Abstract
Successful establishment of infection by bacterial pathogens requires fine-tuning of virulence-related genes. Quorum sensing (QS) is a global regulation process based on the synthesis of, detection of, and response to small diffusible molecules, called N-acyl-homoserine lactones (AHL), in gram-negative bacteria. In numerous species, QS has been shown to regulate genes involved in the establishment of pathogenic interactions with the host. Brucella melitensis produces N-dodecanoyl homoserine lactones (C12-HSL), which down regulate the expression of flagellar genes and of the virB operon (encoding a type IV secretion system), both of which encode surface virulence factors. A QS-related regulator, called VjbR, was identified as a transcriptional activator of these genes. We hypothesized that VjbR mediates the C12-HSL effects described above. vjbR alleles mutated in the region coding for the AHL binding domain were constructed to test this hypothesis. These alleles expressed in trans in a ΔvjbR background behave as constitutive regulators both in vitro and in a cellular model of infection. Interestingly, the resulting B. melitensis strains, unable to respond to AHLs, aggregate spontaneously in liquid culture. Preliminary characterization of these strains showed altered expression of some outer membrane proteins and overproduction of a matrix-forming exopolysaccharide, suggesting for the first time that B. melitensis could form biofilms. Together, these results indicate that QS through VjbR is a major regulatory system of important cell surface structures of Brucella and as such plays a key role in host-pathogen interactions.
Brucella spp. are gram-negative intracellular pathogens belonging to the α-2 proteobacteria group, like Agrobacterium, Rhizobium, and Rickettsia, which also live in close association with a eukaryotic host (46). Bacteria of the genus Brucella are the etiologic agents of brucellosis, a worldwide zoonosis affecting a broad range of mammals and triggering important economic losses (63). Three Brucella species, B. melitensis, B. abortus, and B. suis, are able to infect humans, causing a chronic, debilitating disease with severe, sometimes fatal outcomes.
Brucellae are remarkably well adapted to the intracellular lifestyle, being able to invade and to survive within macrophages and nonprofessional phagocytes (17, 51). This is one of the bases for the still poorly understood chronicity of brucellosis (26). This aptitude relies on the perturbation of vesicular trafficking and the creation of a unique intracellular replication niche derived from the endoplasmic reticulum (7, 8).
Brucella is confronted with very diverse environments and host defenses both in the extracellular milieu and inside host cells. It is thus expected that this pathogen has to sense external and internal signals to achieve successful adaptation throughout its infectious cycle. Among such systems, quorum sensing (QS), stringent response, and signal transduction through two-component regulators have been particularly well studied (for a review, see reference 40). In this study we focused on QS, a communication system used by a large number of bacteria to coordinate gene expression within a population according to population density (30) or limited diffusion in a restricted environment (53). In gram-negative bacteria, this communication system involves the synthesis, release, and subsequent detection of small diffusible molecules or autoinducers (commonly acyl homoserine lactones [AHLs]). As the bacterial population expands, the extracellular concentration of AHLs increases. When the autoinducer concentration reaches a threshold level, AHLs bind to LuxR-type transcriptional regulators comprising an N-terminal AHL binding domain and a C-terminal DNA binding domain containing a helix-turn-helix motif. This interaction leads to conformational changes of the regulator and subsequent modifications of target gene transcription. The phenotypes regulated by QS are as diverse as bioluminescence (37), competence (49), biofilm formation (44), and virulence (50, 75, 78).
We identified two LuxR-type regulators in the sequenced B. melitensis genome (16), the previously described VjbR regulator (BMEI1116) (14) and a second regulator, currently undergoing characterization, called BabR (BMEI1758). Despite several attempts, we were not able to identify an AHL synthetase in B. melitensis. However, we have previously identified N-dodecanoyl-dl-homoserine lactone (C12-HSL) from B. melitensis culture supernatant (73). C12-HSL represses the transcription of the flagellar gene fliF (14) and of the virB operon (73), whereas VjbR is a transcriptional activator of these two surface-associated virulence factors (16). The fliF gene encodes the flagellar MS ring monomer implicated in the establishment of chronic infection (28), while the virB operon encodes a type IV secretion system (TFSS) involved in the control of Brucella-containing vacuole maturation into a replication-permissive organelle (11).
In the current study, we investigated whether VjbR could mediate the C12-HSL repressor effect. To achieve this objective, we used VjbR polypeptides mutated in the N-terminal AHL binding domain of the regulator. These mutant polypeptides behave as signal-independent regulators both in B. melitensis cultures and during cellular infection. Strains expressing these mutated regulators displayed a clumping phenotype that led us to investigate the role of VjbR in the regulation of cell surface components. Our data show that VjbR regulates exopolysaccharide (EPS) synthesis or export and also the production of several outer membrane proteins (Omps), some of which are involved in virulence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All strains and plasmids used in this study are listed in Table 1. Brucella strains were grown with shaking at 37°C in 2YT medium (10% yeast extract, 10 g liter−1 tryptone, 5 g liter−1 NaCl) containing appropriate antibiotics from an initial optical density at 600 nm (OD600) of 0.05.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| B. melitensis strains | ||
| 16M | Wild type, Nalr | A. Macmillan, Central Veterinary Laboratory, Weybridge, United Kingdom |
| CD100 | ΔvjbR::Kanr | 14 |
| CD110 | ΔvjbR::Kanr, pJD27 PvirB Ampr | 14 |
| SB200 | ΔvjbR::Kanromp31::Cmr | This study |
| B. abortus strains | ||
| 2308 | Wild type, Nalr | 54 |
| RMD100 | ΔvjbR::Kanr | R.-M. Delrue, unpublished data |
| E. coli strains | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 lacX74 recA1 endA1 araΔ139 Δ(ara leu)7697 galU galK λ−rps (Strr) nupG | Gibco BRL |
| S17-1(λ pir) | recA thi pro hsdR [res− mod+][RP4::2-Tc::Mu-Km::Tn7] λpir phage lysogen | 61 |
| Plasmids | ||
| pSK-oriT cat | Suicide vector, Cmr | I. Danese and P. Lestrate, unpublished data |
| pSB001 | pSK-oriT cat derivative carying a 500-bp internal fragment of BMEII0844 | This study |
| pJD27 PvirB | PvirB-luxAB, Ampr | 14 |
| pDONR201 | Gateway vector, Kanr | Invitrogen |
| pSB101 | pDONR carrying the PCR product vjbR | This study |
| pSB102 | pDONR carrying the PCR product vjbR(Δ1-180) | This study |
| pSB103 | pDONR carrying the PCR product vjbR(D82A) | This study |
| pMR10-cat | Broad-host-range cloning vector, RK2 OriV Cmr Kanr | A. A. Bourniquel |
| pRH001 | pMR10 derivative gateway destinantion vector, medium copy number, Cmr | 34 |
| pSB201 | pRH001 derivative, Plac-controlled synthesis of VjbR | This study |
| pSB202 | pRH001 derivative, Plac-controlled synthesis of VjbR(Δ1-180) | This study |
| pSB203 | pRH001 derivative, Plac-controlled synthesis of VjbR(D82A) | This study |
| pBBR1-MCS1 | Broad-host-range cloning vector, Cmr | 38 |
| pRH002 | pBBR1-MCS1 derivative gateway destination vector, high copy number, rep, Cmr | 34 |
| pSB301 | pRH002 derivative, Plac-controlled synthesis of VjbR | This study |
| pSB302 | pRH002 derivative, Plac-controlled synthesis of VjbR(Δ1-180) | This study |
| pSB303 | pRH002 derivative, Plac-controlled synthesis of VjbR(D82A) | This study |
| pSB305 | pRH002 derivative, Plac-controlled synthesis of Omp31 (BMEII0844) | This study |
| pRH018 | pRH002 derivative allowing C-terminal fusion with 13Myc tag | 34 |
| pSB401 | pRH018 derivative, Plac-controlled synthesis of VjbR-13Myc | This study |
| pSB402 | pRH018 derivative, Plac-controlled synthesis of VjbR(Δ1-180)-13Myc | This study |
| pSB403 | pRH018 derivative, Plac-controlled synthesis of VjbR(D82A)-13Myc | This study |
Nalr, nalidixic acid resistant; Kanr, kanamycin resistant; Cmr, chloramphenicol resistant; Ampr, ampicillin resistant.
The Escherichia coli DH10B (Gibco BRL), S17-1 (60), and DB3.1 (Invitrogen) strains were grown in Luria-Bertani medium with appropriate antibiotics.
Chloramphenicol, nalidixic acid, and ampicilin were used at 20 μg/ml, 25 μg/ml, and 100 μg/ml, respectively. Synthetic C12-HSL from Fluka was prepared in acetonitrile (ACN) and was added to bacterial growth media at a final concentration of 5 μM. The same volume of ACN was used for a negative control.
Plasmid construction.
DNA manipulations were performed according to standard techniques (1). Restriction enzymes were purchased from Roche, and primers were purchased from Sigma-Aldrich.
Derivatives of the replicative plasmids pRH001 and pRH002 (34) harboring vjbR mutant alleles were constructed using the Gateway technique (Invitrogen). The destination vectors pRH001 and pRH002 harbor a chloramphenicol resistance (cat) marker and the toxic cassette ccdB. This group of genes is flanked by attR1 and attR2 recombination sites. The wild-type (wt) control allele corresponding to the total VjbR protein (amino acids 1 to 260) was amplified with primers VjbR-B1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGAGTCTTGATCTCGTTCAT-3) and VjbR-B2 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACACGAGATGCTGTACCTCG-3′). Gateway primers HTH-B1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGAAGGATGCAAATTCAGTTGCAAG-3′) and VjbR-B2 were used for amplification of the predicted C-terminal DNA binding domain corresponding to amino acids 181 to 260 of VjbR [vjbR(Δ1-180)]. B. melitensis 16M genomic DNA was used as the template for all amplifications. The resulting PCR products (vjbR-wt and vjbR-HTH, respectively) were cloned into pDONR201 (Invitrogen Life Technologies) by the BP reaction as described previously (21). The resulting entry clones pSB101 and pSB102 were confirmed by PCR using primers VjbR-B1 and VjbR-B2 and primers HTH-B1 and VjbR-B2, respectively.
Aspartate 82 of VjbR was mutated into alanine via PCR-based site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene), using pSB101 as the template. The resulting plasmid, pSB103, was sequenced to confirm the mutation using primers VjbR-B1 and VjbR-B2.
Entry clones containing vjbR alleles were used together with the destination vectors pRH001 and pRH002 during Gateway LR reactions as described previously (21). For quantification of the autoinducer response of VjbR mutants, the resulting vectors pSB201, pSB102, and pSB103 were transferred into the CD110 strain (B. melitensis ΔvjbR::Kanr containing a PvirB-luxAB transcriptional fusion) by mating. For a cellular infection experiment the resulting vectors pSB301, pSB302, and pSB303 were transferred into the CD100 strain (B. melitensis ΔvjbR::Kanr). For characterization of the clumping phenotype in B. abortus, the vectors pSB201, pSB202, and pSB203 were transferred into the RMD100 strain (B. abortus ΔvjbR::Kanr).
For construction of ΔvjbR::Kanr omp31::Cmr, an internal fragment of omp31 (BMEII0844) was initially amplified by PCR from B. melitensis 16M genomic DNA with primers F-Omp31 (5′-CTCGGCATTGCGGCTATTTTC-3′) and R-Omp31 (5′-CAGGTTGAACGCAGATTT-3′). The R-Omp31 primer contains a TGA stop codon to avoid production of a functional truncated protein. The 341-bp amplified product was then inserted into the EcoRV-digested pSK-oriT cat vector in the opposite orientation relative to the Plac promoter to avoid expression of the 3′ fragment of the disrupted coding sequences in Brucella. The construct was introduced into B. melitensis 16M (Nalr) from E. coli S17-1 by mating. A single crossover then led to disruption of the wt locus on the chromosome. Integrative mutants were selected on a medium containing kanamycin and nalidixic acid.
Plasmids used to assess mutated polypeptide stability were constructed using the Gateway technology with the pSB101 (pPlac-vjbR), pSB102 [pPlac-vjbR(Δ1-180)], and pSB103 [pPlac-vjbR(D82A)] entry clones and the pRH018 destination vector (34). The resulting plasmids pSB401, pSB402, and pSB403 allowed a C-terminal fusion of the regulator with the 13Myc tag.
The complementation plasmid carrying the omp31 open reading frame under Plac control (pSB305) was constructed from pRH002 and pDONR-BMEII0844 from the ORFeome (21) using the Gateway technology.
Mating.
Mating was performed by mixing equal volumes (100 μl) of liquid cultures of E. coli S17-1 donor cells (OD600 = 0.6) and the B. melitensis 16M Nalr recipient strain (overnight culture) on a 0.22-μm-pore-size filter. The filter was left for 1 h on a 2YT medium plate without antibiotics and then transferred onto a 2YT medium plate containing chloramphenicol and nalidixic acid. After 3 days of incubation at 37°C, the exconjugates were replicated on a 2YT medium plate containing nalidixic acid and chloramphenicol.
Measurement of luciferase activity.
Bacterial strains were grown overnight in 2YT medium with aeration at 37°C. Cultures were centrifuged, and the bacterial pellets were resuspended and washed twice in 2YT medium. For each strain, three 10-ml portions of cultures in 2YT medium (initial OD600 of 0.05) were incubated at 37°C with shaking. After 20 h (PvirB expression peak) the OD600 was measured, and 0.2-ml samples were harvested. N-decanal substrate was added to a final concentration of 0.145 mM (stock concentration, 5.8 mM in 50% ethanol). After 10 min, light production was measured for 5 s using a Microlumat LB96P luminometer (EG and Berthold). Luciferase activity was expressed in relative luminescence units per OD600 unit at a given time point. Measurement was performed in triplicate.
Cellular infections.
Survival of Brucella strains was evaluated in an immortalized cell line of bovine peritoneal macrophages (67) as described previously (15). A ΔvjbR mutant was used as a negative control for replication defects during the cellular infection. C12-HSL was added at a final concentration of 5 μM together with gentamicin. The same volume of ACN, the C12-HSL resuspension solvent, was used as a negative control.
Scanning electron microscopy.
B. melitensis strains were grown overnight in 2YT medium with aeration at 37°C. For each strain three 1-ml portions of cultures in 2YT medium (initial OD600 of 0.05) were incubated at 37°C with shaking in a 24-well plate containing an insert plate with a porous membrane (diameter, 1.0 μm) (BD Falcon). After 24 h, brucellae were fixed for 20 min with 4% paraformaldehyde (PFA), and plates were centrifuged for 10 min at 1,000 rpm. Membranes were cut and dehydrated for 5 min in 25, 50, 75, 95, and 100% ethanol at room temperature. They were finally prepared by critical-point drying, mounted on an aluminum stub, and covered with a thin layer of gold (20 to 30 nm). Examination was carried out with a scanning electron microscope (XL-20; Eindhoven, Philips, The Netherlands) at the Unité Interfacultaire de Microscopie Electronique (University of Namur, Belgium).
EPS staining.
Bacteria in a late-exponential-phase culture (OD600, 1.0) were fixed with 4% PFA for 20 min before staining.
(i) Calcofluor white staining.
For detection of polysaccharides, 1 ml of 0.05% calcofluor white (fluorescent whitener 28; Sigma) was added to 0.1 ml of PFA-fixed cells. Visualization was accomplished with an epifluorescence microscope (Nikon Eclipse E1000) with the appropriate filter sets. Pictures were captured using a Hamamatsu ORCA-ER camera.
(ii) ConA-FITC staining.
For confocal microscopy, 0.1 ml of concanavalin A (ConA)-fluorescein isothiocyanate (FITC) (1 mg/ml) was added to 0.2 ml of PFA-fixed cells. One microliter of propidium iodide (10 mM) was added for visualizing bacteria. After incubation for 30 min in the dark, cells were washed in phosphate-buffered saline (PBS) (pH 8.5), resuspended in 100 μl of the same buffer, and examined immediately using a Leica SP-1 confocal laser scanning microscope.
Dot blot analysis.
Brucellae were grown for 48 h in 2YT medium at 37°C. Crude extracts were prepared as follows. After being washed in fresh 2YT medium, bacteria were concentrated 10-fold in PBS and inactivated for 1 h at 80°C. Equivalent amounts of proteins for each crude extract were used for serial twofold dilutions. Two microliters of each dilution was applied to a nitrocellulose membrane (Hybond; Amersham). Omp immunodetection was performed with the following monoclonal antibodies (MAbs) (10): anti-Omp25 MAb (A68/4B10/F5) at 1/100, anti-Omp31 MAb (A59/10F9/G10) at 1/100, anti-Omp36 MAb (A68/25G5/A5) at 1/100, anti-Omp 89 MAb (A53/10B2) at 1/1,000, anti-Omp10 MAb (A68/7G11/C10) at 1/5, anti-Omp19 MAb (A68/25H10/A5) at 1/5, and anti-Omp16 (A68/08C03/G03) at 1/10. Horseradish peroxidase-conjugated goat anti-mouse antibodies (Amersham) were used at 1/5,000 along with the ECL system (Amersham) to develop blots for chemoluminescence before visualization on film. Dot blots using MAbs specific for Omp16 (PAL lipoprotein) were used as internal loading controls. This protein did not show any change in the conditions tested. Dot blots were quantified using a PhosphorImager. The values used for the graph corresponded to the first dilutions at which differences between samples could be seen.
Polymyxin B test.
Bacterial survival after controlled exposure to polymyxin B (7,870 U/mg; Sigma-Aldrich, Germany) was assayed essentially as described by Sola-Landa et al. (64). Briefly, serial dilutions of polymyxin B prepared in 1 mM HEPES (pH 8.0) were prepared in 96-well microtiter-type plates. Bacteria resuspended at 2 × 104 CFU/ml were dispensed into triplicate rows, and plates were incubated for 1 h at 37°C. Viable bacteria were counted by spreading 20 μl from each well onto 2YT agar. The results were expressed as percentages of survival; 100% corresponded to the control incubated without the peptide.
ELLSA applied to culture supernatants.
A peroxidase-labeled ConA solution stored at −20°C was diluted in PBS containing 0.05% (vol/vol) Tween 20 diluting buffer to obtain a final concentration of 10 μg ml−1. One hundred microliters of the peroxidase-labeled lectin solution was added to wells previously coated for 16 h with 100-μl portions of supernatants of stationary-phase cultures vortexed for 1 min at full speed. At least three parallel experiments per sample dilution were run in each assay. Wells covered with PBS for the same contact time that was used for supernatants were subjected to the same treatment and used to estimate the nonspecific binding in the enzyme-linked lectin sorbent assay (ELLSA) response. Microtiter plates (Maxisorp; Nunc) were placed at room temperature for 1 h to allow the lectin to bind to the polysaccharides. The peroxidase-labeled lectin solution was removed from the wells by inverting the plates and tapping them on absorbent paper. Following five successive washes with 200 μl of diluting buffer to eliminate unbound enzyme conjugate, the linked peroxidase conjugate was visualized following addition of 100 μl of K-Blue as recommended by the manufacturer (Neogen). The reaction was allowed to develop for 15 min in the dark, and the absorbance at 650 nm and 450 nm was measured was a microplate reader.
Statistical analysis.
Anova I was used for data analysis after the homogeneity of variance was tested (Barlett test). Average comparisons were performed by using pairwise Scheffe's test (55). The error bars in figures indicate the 95% confidence intervals of the means (computed from the residual mean square using Student's t test, α = 0.05).
RESULTS
Negative effect of C12-HSL on PvirB expression is mediated by VjbR.
As previously described, the LuxR-type regulator VjbR and C12-HSLs share common targets (14). VjbR is required for virB expression, and C12-HSLs repress the transcription from the virB promoter (PvirB). These observations led to the hypothesis that the C12-HSL repressor effect on the PvirB promoter could be linked to its inhibitory effect on the VjbR regulator. To test this hypothesis, we constructed two vjbR alleles mutated in the predicted AHL binding domain. The structure of TraR (the LuxR-type regulator of Agrobacterium tumefaciens) bound to its autoinducer led to prediction of several conserved amino acids directly involved in the binding of the pheromone (76, 82). These studies suggest that several hydrogen bonds, between the AHL and some conserved amino acids within the AHL binding hydrophobic pocket, are involved in the binding of the AHL. These residues are highly conserved in LuxR-type regulators (76, 82). One of them, Asp70, is conserved in VjbR (Asp82). Mutation of this amino acid has been described to inactivate the AHL binding to LuxR-type regulators (42, 57, 62). Consequently, we constructed the vjbR(D82A) allele encoding replacement of aspartate 82 with alanine. The vjbR(Δ1-180) allele results in the complete deletion of the predicted autoinducer binding domain (14). wt as well as mutant alleles of vjbR were cloned under Plac control into the medium-copy-number plasmid pRH001 to generate pSB201 (pPlac-vjbR), pSB202 [pPlac-vjbR(Δ1-180)], and pSB203 [pPlac-vjbR(D82A)] (Table 1). For the following experiments, the plasmids containing the wt vjbR, vjbR(Δ1-180), and vjbR(D82A) alleles were designated pSBN01, pSBN02, and pSBN03, respectively.
To assess the effect of mutated VjbR regulators on PvirB activity, pSB201, pSB202, and pSB203 were introduced into CD110, a B. melitensis ΔvjbR::Kanr strain containing a PvirB-luxAB transcriptional fusion as a reporter (Table 1). As shown in Fig. 1, the activity of PvirB-luxAB was reduced twofold in the presence of the wt vjbR allele and C12-HSL, analogous to the effect of this signal molecule in the wt strain. We were not able to detect any repression effect on PvirB upon addition of C12-HSL with the vjbR(D82A) or vjbR(Δ1-180) allele. Since both mutant regulators should be unable to bind C12-HSL, these results suggest that the LuxR-type regulator VjbR mediates the repression of PvirB by C12-HSL. As PvirB is insensitive to AHL repression in the presence of the vjbR(D82A) or vjbR(Δ1-180) allele, we propose that the VjbR polypeptides encoded by these alleles are defective in AHL binding and therefore behave like constitutive regulators.
FIG. 1.
Schematic representation of the VjbR mutated polypeptides. The pSB201-encoded wt VjbR polypeptide is shown at the top; the proposed autoinducer (AI) binding region is indicated by a solid bar, and the DNA binding region is indicated by a cross-hatched bar. Mutations in the VjbR polypeptide are indicated in the middle. The D82A substitution is indicated by an arrowhead, and conserved regions of deletants are represented. The relative levels of luciferase activity are indicated on the right. The values are expressed as percentages of the PvirB activity in the B. melitensis CD110 strain containing the pSB201 plasmid (top) grown without C12-HSL. The average PvirB activity in the vjbR mutant was 40%. The values are the means of at least three experiments (the variation coefficients were between 1 and 8%).
VjbR mediates C12-HSL inhibitory effect on B. melitensis intracellular replication.
Since C12-HSLs are known to repress PvirB expression in bacteriological cultures, we tested whether this is also the case during cellular infection. Bovine macrophages were infected with a wt B. melitensis strain in the presence or in the absence of C12-HSL. These signal molecules were added at the beginning of the infection at a final concentration of 5 μM. After 1 h and 48 h of infection the number of intracellular bacteria was evaluated. As shown in Table 2, C12-HSL addition did not affect B. melitensis internalization (log CFU at 1 h postinfection) but strongly reduced its intracellular replication (log CFU at 48 h postinfection). Interestingly, this effect was not observed when C12-HSLs were added at 24 h postinfection (data not shown). These results suggest that perturbation of the QS network impaired intracellular replication or trafficking of the bacteria within macrophages.
TABLE 2.
Intracellular replication of B. melitensis in macrophagesa
| Strain | Conditions | Log CFU/well
|
|
|---|---|---|---|
| 1 h postinfection | 48 h postinfectionb | ||
| wt | ACN | 3.19 ± 0.01 | 5.00 ± 0.12 A |
| C12-HSL | 3.28 ± 0.01 | 3.60 ± 0.23 A | |
| CD100 | ACN | 3.26 ± 0.11 | 2.92 ± 0.07 |
| C12-HSL | 3.07 ± 0.02 | 2.96 ± 0.09 | |
| CD100/pSB301 | ACN | 2.89 ± 0.10 | 3.84 ± 0.07 |
| C12-HSL | 3.01 ± 0.03 | 2.12 ± 0.04 B | |
| CD100/pSB303 | ACN | 2.95 ± 0.03 | 3.82 ± 0.04 B |
| C12-HSL | 2.96 ± 0.01 | 3.82 ± 0.03 | |
Infections were performed in triplicate. At different time points, the cells were lysed and the numbers of intracellular bacteria were determined by plating the cell lysates on agar plates and expressed in log CFU per well ± standard deviations. ACN is the C12-HSL solvent.
Values followed by the same letter were significantly different (P < 0.001).
To assess whether the effect of C12-HSL during infection is also dependent on VjbR, bovine macrophages were infected in the presence or in the absence of C12-HSL with strain CD100/pSB301 (B. melitensis ΔvjbR::Kanr/pPlac-vjbR) or CD100/pSB303 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(D82A)]. B. melitensis 16M and the CD100 vjbR defective strain were used as infection controls.
As shown in Table 2, addition of exogenous C12-HSL to the CD100 (B. melitensis ΔvjbR::Kanr) strain complemented with the wt vjbR allele (CD100/pSB301) reduced the intracellular replication approximately 1.7 log, which is similar to the 1.4 log reduction observed with the wt strain. This repression effect was not observed with strain CD100/pSB303 expressing the mutated allele vjbR(D82A). These results suggest that VjbR mediates the effect of C12-HSL on intracellular replication.
B. melitensis VjbR mutants display a clumping phenotype.
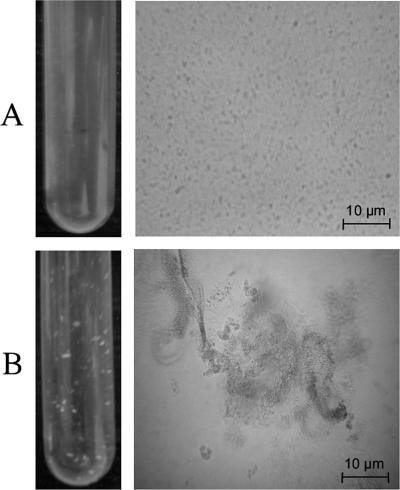
Interestingly, the CD100 strain expressing the vjbR(D82A) or vjbR(Δ1-180) allele exhibits a striking phenotype. As the bacterial cultures reached a high density, cells aggregate and form clumps (Fig. 2).
FIG. 2.
Observation of the clumping phenotype (left panels) and phase-contrast images of B. melitensis (right panels) in exponential growth phase. (A) B. melitensis 16M; (B) strain CD100/pSB203 harboring vjbR(D82A).
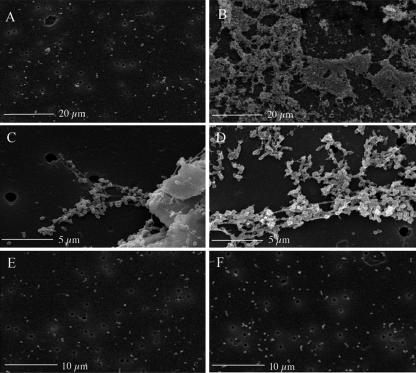
This clumping phenotype was also observed with the CD100 (B. melitensis ΔvjbR::Kanr) strain (clumps were smaller and generally observable only by microscopy), suggesting that various alterations of vjbR may result in this phenotype. This observation leads to the hypothesis that clumping repression in wt B. melitensis could be, via VjbR, under AHL control. Thus, a ΔvjbR strain and a strain unresponsive to AHL both exhibit derepression of the clumping phenotype. In order to characterize this phenotype, we observed aggregates by scanning electron microscopy using culture samples collected at mid-exponential growth phase. As shown in Fig. 3, CD100 (B. melitensis ΔvjbR::Kanr) and CD100/pSB203 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(D82A)] differed greatly from the nonaggregating wt strain. While wt B. melitensis cells were isolated (Fig. 3A), vjbR mutant strains formed large aggregates in which bacteria appear to be embedded within a sticky matrix (Fig. 3B to D). Strain CD100/pSB202 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(Δ1-180)] displayed a similar phenotype (data not shown). To further characterize the clumping phenotype, we focused on the CD100/pSB203 strain as the clumping phenotype is more marked in this strain.
FIG. 3.
Scanning electron micrographs of suspension cultures of Brucella strains. (A) wt B. melitensis strain 16M; (B and C) clumps formed by strain CD100; (D) clumps formed by strain CD100 harboring pSB203; (E) B. abortus 2308; (F) strain RMD100 harboring pSB203.
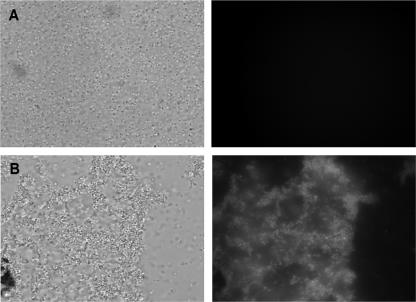
B. melitensis clumps contain EPSs.
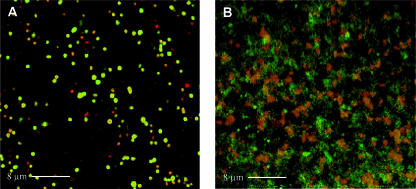
Extracellular matrixes are often composed of EPSs. To test whether EPSs are a major component of the matrix observed in the aggregating strains, culture samples were stained with calcofluor white, a general EPS dye. When calcofluor white was added to the samples, aggregates exhibited a bright fluorescence that was not observed with wt B. melitensis (Fig. 4). This positive staining indicates that bacteria are embedded in a matrix composed at least of a β-linked glucan EPS. We attempted to further characterize the EPS by using the ConA lectin, which is specific for α-mannopyranosyl and α-glucopyranosyl residues (47). Propidium iodide was used to counterstain bacteria red. As shown in Fig. 5, strain CD100 expressing vjbR(D82A) produced a matrix that stained green for ConA-FITC. Staining of the CD100 and CD100/pSB202 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(Δ1-180)] strains gave similar results (data not shown). These results demonstrate that vjbR mutant strains are able to produce EPS containing α-mannopyranosyl and/or α-glucopyranosyl residues as well as a β-linked glucan(s). This EPS is present in large amounts in the CD100/pSB202 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(Δ1-180)] and CD100/pSB203 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(D82A)] strains and seems to be a component of the extracellular matrix of the aggregates.
FIG. 4.
Identification of EPS(s) within aggregates formed by Brucella vjbR mutant strains. Phase-contrast images are shown in the left panels, and calcofluor white staining is shown in the right panels. (A) wt strain; (B) CD100 strain containing the vjbR(D82A) expression plasmid pSB203. Calcofluor white staining of the CD100/pSB202 strain gave similar results.
FIG. 5.
Interactions between FITC-labeled ConA lectin and aggregates formed by Brucella vjbR mutant visualized by confocal laser scanning microscopy. Culture samples of (A) B. melitensis strains 16M and (B) CD100 containing the vjbR(D82A) expression plasmid pSB203 were stained with ConA-FITC (green) and propidium iodide (red).
VjbR-dependent clumping phenotype is not observed in B. abortus 2308.
This study provided the first evidence of the ability of B. melitensis to produce an EPS. To identify genes involved in the synthesis of this polymer, we searched the B. melitensis genome for homologues of genes involved in EPS production in species closely related to Brucella that are able to produce EPS(s) (Agrobacterium tumefaciens, Sinorhizobium meliloti, etc.). Some of these homologues are located in a 25-kb locus that is absent in B. abortus (77). This observation led us to hypothesize that if at least one of these genes is required for the biosynthesis and/or export of EPS, B. abortus should not be able to form aggregates in conditions where B. melitensis does. In order to test this hypothesis, we constitutively expressed AHL-insensitive vjbR alleles described above in strain RMD100, a B. abortus 2308 strain deleted for vjbR (B. abortus VjbR shares 100% identity with B. melitensis VjbR). As illustrated in Fig. 3E and F, neither wt B. abortus 2308 nor the RMD100/pSB203 [B. abortus ΔvjbR::Kanr/pPlac-vjbR(D82A)] strain displayed the typical clumping phenotype. These observations and the homology shared by the genes located in the 25-kb locus described above strongly suggest that one or several genes at this locus are required for EPS synthesis and/or export. Obviously, we cannot rule out the possibility that the absence of a clumping phenotype in B. abortus is due to other differences between B. melitensis and B. abortus.
Implication of QS in the regulation of surface properties in B. melitensis.
VjbR was previously shown to regulate the TFSS VirB and flagella, two surface structures involved in B. melitensis virulence (14). Here we show that VjbR also has a role in the expression of a gene(s) involved in EPS synthesis and/or export. The common feature of the VjbR target genes is their involvement in surface composition. To assess whether other surface components of Brucella are regulated by QS through VjbR, the effect of C12-HSL addition on the abundance of six different Omps was tested by dot blot analysis using specific MAbs (10). Dot blotting was carried out with total B. melitensis extracts from stationary-phase cultures. The same amounts of proteins from all samples were used for dot blot analysis. As shown in Fig. 6A and C, the addition of exogenous C12-HSL led to a slight decrease in Omp10, Omp19, and Omp89 abundance, a strong increase in Omp25 and Omp31 abundance, and a slight increase in Omp36 abundance. To determine if VjbR mediates the effect of the pheromone on Omp production, the same experiment was performed with the vjbR-deficient strain CD100. We observed that in no case was the production of Omps affected by the addition of C12-HSL (Fig. 6B and C). The effect of exogenous C12-HSL is therefore VjbR dependent. As revealed by densitometric quantification (Fig. 6C), the effect of vjbR deletion is in agreement with this observation since the vjbR-deficient strain exhibited decreased production of Omps activated by C12-HSL (Omp25, Omp36, and Omp31) and increased production of Omps repressed by C12-HSL (Omp10, Omp19, and Omp89). These experiments suggest that VjbR is involved in the control of outer membrane composition. Whether this regulation is direct or indirect remains to be determined.
FIG. 6.
QS system of B. melitensis affects the production of several Omps. (A) Dot blot analysis of Omp amounts in the B. melitensis 16M wt strain cultivated with or without exogenous C12-HSL. (B) Dot blot analysis of Omp amounts in the CD100 strain cultivated with or without exogenous C12-HSL. (C) Densitometric quantification of Omps for dot blots in panels A and B. (D) Dot blot analysis of Omp amounts in the B. melitensis 16M wt strain and the CD100 strain complemented with pSB202. Cells were grown in 2YT medium and harvested in stationary phase. Whole-cell extracts were diluted as described in Materials and Methods, and dilutions were subjected to dot blot analysis using different Omp MAbs. ACN, negative control with the solvent (ACN) used for C12-HSL dilution. The data are representative of three experiments.
AHL-independent vjbR mutant strains display a pronounced surface phenotype.
As suggested in this study, VjbR polypeptides mutated in the AHL-binding domain behave like constitutive regulators. We have shown that strains expressing these polypeptides display a clumping phenotype. It was therefore predicted that these strains would also exhibit modified outer membrane properties and probably exhibit modified sensitivity to cationic peptides like polymyxin B. To test this hypothesis, we compared the polymyxin B resistance of the B. melitensis 16M, CD100 (B. melitensis ΔvjbR::Kanr), CD100/pSB201 (B. melitensis ΔvjbR::Kanr/pPlac-vjbR), CD100/pSB202 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(Δ1-180)], and CD100/pSB203 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(D82A)] strains (Fig. 7). B. melitensis is naturally resistant to polymyxin B (43). As shown in Fig. 7, the ΔvjbR CD100 strain is not affected by this cationic polypeptide. Strains CD100/pSB201, CD100/pSB202, and pSB203 expressing wt and mutated vjbR alleles under Plac control display important sensitivity to polymyxin B, suggesting that the overexpression of any form of the VjbR regulator leads to membrane perturbations. We hypothesized that C12-HSL cannot act through VjbR in these strains, either because they express a mutated regulator or because the Plac-controlled overexpression of wt VjbR titrates the C12-HSL effect. To test whether the sensitivity to polymyxin B is correlated to deregulation of Omps, total extracts of CD100/pSB202 [B. melitensis ΔvjbR::Kanr/pPlac-vjbR(Δ1-180)] were subjected to Omp dot blot analysis. As shown in Fig. 6D, strain CD100/pSB202 overexpressed all the Omps tested compared to the wt strain. These results suggest that the sensitivity to polymyxin B might be linked to the overproduction of Omps.
FIG. 7.
Effect of polymixin B on the viability of B. melitensis 16M, CD100, CD100/pSB202, and CD100/pSB203. The data for the wt and CD100 strains are superimposed on the graph. The graph is representative of three distinct experiments.
Omp31 is implied in the clumping phenotype.
One of the Omps regulated by VjbR, Omp31, is encoded in the 25-kb DNA fragment absent in B. abortus. In their characterization of the 25-kb locus, Vizcaino et al. (77) suggested that Omp31 could be involved in the export of unknown polysaccharides, as shown for other bacterial Omps (3, 29, 70, 71). To test whether Omp31 is needed for the export of the B. melitensis EPS identified in this study, we constructed the ΔvjbR::Kanr omp31::Cmr strain SB200 and tested its clumping capacity in 96-well-plate cultures. As shown in Fig. 8A, the CD100 (B. melitensis ΔvjbR::Kanr) strain exhibited large amounts of aggregates in rich bacterial medium, while the wt and SB200 (ΔvjbR::Kanr omp31::Cmr) strains did not. These observations suggest that Omp31 is involved in the aggregation of vjbR mutant strains. To determine if this Omp is involved directly or indirectly in EPS secretion, we used an ELLSA in which a ConA-peroxidase conjugate was incubated with coated stationary-phase culture supernatants. As shown Fig. 8B, the aggregating abilities of the wt, CD100 (B. melitensis ΔvjbR::Kanr), and SB200 (ΔvjbR::Kanr omp31::Cmr) strains were correlated with ConA-peroxidase binding. These results were observed only when cultures were strongly vortexed before supernatants were harvested. EPS detection by ELLSA in strain SB200 was restored by complementation of the omp31 mutation by a plasmid copy of omp31 under the control of the Plac promoter (Fig. 8B). Our observations are in agreement with the proposed role (indirect or not) of Omp31 in EPS export.
FIG. 8.
Omp31 is implicated in the clumping phenotype and EPS production or export in B. melitensis. (A) Clumping phenotypes of the wt strain, vjbR-defective strain CD100, and vjbR/omp31-defective strain SB200 in 96-well 2YT medium cultures. (B) ELLSA with ConA-peroxidase and supernatants of shaken stationary-phase cultures of the wt, CD100, SB200, and SB200/pSB305 strains. Peroxidase activity is represented by the OD450-OD650 value. Significant differences in relation to the wt strain are indicated by an asterisk (P < 0.05).
DISCUSSION
This investigation provided insights into the role of VjbR in AHL-mediated QS in B. melitensis. Evidence that VjbR mediates C12-HSL effects and that this regulator is involved in Omp regulation is described here. We also showed that it is implicated in the regulation of EPS production and/or export.
The VirB TFSS is crucial for the early steps of cellular infection. It reaches an expression peak at 5 h postinfection in B. abortus (60). As described in this study, exogenous C12-HSL represses B. melitensis replication within macrophages when it is added at the beginning of the infection. This inhibition is not observed in the CD100 vjbR-defective strain but is still effective in a babR (BMEI1758)-deficient strain (data not shown), underlying the specificity of VjbR-mediated QS during cellular infection. Addition of signal molecules at 24 h postinfection (data not shown) has no effect on B. melitensis replication, suggesting that if C12-HSLs are produced in vivo by B. melitensis, they could be produced later during the infection, possibly to repress virB when its presence is no longer required. The impairment of B. melitensis replication within macrophages in the presence of C12-HSL could be explained in part by early inactivation of the TFSS VirB. Nevertheless, we cannot exclude the possibility that other virulence factors involved in the establishment of infection are regulated by C12-HSL.
We demonstrated that VjbR mediates the inhibitory effect of C12-HSL on PvirB expression and intracellular replication using two mutated alleles of vjbR. These alleles encode polypeptides unresponsive to signal molecules, thus behaving as constitutive regulators. Our results are in agreement with earlier studies (9, 25, 68, 69) predicting the ability of VjbR Asp82, a highly conserved amino acid in LuxR regulators, to bind AHL.
Interestingly, strains producing the two mutant regulators showed even higher PvirB activity than a strain producing the wt regulator. Immunoblot analysis performed on crude extracts of B. melitensis strains carrying plasmids pSB401, pSB402, and pSB403 revealed that the different VjbR polypeptides tagged with the 13Myc tag were present at similar levels (data not shown). Thus, the effect of the VjbR mutant proteins on PvirB-luxAB is probably not due to their overproduction. We therefore propose that this phenomenon could be the result of the inability of these mutated regulators to mediate the repression effect of AHLs intrinsically produced by B. melitensis (73) that could also repress PvirB-luxAB fusion. Interestingly, Choi and Greenberg described a similar result with a Δ2-162 LuxR mutant (9). These authors suggested that the elevated activity could be explained by the fact that the N-terminal region of LuxR masks the activity of the C-terminal region of the regulator in the absence of autoinducer. However, this model is probably not applicable to VjbR, since the VjbR(D82A) polypeptide also shows greater activity than the wt polypeptide.
VjbR AHL-independent regulators always display exacerbated phenotypes compared to those of wt or CD100 strains (i.e., clumping phenotype, Omp production, and polymyxin B sensitivity). We hypothesize that this behavior could be attributed to the constitutive nature of the mutated regulators.
This investigation shows that mutations of the VjbR LuxR-type regulator lead to a clumping phenotype. The exacerbated clumping phenotype observed in VjbR-AHL-independent strains suggests that AHLs might repress aggregation. The failure of the B. melitensis wt strain to form aggregates in the conditions used in this study could be explained by this hypothesis since AHLs produced by B. melitensis may repress expression of genes involved in clumping.
Bacterial aggregation is one of the initial steps of biofilm formation; thus, the clumping phenotype described in this study suggests for the first time that B. melitensis could be able to form biofilms. Multicellular behavior in bacteria, including biofilm formation (35, 44), competence (39, 41, 49), or coordinated control of virulence factors (14, 65, 81, 83), is often controlled by QS. It is thus interesting that QS is related to aggregation in B. melitensis.
EPS is the major component of biofilm matrixes; consequently, we further characterized aggregates produced by AHL-independent strains. As revealed by calcofluor white staining and a lectin binding assay, aggregates formed by VjbR mutant strains contain an EPS(s) with β-glucan and α-mannopyranosyl and/or α-glucopyranosyl residues. The EPS is currently being characterized (M. Godefroid, unpublished data). The presence of EPS in aggregates is consistent with the hypothesis that the observed clumping phenotype is related to biofilm formation since EPSs are major components of biofilm matrixes (12). However, structures other than EPS, like flagella (36), Omps (4), adhesins (58, 59), or DNA (27, 66, 45, 79), could be involved in B. melitensis aggregation.
The absence of aggregation in B. abortus is most likely due to genomic differences from B. melitensis. We propose that one of the differences could be the deletion in the former organism of a large 25-kb fragment containing the omp31 gene and other genes sharing homology with genes involved in EPS production in other species (A. tumefaciens, S. meliloti, Rhizobium leguminosarum, etc.). Our investigation suggests that Omp31 is involved in EPS export, but this Omp may be involved in other aspects of EPS production. Nevertheless, we cannot exclude the possibility that other differences between these two strains are involved in this phenotype. The ability of some Brucella spp. strains to produce EPS could also contribute to the differences in host preference and disparate virulence of brucellae.
Our investigation demonstrates that other QS-induced surface modifications may occur, as revealed by the drastic modifications in Omp abundance upon C12-HSL addition. While some Omps are detected at a lower rate in the presence of signal molecules (Omp10, Omp16, and Omp19), others are present in greater abundance (Omp25, Omp31 and Omp36). Omp25 and Omp31 are the Omps most represented on outer membrane vesicles (OMV) produced by Brucella (2, 31, 32). This observation is particularly interesting, as Schooling and Beveridge have recently shown that OMV are common constituents of biofilm matrixes (56). These authors suggest that OMV might contribute to biofilm architectures and could be involved in the secretion of several molecules. It will be interesting to further characterize the matrix produced by B. melitensis and to assess the potential role of OMV in this multicellular behavior.
An important conclusion drawn from our investigation is that VjbR is implicated in the regulation of numerous membrane structures in B. melitensis. Besides the previously described role of VjbR in TFSS and flagellum regulation, we report here that it is implicated in the regulation of EPS synthesis and/or export and in outer membrane properties. No S-lipopolysaccharide differences have been revealed by Western blot analysis using S-lipopolysaccharide MAbs (data not shown). Interestingly, numerous attenuated mutants of B. melitensis were previously shown to be altered in membrane composition, like mutants with mutations in the TFSS VirB (15, 48), flagella (28), Omp10 and Omp19 (74), Omp25 (22, 23, 24), and the BvrR/BvrS two-component system (64). BvrR/BvrS transpositional mutants show Omp25 and Omp3b underproduction (33) and high susceptibility to polymyxin B (64). These observations led to the hypothesis that the BvrR/BvrS two-component system is involved in cell envelope changes required for adaptation to the intracellular environment. Here we describe a second system involved in outer membrane regulation.
We report for the first time evidence that B. melitensis could be able to form a biofilm. This aptitude could have several advantages in the host, including (i) protection against host defenses (18, 19, 52), (ii) adhesion to host cells and surfaces (5, 6), and (iii) protection against acidity (84), among others. Furthermore, biofilms have been shown to be implicated in several chronic infections, mediating persistence of pathogens despite host defenses or antibiotic treatments (13, 20). Brucellosis is a chronic infection, and the ability of B. melitensis to produce an EPS(s) demonstrated in this work could be a cause of the still unsolved persistence of this pathogen within the host. Besides, B. melitensis is able to survive for several weeks on inert surfaces (http://www.fao.org/ag/againfo/subjects/fr/health/diseases-cards/brucellosi-bo.html); thus, a possible role for the biofilm outside the host could be protection against desiccation, as described for Salmonella enterica serovar Typhimurium (80) and for Nostoc commune (72).
Acknowledgments
We are grateful to C. Didembourg for helpful technical assistance, B. Nkengfac for relevant English corrections, and E. Depiereux for statistical analysis of our results. We are grateful to R. M. Delrue for the construction of plasmids pSB101 and pSB102 and for helpful discussions. We thank past and present members of the Brucella team of the URBM for fruitful discussions. We also thank the Unité Inter-facultaire de Microscopie Electronique and the Unité de Recherche en Biologie Cellulaire (University of Namur, Belgium) for help with use of the scanning electron microscope and the confocal microscope, respectively.
This work was supported by the Commission of the European Communities (contract QLK2-CT-2002-00918). S. Uzureau, M. Godefroid, C. Deschamps, and J. Lemaire hold a specialization grant from the Fonds pour la Formation à la Recherche dans l'Industrie et l'Agriculture (FRIA).
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. E. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Green Publishing Associates, New York, NY.
- 2.Boigegrain, R. A., I. Salhi, M. T. Alvarez-Martinez, J. Machold, Y. Fedon, M. Arpagaus, C. Weise, M. Rittig, and B. Rouot. 2004. Release of periplasmic proteins of Brucella suis upon acidic shock involves the outer membrane protein Omp25. Infect. Immun. 72:5693-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bugert, P., and K. Geider. 1995. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol. Microbiol. 15:917-933. [DOI] [PubMed] [Google Scholar]
- 4.Burdman, S., E. Jurkevitch, B. Schwartsburd, and Y. Okon. 1999. Involvement of outer-membrane proteins in the aggregation of Azospirillum brasilense. Microbiology 145:1145-1152. [DOI] [PubMed] [Google Scholar]
- 5.Castaneda-Roldan, E. I., F. Avelino-Flores, M. Dall'Agnol, E. Freer, L. Cedillo, J. Dornand, and J. A. Giron. 2004. Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell. Microbiol. 6:435-445. [DOI] [PubMed] [Google Scholar]
- 6.Castaneda-Roldan, E. I., S. Ouahrani-Bettache, Z. Saldana, F. Avelino, M. A. Rendon, J. Dornand, and J. A. Giron. 2006. Characterization of SP41, a surface protein of Brucella associated with adherence and invasion of host epithelial cells. Cell. Microbiol. 8:1877-1887. [DOI] [PubMed] [Google Scholar]
- 7.Celli, J. 2006. Surviving inside a macrophage: the many ways of Brucella. Res. Microbiol. 157:93-98. [DOI] [PubMed] [Google Scholar]
- 8.Celli, J., and J. P. Gorvel. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 7:93-97. [DOI] [PubMed] [Google Scholar]
- 9.Choi, S. H., and E. P. Greenberg. 1991. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. USA 88:11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., P. de Wergifosse, G. Dubray, and J. N. Limet. 1990. Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect. Immun. 58:3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159-168. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 14.Delrue, R. M., C. Deschamps, S. Leonard, C. Nijskens, I. Danese, J. M. Schaus, S. Bonnot, J. Ferooz, A. Tibor, X. De Bolle, and J. J. Letesson. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151-1161. [DOI] [PubMed] [Google Scholar]
- 15.Delrue, R. M., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. P. Gorvel, and J. J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 16.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 58:2320-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Haeze, W., J. Glushka, R. De Rycke, M. Holsters, and R. W. Carlson. 2004. Structural characterization of extracellular polysaccharides of Azorhizobium caulinodans and importance for nodule initiation on Sesbania rostrata. Mol. Microbiol. 52:485-500. [DOI] [PubMed] [Google Scholar]
- 19.D'Haeze, W., and M. Holsters. 2004. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 12:555-561. [DOI] [PubMed] [Google Scholar]
- 20.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dricot, A., J. F. Rual, P. Lamesch, N. Bertin, D. Dupuy, T. Hao, C. Lambert, R. Hallez, J. M. Delroisse, J. Vandenhaute, I. Lopez-Goni, I. Moriyon, J. M. Garcia-Lobo, F. J. Sangari, A. P. Macmillan, S. J. Cutler, A. M. Whatmore, S. Bozak, R. Sequerra, L. Doucette-Stamm, M. Vidal, D. E. Hill, J. J. Letesson, and X. De Bolle. 2004. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 14:2201-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edmonds, M. D., A. Cloeckaert, N. J. Booth, W. T. Fulton, S. D. Hagius, J. V. Walker, and P. H. Elzer. 2001. Attenuation of a Brucella abortus mutant lacking a major 25 kDa outer membrane protein in cattle. Am. J. Vet. Res. 62:1461-1466. [DOI] [PubMed] [Google Scholar]
- 23.Edmonds, M. D., A. Cloeckaert, and P. H. Elzer. 2002. Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 88:205-221. [DOI] [PubMed] [Google Scholar]
- 24.Edmonds, M. D., A. Cloeckaert, S. D. Hagius, L. E. Samartino, W. T. Fulton, J. V. Walker, F. M. Enright, N. J. Booth, and P. H. Elzer. 2002. Pathogenicity and protective activity in pregnant goats of a Brucella melitensis Deltaomp25 deletion mutant. Res. Vet. Sci. 72:235-239. [DOI] [PubMed] [Google Scholar]
- 25.Egland, K. A., and E. P. Greenberg. 2000. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J. Bacteriol. 182:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ficht, T. A. 2003. Intracellular survival of Brucella: defining the link with persistence. Vet. Microbiol. 92:213-223. [DOI] [PubMed] [Google Scholar]
- 27.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fretin, D., A. Fauconnier, S. Kohler, S. Halling, S. Leonard, C. Nijskens, J. Ferooz, P. Lestrate, R. M. Delrue, I. Danese, J. Vandenhaute, A. Tibor, X. DeBolle, and J. J. Letesson. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell. Microbiol. 7:687-698. [DOI] [PubMed] [Google Scholar]
- 29.Frosch, M., D. Muller, K. Bousset, and A. Muller. 1992. Conserved outer membrane protein of Neisseria meningitidis involved in capsule expression. Infect. Immun. 60:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gamazo, C., and I. Moriyon. 1987. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect. Immun. 55:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamazo, C., A. J. Winter, I. Moriyon, J. I. Riezu-Boj, J. M. Blasco, and R. Diaz. 1989. Comparative analyses of proteins extracted by hot saline or released spontaneously into outer membrane blebs from field strains of Brucella ovis and Brucella melitensis. Infect. Immun. 57:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guzman-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. P. Gorvel, I. Moriyon, E. Moreno, and I. Lopez-Goni. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallez, R., J. J. Letesson, J. Vandenhaute, and X. De Bolle. 2007. Gateway-based destination vectors for functional analyses of bacterial ORFeomes: application to the Min system in Brucella abortus. Appl. Environ. Microbiol. 73:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 36.Jeon, B., K. Itoh, N. Misawa, and S. Ryu. 2003. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol. Immunol. 47:833-839. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan, H. B., and E. P. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 39.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letesson, J. J., and X. De Bolle. 2004. Brucella—molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 41.Luo, P., H. Li, and D. A. Morrison. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50:623-633. [DOI] [PubMed] [Google Scholar]
- 42.Luo, Z. Q., A. J. Smyth, P. Gao, Y. Qin, and S. K. Farrand. 2003. Mutational analysis of TraR. Correlating function with molecular structure of a quorum-sensing transcriptional activator. J. Biol. Chem. 278:13173-13182. [DOI] [PubMed] [Google Scholar]
- 43.Martinez de Tejada, G., J. Pizarro-Cerda, E. Moreno, and I. Moriyon. 1995. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect. Immun. 63:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLean, R. J., M. Whiteley, D. J. Stickler, and W. C. Fuqua. 1997. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154:259-263. [DOI] [PubMed] [Google Scholar]
- 45.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 46.Moreno, E., E. Stackebrandt, M. Dorsch, J. Wolters, M. Busch, and H. Mayer. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172:3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naismith, J. H., and R. A. Field. 1996. Structural basis of trimannoside recognition by concanavalin A. J. Biol. Chem. 271:972-976. [DOI] [PubMed] [Google Scholar]
- 48.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 49.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 50.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pizarro-Cerda, J., S. Meresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reckseidler-Zenteno, S. L., R. DeVinney, and D. E. Woods. 2005. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect. Immun. 73:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365-370. [DOI] [PubMed] [Google Scholar]
- 54.Sangari, F., and J. Aguero. 1991. Mutagenesis of Brucella abortus: comparative efficiency of three transposon delivery systems. Microb. Pathog. 11:443-446. [DOI] [PubMed] [Google Scholar]
- 55.Scheffé, H. 1959. The analysis of variance. John Wiley and Sons, New York, NY.
- 56.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shadel, G. S., R. Young, and T. O. Baldwin. 1990. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J. Bacteriol. 172:3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sieira, R., D. J. Comerci, L. I. Pietrasanta, and R. A. Ugalde. 2004. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 54:808-822. [DOI] [PubMed] [Google Scholar]
- 61.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:783-791. [Google Scholar]
- 62.Slock, J., D. VanRiet, D. Kolibachuk, and E. P. Greenberg. 1990. Critical regions of the Vibrio fischeri LuxR protein defined by mutational analysis. J. Bacteriol. 172:3974-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, L. D., and T. A. Ficht. 1990. Pathogenesis of Brucella. Crit. Rev. Microbiol. 17:209-230. [DOI] [PubMed] [Google Scholar]
- 64.Sola-Landa, A., J. Pizarro-Cerda, M. J. Grillo, E. Moreno, I. Moriyon, J. M. Blasco, J. P. Gorvel, and I. Lopez-Goni. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 65.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9:133-137. [DOI] [PubMed] [Google Scholar]
- 67.Stabel, J. R., and T. J. Stabel. 1995. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet. Immunol. Immunopathol. 45:211-220. [DOI] [PubMed] [Google Scholar]
- 68.Stevens, A. M., and E. P. Greenberg. 1997. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J. Bacteriol. 179:557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 91:12619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stevenson, G., R. Lan, and P. R. Reeves. 2000. The colanic acid gene cluster of Salmonella enterica has a complex history. FEMS Microbiol. Lett. 191:11-16. [DOI] [PubMed] [Google Scholar]
- 72.Tamaru, Y., Y. Takani, T. Yoshida, and T. Sakamoto. 2005. Crucial role of extracellular polysaccharides in desiccation and freezing tolerance in the terrestrial cyanobacterium Nostoc commune. Appl. Environ. Microbiol. 71:7327-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 70:3004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tibor, A., V. Wansard, V. Bielartz, R. M. Delrue, I. Danese, P. Michel, K. Walravens, J. Godfroid, and J. J. Letesson. 2002. Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 70:5540-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ulrich, R. L., D. Deshazer, H. B. Hines, and J. A. Jeddeloh. 2004. Quorum sensing: a transcriptional regulatory system involved in the pathogenicity of Burkholderia mallei. Infect. Immun. 72:6589-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vizcaino, N., A. Cloeckaert, M. S. Zygmunt, and L. Fernandez-Lago. 2001. Characterization of a Brucella species 25-kilobase DNA fragment deleted from Brucella abortus reveals a large gene cluster related to the synthesis of a polysaccharide. Infect. Immun. 69:6738-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.von Bodman, S. B., D. R. Majerczak, and D. L. Coplin. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. USA 95:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 80.White, A. P., D. L. Gibson, W. Kim, W. W. Kay, and M. G. Surette. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, R. G., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 83.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu, M., S. Takenaka, M. Sato, and E. Hoshino. 2001. Influence of starvation and biofilm formation on acid resistance of Streptococcus mutans. Oral Microbiol. Immunol. 16:24-27. [DOI] [PubMed] [Google Scholar]