Abstract
The ulcer-causing pathogen Helicobacter pylori uses directed motility, or chemotaxis, to both colonize the stomach and promote disease development. Previous work showed that mutants lacking the TlpB chemoreceptor, one of the receptors predicted to drive chemotaxis, led to less inflammation in the gerbil stomach than did the wild type. Here we expanded these findings and examined the effects on inflammation of completely nonchemotactic mutants and mutants lacking each chemoreceptor. Of note, all mutants colonized mice to the same levels as did wild-type H. pylori. Infection by completely nonchemotactic mutants (cheW or cheY) resulted in significantly less inflammation after both 3 and 6 months of infection. Mutants lacking either the TlpA or TlpB H. pylori chemotaxis receptors also had alterations in inflammation severity, while mutants lacking either of the other two chemoreceptors (TlpC and HylB) behaved like the wild type. Fully nonchemotactic and chemoreceptor mutants adhered to cultured gastric epithelial cells and caused cellular release of the chemokine interleukin-8 in vitro similar to the release caused by the wild type. The situation appeared to be different in the stomach. Using silver-stained histological sections, we found that nonchemotactic cheY or cheW mutants were less likely than the wild type to be intimately associated with the cells of the gastric mucosa, although there was not a strict correlation between intimate association and inflammation. Because others have shown that in vivo adherence promotes inflammation, we propose a model in which H. pylori uses chemotaxis to guide it to a productive interaction with the stomach epithelium.
Chemotaxis, the ability of microorganisms to move in response to chemical cues, is widespread within the prokaryotic world, but its role in mammalian infection is not well understood (34, 48). In some pathogens, nonchemotactic mutants have altered animal colonization, but the underlying cause of this phenotype is not known. Helicobacter pylori has become a model organism for understanding the biological contributions of chemotaxis to infection. This microbe chronically infects gastric tissue, which can lead to peptic ulcers, gastric adenocarcinoma, and mucosa-associated lymphoid tumors (42).
Motility is known to be critical for H. pylori infection of piglet, mouse, and gerbil stomachs (19, 21, 39, 47). Similarly, mutants that lack directed motility (nonchemotactic mutants) are attenuated, but to various degrees depending on the infection model (19, 24, 40, 62). These results are usually interpreted to mean that chemotaxis promotes growth in the stomach. Recent work has suggested that chemotaxis also plays roles in processes other than colonization (40). In particular, it was found that mutants lacking one of the chemotaxis pathway receptors, TlpB, colonized gerbils to wild-type levels but caused less inflammation.
Chemotaxis has been extensively studied in Escherichia coli (4, 8). In this microbe, chemoreceptors sense environmental cues, such as amino acids, and transmit this ligand-binding information to a signal transduction cascade that affects flagellar rotation. The core signal transduction proteins consist of the CheW receptor-kinase coupling/adapter protein, the CheA kinase, and the CheY response regulator. CheY interacts with the flagellar motor in its phosphorylated state. When an attractant ligand binds to the chemoreceptor, the kinase activity of CheA is diminished and nonphosphorylated CheY predominates. In this form, CheY fails to interact with the flagellar motor, the flagella rotate counterclockwise, and the bacteria swim. When no ligand is bound to the chemoreceptor, the CheA kinase is active, and CheY is phosphorylated (CheY∼P) and interacts with the flagellar motor. In the presence of CheY∼P, the flagellar motor rotates clockwise, and the bacteria randomly reorient in a behavior called tumbling.
H. pylori chemotaxis appears to have some similar and some distinct features compared with E. coli chemotaxis (2, 46, 63). H. pylori has four chemoreceptors: TlpA (HP099), TlpB (HP0103), TlpC (HP0082), and HylB (HP0599). In addition, it has the core signaling components CheW, CheA, and CheY, and mutants lacking each of these have the predicted chemotaxis deficiencies (7, 24, 49). H. pylori furthermore has three proteins that are hybrids of CheW and CheY, called CheVs. While Bacillus subtilis CheV is redundant with CheW (51), none of the H. pylori CheVs is redundant with its CheW (49), suggesting that H. pylori may have a number of unique features in its chemotaxis pathway. Finally, it was recently shown that H. pylori contains a remote homolog of CheZ, a protein that accelerates the dephosphorylation of CheY∼P (61).
Two of the four H. pylori chemoreceptors, TlpA and TlpC, are important for mouse colonization in a coinfection model (3) but dispensable for occupation of the gastric niche when there is only one H. pylori strain present (3, 16). TlpB mutants have no defect for gerbil infection in either a single-strain or coinfection model (40) but recently were reported to have a defect for single-strain infection of interleukin-12 (IL-12) knockout mice (16). HylB mutants behave similar to TlpA and TlpC mutants (S. M. Williams and K. M. Ottemann, unpublished data). There is experimental evidence that TlpA senses arginine and bicarbonate (11) (see Discussion) and that TlpB senses protons (16), but the ligands of the other receptors remain unknown.
An important part of the H. pylori disease process is the ability to cause inflammation, a process believed to underlie all H. pylori-induced pathology. Inflammation in the gastric mucosa is caused by the production of proinflammatory cytokines and chemokines (22, 41, 57, 66). Several cells produce these mediators, including gastric epithelial cells and immune cells. Bacterial molecules that stimulate cytokine/chemokine manufacture are known from studies of H. pylori or the related organism Helicobacter felis. For example, proteins encoded by the cag pathogenicity island (Cag PAI) trigger epithelial cells to secrete the proinflammatory mediators IL-8, IL-6, and tumor necrosis factor alpha (66). The same chemokines and cytokines are released by macrophages confronted with H. pylori urease, while a different protein, OipA, has been shown to cause cultured gastric epithelial cells to produce IL-8 (68). Neutrophil-activating protein activates neutrophils such that they produce more reactive oxygen species and thus enhance inflammation. Helicobacter proteins that directly stimulate Th-1 T-helper cells are not yet known, although it is well known that these cells are critical for the development of gastritis (17, 52). The strain used here, SS1, induces a strong inflammatory response in both mice and gerbils (18, 23, 39, 40, 64) but is proposed to have an incomplete Cag PAI (14).
Previous work revealed that H. pylori mutants lacking the TlpB chemoreceptor colonize gerbils to wild-type levels but the animals have decreased inflammation. There are other mutants that share this phenotype. The best understood are mutants lacking the Cag PAI (1, 31, 45), which lose the ability to trigger secretion of proinflammatory cytokines and chemokines from gastric epithelial cells (42). Here we expand the study of the chemotaxis-inflammation connection by employing a mouse model. Because nonchemotactic mutants (e.g., mutants lacking cheW, cheY, or cheA [62]) infect mice but not gerbils, we are able to determine that this bacterial process in general influences inflammation and also that one other H. pylori chemoreceptor aids this immune response. We also find that in vivo adherence is lessened in chemotaxis mutants, and we suggest that this lack of intimate contact with the gastric epithelium underlies the lessened inflammation.
MATERIALS AND METHODS
H. pylori strains, growth conditions, and general molecular biology.
For these studies we used H. pylori strains SS1 and G27 (Table 1) and isogenic mutants of these strains. To culture H. pylori, we used either Columbia blood agar (Difco) plates with 5% defibrinated horse blood (Hemostat Labs, Davis, CA), 50 μg/ml cycloheximide, and 0.2% (wt/vol) β-cyclodextrin (Sigma) (CHBA) or brucella broth (Difco) with 10% fetal bovine serum (FBS). Cultures were incubated at 37°C under 7 to 10% O2, 10% CO2, and 80 to 83% N2. All antibiotics were from Sigma or ISC Bioexpress. For selection of H. pylori mutants, chloramphenicol was used at 5 to 10 μg/ml, and kanamycin was used at 15 μg/ml. For selection of plasmid-bearing E. coli we used chloramphenicol at 20 μg/ml, kanamycin at 30 μg/ml, and ampicillin at 100 μg/ml.
TABLE 1.
Strains used in this study
| Strain | Characteristics | Chemotaxis phenotype | Gene in 26695a | Reference(s) and/or source |
|---|---|---|---|---|
| H. pylori SS1 | Wild type, type I | Che+ | 37; Janie O'Rourke | |
| H. pylori SS1 cheY | ΔcheY::cat-102, Cmrb | Che− | HP1067 | 40, 62 |
| H. pylori SS1 cheW | ΔcheW::aphA3, Kmr | Che− | HP0391 | 62 |
| H. pylori SS1 tlpB::cat-2 | tlpB::cat-2, Cmr | Che+ | HP0103 | This study |
| H. pylori SS1 ΔtlpB::cat | ΔtlpB::cat, Cmrc | Che+ | HP0103 | 40 |
| H. pylori SS1 ΔtlpA::cat | ΔtlpA::cat-1, Cmr | Che+ | HP0099 | 3 |
| H. pylori SS1 ΔtlpC::cat | ΔtlpC::cat-1, Cmr | Che+ | HP0082 | 3 |
| H. pylori SS1 ΔhylB::cat | ΔhylB::cat-D1, Cmr | Che+/− | HP0599 | This study |
| H. pylori G27 | Wild type, type I | Che+ | 10; Nina Salama | |
| H. pylori G27 tlpB::cat-2 | tlpB::cat-2, Cmr | Che+ | HP0103 | This study |
| H. pylori G27 ΔtlpB::cat | ΔtlpB::cat, Cmr | Che+ | HP0103 | This study |
| E. coli DH10B | Cloning strain; F−mcrA Δ(mrr-hsdRMS-mcrBC) Δ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Lab stock | ||
| E. coli BL21 | Overexpression strain; E. coli B F−ompT gal (dcm) (lon) hsdSB (rB− mB−) | Novagen |
Plasmids were prepared using QIAGEN kits, and genomic DNA was prepared using either Wizard genomic kits (Promega) or DNeasy (QIAGEN). All restriction and DNA modification enzymes were from New England Biolabs or Invitrogen. Amplification of DNA was carried out using Pfu-Turbo polymerase (Stratagene) or Taq polymerase (generous gift from D. Kellogg). DNA sequencing was performed by the University of California at Berkeley sequencing facility.
Assessment of motility and chemotaxis.
Motility was determined by phase-microscopic inspection of cultures and using soft-agar plates composed of 0.35% agar in brucella broth, 2.5% FBS, and H. pylori-selective antibiotics (5 μg/ml trimethoprim, 8 μg/ml amphotericin B, 10 μg/ml vancomycin, 50 μg/ml cycloheximide, 5 μg/ml cefsulodin, 2.5 U/ml polymyxin B), as described previously (47).
Cloning and mutagenesis of tlpB to create tlpB::cat-2.
For creation of tlpB::cat-2, the tlpB gene (HP0103) with ∼50-bp flanking sequences was amplified from H. pylori G27 genomic DNA using primers HPTlpB1 (5′-CTCTGGATCCCCCGTTGTTGGAAAAATTG-3′; the underlined region indicates homology) and HPTlpB3 (5′-TGGAAGCTTGCACTTGTTTGTCTAAATTC-3′) and Pfu polymerase. All primers for cloning and mutagenesis were designed from the H. pylori 26695 genome sequence (63). The 2.2-kb PCR product was treated with Taq polymerase to add an overhanging T and then ligated with pCR2.1 using T4 DNA ligase (Invitrogen) to create pKO105. This plasmid was verified by restriction digestion and sequencing. The tlpB insert was then removed by EcoRI digestion and ligated with EcoRI-cut pUC19 (69) using T4 DNA ligase to create pKO115. Restriction enzyme analysis was used to confirm the construction of pKO115.
To create the tlpB::cat-2 mutant, pKO115 was digested with the restriction enzyme Bpu11021, which cuts about two-thirds into tlpB. The cut DNA was rendered blunt by treatment with T4 DNA polymerase, and the 5′ phosphates were removed with calf intestinal phosphatase. pBS-cat (54) was then digested with KpnI and SacI to release a 0.8-kb cat fragment. The cat fragment was rendered blunt as described above and gel purified. The purified fragments were ligated together using T4 DNA ligase to create plasmid pTC1 (pUC19::tlpB::cat-2). pTC1 was screened by restriction enzyme digestion to verify that the cat gene was correctly inserted into tlpB.
Cloning and mutagenesis of hylB.
hylB (HP0599) and flanking sequences were amplified from H. pylori SS1 genomic DNA using primers hylB3 (5′-GCATGGTTGCCTTGGGG) and hylB4 (5′-CGGCAAGAATGCTAGCGG). The resulting 2.2-kb PCR fragment was cloned into pBad18 (27) cut with SmaI to generate pL30A2. A deletion was made in the hylB coding sequence by using inverse PCR to amplify the plasmid backbone and hylB flanking sequences with primers hylB10 (5′-GGTGGCATTCCTTATTTAAATTTG) and hylB11 (5′-TGATTCCACTCAATGAAGTGTTTTG), resulting in a 1,300-bp deletion of the entire hylB open reading frame and a product that was 5,530 bp long. An 817-bp fragment containing the chloramphenicol resistance gene (cat) was cut from pBS-cat (54) with NotI and PstI and rendered blunt ended with T4 polymerase, and the resulting product was ligated into the hylB-deleted PCR product to generate pL30A2cat2.
Transformation of H. pylori to create tlpB::cat-2 and ΔhylB::cat-1 mutants.
Plasmids pTC1 and pL30A2cat2 were used individually to transform H. pylori G27 or SS1 to chloramphenicol resistance, using natural transformation as described previously (47). Chloramphenicol-resistant colonies were colony purified twice, and the location of the insertion was verified by PCR amplification of chromosomal DNA from the mutants, using primers HPTlpB1 and HPTlpB3 for tlpB mutants and primers hylB5 (5′-TAAATGGGCCAAAGTCAAAG) and hylB6 (5′-GCTGGGTATAGCATTGATGATAAT) for hylB mutants (data not shown). Additionally, we performed Southern blotting of genomic DNA digested with HindIII to verify that a single copy of cat was present in each H. pylori mutant, using a probe complementary to the cat gene labeled with an AlkPhos Direct kit (Amersham Pharmacia) (data not shown).
Western blotting and chemoreceptor antibody creation.
Total cell proteins were prepared from H. pylori cultured on Columbia horse blood agar plates for 2 days by resuspending and lysing the cells in 2× Laemmli sample buffer (6). Samples were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, transferred to immunoblot polyvinylidene difluoride membranes (Bio-Rad), and incubated with a 1:5,000 dilution of anti-glutathione transferase (GST)-TlpA22. For visualization, the blots were incubated with the secondary antibody goat anti-rabbit conjugated to horseradish peroxidase (Santa Cruz Biotech) at a dilution of 1:2,000, followed by incubation with luminol, p-coumoric acid, and hydrogen peroxide. Luminescent blots were visualized by exposure to Biomax Light film (Kodak).
The GST-TlpA22 antibody is a rabbit polyclonal antibody directed toward a GST-TlpA(cyto) fusion protein. GST-TlpA(cyto) was made by cloning the portion of tlpA that encodes amino acids 358 to 666 of the TlpA cytoplasmic domain into pGEX-4T.1 (Amersham) to create an N-terminal GST fusion in plasmid pTA22. This region of TlpA was chosen because it encompasses the conserved region of chemoreceptors. GST-TlpA(cyto) was overexpressed in E. coli BL21, and the cells were lysed by grinding them under liquid N2. The protein was then purified by glutathione chromatography (Sigma G4510 glutathione column), eluted using glutathione, dialyzed, and then concentrated using a 3,500-molecular-weight cutoff Centricon concentrator (Millipore). Purified protein was sent to Animal Pharm Inc. for rabbit inoculation and serum isolation.
Mouse infections.
All animal protocols were approved by the Institutional Animal Use and Care Committee. H. pylori SS1 and its isogenic mutants were grown overnight in shaking brucella broth plus 10% FBS and used for mouse infection as described previously (47). The mice used were 4- to 6-week-old female FVB/N mice (Charles River) or 4- to 6-week-old female C57BL/6 mice (Taconic). After the infection period, one half of each stomach was homogenized with a sterile pestle in brucella broth plus 10% FBS and plated to determine the number of CFU/gram of stomach as described previously (62); the other half was used for histology (see below). To obtain coinfection data, the stomach homogenates were plated on two media that differentiate between the two strains, as described previously (62). Ratios are given as mutant/wild type.
Histology and pathology.
After mouse sacrifice, one half of each stomach was placed in a histology cassette with a sponge (Fisher) and fixed using buffered formalin (Fisher). The tissue was embedded in paraffin, sectioned (5 μm), stained with hematoxylin and eosin, and evaluated in a blind fashion by a pathologist (J. E. Carter). Each slide was read twice to ensure reproducibility, and identical grades were obtained in all cases. Grading was conducted by two established methods. First, lymphocytic infiltration was scored using the method of Eaton et al. (20). The scores are as follows: 0, no infiltrate; 1, mild, multifocal infiltration; 2, mild, widespread infiltration; 3, mild, widespread and moderate, multifocal infiltration; 4, moderate, widespread infiltration; and 5, moderate, widespread and severe, multifocal infiltration. Neutrophil infiltration was scored as present or absent. Gastric atrophy was assessed using the method of Rugge et al. (53). In general terms, atrophy refers to the shrinkage of cells in a histologic population by loss of cell substance. We used the definition of Rugge et al., in which atrophy is defined as the loss of gastric glands histologically specific to the area of the gastric mucosa being sampled. This definition allows division of atrophy into two categories: atrophy associated with metaplasia, in which gastric glands in one portion of the stomach are lost but are replaced by gastric glands from a histologically separate region of the gastric mucosa, and atrophy without metaplasia, in which gastric glands in one portion of the stomach are lost without replacement by other types of gastric glands and fibrosis of the lamina propria is seen. Categorization of a specimen as negative for atrophy meant that it lacked atrophy associated with metaplasia and atrophy without metaplasia and was indeterminate for atrophy if inflammation obscured histologic classification.
To assess the proximity of the H. pylori to the cells, sections were scored by a researcher (S. M. Williams) who was blind to the identity of the infecting strain. H. pylori in the glands of the corpus or antrum was scored as either (i) “touching,” with the majority of the bacterium touching the gastric cell surface; (ii) “near,” with the bacterium within one bacterial cell length of the gastric cell or only a small part of the bacterium touching the cell; or (iii) “distant,” with the bacterium completely in the center of the gland or in the superficial mucus and not near the cells. Bacterial cells were usually found in groups, within the gastric glands. If there were adherent or nearby cells in a gland, the group was scored as touching/near regardless of whether some of the bacteria were far. A group was scored as distant only if no members were adherent or nearby.
Cytokine array assay.
AGS (ATCC CRL-1739 [56]) or NCI-N87 (CRL-5822 [13]) human gastric epithelial cells were cultured according to the ATCC guidelines in Dulbecco's modified Eagle's medium (DMEM). To assay cytokine and chemokine production, cells were seeded at 2 × 105 cells/well in 24-well tissue culture plates and incubated for 24 h. After this time, H. pylori, cultured for 24 h on CHBA, was scraped from a plate and resuspended in sterile DMEM or phosphate-buffered saline. The bacterial concentration was determined by determining the optical density at 600 nm, and a volume equivalent to 2 × 107 bacteria was added to each well in fresh tissue culture medium to give a multiplicity of infection of 100. After 22 to 24 h, the culture supernatant was removed, centrifuged to remove any bacteria, and stored at −80°C until it was assayed (less than 1 week). This supernatant was applied to a Human Cytokine Array I (RayBiotech, Inc.) by following the manufacturer's protocols. This array tests for granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, Gro, Gro-α, IL-1α, IL-2, IL-3, IL-5, IL-6, IL-7, IL-8, IL-10, IL-13, IL-15, gamma interferon, monocyte chemoattractant protein 1 (MCP1), MCP2, MCP3, MIG, RANTES, transforming growth factor β, and tumor necrosis factor beta. The results were visualized by exposing the blots to Biomax Light film (Kodak).
In vitro adherence assay.
AGS cells (ATCC CRL-1739 [56]) were cultured according to ATCC recommendations in DMEM and were prepared and inoculated with H. pylori as described above for the cytokine arrays. The cells were incubated with bacteria for 2 to 3 h and then washed extensively with phosphate-buffered saline, as described by Segal et al. (56). AGS cells along with attached H. pylori were removed from the plates with trypsin-EDTA, and the H. pylori was plated on CHBA plates to determine the number of associated microbes.
Statistical analysis of data.
Inflammatory scores and mouse colonization levels were analyzed statistically using a two-tailed t test (unpaired) at http://www.graphpad.com/quickcalcs/ttest1.cfm. P values of <0.05 were considered significant. H. pylori-gastric epithelium interactions were analyzed using the chi-square test for categorical data. Briefly, all the bacteria in each class (touching, near, and distant) were summed, and an expected frequency table was generated. We then compared our data to the expected table.
RESULTS
Bacterial chemotaxis promotes inflammation in mice.
In our previous studies, we found that H. pylori lacking the TlpB chemoreceptor was defective for causing inflammation in gerbils (40). We decided to expand those studies to investigate whether chemotaxis in general was important for triggering an inflammatory response, as well as whether other chemoreceptor mutants have the same phenotype as mutants lacking TlpB. Unlike the TlpB mutants, nonchemotactic mutants (e.g., mutants lacking cheY) do not infect gerbils at all (40), so we could not analyze such mutants in the gerbil model. Nonchemotactic mutants do, however, infect mice, so we decided to switch to this animal model to analyze the link between chemotaxis and inflammation. Previous work has shown that C57BL/6 mice develop inflammation in response to H. pylori infection (37), so we infected mice of this strain with previously characterized nonchemotactic SS1 mutants lacking either cheY or cheW (7, 40, 62). Each of these mutants was generated by deletion of most of the open reading frame and replacement with a nonpolar antibiotic resistance cassette. The cheY mutation has been complemented previously (40, 62). This analysis found that all cheY-associated phenotypes were corrected by the complementation, suggesting the cheY mutation is responsible for the defects. The cheW gene is predicted to be at the end of its operon, and thus this mutation is likely nonpolar (http://www.microbesonline.org). Both of these mutants swim without changing direction and are thus not chemotactic in laboratory assays. These nonchemotactic mutants do not have a mouse colonization defect at later time points during infection (>1 month) when they are the sole infecting strains (62). We carried out two separate infections, allowing one to proceed for 3 months and the other to proceed for 6 months. For both of these experiments, 4- to 6-week-old female C57BL/6 mice were infected with ∼5 × 107 CFU.
After 3 months of infection, we found that the nonchemotactic mutant (cheY) caused less inflammation than did wild-type H. pylori SS1. For this time point we did not analyze the cheW mutant. The average inflammatory score for the cheY mutant was 0.80 ± 0.45, while that for the wild type was 1.4 ± 1.1 (Table 2 and Fig. 1A). Although the scores of mice infected with the cheY mutant were lower than those of wild-type-infected mice, the difference between the two was not significant (P = 0.3). There was no difference in colonization levels between the wild type and the cheY mutant (Fig. 1B), suggesting that differences in bacterial load were not the cause of the altered inflammation levels. Furthermore, we found that the cheY mutant and wild type were distributed similarly between the antrum and corpus (data not shown), indicating that this mutant colonizes the stomach as fully as the wild type at this time point.
TABLE 2.
Histology and numbers of CFU for stomachs from mice infected for 3 months with SS1 and isogenic chemotaxis mutantsa
| Infecting strain | Log CFU/g stomach | Inflammation grade | Presence of neutrophils |
|---|---|---|---|
| Wild-type SS1 | |||
| S1 | 6.57 | 3 | No |
| S2 | 7.44 | 0 | No |
| S3 | 5.51 | 1 | Yes |
| S4 | 4.6 | 1 | Yes |
| S5 | 6.3 | 2 | Yes |
| Avg | 6.82 | 1.4 ± 0.51b | |
| SS1 ΔcheY::cat-102 | |||
| Y1 | 5.05 | 1 | Yes |
| Y2 | 6.31 | 1 | Yes |
| Y3 | 7.03 | 0 | No |
| Y4 | 5.40 | 1 | Yes |
| Y5 | 5.44 | 1 | Yes |
| Avg | 6.42 | 0.8 ± 0.20 | |
| SS1 ΔtlpA::cat-1 | |||
| A1 | 7.22 | 1 | Yes |
| A2 | 4.96 | 1 | Yes |
| A3 | 5.65 | 1 | Yes |
| A4 | 6.89 | 0 | No |
| A5 | 5.72 | 1 | Yes |
| Avg | 6.7 | 0.8 ± 0.2 | |
| SS1 ΔtlpB::cat | Yes | ||
| B1 | 7.15 | 1 | Yes |
| B2 | 6.09 | 1 | Yes |
| B3 | 5.12 | 1 | Yes |
| B4 | 7.27 | 1 | No |
| B5 | 4.80 | 1 | Yes |
| Avg | 6.83 | 1.0 ± 0 | |
| SS1 ΔtlpC::cat-1 | |||
| C1 | 5.57 | 1 | Yes |
| C2 | 5.83 | 3 | Yes |
| C3 | 5.65 | 2 | Yes |
| C4 | 5.07 | 1 | Yes |
| C5 | 6.46 | 1 | Yes |
| Avg | 5.96 | 1.6 ± 0.4 | |
| SS1 ΔhylB::cat-D1 | |||
| H1 | 5.52 | 1 | No |
| H2 | 6.41 | 1 | Yes |
| H3 | 5.97 | 2 | Yes |
| H4 | 5.94 | 3 | Yes |
| H5 | 7.39 | 1 | Yes |
| Avg | 6.77 | 1.6 ± 0.4 | |
| Uninfected | |||
| U1 | 0 | 0 | No |
| U2 | 0 | 0 | No |
C57BL/6 mice were used for these infections. All grading was done using a stomach sample that included both the antrum and the corpus. All mice were also graded for gastric atrophy and had a score of 0.
Average ± standard error of the mean.
FIG. 1.
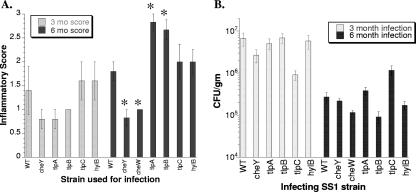
(A) Inflammatory scores of mouse stomachs infected with H. pylori SS1 or its isogenic chemotaxis mutants. Mouse stomach sections were stained with hematoxylin and eosin and graded for the amount of lymphocyte infiltration as described in Materials and Methods. Mice were infected for 3 months or 6 months with the strains indicated below the bars. In all experiments C57BL/6 mice were used. Five mice were used for each group for the 3-month infections. For the 6-month infections, six mice were used for all groups except the wild-type and ΔtlpB::cat groups (five mice). Uninfected mice for both time points gave inflammatory scores of zero. The error bars indicate the standard errors of the means. An asterisk indicates that there is a statistically significant difference between the inflammatory score for a strain and the score for mice infected with the wild type for the same length of time. The data are also shown in Tables 2 and 3. WT, wild type. (B) CFU per gram of mouse stomach. The numbers of viable bacteria were determined after 3 or 6 months of infection, using samples described above. At each time point, there is no difference between the number of wild-type bacterial cells and the number of cells of any of the mutants. The error bars indicate the standard errors of the means.
Although the data suggested that nonchemotactic cheY mutants cause less inflammation, the levels of inflammation caused by wild-type H. pylori were low and variable. Because others had shown that longer H. pylori infections result in higher inflammatory scores, we repeated the experiment and allowed infection to proceed for 6 months. In addition, we used two different nonchemotactic mutants, one lacking cheY and the other lacking cheW. Consistent with the 3-month infection, both nonchemotactic mutants caused significantly less inflammation than did wild-type H. pylori (Table 3 and Fig. 1A) but colonized to similar levels (Fig. 1B). These findings thus suggest that bacterial chemotaxis enhances H. pylori-triggered inflammation.
TABLE 3.
Histology and numbers of CFU for stomachs from mice infected for 6 months with SS1 and isogenic chemotaxis mutantsa
| Infecting strain | Log CFU/g stomach | Inflammation grade | Presence of neutrophils |
|---|---|---|---|
| Wild-type SS1 | |||
| S1 | 4.96 | 1 | Yes |
| S2 | 5.01 | 2 | Yes |
| S3 | 5.17 | 2 | Yes |
| S4 | 4.85 | 2 | Yes |
| S5 | 5.97 | 2 | Yes |
| Avg | 5.40 | 1.8 ± 0.20b | |
| SS1 ΔcheY::cat-102 | |||
| Y1 | 4.79 | 1 | Yes |
| Y2 | 4.86 | 1 | Yes |
| Y3 | 5.34 | 0 | No |
| Y4 | 5.34 | 1 | Yes |
| Y5 | 5.39 | 1 | No |
| Y6 | 5.69 | 1 | Yes |
| Avg | 5.3 | 0.8 ± 0.17 | |
| SS1 ΔcheW::kan | |||
| W1 | 4.53 | 1 | No |
| W2 | 5.01 | 1 | No |
| W3 | 4.93 | 1 | No |
| W4 | 5.22 | 1 | No |
| W5 | 5.39 | 1 | No |
| W6 | 4.77 | 1 | No |
| Avg | 5.06 | 1.0 ± 0 | |
| SS1 ΔtlpA::cat-1 | |||
| A1 | 6.12 | 3 | Yes |
| A2 | 5.37 | 3 | Yes |
| A3 | 4.63 | 3 | Yes |
| A4 | 5.59 | 4 | Yes |
| A5 | 5.43 | 3 | Yes |
| A6 | 4.45 | 3 | Yes |
| Avg | 5.58 | 2.83 ± 0.17 | |
| SS1 ΔtlpB::cat | Yes | ||
| B1 | 3.53 | 3 | Yes |
| B2 | 4.74 | 3 | Yes |
| B3 | 3.94 | 2 | Yes |
| B4 | 5.54 | 2 | Yes |
| B5 | 4.59 | 3 | Yes |
| Avg | 4.99 | 2.7 ± 0.37 | |
| SS1 ΔtlpC::cat-1 | |||
| C1 | 4.86 | 3 | Yes |
| C2 | 6.08 | 2 | Yes |
| C3 | 4.78 | 3 | Yes |
| C4 | 6.70 | 2 | Yes |
| C5 | 5.66 | 1 | No |
| C6 | 5.32 | 1 | No |
| Avg | 6.07 | 2.0 ± 0.37 | |
| SS1 ΔhylB::cat-D1 | |||
| H1 | 4.6 | 2 | Yes |
| H2 | 5.09 | 2 | Yes |
| H3 | <2.40 | 2 | Yes |
| H4 | 5.16 | 1 | Yes |
| H5 | 4.38 | 2 | Yes |
| H6 | 5.72 | 3 | Yes |
| Avg | 5.23 | 2.0 ± 0.26 | |
| Uninfected | |||
| U1 | 0 | 0 | No |
| U2 | 0 | 0 | No |
| U3 | 0 | 0 | No |
| Avg | 0 |
C57BL/6 mice were used for these infections. All grading was done using a stomach sample that included both the antrum and the corpus. All mice were also graded for gastric atrophy and had a score of 0.
Average ± standard error of the mean.
Two H. pylori chemoreceptors modulate inflammation.
The results presented above suggest that proper chemotaxis is important for the development of inflammation, because nonchemotactic cheY or cheW mutants cause less-severe inflammation (Tables 2 and 3 and Fig. 1). Previous work in gerbils had shown that one chemoreceptor, TlpB, was important for inflammation in that animal model. We were curious whether other H. pylori chemoreceptor mutants have similar phenotypes, so we analyzed inflammation in mice infected for 3 or 6 months with H. pylori SS1 mutants lacking tlpA, tlpB, tlpC, or hylB. Each of these mutants was constructed by replacing most or all of the target gene with a cat gene that can be slightly polar due to its very weak transcriptional terminator (A. Castillo and K. M. Ottemann, unpublished data). We have shown previously that the gene downstream of tlpB is transcribed at wild-type levels, suggesting that the tlpB mutation is not polar (40). This finding is consistent with predictions that tlpB is in a single-gene operon (http://www.microbesonline.org). tlpC is similarly predicted to be the only gene in its operon. The hylB and tlpA mutations may be polar because these genes are predicted to be in three- and two-gene operons, respectively. Each of these mutations results in loss of a specific protein recognized by an anti-chemoreceptor antibody of the correct molecular weight (Fig. 2). H. pylori lacking tlpA, tlpB, or tlpC does not have any detectable in vitro phenotype (3, 40), while mutants lacking hylB have a modest defect in the soft-agar assay commonly used for H. pylori chemotaxis (unpublished data). tlpA, tlpC, and hylB mutants have defects for mouse colonization when they are in competition with the wild type (3; data not shown). We found that H. pylori mutants lacking tlpA behaved similar to the mutants lacking tlpB. After 3 months of infection, C57BL/6 mice infected with tlpA and tlpB mutants had less inflammation than mice infected with the wild type, although the difference was not statistically significant (Fig. 1A and Table 2). After 6 months of infection, tlpA and tlpB mutants caused enhanced inflammation that was statistically significant (Fig. 1A and Table 3). tlpC and hylB mutants displayed phenotypes different from those of tlpA and tlpB mutants. These mutants had inflammation scores that were very similar to those of the wild type at both 3 and 6 months (Fig. 1). All H. pylori chemoreceptor mutants colonized to wild-type levels (Fig. 1B). Thus, it appears that some chemoreceptors influence inflammation, while others do not, suggesting that each chemoreceptor may have evolved to have different roles in the disease process.
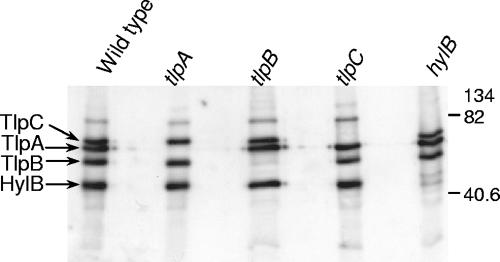
FIG. 2.
Western analysis of H. pylori SS1 and its isogenic chemoreceptor mutants, showing that each mutant lacks one chemoreceptor. Total bacterial proteins were collected from 2-day CHBA cultures. Proteins were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred to polyvinylidene difluoride membranes. Transferred proteins were incubated with 1:5,000 anti-GST-TlpA22, which reacts to the conserved domain that is shared between all chemoreceptors. The strains are indicated above the lanes, and SS1 ΔtlpB::cat was used for this analysis. The position of each chemoreceptor is shown on the left, and the molecular masses (in kilodaltons) are given at the right. TlpC is predicted to be a 75.2-kDa protein, TlpA is predicted to be a 74.5-kDa protein, TlpB is predicted to be a 62.8-kDa protein, and HylB is predicted to be a 48.4-kDa protein; the sizes determined in this study agree well with these predictions.
Taken together, these results suggest that proper chemotaxis is important for development of inflammation. In particular, the H. pylori tlpA and tlpB chemoreceptors influence how inflammation develops in mice. TlpB mutants also had inflammation phenotypes in gerbils after 1 month of infection; they caused less inflammation, and the immune cell population was skewed toward neutrophils relative to the wild-type immune cell population (40).
Mutants lacking tlpB colonize mice to wild-type levels.
Previous work had shown that chemotaxis is not required for colonization of mice when there is only one H. pylori strain infecting the animals (62). Consistent with this notion, we also found that H. pylori can lack its individual chemoreceptors and achieve wild-type colonization in mice (Fig. 1B) (3) and gerbils (40). Recently, Croxen et al. reported that H. pylori SS1 tlpB mutants did have a mouse colonization defect (16). Because decreased colonization could underlie the decreased inflammation of tlpB mutants, we decided to analyze whether tlpB mutants had any colonization defects. For these analyses we used two independent tlpB mutants. One, used in the experiments described above and in our previous work (40), has almost the entire tlpB gene deleted and replaced by cat. This mutant is called ΔtlpB::cat. The second tlpB mutant, tlpB::cat-2, has the cat gene inserted at a BpuII0121 restriction site within tlpB, corresponding to codon 380 out of 565 codons. This insertion disrupts the portion of TlpB predicted to interact with the downstream signaling protein CheW (residues 303 to 564) and is likely a null mutation. Both tlpB alleles were generated in H. pylori strains SS1 and G27 and analyzed for in vitro chemotaxis and the presence of the TlpB protein.
Loss of TlpB in any strain, using either insertion mutant, did not significantly affect chemotaxis as measured using the soft-agar plate assay. In the SS1 and G27 H. pylori strain backgrounds, the tlpB mutations resulted in soft-agar migration rates ranging from 97% to 119% of those of the corresponding wild type (data not shown). These phenotypes were the same as those reported previously for SS1 ΔtlpB::cat (40). As shown in Fig. 2, mutation of tlpB resulted in elimination of a 63-kDa band, supporting the conclusion that the tlpB mutant does not have the TlpB protein.
We next examined the role of the TlpB chemoreceptor in mouse infection, using both single-strain infections and competition infections. For both types of infections we used 4- to 6-week-old FVB/N mice and allowed the infection to proceed for 2 weeks. We commonly use this mouse strain for colonization studies and have found that H. pylori infects it and C57BL/6 mice similarly (data not shown). After 2 weeks, we determined the number of H. pylori cells in the stomach. In the single-strain infections, both tlpB mutants infected to levels that were indistinguishable from the levels obtained with the wild type, suggesting that the TlpB chemoreceptor is not needed for infection of mice (Table 4). This result is similar to that in gerbils (40). We also carried out competition experiments because these types of experiments can reveal subtle colonization defects (3, 54). For these experiments, mice were coinfected with a mixture of equal amounts of the wild type and either the SS1 ΔtlpB::cat or tlpB::cat-2 mutant. After 2 weeks of infection, there were similar numbers of wild-type and mutant cells (Table 5), and in some coinfected animals there was a slight decrease in the number of wild-type bacteria relative to the number when the wild type was used alone, although we do not know the basis for this phenotype (Table 5). Taken together with the 3- and 6-month infection results, these findings indicate that the TlpB chemoreceptor is not required for bacterial growth or maintenance in the rodent stomach.
TABLE 4.
Single-strain experiments with wild-type SS1, tlpB mutants, and FVB/N micea
| Strain | N | 106 CFU/g (avg ± SEM) |
|---|---|---|
| SS1 ΔtlpB::cat | 5 | 3.73 ± 0.79 |
| SS1 tlpB::cat-2 | 4 | 4.41 ± 0.34 |
| SS1 wild type | 8 | 2.77 ± 0.42 |
The inoculating doses were between 4 × 106 and 1.7 × 108 CFU. All infections proceeded for 2 weeks, and we detected bacteria in all mice that were infected. N is the number of infected mice in each experiment.
TABLE 5.
Competition experiments with wild-type SS1, tlpB mutants, and FVB/N micea
| Expt | N | Strains | Input ratio | Output ratio | Competitive index | Strain | Input (CFU/g) | Output (CFU/g)
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse 1 | Mouse 2 | Mouse 3 | Mouse 4 | Mouse 5 | ||||||||
| 1 | 2 | SS1 tlpB::cat-2 + wild type | 1.5 | 0.45 | 0.3 | Mutant | 2.5 × 107 | 7.3 × 105 | 3.8 × 105 | |||
| Wild type | 3.8 × 107 | 1.2 × 106 | 1.2 × 106 | |||||||||
| 2 | 5 | SS1 ΔtlpB::cat + wild type | 1.3 | 9.0 | 7.0 | Mutant | 2.0 × 108 | 3.4 × 106 | 6.0 × 106 | 7.6 × 106 | 6.5 × 106 | 3.2 × 106 |
| Wild type | 1.5 × 108 | 1.1 × 106 | 1.7 × 106 | 4.7 × 105 | 2.9 × 105 | 1.3 × 106 | ||||||
| 3 | 3 | SS1 ΔtlpB::cat + wild type | 0.6 | 2 | 3.3 | Mutant | 4 × 106 | 1.9 × 106 | 4.2 × 106 | 2.5 × 106 | ||
| Wild type | 6.7 × 106 | 4.2 × 105 | 3.6 × 106 | 7.6 × 106 | ||||||||
The inoculating doses were between 4 × 106 and 1.7 × 108 CFU. All infections proceeded for 2 weeks, and we detected bacteria in all mice that were infected. N is the number of infected mice in each experiment. All ratios are mutant/wild type ratios. The competitive index is defined as output ratio/input ratio. The input is the number of CFU that each mouse was infected with. The output is the number of CFU/g stomach found in each mouse after 2 weeks of infection.
Chemotaxis mutants and wild-type H. pylori both adhere to and trigger IL-8 release from cultured gastric cells in vitro.
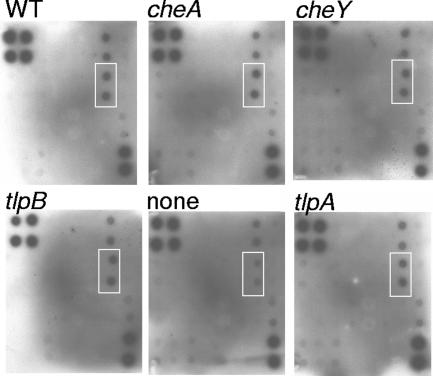
We observed consistently that nonchemotactic H. pylori SS1 cheY or cheW mutants do not cause as much inflammation as wild-type H. pylori SS1. One of the ways that H. pylori leads to inflammation is by adhering to the gastric epithelial cells (26). We examined two aspects of H. pylori-epithelial cell interactions: adherence and the ability to stimulate the cells to release cytokines and chemokines. For assessing in vitro adherence, we cocultured H. pylori with the gastric epithelial cell line AGS. This cell line has been used extensively for similar studies (10, 35, 56). After allowing 2 h for the bacteria to interact with the cells, we washed the cells extensively and then determined the number of bacteria that remained adherent to the cells. We found that there was not a significant difference between the numbers of wild-type H. pylori cells and the numbers of mutant H. pylori cells adhering to the AGS cells (cheY, tlpA, and tlpB mutants were tested) (data not shown). As stated above, H. pylori promotes inflammation in part by interacting with epithelial cells and causing them to release the chemokine IL-8. This response is mediated largely through the bacterial Cag PAI, which encodes a type IV secretion apparatus that sends both peptidoglycan and the CagA protein into eukaryotic cells (9, 65). Both of these factors cause IL-8 production (9, 25, 36). H. pylori Cag PAI-deficient mutants cause less inflammation in animals and less IL-8 release from cultured gastric epithelial cells, consistent with a strong link between these two processes (1, 15). We thus determined whether our cheW, cheY, tlpA, and tlpB mutants caused cultured gastric cells to release IL-8 and other cytokines by coculturing gastric AGS cells with wild-type or mutant H. pylori. We then analyzed the cell supernatant for several cytokines and chemokines using a cytokine array that functions like several simultaneous immunoblots. We found that all H. pylori strains, including the wild type and the isogenic chemotaxis mutants, resulted in comparable levels of IL-8 release (Fig. 3). Similar results were obtained with nonmotile and tlpB mutants of a more robust IL-8-inducing H. pylori strain, G27 (Data not shown). In addition, there were no differences in the release of any of the other cytokines tested, although most cytokines were not detected even with wild-type H. pylori. An exception was Gro (the two spots above the IL-8 samples in Fig. 3). Taken together, these findings suggest that nonchemotactic H. pylori interacts normally in vitro with cultured gastric cells.
FIG. 3.
Cytokine/chemokine release from AGS cells. AGS cells were incubated with H. pylori wild-type strain SS1 and its isogenic mutants as indicated above the panels. Culture supernatant was collected and assayed using a cytokine array. The six panels show representative arrays from H. pylori SS1 strains. The white box on each array encompasses the IL-8 spots; the two spots above these spots, positive in every sample, are Gro. The group of four spots at the top left and the group of two spots at the bottom right are positive controls. The assays were repeated three or four times for each H. pylori strain; the results of one representative assay are shown.
cheY and cheW mutants are not as close to the gastric epithelium as wild-type H. pylori.
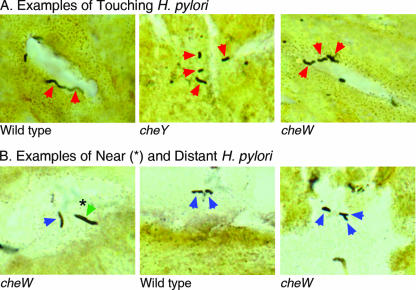
In vivo, inflammation is enhanced by bacterium-host cell interactions. This connection was shown in an elegant series of experiments by using transgenic mice that expressed the H. pylori adhesin Lewis B (LeB). These LeB mice had a greater number of H. pylori cells adhering and, concomitantly, a stronger inflammatory response than their wild-type siblings, strongly suggesting that bacterial adherence is a key factor for promoting inflammation (26). We thus decided to examine whether our various chemotaxis mutants had defects in interactions with gastric epithelial cells in vivo. We carried out this analysis in a blind fashion by examining Warthin-Starry-stained gastric tissue sections of H. pylori-infected mice for the bacterial position relative to the gastric gland surface. We used tissue sections from the same 6-month-infected C57BL/6 mice that were scored for inflammation (Fig. 1 and Table 3). We placed all visible bacteria with Helicobacter-like morphology in a stomach section into one of three classes, based on the nature of the bacterial contact with the gastric epithelial surface. These classes were (i) touching, (ii) near, and (iii) distant (see Materials and Methods) (Fig. 4). Using this rating system, we found there was a statistically significant difference in the distributions within each category between the mutants and the wild type in mice infected for 6 months (Table 6). For all nonchemotactic mutants (cheW, cheY, tlpA, and tlpB) 11 to 21% of the enumerated cells were in the superficial mucous layer, while only 3% of wild-type cells were found in this portion of the stomach. In contrast, 88% of the wild-type cells were scored as “touching,” while only 53 to 74% of the mutant cells were scored similarly. These observations are consistent with a model in which chemotaxis facilitates H. pylori-epithelium interactions. cheW and cheY nonchemotactic mutants have decreased cell interactions, and the mutants result in reduced inflammation. Although the tlpA and tlpB mutants do not fit as nicely into this model, we speculate that they may have differential interactions with a particular cell type that is difficult to detect in this assay.
FIG. 4.
Examples of H. pylori-gastric epithelium interactions observed in infected mouse stomach sections. Stomach samples from C57BL/6 mice infected for 6 months (same mice as those described in Fig. 1 and Tables 3 and 6) were stained with Warthin-Starry stain and visualized using light microscopy with a ×100 oil immersion objective. Bacteria stain dark brown, the epithelium stains gold, and the mucus stains light yellow-clear. A grader who was blinded to the identity of the infecting strain scored every visible bacterium with morphology appropriate for H. pylori as either touching, near, or distant, as described in the text. (A) Examples of bacteria scored as touching (indicated by arrowheads) for each bacterial strain (H. pylori wild-type strain SS1, SS1 cheY, or SS1 cheW). (B) Examples of bacteria scored as near and bacteria scored as distant (indicated by green and blue arrowheads, respectively).
TABLE 6.
Categorization of H. pylori-gastric epithelium interactions in C57BL/6 mice infected for 6 monthsa
| Strain | % Touching | % Near | % Distant | N |
|---|---|---|---|---|
| Wild-type SS1 | 87.6 | 9.0 | 3.4 | 276 |
| SS1 cheW | 53.4 | 24.9 | 21.7 | 178 |
| SS1 cheY | 70.6 | 17.9 | 11.4 | 243 |
| SS1 tlpA | 74.2 | 14.6 | 11.2 | 474 |
| SS1 tlpB | 63.0 | 22.2 | 14.8 | 1,005 |
The mouse samples were the same samples that were used for the experiments whose results are shown in Fig. 1 and Table 3. Categories are defined in the text. N is the number of bacteria examined; for each strain five or six mouse stomach sections (each from an individual mouse) were examined, and all visible helical bacteria were placed into one of the three categories. The data were analyzed using a chi-square test to compare the distribution between the three categories to a distribution that would occur randomly. The analysis indicated that the differences in frequency observed between groups have a very low probability of being generated by random chance (P < 0.00001) from a single theoretical frequency distribution.
DISCUSSION
The major finding of this study is that H. pylori chemotaxis has a profound influence on inflammation. This finding was presaged by studies showing that tlpB mutants cause less inflammation in gerbils, but we were unable to analyze the inflammatory response to totally nonchemotactic mutants (Che−) because they do not infect gerbils (40). Che− mutants do, in contrast, infect mice, and so these types of mutants were assessed for the ability to trigger inflammation. We found that two independent Che− mutants, lacking cheY or cheW, result in significantly less mouse stomach inflammation but colonize this organ to wild-type levels. This result suggests that these mutants have a specific defect in cell interactions that cause inflammation.
We carried out several experiments to investigate why nonchemotactic cheW or cheY mutants cause lessened inflammation. In tissue sections of infected mouse stomachs, we analyzed the interactions of Che− and wild-type H. pylori with the gastric epithelium. We found that Che− mutants are less closely cell associated, on average, than wild-type H. pylori is. The decreased intimate association may underlie the diminished inflammation, as other experiments have shown that bacterial adherence to gastric cells enhances inflammation (26). Thus, one possible model is that chemotaxis guides H. pylori to the epithelial cells and facilitates intimate bacterium-cell association. There are other possibilities, however, that are consistent with our findings. One possibility is that chemotaxis guides H. pylori to a particular niche and in that niche H. pylori expresses certain adhesins or proinflammatory molecules. In this model, the inflammation defect is due to Che− H. pylori not expressing certain proteins, rather than chemotaxis per se being important for bacterium-cell contact. This model is consistent with findings for Vibrio cholerae, in which nonchemotactic mutants misexpress certain virulence factors (38). Another possibility is that the dysregulated flagella of Che− mutants result in poor attachment. In the Che− mutants used here, flagella rotate continuously in the swim/counterclockwise direction (unpublished observations). There are other examples in which flagella locked into one rotational mode affect bacterium-cell interactions, such as Salmonella enterica serovar Typhimurium (32). In this case, swim/counterclockwise-locked flagellum mutants invaded cultured cells better than the wild type; mutants with tumble/clockwise-locked or paralyzed flagella invaded less well than the wild type, and totally aflagellate bacteria invaded similar to the wild type. These studies were interpreted to mean that the orientation of the flagella around the bacteria influences how the bacteria interact with cells. Salmonella flagella are peritrichous, and the flagella could whirl around the bacterium and block adhesins. H. pylori, in contrast, has polar flagella, and the whirling flagellar mass appears to be localized at one end of the cell (unpublished observations). Thus, it seems unlikely that a polar flagellar structure would block a significant portion of the bacterial cell surface. Consistent with this idea, we and others have found that H. pylori with paralyzed flagella adhere and cause IL-8 release similar to the wild type (12, 30; data not shown). Intriguingly, there is a link between adherence and chemotaxis in a relative of H. pylori, Campylobacter jejuni. In this case, however, mutants lacking cheY are hyperadherent (70), suggesting that this system is opposite that of H. pylori.
An H. pylori virulence factor known to contribute to severity of inflammation is the Cag PAI (10, 42). The Cag PAI encodes a type IV secretion system that delivers the CagA protein (5, 44, 55, 60) and peptidoglycan (65) to the mammalian cells. Both of these factors cause the mammalian cells to produce the IL-8 chemokine (9, 25, 65). The Cag PAI is not needed for animal colonization (18, 45) but is needed for full inflammation (45, 67). There is some controversy about whether the strain used here, SS1, contains a functional Cag PAI (14). Some studies have shown that SS1 causes low production of IL-8 relative to other strains and that mutation of the Cag PAI does not decrease the amount of IL-8 further (18, 64), while other studies have shown that SS1 does lead to significant IL-8 production (67, 68). Yamoaka et al. (67) discussed the variability in strains called SS1, indicating that some elicit IL-8 and others do not. The SS1 strain used here came directly from Adrian Lee and Janie O'Rourke and has been minimally passaged in the laboratory, suggesting that it likely retains any original functions. Furthermore, we show here that our SS1 does elicit at least some IL-8 production. The role of IL-8 induction in mouse infection is also unclear. Mice lack the gene for IL-8 and instead have functional homologs, MIP-2 and KC, that act through the same cellular receptor (58). H. pylori does cause the upregulation of MIP-2 and KC, and this response appears to be Cag PAI dependent (43, 68). Taken together, these studies suggest that H. pylori strain SS1 does upregulate IL-8, but whether this response is Cag dependent remains to be determined.
Intriguingly, S. enterica serovar Typhimurium Che− mutants have an inflammation deficit similar to the deficit that we report here for Che− H. pylori (59). In a mouse colitis model, serovar Typhimurium mutants lacking cheY infect the cecum and mesentery lymph nodes similar to the wild type but cause less intestinal inflammation than the wild type. The inflammatory defect of Che− mutants was similar to the defect found with aflagellate mutants. The latter mutants were analyzed for bacterial position relative to the intestinal epithelial surface in infected mouse colons. Aflagellate mutants were, on average, further from the cell surface than the wild type. Taken together, these results suggest that chemotactic motility drives serovar Typhimurium to the intestinal epithelial surface and this interaction in turn promotes inflammation. This phenotype is similar to the one that we describe for H. pylori and suggests that chemotaxis plays similar roles in the infection cycles of these microbes.
Two of the four H. pylori chemoreceptors, TlpB and TlpA, modulate inflammation. Interestingly, mutants lacking these chemoreceptors initially cause somewhat lower inflammatory scores, but at later time points mice infected with these mutants have significantly higher inflammatory scores. One possibility is that these mutants have delayed inflammation kinetics such that they have “caught up” to the wild type by 6 months. Even at 6 months, however, these mutants still seem to be removed from the gastric epithelial surface. Although we do not yet understand this phenotype, it should be noted that our method for monitoring the bacterium-epithelial distance is crude. Most of the Che− tlpA and tlpB mutants are close to the gastric epithelium in toto, but we do not know if the mutants home to different cells than the wild type. TlpB has been suggested to sense protons, as mutants lacking the TlpB gene lose a characteristic swimming response to acid (16). The same workers found that H. pylori SS1 tlpB mutants have a colonization defect in IL-12-deficient mice. IL-12 is a proinflammatory cytokine, and mice lacking this protein are extrapermissive for H. pylori infection (29). It is not yet clear why tlpB mutants have a colonization defect in this model and not in normal mice, although possibly the lack of an inflammatory response makes tlpB mutants less able to survive. Recruitment of other T-cell populations found to be regulators of inflammation and bacterial colonization may be disrupted in this mouse strain (50). TlpA is required for colonization of wild-type mice in a competition infection model (3). Using H. pylori strain 26695, some workers have suggested that TlpA senses both arginine and bicarbonate (11). H. pylori strain 26695 is typically nonmotile (33), and Cerda et al. (11) gave no information about whether the strain was treated to yield a motile variant. We have been unable to demonstrate chemotaxis toward arginine using H. pylori strain SS1 and several chemotaxis assays (28, 71; K. M. Ottemann and T. M. Andermann, unpublished data).
In summary, we have shown that proper chemotaxis enhances H. pylori-triggered inflammation. Chronic inflammation is the source of clinical disease in H. pylori infection, and so a better understanding of this process may help us design therapies that can thwart this pathological response. Our data furthermore highlight the finding that two of the four H. pylori chemoreceptors direct this response, suggesting that H. pylori senses specific host molecules in order to modulate inflammation. We also observed that nonchemotactic mutants are less intimate with host cells than wild-type H. pylori is. Because adherence enhances gastritis, this finding suggests a model in which chemotaxis guides H. pylori to adhere to cells of the gastric epithelium, thereby triggering an inflammatory response to the pathogen.
Acknowledgments
We sincerely thank Marla Hill for assistance with the animal experiments, Tommy Williams for suggestions on statistical analysis of categorical data, and members of the Ottemann lab for comments on the manuscript.
This work was supported by Public Health Service grants R01 CA101931 (to D.J.M.) and RO1 AI050000 (to K.M.O.) from the National Institutes of Health.
Editor: A. Camilli
Footnotes
Published ahead of print on 21 May 2007.
REFERENCES
- 1.Akanuma, M., S. Maeda, K. Ogura, Y. Mitsuno, Y. Hirata, T. Ikenoue, M. Otsuka, T. Watanabe, Y. Yamaji, H. Yoshida, T. Kawabe, Y. Shiratori, and M. Omata. 2002. The evaluation of putative virulence factors of Helicobacter pylori for gastroduodenal disease by use of a short-term Mongolian gerbil infection model. J. Infect. Dis. 185:341-347. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. Dejonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merber, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Andermann, T. M., Y.-T. Chen, and K. M. Ottemann. 2002. Two predicted chemoreceptors promote Helicobacter pylori infection. Infect. Immun. 70:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 5.Asahi, M., T. Azuma, S. Ito, Y. Ito, H. Suto, Y. Nagai, M. Tsubokawa, Y. Tohyama, S. Maeda, M. Omata, T. Suzuki, and C. Sasakawa. 2000. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 191:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley and Sons, Hoboken, NJ.
- 7.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179:4676-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-522. [DOI] [PubMed] [Google Scholar]
- 9.Brandt, S., T. Kwok, R. Hartig, W. Konig, and S. Backert. 2005. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc. Natl. Acad. Sci. USA 102:9300-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerda, O., A. Rivas, and H. Toldeo. 2003. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224:175-181. [DOI] [PubMed] [Google Scholar]
- 12.Clyne, M., T. O'Croinin, S. Suerbaum, C. Josenhans, and B. Drumm. 2000. Adherence of isogenic flagellum-negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect. Immun. 68:4335-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conlin, V. S., S. B. Curtis, Y. Zhao, E. D. Moore, V. C. Smith, R. M. Meloche, B. B. Finlay, and A. M. Buchan. 2004. Helicobacter pylori infection targets adherens junction regulatory proteins and results in increased rates of migration in human gastric epithelial cells. Infect. Immun. 72:5181-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crabtree, J., R. Ferrero, and J. Kusters. 2002. The mouse colonizing Helicobacter pylori strain SS1 may lack a functional cag pathogenicity island. Helicobacter 7:139-140. [DOI] [PubMed] [Google Scholar]
- 15.Crabtree, J. E., A. Covacci, S. M. Farmery, Z. Xiang, D. S. Tompkins, S. Perry, and I. J. D. Lindley. 1995. Helicobacter pylori induced interleukin-8 expression in gastric epithelial cells is associated with CagA positive phenotype. J. Clin. Pathol. 48:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croxen, M. A., G. Sisson, R. Melano, and P. S. Hoffman. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188:2656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton, K. A., L. H. Benson, J. Haeger, and B. M. Gray. 2006. Role of transcription factor T-bet expression by CD4+ cells in gastritis due to Helicobacter pylori in mice. Infect. Immun. 74:4673-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eaton, K. A., D. Kersulyte, M. Mefford, S. J. Danon, S. Krakowka, and D. E. Berg. 2001. Role of Helicobacter pylori cag region genes in colonization and gastritis in two animal models. Infect. Immun. 69:2902-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 20.Eaton, K. A., M. J. Radin, and S. Krakowka. 1995. An animal model of gastric ulcer due to bacterial gastritis in mice. Vet. Pathol. 32:489-497. [DOI] [PubMed] [Google Scholar]
- 21.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrero, R. L., and J. G. Fox. 2001. In vivo modeling of Helicobacter-associated gastrointestinal diseases, p. 565-582. In H. L. T. Mobley, G. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed]
- 23.Ferrero, R. L., J. M. Thiberge, M. Huerre, and A. Labigne. 1998. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect. Immun. 66:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girardin, S. E., I. G. Boneca, L. A. Carneiro, A. Antignac, M. Jehanno, J. Viala, K. Tedin, M. K. Taha, A. Labigne, U. Zahringer, A. J. Coyle, P. S. DiStefano, J. Bertin, P. J. Sansonetti, and D. J. Philpott. 2003. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300:1584-1587. [DOI] [PubMed] [Google Scholar]
- 26.Guruge, J. L., P. G. Falk, R. G. Lorenz, M. Dans, H.-P. Wirth, M. J. Blaser, D. E. Berg, and J. I. Gordon. 1998. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 95:3925-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkins, A. C., and C. S. Harwood. 2002. Chemotaxis of Ralstonia eutropha JMP134(pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Appl. Environ. Microbiol. 69:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman, P. S., N. Vats, D. Hutchison, J. Butler, K. Chisholm, G. Sisson, A. Raudonikiene, J. S. Marshall, and S. J. Veldhuyzen van Zanten. 2003. Development of an interleukin-12-deficient mouse model that is permissive for colonization by a motile KE26695 strain of Helicobacter pylori. Infect. Immun. 71:2534-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, J., P. W. O'Toole, P. Doig, and T. Trust. 1995. Stimulation of interleukin-8 production in epithelial cell lines by Helicobacter pylori. Infect. Immun. 63:1732-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Israel, D. A., N. Salama, C. N. Arnold, S. F. Moss, T. Ando, H.-P. Wirth, K. T. Tham, M. Camorlinga, M. J. Blaser, S. Falkow, and R. M. Peek. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Investig. 107:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Josenhans, C., K. A. Eaton, T. Thevenot, and S. Suerbaum. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 68:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 35.Kim, N., E. A. Marcus, Y. Wen, D. L. Weeks, D. R. Scott, H. C. Jung, I. S. Song, and G. Sachs. 2004. Genes of Helicobacter pylori regulated by attachment to AGS cells. Infect. Immun. 72:2358-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, S. Y., Y. C. Lee, H. K. Kim, and M. J. Blaser. 2006. Helicobacter pylori CagA transfection of gastric epithelial cells induces interleukin-8. Cell. Microbiol. 8:97-106. [DOI] [PubMed] [Google Scholar]
- 37.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 38.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGee, D. J., C. Coker, T. L. Testerman, J. M. Harro, S. V. Gibson, and H. L. Mobley. 2002. The Helicobacter pylori flbA flagellar biosynthesis and regulatory gene is required for motility and virulence and modulates urease of H. pylori and Proteus mirabilis. J. Med. Microbiol. 51:958-970. [DOI] [PubMed] [Google Scholar]
- 40.McGee, D. J., M. L. Langford, E. L. Watson, J. E. Carter, Y.-T. Chen, and K. M. Ottemann. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michetti, P., and A. M. Svennerholm. 2003. Helicobacter pylori—inflammation, immunity and vaccines. Helicobacter 8:31-35. [DOI] [PubMed] [Google Scholar]
- 42.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 43.Obonyo, M., D. G. Guiney, J. Fierer, and S. P. Cole. 2003. Interactions between inducible nitric oxide and other inflammatory mediators during Helicobacter pylori infection. Helicobacter 8:495-502. [DOI] [PubMed] [Google Scholar]
- 44.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 45.Ogura, K., S. Maeda, M. Nakao, T. Watanabe, M. Tada, T. Kyutoku, H. Yoshida, Y. Shiratori, and M. Omata. 2000. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J. Exp. Med. 192:1601-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Toole, P. W., M. C. Lane, and S. Porwollik. 2000. Helicobacter pylori motility. Microbes Infect. 2:1207-1214. [DOI] [PubMed] [Google Scholar]
- 47.Ottemann, K. M., and A. Lowenthal. 2002. Helicobacter pylori uses motility for both initial colonization and to attain robust infection. Infect. Immun. 70:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 49.Pittman, M. S., M. Goodwin, and D. J. Kelly. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493-2504. [DOI] [PubMed] [Google Scholar]
- 50.Rad, R., L. Brenner, S. Bauer, S. Schwendy, L. Layland, C. P. da Costa, W. Reindl, A. Dossumbekova, M. Friedrich, D. Saur, H. Wagner, R. M. Schmid, and C. Prinz. 2006. CD25+/Foxp3+ T cells regulate gastric inflammation and Helicobacter pylori colonization in vivo. Gastroenterology 131:525-537. [DOI] [PubMed] [Google Scholar]
- 51.Rosario, M. M. L., K. L. Fredrick, G. W. Ordal, and J. D. Helmann. 1994. Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs. J. Bacteriol. 176:2736-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth, K. A., S. B. Kapadia, S. M. Martin, and R. G. Lorenz. 1999. Cellular immune responses are essential for the development of Helicobacter felis-associated gastric pathology. J. Immunol. 163:1490-1497. [PubMed] [Google Scholar]
- 53.Rugge, M., P. Correa, M. F. Dixon, R. Fiocca, T. Hattori, J. Lechago, G. Leandro, A. B. Price, P. Sipponen, E. Solcia, H. Watanabe, and R. M. Genta. 2002. Gastric mucosal atrophy: interobserver consistency using new criteria for classification and grading. Aliment. Pharmacol. Ther. 16:1249-1259. [DOI] [PubMed] [Google Scholar]
- 54.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. The vacuolating cytotoxin of Helicobacter pylori plays a role during colonization of a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segal, E. D., S. Falkow, and L. S. Tompkins. 1996. Helicobacter pylori attachment to gastric cells induces cytoskeletal rearrangements and tyrosine phosphorylation of host cell proteins. Proc. Natl. Acad. Sci. USA 93:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimoyama, T., and J. E. Crabtree. 1998. Bacterial factors and immune pathogenesis in Helicobacter pylori infection. Gut 43(Suppl.):S2-S5. [PMC free article] [PubMed] [Google Scholar]
- 58.Singer, M., and P. J. Sansonetti. 2004. IL-8 is a key chemokine regulating neutrophil recruitment in a new mouse model of Shigella-induced colitis. J. Immunol. 173:4197-4206. [DOI] [PubMed] [Google Scholar]
- 59.Stecher, B., S. Hapfelmeier, C. Muller, M. Kremer, T. Stallmach, and W. D. Hardt. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:4138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA 97:1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terry, K., A. C. Go, and K. M. Ottemann. 2006. Proteomic mapping of a suppressor of nonchemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol. Microbiol. 61:871-882. [DOI] [PubMed] [Google Scholar]
- 62.Terry, K., S. M. Williams, L. Connolly, and K. M. Ottemann. 2005. Chemotaxis plays multiple roles in Helicobacter pylori animal infection. Infect. Immun. 73:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Lftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Tizegerald, N. Lee, M. D. Adams, E. K. Kichey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Person, J. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Wathey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 64.van Doorn, N. E., F. Namavar, M. Sparrius, J. Stoof, E. P. van Rees, L. J. van Doorn, and C. M. Vandenbroucke-Grauls. 1999. Helicobacter pylori-associated gastritis in mice is host and strain specific. Infect. Immun. 67:3040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viala, J., C. Chaput, I. G. Boneca, A. Cardona, S. E. Girardin, A. P. Moran, R. Athman, S. Memet, M. R. Huerre, A. J. Coyle, P. S. DiStefano, P. J. Sansonetti, A. Labigne, J. Bertin, D. J. Philpott, and R. L. Ferrero. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5:1166-1174. [DOI] [PubMed] [Google Scholar]
- 66.Wang, J., T. G. Blanchard, and P. B. Ernst. 2001. Host inflammatory response to infection, p. 471-480. In H. L. T. Mobley, G. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC.
- 67.Yamaoka, Y., M. Kita, T. Kodama, S. Imamura, T. Ohno, N. Sawai, A. Ishimaru, J. Imanishi, and D. Y. Graham. 2002. Helicobacter pylori infection in mice: role of outer membrane proteins in colonization and inflammation. Gastroenterology 123:1992-2004. [DOI] [PubMed] [Google Scholar]
- 68.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 70.Yao, R., D. H. Burr, and P. Guerry. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23:1021-1031. [DOI] [PubMed] [Google Scholar]
- 71.Yu, H. S., and M. Alam. 1997. An agarose-in-plug bridge method to study chemotaxis in the Archaeon Halobacterium salinarum. FEMS Microbiol. Lett. 156:265-269. [DOI] [PubMed] [Google Scholar]