Abstract
We isolated the c rings of F-ATP synthases from eight cyanobacterial strains belonging to four different taxonomic classes (Chroococcales, Nostocales, Oscillatoriales, and Gloeobacteria). These c rings showed different mobilities on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), probably reflecting their molecular masses. This supposition was validated with the previously characterized c11, c14, and c15 rings, which migrated on SDS-PAGE in proportion to their molecular masses. Hence, the masses of the cyanobacterial c rings can conveniently be deduced from their electrophoretic mobilities and, together with the masses of the c monomers, allow the calculation of the c ring stoichiometries. The method is a simple and fast way to determine stoichiometries of SDS-stable c rings and hence a convenient means to unambiguously determine the ion-to-ATP ratio, a parameter reflecting the bioenergetic efficacy of F-ATP synthases. AFM imaging was used to prove the accuracy of the method and confirmed that the c ring of Synechococcus elongatus SAG 89.79 is a tridecameric oligomer. Despite the high conservation of the c-subunit sequences from cyanobacterial strains from various environmental groups, the stoichiometries of their c rings varied between c13 and c15. This systematic study of the c-ring stoichiometries suggests that variability of c-ring sizes might represent an adaptation of the individual cyanobacterial species to their particular environmental and physiological conditions. Furthermore, the two new examples of c15 rings underline once more that an F1/Fo symmetry mismatch is not an obligatory feature of all F-ATP synthases.
Conversion of an electrochemical ion gradient into the universal energy currency ATP is performed by the enzyme F1Fo ATP synthase. The protein complex, which synthesizes ATP from ADP and phosphate, consists of two rotary motors, F1 and Fo, which are connected by a central and a peripheral stalk to exchange energy with each other. During ATP synthesis, the translocation of H+ or Na+ through the membrane-embedded Fo motor induces torque, which enables the F1 motor to act as an ATP generator (4, 5, 7).
The Fo motor consists of the stator subunits a and b2 and the rotating c ring. The c ring is a cylindrical assembly of helical hairpin subunits with their loops located on the cytoplasmic side and interacting with the γ and ɛ subunits of the F1 motor. The oligomeric c-ring assemblies consist of an inner ring of α-helices surrounding a central phospholipid-containing cavity (27) and an outer ring of α-helices that contacts the membrane phospholipids and/or subunit a (11, 43). Each c subunit harbors an ion binding site in the middle of the membrane (39). According to the crystal structure of the Na+-translocating c ring from Ilyobacter tartaricus, residues on the inner and outer helices of one c subunit and the outer helix of the neighboring c subunit contribute to one ion binding site per subunit (20). A number of c-ring preparations from various species revealed a remarkable flexibility in the c-ring stoichiometry, ranging from 10 subunits in Saccharomyces cerevisiae (36), Escherichia coli (12), and Bacillus sp. strain PS3 (23); 11 subunits in I. tartaricus (35), Propionigenium modestum (19), and Clostridium paradoxum (18); and 14 subunits in chloroplasts (22, 34) to 15 subunits in the alkaliphilic cyanobacterium Arthrospira sp. strain PCC 9438 (28). Furthermore, data obtained via bioinformatics suggested a single polypeptide ring consisting of 13 fused subunit c hairpins in the ATP synthase from Methanopyrus kandleri (15).
The number of c subunits in the oligomeric ring (n) has important physiological implications, because it defines the number of ions translocated per synthesis of three molecules of ATP in the β3 subunits of the F1 complex in a complete 360° rotation (9). Accordingly, the “cn-to-β3 ratio” represents the theoretical ion-to-ATP ratio and is equal to the number of c subunits divided by three. Until now, the number of c subunits in the ring has been determined by time-consuming methods, such as structure analysis by X-ray crystallography (20), electron crystallography (40), and atomic force microscopy (AFM) (35) or the biochemical analysis of genetically fused c subunits (12, 13, 23).
We show here with c rings of known oligomeric composition that their relative electrophoretic mobilities and molecular masses are proportional. Using this approach, the unknown c ring from Synechococcus elongatus SAG 89.79 F-ATP synthase was found to be a tridecamer. Indeed, high-resolution AFM topographs resolve the structural assembly of c subunits into c13 rings. With this new method, we determined the c-ring stoichiometries from eight different cyanobacterial species of four different taxonomic classes (Chroococcales, Nostocales, Oscillatoriales, and Gloeobacteria) and from different environmental groups (freshwater, thermal springs, soda lakes, and calcareous rocks). Although, the sequences of these c subunits are very similar, their oligomeric states range from 13 to 15.
MATERIALS AND METHODS
Strains.
The strains used in this study were Arthrospira sp. strain PCC 9438 (28); Arthrospira sp. strain PCC 9108, Synechocystis sp. strain PCC 6803, Anabaena sp. strain PCC 7120, Synechococcus sp. strain PCC 6716, S. elongatus PCC 6301, S. elongatus PCC 7942, and Gloeobacter violaceus PCC 7421 (obtained from the Pasteur Culture Collection of Cyanobacteria [PCC], Paris, France); and S. elongatus SAG 89.79 (obtained from the Culture Collection of Algae [SAG] at the University of Göttingen, Göttingen, Germany).
Growth conditions.
Arthrospira sp. strain PCC 9438 and Arthrospira sp. strain PCC 9108 cells were grown in Zarrouk's medium (42). Synechococcus sp. strain PCC 6716, Synechocystis sp. strain PCC 6803, Anabaena sp. strain PCC 7120, S. elongatus PCC 6301, S. elongatus PCC 7942, G. violaceus PCC 7421, and S. elongatus SAG 89.79 were grown in BG-11 medium as described previously (29). G. violaceus PCC 7421 was grown in BG-11 at 25°C in dim light as described previously (3). Cells were harvested either by centrifugation or by filtration in the mid-log phase at a density of about 5 μg chlorophyll per ml of culture and frozen in liquid nitrogen for storage. Large-scale cell cultivation of all strains (except G. violaceus PCC 7421) was performed in 10-liter bottles at 30°C under continuous illumination (white light; 60 μmol photons m−2 s−1) and continuous aeration with filtered air.
Isolation of thylakoid membranes from cyanobacterial strains.
Purification of thylakoid membranes was performed as described previously (28) with the following modifications. One gram of cells (wet weight) was suspended in 2 ml buffer A (20 mM Tris-HCl, pH 8.0, 5 mM EDTA supplemented with 30 mM dithiothreitol, 0.1 mM diisopropylfluorophosphate, and one spatula tip of DNase I) and passed twice through a French pressure cell at 8.3 × 104 kPa. Unbroken cells and cell debris were removed by low-speed centrifugation (5,000 × g; 10 min; 4°C). The thylakoid membrane fractions were sedimented by ultracentrifugation for 45 min at 200,000 × g at 4°C as a dense (green) pellet at the bottom of the centrifuge tubes and collected, avoiding mixing it with a greasy yellowish pellet (plasma and outer membranes) on the top.
Preparation of c rings from cyanobacterial thylakoid membranes and spinach chloroplasts.
Purification of the c rings from membranes was performed as described previously (17) with modifications. The obtained thylakoid membranes and spinach chloroplasts were resuspended in 2 ml of buffer A containing 1% N-lauroylsarcosine and incubated for 10 min at 65°C to solubilize the membranes. The mixture was centrifuged for 45 min at 200,000 × g (25°C), and (NH4)2SO4 at 65% (wt/vol) saturation was added to the supernatant to precipitate contaminating membrane proteins. After 20 min of incubation at 20°C, the samples were centrifuged for 20 min at 39,000 × g. Supernatants containing the c oligomers were passed through a 0.22-μm polyvinylidene difluoride (PVDF) membrane and dialyzed against 5 liters of 10 mM Tris-HCl buffer (pH 8.0) using a dialysis membrane with a molecular mass cutoff (MMCO) of 6,000 Da. c-ring samples were subsequently concentrated by ultrafiltration (MMCO, 10,000 Da) (Centricon tubes; YM-10; Millipore, Billerica, MA) to a protein concentration of 1 mg/ml. The protein concentration was determined according to the bicinchoninic acid method (Pierce). To improve the purity of these c-ring preparations, 1% N-lauroylsarcosine was added again to the samples, and the precipitation procedure with (NH4)2SO4 at 65% (wt/vol) saturation was repeated. After dialysis against 5 liters of 10 mM Tris-HCl, pH 8.0, the protein samples were concentrated to 1 mg protein/ml by ultrafiltration with Centricon tubes (YM-10) and applied on top of a density gradient (5 ml) of 5 to 30% sucrose in 5% steps containing 20 mM Tris-HCl, pH 8.0, and 0.05% (wt/vol) dodecylmaltoside. After ultracentrifugation (4°C; 16 h; 150,000 × g), fractions of 0.5 ml were collected from the bottom and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). c-oligomer-containing samples were pooled and dialyzed (MMCO, 6,000 Da) against 10 mM Tris-HCl, pH 8.0, and concentrated to 1 mg protein/ml by ultrafiltration. Dodecylmaltoside or octylglucoside was added to a final concentration of 0.05% or 1%, respectively. For storage, the samples were sterilized through a PVDF membrane (0.22 μm) and kept at +4°C.
SDS-PAGE.
Dual vertical Mini-Gel systems (113 by 100 mm) from C.B.S. Scientific (Del Mar, CA) were used for all gels presented in this work. SDS gels were prepared as described previously (30). Separation gels contained 13.2% acrylamide, and stacking gels contained 3.8% acrylamide. Prior to being loaded, the samples were mixed 1:1 (vol/vol) with loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 12% glycerol, 0.01% Coomassie blue G, 0.01% bromphenol blue) and incubated for 5 min at 65°C. During the run, the gels were permanently cooled with water (+4°C). The current was 30 mA for the first 30 min and then increased to 40 mA (per gel). At the end of the run, the gels were stained with silver as described previously (26).
Mass determination of cyanobacterial c subunits.
The masses of the different cyanobacterial c subunits were determined by mass spectroscopy (MS). The c subunit was extracted into organic solvents as described previously (16) either directly from cells or from purified c-ring samples. The organic c-ring samples were dried and stored at −80°C. Immediately prior to the measurement, the dry pellets were redissolved in 20 μl of chloroform-methanol mixture (1:1 [vol/vol]) and spotted onto a layer of matrix (dihydroxybenzoic acid), which had been deposited before from 10% stock solution in acetonitrile-water (2:1 [vol/vol]) supplemented with 0.1% trifluoroacetic acid. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis was performed as described previously (39).
De novo sequencing of c subunits from Arthrospira sp. strain PCC 9438, Arthrospira sp. strain PCC 9108, and S. elongatus SAG 89.79.
Total amino acid composition analysis of purified c subunits from Arthrospira sp. strain PCC 9438 and S. elongatus SAG 89.79 was conducted as follows. Dried samples from three time periods (24, 72, and 96 h) were hydrolyzed with gaseous 6 M HCl at 110°C under argon. The hydrolysates were derivatized with the AccqTag Ultra reagent (Waters), and amino acid derivatives were separated on an Acquity UPLC according to the manufacturer's instructions. The first 31 amino acids from the N termini of the protein sequences of the c subunits from Arthrospira sp. strain PCC 9438 and S. elongatus SAG 89.79 were obtained by Edman sequencing of the purified c subunits in organic solvents. After electrophoresis, the gel was blotted onto a PVDF membrane (0.2 μm). The Coomassie blue-stained band was cut out from the membrane. A deformylation reaction (41) was performed only for the c subunit from Arthrospira sp. strain PCC 9438. All single bands were analyzed on a protein/peptide sequencer (492cLC Procise; Applied Biosystems, Foster City, CA) with an on-line Amino Acid Analyzer (140C PTH; Perkin Elmer, Applied Biosystems, Foster City, CA), using a slightly modified pulsed-liquid program provided by the source company. Fragmentation of the cyanobacterial c subunit for the peptide mass fingerprint analysis was conducted by tryptic digestion (trypsin, 1:20 [wt/wt]) for 16 h at 37°C in 50 mM (NH4)HCO3, pH 8.5, 10 mM dithiothreitol, and 20% CH3CN. The peptide fragments were subjected to MALDI-TOF using an Ultraflex TOF/TOF II mass spectrometer equipped with the control and analysis software Compass v. 1.2 (Bruker Daltonics, Bremen, Germany), followed by data analysis with the Mascot software package. The putative c-subunit sequences were deduced in a combinatory approach considering the positions of the conserved amino acid residues and fitted with the MS data. The primary structures of the c subunits from Arthrospira sp. strain PCC 9438 and Arthrospira sp. strain PCC 9108 were considered to be identical, since the obtained masses of the c subunits and the peptide fragments thereof were found to be the same. The putative sequences of the c subunits from Arthrospira sp. strain PCC 9438 and Arthrospira sp. strain PCC 9108 were found to be identical to the translated sequence of the atpH gene from Arthrospira sp. (C.-C. Zhang, Marseille, France, personal communication) except for one D-to-A residue exchange at position 8 of the protein sequence.
Estimation of molecular masses of c oligomers from SDS-PAGE.
The mobilities the c rings on SDS-PAGE were measured as the distance between the border between stacking and separation gels and the front line of the protein band. The relative mobilities (Rm) were calculated according to the following formula: Rm = [Δx(csample) − Δx(cmarker)]/Δx(cmarker), where Δx(csample) is the mobility of the examined c-ring band in millimeters and Δx(cmarker) is the mobility of the marker protein bovine serum albumin (66.2 kDa).
Reconstitution of c rings from S. elongatus SAG 89.79.
c-ring samples from S. elongatus SAG 89.79 (1 mg/ml) were mixed with 1-palmitoyl-2-oleoyl-phosphatidylcholine at a lipid-to-protein ratio of 1 (wt/wt) to yield 2% (wt/vol) dodecylmaltoside. Dialysis was performed for 20 days at 25°C against 50 ml buffer containing 10 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 3 mM NaN3 using 10-kDa-cutoff dialysis membranes.
AFM.
An atomic force microscope equipped with a 100-μm X-Y piezo scanner was optimized for observing single molecules in buffer solution (Nanoscope IIIa; DI-Veeco). Silicon nitride AFM cantilevers (100 μm long; Olympus, Tokyo, Japan) exhibited nominal spring constants of ∼0.9 N/m. To adsorb the protein membranes, 30 μl of the sample buffer (20 mM Tris-HCl, 300 mM NaCl, 0.02% NaN3, and 1% glycerol, pH 8.0) was placed onto freshly cleaved mica for ∼15 min. After this, the sample was rinsed with 20 mM Tris-HCl, 150 mM KCl, 1% glycerol, pH 8.0, to remove weakly attached material. Contact mode AFM topographs were recorded in this buffer at 24°C with a stylus loading force of <100 pN and a line frequency of 5 to 6 Hz. No differences between topographs recorded in trace and in retrace directions were observed, indicating that the scanning process did not influence the appearance of the sample.
Image processing.
Individual c rings of the AFM topographs were selected manually and subjected to reference-free alignment and averaging using the SEMPER image-processing system (Synoptics Ltd., United Kingdom). To assess the ring symmetry, the rotational-power spectrum of the averaged image was calculated using SEMPER. Alternatively, the stoichiometries of individual c rings were calculated and then averaged (data not shown).
RESULTS
Sequence comparison of selected c subunits from cyanobacteria and plant chloroplast ATP synthases.
We compared the protein sequences of subunits c (atpH) in the atp1 operons of a selected group of cyanobacteria (Fig. 1). The cyanobacterial c-subunit sequences were available in full length from public databases or were sequenced in this work (Arthrospira sp. strain PCC 9438, Arthrospira sp. strain PCC 9108, and S. elongatus SAG 89.79). The data summarized in Fig. 1 show a very high sequence identity among the cyanobacterial c subunits (84.5% on average; range, 77.1 to 96.2%). Significant differences were observed only between amino acids 1 to 10 and 23 to 41 (I. tartaricus numbering). Furthermore, a conserved exchange at position 16, three exchanges at position 66, and some conserved variations at positions 73 to 77 and in the amino acids at the end of the sequence can be observed.
FIG. 1.
Alignment of c-subunit sequences from ATP synthases from cyanobacteria, chloroplasts, and selected bacteria used in this study. Individual sequences were aligned on their highly conserved cytoplasmic loops (marked in black). To save space, the species names are abbreviated (for the full names, see Table 1). The numbering is according to the sequence of I. tartaricus. In this sequence, the following amino acids were marked in black: the GXGXGXGXG motif (40) and the sodium binding motif (Q32, V63, E65, S66) (20). c-ring stoichiometries (St) are shown on the right; the known ones are highlighted in black. Except for sequences of Arthrospira sp. strain PCC 9438, Arthrospira sp. strain PCC 9108, and S. elongatus SAG 89.79, the translated protein sequences of the cyanobacterial c subunits were available in full length from Cyanobase (http://www.bacteria.kazusa.or.jp/cyanobase/) and Swiss-Prot (http://expasy.org/) databases. The putative protein sequences of Arthrospira sp. strain PCC 9438, Arthrospira sp. strain PCC 9108, and S. elongatus SAG 89.79 were deduced from de novo sequencing, amino acid analysis, and MS of peptide fragments as described in Materials and Methods.
Purification of the c rings from several cyanobacterial species.
c rings from thylakoid membranes from several cyanobacterial strains were purified by the protocol developed for the c ring of I. tartaricus ATP synthase (19). All purified c rings were resistant to incubation for 5 min at 65°C in the presence of 2% SDS. SDS-PAGE (Fig. 2) showed the isolated c rings in the upper part of the gel and the monomeric c subunits migrating near the gel front. The putative c-ring bands were confirmed by trichloroacetic acid treatment (18), which dissociated all the c oligomers into their monomeric units (not shown).
FIG. 2.
SDS-PAGE of c rings purified from various bacterial strains. Aliquots of 0.5 to 2 μg of the isolated c-ring samples were subjected to SDS-PAGE as described in Materials and Methods. Lanes 1 to 4, migration of c rings with known oligomeric compositions (c11 to c15, as indicated). The masses of these rings are indicated between lanes 4 and 5. Lanes 5 to 11, migration of c rings from cyanobacterial strains, except the one from G. violaceus PCC 7421. Bands corresponding to oligomeric c rings are indicated on the right (cn). Lanes 1 to 7 and 8 to 11 were taken from two separate gels; they have different molecular mass marker scales on the left and right, respectively. Lanes: 1, c11 (C. paradoxum; Cp); 2, c11 (I. tartaricus; It); 3, c14 (spinach chloroplasts); 4, c15 (Arthrospira sp. strain PCC 9438); 5, cn (S. elongatus PCC 6301); 6, cn (Synechocystis sp. strain PCC 6803); 7, cn (S. elongatus SAG 89.79); 8, cn (Arthrospira sp. strain PCC 9108); 9, cn (Anabaena sp. strain PCC 7120); 10, cn (7942); 11, cn (Synechococcus sp. strain PCC 6716).
Determination of molecular masses of the monomeric c subunits by MS-MALDI.
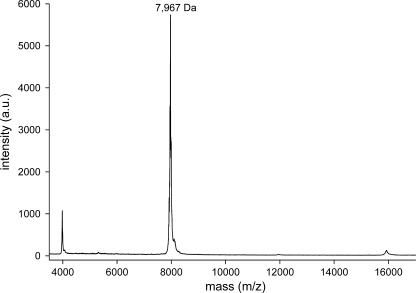
To calculate the oligomeric states of the c rings from their apparent masses (see below), the masses of the monomeric c subunits must be known. The c subunits were therefore extracted with chloroform-methanol and then subjected to MS-MALDI. A sample spectrum of subunit c from S. elongatus SAG 89.79 is shown in Fig. 3. The masses were in the range of 8,000 Da, as expected for monomeric c subunits, but deviated from the masses calculated from the amino acid sequences by +16 to +60 Da. In previous investigations with the c subunits of spinach chloroplasts, similar mass increases were attributed to N-terminal formylation (33) or to oxidation reactions (28). By combining this information with the MS data obtained, the measured masses fit well with the masses calculated from the amino acid sequences of the corresponding c subunits. Putative modifications of the various c subunits are summarized in Table 1.
FIG. 3.
MALDI-TOF mass spectrum of subunit c from S. elongatus SAG 89.79 extracted with chloroform-methanol (1:1). The identified masses are indicated in Da. The samples were prepared as described in Materials and Methods. a.u., arbitrary units.
TABLE 1.
Properties of the c rings used in this study
| Species | Mass(es) of c monomer (Da)
|
Modificatione
|
Rm | Estimated c-ring mass (kDa) | Proposed stoichiometry | Theoretical mass of c oligomer (kDa) (modifications included) | ||
|---|---|---|---|---|---|---|---|---|
| Theoretical | Detected | Formylation | Oxidation | |||||
| Arthrospira sp. strain PCC 9438 | 8,181 | 8,207/8,224 | + | + | 0.161 ± 0.034 | 122.03 ± 1.28 | 15 | 123.12 |
| Arthrospira sp. strain PCC 9108 | 8,181 | 8,208/8,228 | + | + | 0.152 ± 0.026 | 122.35 ± 0.98 | 15 | 123.12 |
| Synechocystis sp. strain PCC 6803 | 7,968 | 7,993/8,014 | + | + | 0.39 ± 0.036 | 113.42 ± 1.35 | 14 | 111.93 |
| Anabaena sp. strain PCC 7120 | 8,002 | 8,027/8,042 | + | + | 0.389 ± 0.043 | 113.45 ± 1.62 | 14 | 112.41 |
| S. elongatus SAG 89.79 | 7,956 | 7,967d | − | + | 0.601 ± 0.059 | 105.5 ± 2.22 | 13 | 103.57 |
| S. elongatus PCC 6301 | 7,967 | 7,998/8,012 | + | + | 0.399 ± 0.05 | 113.07 ± 1.87 | 14 | 111.93 |
| Synechococcus sp. strain PCC 6716 | 8,180 | 8,205/8,244 | + | + | 0.376 ± 0.039 | 113.96 ± 1.47 | 14 | 114.9 |
| S. elongatus PCC 7942 | 7,967 | 7,967/7,985/7,998/8,012 | −/+ | + | 0.399 ± 0.038 | 113.09 ± 1.43 | 14 | 111.93 |
| G. violaceus PCC 7421 | 8,196 | 8,198/8,214/8,242 | −/+ | + | 0.155 ± 0.021 | 122.25 ± 0.79 | 15 | 122.94 |
| Spinach chloroplast | 7,974 | 8,003a | + | ND | 0.385 ± 0.053 | 113.59 ± 2.01 | 14 | 112.04 |
| C. paradoxum | 8,257 | 8,266b | − | ND | 0.995 ± 0.093 | 90.66 ± 3.5 | 11 | 90.93 |
| I. tartaricus | 8,795 | 8,796c | − | ND | 0.842 ± 0.07 | 96.44 ± 2.63 | 11 | 96.76 |
Estimation of c-ring masses from electrophoretic mobilities.
All c rings from different sources migrate on SDS-PAGE according to their molecular masses (18, 22, 23, 28). However, the electrophoretic mobilities of c rings are approximately two times faster than those of soluble proteins, as was also observed for other hydrophobic membrane proteins (8). Therefore, the c-ring masses cannot be determined by using commercial SDS gel markers. However, within the family of isolated c rings, the electrophoretic mobilities correlate linearly with their molecular masses (Fig. 2, lanes 1 to 4). Hence, by using c rings of known masses as standards, the molecular mass of an unknown c ring may be determined by its relative electrophoretic mobility. The method is shown in Fig. 4 for the determination of the mass of the c ring from S. elongatus SAG 89.79. The mobility of this c ring was compared with those of the c rings of known masses (Fig. 4A). It migrated between the I. tartaricus c11 ring (96.76 kDa) and the chloroplast c14 ring (112.04 kDa), with an apparent molecular mass of 105 ± 2.2 kDa derived from this analysis. As c-ring mobilities slightly varied in different experiments, an accurate determination of the molecular mass of an unknown c ring required calibration with at least three c rings of known masses in several parallel runs and a subsequent regression analysis (Fig. 4B). By following this scheme, we determined the molecular masses of the other cyanobacterial c rings (Table 1). In agreement with the different mobilities of c rings and the molecular masses of the corresponding c monomers, the following c-ring stoichiometries have been calculated: 15-mers for Arthrospira sp. strain PCC 9108 and G. violaceus PCC 7421; 14-mers for the five strains Synechocystis sp. strain PCC 6803, Anabaena sp. strain PCC 7120, Synechococcus sp. strain PCC 6716, S. elongatus PCC 7942, and S. elongatus PCC 6301; and a 13-meric c ring for S. elongatus SAG 89.79.
FIG. 4.
Estimation of the molecular mass of the c ring of S. elongatus SAG 89.79 from its relative mobility on SDS-PAGE. (A) The c rings from C. paradoxum (Cp; lane 1), I. tartaricus (It; lane 2), spinach chloroplasts (lane 4), and Arthrospira sp. strain PCC 9438 (lane 5), with molecular masses shown on the left side of the gel, were used for calibration. The relative mobilities of individual c rings were determined as shown by the dashed lines (see Materials and Methods). The oligomeric c-ring compositions are shown on the right (c11 to c15). Lane 3 shows the migration of the c ring from S. elongatus SAG 89.79. The dashed line at the top of the gel represents the border between stacking and separation gels. A scale bar indicating the different migration lengths of the c rings and bovine serum albumin (66.2 kDa) is on the left. (B) Estimation of c-ring molecular masses from the relative mobilities. The molecular masses (kDa) of the different c rings were plotted against their relative mobilities as determined in panel A. ▴, from left to right, c rings from Arthrospira sp. strain PCC 9438, spinach chloroplasts, I. tartaricus, and C. paradoxum; •, c ring from S. elongatus SAG 89.79; ○, calculated relative mobility of the c ring from S. elongatus SAG 89.79, assuming a c13 stoichiometry. The data represent average values and standard deviations (indicated by error bars) from 12 independent experiments. The dashed line represents a linear-regression trend line, and the confidence intervals (P = 0.05) are displayed with dotted lines.
High-resolution AFM of the S. elongatus SAG 89.79 c ring.
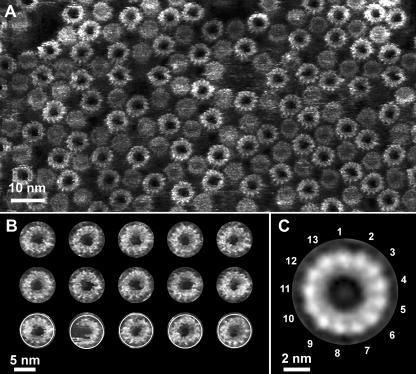
The above analysis suggested that the F-ATP synthase of S. elongatus SAG 89.79 has a c13 ring. To prove this observation structurally, the rings were reconstituted into a lipid bilayer and imaged by AFM. High-resolution topographs (Fig. 5A) showed a densely packed assembly of c rings with two different diameters. Since transmembrane c rings exhibit ends of two different sizes (35), the AFM topograph suggested that the rings were oriented alternating up and down in the bilayer. Closer inspection of individual c rings allowed their numbers of subunits to be directly counted from the unprocessed topographs (Fig. 5B). It appeared that every c ring was assembled from 13 subunits. Also, reference-free single-molecule averaging showed that indeed every class of c rings was composed of 13 subunits. The major average class (Fig. 5C) showed the circular arrangement of c subunits in detail. On average, the wide end of the c ring protruded 1.5 ± 0.2 nm from the lipid bilayer and had outer and inner diameters of 6.6 ± 0.5 nm and 5.5 ± 0.5 nm (n = 40), respectively.
FIG. 5.
High-resolution AFM of the S. elongatus SAG 89.79 c ring. (A) Topograph showing the densely packed c-ring assembly after its reconstitution into the lipid bilayer. The wide and narrow ends of c rings suggest their alternating orientations in the membrane. (B) Gallery of single rings showing their c-subunit arrangements and stoichiometries. (C) Reference-free single-particle average of c rings (n = 131). Independent of the classes found by averaging, the ring stoichiometry was always 13. Topographs were recorded in buffer solution and exhibited gray levels corresponding to a vertical scale of 2.0 nm.
DISCUSSION
We report on c-subunit rings from F-ATP synthases of eight different cyanobacterial strains isolated from their thylakoid membranes. These c rings were stable in SDS, allowing us to determine their electrophoretic mobilities in SDS-PAGE. As the quaternary structures of the c rings from F-ATP synthases (and the k ring from the V-type ATPase in Enterococcus hirae) appear to be highly conserved (20, 25, 36), their relative electrophoretic mobilities were expected to be proportional to their relative molecular masses. With several c rings of known molecular masses, this proposition could be verified and applied to determine the masses of cyanobacterial c rings. The accuracy allows the calculation of their oligomeric compositions by the use of c-subunit masses revealed from MS-MALDI. Based on this analysis, the cyanobacterial c rings investigated were composed of 13 (1×), 14 (5×), and 15 (2×) monomers. Hence, the cyanobacterial c rings exhibited different sizes, and they were generally larger than their bacterial and mitochondrial counterparts, having 10 or 11 subunits, but were more closely related to the c14 ring of chloroplasts. While the ATP synthases with small c rings have a low H+ (Na+)-to-ATP ratio, they can perform ATP synthesis at high efficiency at a sufficiently high ion motive force. ATP synthases with larger c rings have a high H+ (Na+)-to-ATP ratio and enable ATP synthesis at lower ion motive forces, albeit less efficiently. Furthermore, two more examples of c15 rings further support the idea that a symmetry mismatch between the F1 and Fo motors (36) is not obligatory for all F-ATP synthases (28).
To put our fast and simple analysis method on a solid structural basis, we reconstituted the c ring of S. elongatus SAG 89.79 F-ATP synthase and performed high-resolution AFM. The topographs show an assembly of c subunits into a c13 ring. A monomeric rotor consisting of one single polypeptide with 13 helical hairpins has been postulated on the basis of a tridecamerization of the uncE/atpE equivalent ntpK gene in the archaeon M. kandleri (15), but such a gene product has not been biochemically characterized or structurally visualized yet.
The range of the cyanobacterial strains used here includes representatives of the major classes of cyanobacteria (Chroococcales, Nostocales, Oscillatoriales, and Gloeobacteria), with the exception of Prochlorophyta. Rings with the most frequent c14 stoichiometry are found in four mesophilic freshwater cyanobacteria (Synechocystis sp. strain PCC 6803, Anabaena sp. strain PCC 7120, S. elongatus PCC 6301, and S. elongatus PCC 7942) and in Synechococcus sp. strain PCC 6716. The last, and the c13 ring of S. elongatus SAG 89.79, was found in strains isolated as natural inhabitants from thermal springs, which could resist elevated temperatures (>40°C). Two of three c15 rings were found in species living in extreme alkaline environments (Arthrospira sp. strains PCC 9438 and 9108 [reference 28 and this study, respectively]). In vitro experiments indicated that ATP synthesis in Arthrospira sp. strain PCC 9438 (c15) required a lower proton motive force and a higher H+/ATP ratio than ATP synthesis in the two strains Synechococcus sp. strain PCC 6716 (c14) and Synechocystis sp. strain PCC 6803 (c14) (2, 38). One c15 ring was found in G. violaceus PCC 7421, isolated from calcareous rocks, a unique cyanobacterium lacking internal membranes and characterized by very slow growth, low capacity for maintaining pH homeostasis in the cytoplasm, and low delta pH across the plasma membrane (3). It must therefore be taken into account that the 13-meric and the 15-meric rings may represent strain-specific adaptations to various physiological conditions imposed by their natural habitats, e.g., elevated temperature and/or alkaline pH.
What determines the sizes of the c rings?
Cyanobacterial c subunits from different organisms show highly conserved sequences but assemble into rings of variable stoichiometries. The question therefore arises as to whether these c rings would occasionally change their sizes depending on external living conditions, such as the carbon source, as has been suggested for the c ring from E. coli (31). A variation of the c-ring size has been experimentally assessed by genetically engineering the c-ring size in E. coli from 10 to 12 hairpins, and in both cases, an active ATP synthase was reported (12, 13). Though, the c-ring size of E. coli has not yet been determined by structural means, these data imply a certain flexibility of the c-ring size, at least within the E. coli ATP synthase. In great contrast to this, the number of c subunits in the ring of the thermophilic bacterium Bacillus sp. strain PS3 is restricted to 10, because only marginal ATP synthesis rates have been found with all ATP synthases containing c rings deviating from c10 (23). In support of this, recent structural data for the c rings suggest a fixed stoichiometry for a given species rather than a flexible one. AFM images of I. tartaricus c11 rings, plant chloroplast c14 rings, and c15 rings from Arthrospira sp. strain PCC 9438 showed that c rings occasionally lack individual subunits. However, instead of closing this gap immediately by collapsing into smaller rings, these incomplete oligomers retain the same shape and diameter as observed for their complete c rings (24, 28). This indicates strongly that the c11, c14, and c15 stoichiometries of these rings are intrinsic properties of the c subunits determined by the primary structure of the protein. This finding is also supported by the observation of annular structures after isolation of monomeric c subunits from E. coli (1). Furthermore, subunit c from I. tartaricus, when synthesized in E. coli, is correctly assembled into c11 rings (21) despite the preferred c10 stoichiometry of the native E. coli c ring (12). Hence, if the size is defined entirely by the sequence of subunit c, cyanobacterial c rings with their variable sizes and highly conserved primary structures (Fig. 1) may help to identify amino acids determining the c-ring stoichiometries. However, an expression system that allows genetic modification and expression of cyanobacterial (or plant) c rings is necessary to investigate this type of question, and it remains to be established.
Structural considerations.
It is not obvious from the structure of the c11 ring of I. tartaricus (20) how differences close to the N termini of the various cyanobacterial c subunits may have an impact on the ring size. However, additional asparagines at position 4 of the c subunit occur in the sequences of the 15-meric rings from Arthrospira sp. strain PCC 9438 and Arthrospira sp. strain PCC 9108. Another suitable candidate to induce stoichiometry variations may be found in the differing amino acid sequences at positions 23 to 41. It was observed earlier that the GXGXGXG motif positioned in this region leads to a very tight packing of the inner ring of α-helices (40). Hence, variations at these positions might well influence the packing of neighboring helices (6) and therefore influence the ring size. Furthermore, membrane insertion chaperones, such as YidC (14, 37), or the role of the ATP synthase i subunit, which is still not well understood (10, 32), as well as modifications like the N-terminal formylation of some c subunits as observed in this work may have an influence.
Acknowledgments
We thank Christoph von Ballmoos (ETH, Zurich, Switzerland) for providing spinach chloroplast thylakoid membranes and assisting with MS determinations, Janet Vonck (MPI-BP, Frankfurt, Germany) for EM analysis, Cheng-Cai Zhang (IBSM-CNRS, Marseille, France) and Ju-Yuan Zhang (Huazhong Agricultural University, Wuhan, China) for providing access to the DNA sequence of the atp operons from Arthrospira sp., and Birgit Roth Zgraggen for amino acid analyses.
This work was supported by the ETH Research Commission, the European Union, and the DFG.
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Arechaga, I., P. J. Butler, and J. E. Walker. 2002. Self-assembly of ATP synthase subunit c rings. FEBS Lett. 515:189-193. [DOI] [PubMed] [Google Scholar]
- 2.Bakels, R. H., H. S. van Walraven, K. Krab, M. J. Scholts, and R. Kraayenhof. 1993. On the activation mechanism of the H+-ATP synthase and unusual thermodynamic properties in the alkalophilic cyanobacterium Spirulina platensis. Eur. J. Biochem. 213:957-964. [DOI] [PubMed] [Google Scholar]
- 3.Belkin, S., R. J. Mehlhorn, and L. Packer. 1987. Proton gradients in intact cyanobacteria. Plant Physiol. 84:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, P. D. 1997. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 66:717-749. [DOI] [PubMed] [Google Scholar]
- 5.Capaldi, R. A., and R. Aggeler. 2002. Mechanism of the F1Fo-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27:154-160. [DOI] [PubMed] [Google Scholar]
- 6.Curran, A. R., and D. M. Engelman. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13:412-417. [DOI] [PubMed] [Google Scholar]
- 7.Dimroth, P., C. von Ballmoos, T. Meier, and G. Kaim. 2003. Electrical power fuels rotary ATP synthase. Structure 11:1469-1473. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, M. J., and S. J. Bradd. 1993. Separation and analysis of membrane proteins by SDS-polyacrylamide gel electrophoresis. Methods Mol. Biol. 19:203-210. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, S. J. 2000. ATP synthase: what dictates the size of a ring? Curr. Biol. 10:R804-R808. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, D. K., and W. S. Brusilow. 1995. Effects of the uncI gene on expression of uncB, the gene coding for the a subunit of the F1Fo ATPase of Escherichia coli. FEBS Lett. 371:127-131. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, W., and R. H. Fillingame. 1998. Interacting helical faces of subunits a and c in the F1Fo ATP synthase of Escherichia coli defined by disulfide cross-linking. Proc. Natl. Acad. Sci. USA 95:6607-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, W., J. Hermolin, and R. H. Fillingame. 2001. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl. Acad. Sci. USA 98:4966-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, P. C., and R. H. Fillingame. 1998. Genetic fusions of subunit c in the Fo sector of H+-transporting ATP synthase. Functional dimers and trimers and determination of stoichiometry by cross-linking analysis. J. Biol. Chem. 273:29701-29705. [DOI] [PubMed] [Google Scholar]
- 14.Kol, S., B. R. Turrell, J. de Keyzer, M. van der Laan, N. Nouwen, and A. J. Driessen. 2006. YidC-mediated membrane insertion of assembly mutants of subunit c of the F1Fo ATPase. J. Biol. Chem. 281:29762-29768. [DOI] [PubMed] [Google Scholar]
- 15.Lolkema, J. S., and E. J. Boekema. 2003. The A-type ATP synthase subunit K of Methanopyrus kandleri is deduced from its sequence to form a monomeric rotor comprising 13 hairpin domains. FEBS Lett. 543:47-50. [DOI] [PubMed] [Google Scholar]
- 16.Matthey, U., G. Kaim, and P. Dimroth. 1997. Subunit c from the sodium-ion-translocating F1Fo-ATPase of Propionigenium modestum—production, purification and properties of the protein in dodecylsulfate solution. Eur. J. Biochem. 247:820-825. [DOI] [PubMed] [Google Scholar]
- 17.Meier, T., and P. Dimroth. 2002. Intersubunit bridging by Na+ ions as a rationale for the unusual stability of the c-rings of Na+-translocating F1Fo ATP synthases. EMBO Rep. 3:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meier, T., S. A. Ferguson, G. M. Cook, P. Dimroth, and J. Vonck. 2006. Structural investigations of the membrane-embedded rotor ring of the F-ATPase from Clostridium paradoxum. J. Bacteriol. 188:7759-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier, T., U. Matthey, C. von Ballmoos, J. Vonck, T. Krug von Nidda, W. Kühlbrandt, and P. Dimroth. 2003. Evidence for structural integrity in the undecameric c-rings isolated from sodium ATP synthases. J. Mol. Biol. 325:389-397. [DOI] [PubMed] [Google Scholar]
- 20.Meier, T., P. Polzer, K. Diederichs, W. Welte, and P. Dimroth. 2005. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308:659-662. [DOI] [PubMed] [Google Scholar]
- 21.Meier, T., J. Yu, T. Raschle, F. Henzen, P. Dimroth, and D. J. Müller. 2005. Structural evidence for a constant c11 ring stoichiometry in the sodium F-ATP synthase. FEBS J. 272:5474-5483. [DOI] [PubMed] [Google Scholar]
- 22.Meyer zu Tittingdorf, J. M., S. Rexroth, E. Schäfer, R. Schlichting, C. Giersch, N. A. Dencher, and H. Seelert. 2004. The stoichiometry of the chloroplast ATP synthase oligomer III in Chlamydomonas reinhardtii is not affected by the metabolic state. Biochim. Biophys. Acta 1659:92-99. [DOI] [PubMed] [Google Scholar]
- 23.Mitome, N., T. Suzuki, S. Hayashi, and M. Yoshida. 2004. Thermophilic ATP synthase has a decamer c-ring: indication of noninteger 10:3 H+/ATP ratio and permissive elastic coupling. Proc. Natl. Acad. Sci. USA 101:12159-12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller, D. J., N. A. Dencher, T. Meier, P. Dimroth, K. Suda, H. Stahlberg, A. Engel, H. Seelert, and U. Matthey. 2001. ATP synthase: constrained stoichiometry of the transmembrane rotor. FEBS Lett. 504:219-222. [DOI] [PubMed] [Google Scholar]
- 25.Murata, T., I. Yamato, Y. Kakinuma, A. G. Leslie, and J. E. Walker. 2005. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science 308:654-659. [DOI] [PubMed] [Google Scholar]
- 26.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239-242. [DOI] [PubMed] [Google Scholar]
- 27.Oberfeld, B., J. Brunner, and P. Dimroth. 2006. Phospholipids occupy the internal lumen of the c ring of the ATP synthase of Escherichia coli. Biochemistry 45:1841-1851. [DOI] [PubMed] [Google Scholar]
- 28.Pogoryelov, D., J. Yu, T. Meier, J. Vonck, P. Dimroth, and D. J. Müller. 2005. The c15 ring of the Spirulina platensis F-ATP synthase: F1/Fo symmetry mismatch is not obligatory. EMBO Rep. 6:1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rippka, R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 167:3-27. [DOI] [PubMed] [Google Scholar]
- 30.Schägger, H., and G. Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 31.Schemidt, R. A., J. Qu, J. R. Williams, and W. S. Brusilow. 1998. Effects of carbon source on expression of Fo genes and on the stoichiometry of the c subunit in the F1Fo ATPase of Escherichia coli. J. Bacteriol. 180:3205-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneppe, B., G. Deckers-Hebestreit, and K. Altendorf. 1990. Overproduction and purification of the uncI gene product of the ATP synthase of Escherichia coli. J. Biol. Chem. 265:389-395. [PubMed] [Google Scholar]
- 33.Seelert, H., N. A. Dencher, and D. J. Müller. 2003. Fourteen protomers compose the oligomer III of the proton-rotor in spinach chloroplast ATP synthase. J. Mol. Biol. 333:337-344. [DOI] [PubMed] [Google Scholar]
- 34.Seelert, H., A. Poetsch, N. A. Dencher, A. Engel, H. Stahlberg, and D. J. Müller. 2000. Proton-powered turbine of a plant motor. Nature 405:418-419. [DOI] [PubMed] [Google Scholar]
- 35.Stahlberg, H., D. J. Müller, K. Suda, D. Fotiadis, A. Engel, T. Meier, U. Matthey, and P. Dimroth. 2001. Bacterial Na+-ATP synthase has an undecameric rotor. EMBO Rep. 2:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock, D., A. G. Leslie, and J. E. Walker. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700-1705. [DOI] [PubMed] [Google Scholar]
- 37.van der Laan, M., P. Bechtluft, S. Kol, N. Nouwen, and A. J. Driessen. 2004. F1Fo ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Walraven, H. S., H. Strotmann, O. Schwarz, and B. Rumberg. 1996. The H+/ATP coupling ratio of the ATP synthase from thiol-modulated chloroplasts and two cyanobacterial strains is four. FEBS Lett. 379:309-313. [DOI] [PubMed] [Google Scholar]
- 39.von Ballmoos, C., Y. Appoldt, J. Brunner, T. Granier, A. Vasella, and P. Dimroth. 2002. Membrane topography of the coupling ion binding site in Na+-translocating F1Fo ATP synthase. J. Biol. Chem. 277:3504-3510. [DOI] [PubMed] [Google Scholar]
- 40.Vonck, J., T. Krug von Nidda, T. Meier, U. Matthey, D. J. Mills, W. Kühlbrandt, and P. Dimroth. 2002. Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J. Mol. Biol. 321:307-316. [DOI] [PubMed] [Google Scholar]
- 41.Zakharov, S. D., R. G. Ewy, and R. A. Dilley. 1993. Subunit III of the chloroplast ATP-synthase can form a Ca2+-binding site on the lumenal side of the thylakoid membrane. FEBS Lett. 336:95-99. [DOI] [PubMed] [Google Scholar]
- 42.Zarrouk, C. 1966. Contribution à l'étude d'une cyanophycée. Influence de divers facteurs physiques et chimiques sur la croissance et la photosynthèse de Spirulina maxima. Ph.D. thesis. Université de Paris, Paris, France.
- 43.Zhang, D., and S. B. Vik. 2003. Helix packing in subunit a of the Escherichia coli ATP synthase as determined by chemical labeling and proteolysis of the cysteine-substituted protein. Biochemistry 42:331-337. [DOI] [PubMed] [Google Scholar]