prp40p. In addition, we have identified the homolog of the human U1-C protein and four novel specific proteins of molecular masses of 52, 55, 57, and 77 kDa. The corresponding genes of all of them were found in the data base.
MATERIALS AND METHODS
Preparation of Extracts, Immunoaffinity, Affinity, Mono-Q Chromatography, and Gycerol Gradients.
Yeast strain BSY283 (MATa, ade2, arg4, leu2-3, 112, trp1-289, ura3-52, snp1::LEU2) carrying plasmid pBS357 (SNP1 with N-terminal hexahistidine tag, CEN, TRP1) was grown at 30°C in 16 liters of YPD medium (1% yeast extract/2% peptone/2% dextrose) to OD600 = 3. Whole cell extracts were prepared as described (12). Extracts were dialyzed against buffer D200 [20 mM Hepes-KOH (pH 7.9)/200 mM KCl/8% glycerol/0.5 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/4 μg/ml pepstatin]. For the purification of snRNPs, anti-m3G chromatography was performed as described (12), except that EDTA was omitted from all buffers. Nickel nitrilotriacetic acid (Ni-NTA) affinity chromatography was performed as follows. The salt concentration of the snRNP eluate was raised to 300 mM KCl. Total snRNPs were passed over a 500-μl bed volume Ni-NTA–agarose column, preequilibrated with buffer D300 (as buffer D200, but containing 300 mM KCl and 10 mM 2-mercaptoethanol instead of DTT). After washing with 12 bed volumes of buffer D300 containing 0.8 mM imidazole and with 8 bed volumes of buffer D300 containing 8 mM imidazole, U1 snRNP was eluted with buffer D300 containing 40 mM imidazole, yielding 10–20 μg of U1 snRNP.
Mono-Q FPLC was performed as described (20). The flow-through of the Ni-NTA column was diluted with buffer DE0 [20 mM Tris·HCl (pH 6.8)/1.5 mM MgCl2/0.5 mM DTT/0.5 mM PMSF] to a final concentration of 100 mM KCl. After loading onto a 100-μl Mono-Q column (Pharmacia), elution was performed using a three-step salt gradient from 50 to 1000 mM KCl in buffer DE. 100-μl fractions were collected.
Glycerol gradient centrifugation was performed as described (12), except that all buffers contained 0.01% Nonidet P-40. About 10 μg of U1 snRNPs were loaded on a linear 10–30% (wt/vol) glycerol gradient. After centrifugation, 500-μl fractions were taken from the top, and proteins and RNA were analyzed by gel electrophoresis. Identical gradients loaded with total yeast RNA and/or with T7 U1 RNA as sedimentation markers were run for comparison.
In Gel Digestion of snRNP Proteins.
A Ni-NTA fraction containing about 5–10 μg of proteins was extracted using phenol/chloroform/isoamylalcohol (50:49:1); proteins were precipitated from the organic phase with acetone, fractionated by 12% SDS/PAGE, and stained with colloidal Coomassie blue (Sigma). The same was done for fractions containing core U2 snRNP. In gel digestion and subsequent extraction of the peptides was carried out as described previously (8, 21). The protein bands were excised, washed, in gel-reduced, and alkylated, followed by in gel digestion with trypsin (Boehringer Mannheim, sequencing grade). The resulting peptides were extracted in several steps and dried down in a vacuum centrifuge.
Nanoelectrospray Mass Spectrometry.
The peptide mixture was desalted in a capillary needle as described (7, 8). The unseparated peptide mixture was step-eluted with a total volume of 0.5–1 μl of 50% methanol/5% formic acid directly into the spraying needle of the nano electrospray ion source developed in our group (6, 7). All experiments were carried out on a Sciex API III triple quadrupole instrument (Sciex, Perkin–Elmer). Sequencing time per peptide mixture was 30–60 min on a sample volume of about 0.5–1 μl.
Database and Homology Search.
peptidesearch (5), a program for correlating mass spectrometric information with sequence data bases, was used to search peptide sequence tags in a comprehensive nonredundant data base, which currently contains about 200,000 entries. No constraints on protein molecular weight or species of origin were specified. Homology searches were performed using blitz, blast, and fasta.
RESULTS
Purification of U1 snRNPs.
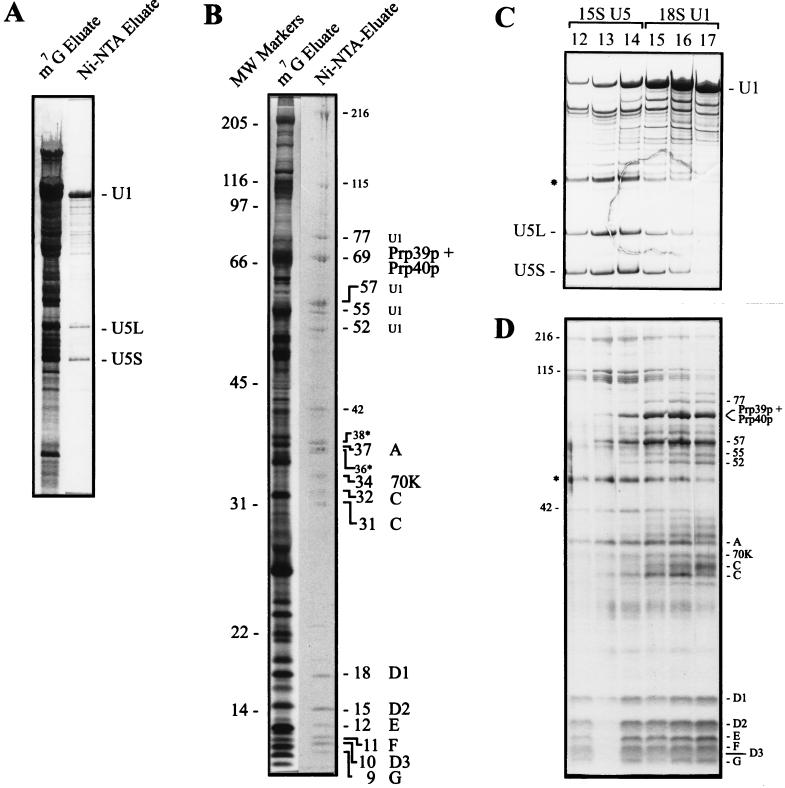
U1 snRNPs were highly enriched by a two-step purification. In the first step, all of the m3G-capped snRNPs (including nucleolar snRNPs) were isolated from whole cell extracts by anti-m3G immunoaffinity chromatography as described previously (12), except that a strain was used that expresses a histidine-tagged U1-70K (Snp1p) protein. From this mixture of snRNPs, U1 snRNP was specifically purified by Ni-NTA chromatography. Fig. 1A shows the RNA analysis of the Ni-NTA column. The U1 snRNA is by far the predominant RNA species in the fraction eluted with 40 mM imidazole. In addition to U1 RNA, U5 RNA is also detected, however, to a much lesser extent, as compared with the U1 RNA. Co-isolation of some U5 snRNP with the U1 snRNP on the Ni-NTA column may be due either to an U1–U5 interaction or to the presence of a histidine-rich sequence in one of the U5 specific snRNP proteins. The Ni-NTA eluate exhibits 20 protein bands that have the apparent molecular weights listed in Fig. 1B. When the Ni-NTA eluate was further subjected to glycerol gradient centrifugation, the U1 and U5 snRNP particles could be separated at least in part (Fig. 1C, RNA analysis), resulting in a pure U1 snRNP preparation (fractions 16 and 17, Fig. 1C). Fig. 1D shows the protein composition of these snRNP fractions. We observed 15 proteins that clearly co-sediment with the yeast U1 RNA. These 15 proteins have apparent molecular masses of 9, 10, 11, 12, 15, 18, 31, 32, 34, 37, 52, 55, 57, 69, and 77 kDa. Fraction 13 and 14 contained mainly U5 RNA and U5 proteins (Fig. 1 C and D). The two proteins of 36 and 38 kDa (Fig. 1B, asterisks) were found at the top of the gradient (data not shown). Thus, they are likely contaminants.
Figure 1.
Purification of U1 snRNPs from S. cerevisiae. (A) Silver staining of snRNAs eluted from anti-m3G-cap (m7G eluate) and Ni-NTA affinity columns (Ni-NTA eluate). The positions of U1 and U5 (long and short form) RNAs are indicated. (B) Proteins of the total snRNPs (m7G eluate) were stained with silver. The proteins of the Ni-NTA eluate were stained with Coomassie blue and then partially sequenced by mass spectrometry. We estimate the protein amounts to be between 5 and less than 1 picomole. Proteins of 216, 115, and 42 kDa are associated with U5 snRNPs (see gradient centrifugation, C and D). The two proteins of 36 and 38 kDa (asterisks) are contaminants. All of the other proteins are U1 snRNP-associated. The band of 69 kDa contained two U1 specific proteins, Prp39p and Prp40p. The U1-C protein was identified in two bands of 32 and 31 kDa. Six yeast common snRNP proteins were identified in the low molecular weight region between 9 and 18 kDa. Molecular weights are indicated in kilodaltons. (C) U1 and U5 snRNPs were separated on a glycerol gradient, and the snRNAs of the corresponding fractions were stained with silver. U1 and U5 snRNAs are indicated. 18S U1 snRNP peaks in fractions 16 and 17, whereas 15S U5 snRNP peaks in fractions 13 and 14. The RNA in the U3 region (asterisk), which co-migrates with U5, was not present in the preparation used for the sequencing of proteins. (D) Proteins of separated U1 and U5 snRNPs were stained with silver. Proteins of 216, 115, and 42 kDa co-migrate with U5 RNA. All of the other proteins co-migrate with U1 RNA. The common proteins of molecular masses ranging from 9 to 18 kDa co-migrate with U1 as well as U5 RNAs. U1 snRNP proteins are indicated on the right, U5 proteins on the left. The band at 49 kDa (asterisk) is a RNase inhibitor.
Identification of Yeast snRNP Proteins by Nanoelectrospray Mass Spectrometry and Detection of Their Genes by Database Search.
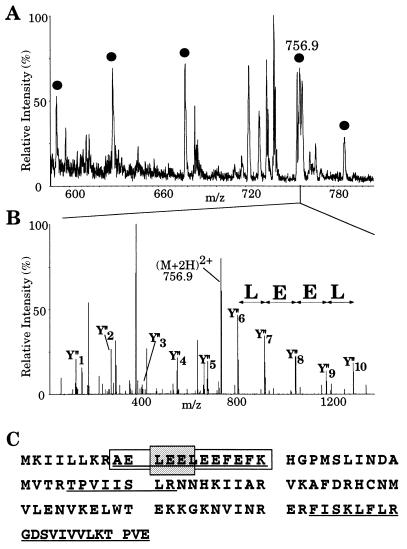
As a source for nanoelectrospray mass spectrometry of the U1 snRNP proteins we used the Ni-NTA eluate shown in Fig. 1B, which contained the U1 as well as some of the U5 proteins. All of the 20 proteins visualized by Coomassie blue staining were partially sequenced by nanoelectrospray tandem mass spectrometry and their genes identified by database search (Table 1). The protein bands were excised from the gel and digested in gel with trypsin after reduction and alkylation of cysteine residues. The resulting peptides were extracted and desalted in a single step. The unseparated peptide mixture was then analyzed by nanoelectrospray mass spectrometry (6, 7) on a triple quadrupole mass spectrometer. For each protein digest, a mass spectrum of the peptide mixture or “peptide mass map” was acquired by scanning the first quadrupole Q1 (Fig. 2A). Then peptide ion peaks of interest were selected in turn by Q1 and fragmented in the collision cell Q2, and the fragments were separated by the third quadrupole Q3. For each peptide a tandem mass spectrum is thus obtained which contains “nested” sets of fragments produced by cleavage at adjacent peptide bonds (one cleavage per peptide ion) and which differ in mass by the molecular weight of one amino acid residue. As shown in Fig. 2 B and C, it is straightforward to construct a peptide sequence tag (5) from the tandem mass spectrum, which is sufficient to retrieve the peptide uniquely from the database. The complete peptide sequence is verified by comparing the tandem mass spectrum with the predicted one for the retrieved sequence. Since several peptides are fragmented for each protein, identifications by this method are certain rather than probable.
Table 1.
Identification of yeast U1 snRNP proteins
| Mr (app.) (kDa) | Mr (calc.) (kDa) | Peptides sequenced | Protein name | Corresponding gene | Accession number |
|---|---|---|---|---|---|
| 69 | 75.1 | 7 | Prp39p | PRP39 | L29224L29224 |
| 69 | 69.1 | 3 | Prp40p | PRP40 | P33203P33203 |
| 37 | 34.4 | 3 | Mud1p | MUD1 | P32605P32605 |
| 34 | 34.4 | 2 | Snp1p | SNP1 | X59986X59986 |
| 31 | 27.0 | 3 | U1-C | SCL8003 17 | U17243U17243 |
| 18 | 16.3 | 3 | Sm D1 | SMD1 | Q02260Q02260 |
| 15 | 12.1 | 5 | Sm D2 | SCL9328 3 | U17245U17245 |
| 12 | 10.4 | 1* | Sm E | SC55020 15 | U55020U55020 |
| 11 | 9.6 | 2 | Sm F | SCL9705 11 | U25842U25842 |
| 10 | 11.2 | 4 | Sm D3 | SMD3 | P43321P43321 |
| 9 | 8.5 | 2 | Sm G | SNP2 | P40204P40204 |
The identity of this peptide was further ascertained by methylation and 18O-labeling.
Figure 2.
Identification of the 15 kDa band of Fig. 1B by mass spectrometry. (A) Part of the mass spectrum of the peptide mixture obtained after digestion of the protein band. (B) Fragmentation of the peak at m/z 756.9 in part A. Mass spectrum of the fragments obtained by scanning the third quadrupole Q3. The prominent fragments in the high molecular weight region are spaced by amino acid molecular weights and reveal the partial sequence LEEL as shown by the arrows (note that the isobaric amino acids leucine and isoleucine are indistinguishable by our method). The sequence stretch, together with its starting mass, its end mass, and the molecular weight of the peptide are entered into a database searching program (peptidesearch) where they are converted to a peptide sequence tag. Search of the tag in a nonredundant database uniquely retrieves the sequence AELEELEEFEFK from gene L9328.5 (GenBank). The predicted C-terminal or Y" ion fragments (4, 22) of this peptide are marked in the spectrum and verify the complete sequence. (C) Protein sequence of gene L9328.5. Five peptides were partially sequenced that identified peptides from the gene as indicated by underlining (those within the mass range are also marked by bullets in A). The partial sequence deduced from B is in gray and the complete peptide is boxed.
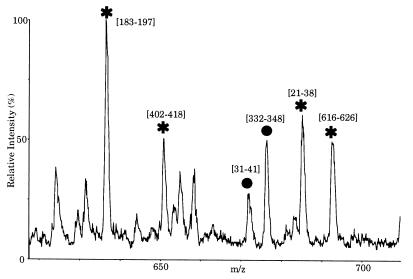
In this way all of the proteins of Fig. 1B have been identified unequivocally. This method also allows the identification of a mixture of two proteins which co-migrate in one single band (Fig. 3). This is shown for the identification of the U1 specific proteins Prp39p and Prp40p, which co-migrated in the 69-kDa band (see Figs. 1B and 3 below).
Figure 3.
Identification of Prp39p and Prp40p in the 69 kDa band of Fig. 1B. Part of the tandem mass spectrum of peptides extracted from the band. The marked peaks have been sequenced, and their peptide sequence tags searched in the data base. They mapped to the indicated sequence range in Prp39p (stars) or Prp40p (bullets). The sequenced peptides cover 106 and 41 amino acids for Prp39p and Prp40p, respectively; thus identification of both proteins is certain.
The Sm Proteins D1, D2, D3, E, F, and G but not B/B′ Are Found in Yeast U1 and U2 snRNPs.
Fig. 1D shows that the low molecular weight proteins ranging from 9 to 18 kDa co-sediment, after glycerol gradient centrifugation, with U1 as well as with U5 snRNAs, making them candidates for the common Sm proteins. These six proteins were sequenced as described above and identified as the yeast homologs of the human Sm proteins D1, D2, D3, E, F, and G (12–19, 23, 24). The yeast homologs of D2 and E were identified here for the first time. As it was shown previously for D1, D3, F, and G (17–19, 24, 25), D2 and E are also evolutionarily highly conserved. Fig. 4 A and B show an alignment between the human and yeast E and D2 Sm proteins, respectively. The yeast Sm E and D2 have apparent masses of 12 and 15 kDa and share 46.7 and 58.3% identity with their human counterpart, respectively. Both proteins possess high homology in the two regions termed Sm motifs 1 and 2, which are found in all of the known Sm proteins from diverse organisms (19, 24, 25).
Figure 4.
(A) Alignment of the yeast and human proteins Sm E. The Sm motifs 1 (boxed in light gray) and Sm motif 2 (boxed in dark gray) are indicated. (B) Alignment of the yeast and human proteins Sm D2. (C) Alignment of yeast and human U1-C proteins. The four metal binding residues of a putative zinc finger motif are boxed in gray. Printed in boldface letters are the peptides which were sequenced by mass spectrometry. SC, S. cerevisiae; HS, Homo sapiens.
Considering that the yeast Sm proteins D1, D2, D3, E, F, and G were identified in purified U1 snRNPs, it was unexpected that no yeast Sm B or B′ proteins were detected. By searching the database for homologs of the human B/B′, we found a putative yeast B/B′ Sm protein. However, if present, this protein must be loosely associated with the yeast snRNPs and thus dissociates at the salt concentration (300 mM) used during fractionation.
To investigate whether all of the yeast snRNP preparations lacked Sm B/B′ or whether this was a peculiarity of the U1 snRNP, we purified yeast core U2 snRNPs in three chromatographic steps (see Materials and Methods). Core U2 snRNPs were isolated from the Ni-NTA flow-through by Mono-Q anion-exchange chromatography at about 600 mM KCl. Again, six proteins in the molecular weight range of 9 to 18 kDa were co-isolated with the U2 RNA (data not shown). The Sm proteins are known to be stably associated with the human snRNAs, even at high salt concentrations. Sequencing of the proteins from the yeast core U2 snRNP preparation revealed that they were identical to the Sm proteins of the yeast U1 snRNP. Again the yeast Sm B/B′ homolog was not detected. We conclude that B/B′ is absent from all of our snRNP preparations, raising the question whether this protein is lost during fractionation of snRNPs or whether it is not associated at all with the yeast snRNPs.
All of the Known Yeast U1-Specific Proteins, Including U1-C and Four Novel Proteins, Are Present in Purified Yeast U1 snRNPs.
All of the known U1 specific proteins previously identified by genetic means, namely U1-70K (Snp1p), U1-A (Mud1p), Prp39p, and Prp40p (13–16), were identified in purified yeast U1 snRNPs. These were found in the protein bands of apparent molecular masses of 34 kDa (U1-70K), 37 kDa (U1-A), and 69 kDa (Prp39p and Prp40p, which co-migrated in the same band) (Figs. 1 B and D and Fig. 3). In addition to the 70K and A proteins, the human U1 snRNP contains a third U1 specific protein, U1-C (11). We identified a homolog of the human U1-C also in yeast U1 snRNP particles (Fig. 1 B and D). The yeast U1-C migrates as a double band of molecular masses of 31 and 32 kDa. We do not know whether this doublet is due to degradation of the U1-C protein or to posttranslational modifications. In this respect, it is interesting to note that the human U1-C protein shows, reproducibly, a similar double band after gel electrophoresis (11). Fig. 4C shows an alignment of human and yeast U1-C proteins. The N-terminal 40 amino acids of the two C proteins show 50% identity. Interestingly, the yeast protein contains the same zinc finger motif of the CC—HH type with the corresponding amino acids in identical positions (ref. 26; Fig. 4). The C-terminal parts of the two proteins show almost no homology.
Four novel proteins with molecular masses of 52, 55, 57, and 77 kDa were similarly sequenced and identified in the database (Fig. 1 B and D). These proteins co-migrate exclusively with U1 but not with U5 snRNAs after glycerol gradient fractionation (Fig. 1D, fractions 15, 16, and 17). Thus, they are good candidates for additional U1 specific proteins. However, even though they co-elute with the U1 RNA from Mono-Q columns (data not shown), their specific association with U1 snRNPs will have to be demonstrated by additional techniques.
DISCUSSION
The recent advances in mass spectrometry combined with the availability of completely sequenced genomes open new exciting possibilities in the analysis of protein-containing complexes, as demonstrated here with the S. cerevisiae U1 snRNP. Mass spectrometry combined with database searching was applied here for the first time to characterize the components of a multiprotein complex.
Isolation of the yeast U1 snRNP was performed successfully, despite the low abundance of snRNPs in yeast. All of the proteins associated with yeast U1 RNA and their corresponding genes were identified by nanoelectrospray mass spectrometry combined with peptide sequence tag searching. The extremely low flow rate of the nanoelectrospray ion source allowed collection of sequence information of low amounts of peptides out of the unseparated digest mixture, which, together with the mass information of the peptides, yielded reliable identification of the proteins. The lack of any chromatographic procedures for the separation of the peptides makes this method fast and robust, which allowed us to analyze the whole complex in a few weeks. In no case did additional analytical techniques have to be employed for the certain identification of protein bands.
Similar to its human counterpart, the yeast U1 snRNP is composed of common and specific proteins. We have identified unequivocally only six yeast common proteins: D1, D2, D3, E, F, and G as compared with the human counterpart, which contains two additional Sm proteins: B and B′ (11). Of these, D2 and E were identified as U1 components for the first time, while the others were identified previously by other means (17–19). Since D2 and E were also identified in purified U2 snRNPs, it is likely that they are associated with the other m3G-capped yeast snRNAs as well (i.e. U4 and U5; Fig. 1D). The identification of these six proteins in yeast snRNPs is shown here for the first time by a direct biochemical approach. Like D1, D3, F, and G, D2, and E are evolutionarily highly conserved (about 50% identity with their human counterparts; Fig. 4 A and B). Considering the structural conservation of the six Sm proteins, it is surprising that we were unable to identify a yeast homolog of the vertebrate Sm B/B′ protein. We exclude that the yeast B/B′ was not detected because of possible co-migration in the gel with one of the other U1 proteins, since the mass spectrometry technique would have been able to detect the B/B′ candidate even at a molar ratio of 1:5. As a positive control, we have been able to identify Prp39p and Prp40p in the same protein band (Fig. 3). We cannot finally answer the question whether yeast U1 snRNPs contain a counterpart of the human B/B′ protein or whether B/B′ is only loosely associated with U1 snRNPs and dissociates from the particle during the biochemical fractionation. We have identified a candidate for a yeast B/B′ counterpart in the data base (22.4 kDa, accession no. P40460P40460, data not shown), however, we do not know yet whether the putative Sm B/B′ is a bona fide yeast homolog. Additional experiments, such as tagging of its gene, would be required to show co-immunoprecipitation of the putative yeast Sm B/B′ with snRNAs.
Four of the nine specific proteins detected were known components of the yeast U1 snRNP: U1-70K, U1-A, Prp39p, and Prp40p (13–16). Here we also identify the yeast U1 specific protein C. Yeast U1-C is 50% identical to the human counterpart in the N-terminal 40 amino acids. This part contains a zinc finger-like domain of the CC—HH type, which was shown to be necessary and sufficient for incorporation of the protein into human U1 snRNP (26). The C-terminal region of the yeast U1-C shows almost no homology, except for a number of seven to eight proline residues located at regular intervals in the yeast and human proteins. However, the homology in this region is very low, suggesting different functions of the two regions in yeast and man.
In conclusion, the yeast U1 snRNPs not only contain the previously identified U1 specific proteins, but also four novel specific proteins that are absent or as yet remain to be identified in metazoans. Although the genes corresponding to all of the novel proteins of the U1 snRNP were identified, they will not be presented here. This will await additional experiments, such as tagging and immunoprecipitation studies, to further characterize the association of these gene products with U1 RNA.
Data presented here and in our earlier studies suggest that the large 18S yeast U1 snRNP could represent a preassembled complex required for commitment of pre-mRNA to splicing (ref. 12 and references therein). The differences in the biochemical composition and shape of the yeast and the human U1 snRNP could reflect functional differences at early steps of spliceosome assembly. Since the human U1 snRNP is required for commitment to splicing of a plethora of distinct pre-mRNAs, it may need more flexibility and therefore need to recruit specific factors depending on the particular task to be performed. It would be interesting to identify the vertebrate homologs of the species-specific yeast U1 proteins (Prp39p and Prp40p) and to understand whether they play a role as non-snRNP associated factors in human spliceosomes.
The analysis of the yeast U1 snRNP proteins would have been very difficult or even impossible with other methods currently available. Edman degradation would not have been successful, since at least two proteins, Sm F and U1-A, proved to be N-terminally blocked and since the amount of material available would have been insufficient. Another approach is the identification of gel-isolated proteins by their peptide map, where low picomole amounts of protein are required. This approach has been successfully used for the large scale identification of proteins from yeast (2). However, in the case of the U1 snRNP, this method would also have failed for many proteins, because the digestion of small proteins yields only very few peptides (which were here in some cases also modified) and some of the bands were mixtures, making certain identification difficult.
The identification of the genes of yeast U1 snRNP proteins allows in vivo studies of the functions of the individual gene products to be performed. The cloning of the corresponding genes is straightforward once they have been identified and amplified from yeast genomic DNA by PCR. In vivo studies of the effects of introduced mutations into yeast U1 snRNP genes should yield a more complete picture of early steps of the splicing process and allow the differences between man and yeast to be clarified.
In addition to a characterized genome, our approach for the identification of the components of multiprotein complexes only requires a biochemical “handle” to purify the complex prior to gel separation. The histidine tagging of one of the components of a particular complex would be one strategy for the isolation of other such complexes from yeast and other organisms in which genetics can be performed easily, provided that at least one of the components is known. If at least 0.1–0.5 pmol of such a complex can be separated by gel electrophoresis, it should be possible to rapidly characterize its protein components by the techniques demonstrated here.
Acknowledgments
We thank the other members of the Protein & Peptide group, especially Drs. Matthias Wilm and Andrej Shevchenko for establishing many of the experimental techniques used here and Anna Shevchenko for expert sample preparation. We thank Dr. C. Marshallsay for critically reading the manuscript. This work was supported by a grant of the German Technology ministry (BMBF) to M.M. and by the Deutsche Forschungsgemeinschaft (SFB272) and the Fonds der Chemischen Industrie to R.L.
Footnotes
Abbreviations: snRNP, small nuclear ribonucleoprotein particle; Ni-NTA, nickel nitrilotriacetic acid.
References
- 1.Patterson S D, Aebersold R. Electrophoresis. 1995;16:791–814. doi: 10.1002/elps.11501601299. [DOI] [PubMed] [Google Scholar]
- 2.Shevchenko A, Wilm M, Vorm O, Jensen O, Podtelejnikov A, Mortensen P, Shevchenko A, Sagliocco F, Boucherie H, Mann M. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fenn J B, Mann M, Meng C K, Wong S F, Whitehouse C M. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 4.Mann M, Wilm M. Trends Biochem Sci. 1995;20:219–223. doi: 10.1016/s0968-0004(00)89019-2. [DOI] [PubMed] [Google Scholar]
- 5.Mann M, Wilm M. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 6.Wilm M, Mann M. Int J Mass Spectrom Ion Processes. 1994;136:167–180. [Google Scholar]
- 7.Wilm M, Mann M. Anal Chem. 1996;66:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 8.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Nature (London) 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 9.Rymond B C, Rosbash M. In: The Molecular and Cellular Biology of the Yeast Saccaromyces. Jones E W, Pringle J R, Broach J R, editors. I. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 143–192. [Google Scholar]
- 10.Moore M J, Query C C, Sharp P A. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–357. [Google Scholar]
- 11.Will C L, Fabrizio P, Lührmann R. In: Nucleic Acids and Molecular Biology. Eckstein F, Lilley D M J, editors. Vol. 9. Berlin: Springer Verlag; 1995. pp. 342–372. [Google Scholar]
- 12.Fabrizio P, Esser S, Kastner B, Lührmann R. Science. 1994;264:261–265. doi: 10.1126/science.8146658. [DOI] [PubMed] [Google Scholar]
- 13.Smith V, Barrell B G. EMBO J. 1991;10:2627–2634. doi: 10.1002/j.1460-2075.1991.tb07805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao X C, Tang J, Rosbash M. Genes Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart S, Rymond B. Mol Cell Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao H-Y, Siliciano P G. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rymond B C. Proc Natl Acad Sci USA. 1993;90:848–852. doi: 10.1073/pnas.90.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy J, Zheng B, Rymond B C, Woolford J L. Mol Cell Biol. 1995;15:445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Séraphin B. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lübben B, Fabrizio P, Lührmann R. J Biol Chem. 1995;270:11549–11554. doi: 10.1074/jbc.270.19.11549. [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 22.Roepstorff P, Fohlman J. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 23.Lehmeier T, Raker V, Hermann H, Lührmann R. Proc Natl Acad Sci USA. 1994;91:12317–12321. doi: 10.1073/pnas.91.25.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Lührmann R. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper M, Johnston L H, Beggs J D. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelissen R L H, Will C L, van Venrooij W J, Lührmann R. EMBO J. 1994;13:4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]