Abstract
Native Americans derive from a small number of Asian founders who likely arrived to the Americas via Beringia. However, additional details about the intial colonization of the Americas remain unclear. To investigate the pioneering phase in the Americas we analyzed a total of 623 complete mtDNAs from the Americas and Asia, including 20 new complete mtDNAs from the Americas and seven from Asia. This sequence data was used to direct high-resolution genotyping from 20 American and 26 Asian populations. Here we describe more genetic diversity within the founder population than was previously reported. The newly resolved phylogenetic structure suggests that ancestors of Native Americans paused when they reached Beringia, during which time New World founder lineages differentiated from their Asian sister-clades. This pause in movement was followed by a swift migration southward that distributed the founder types all the way to South America. The data also suggest more recent bi-directional gene flow between Siberia and the North American Arctic.
Introduction
The mitochondrial DNA haplogroup nomenclature that is widely used today in population and medical genetics, forensic science, and in other interdisciplinary studies, traces back to the analysis of Native American populations by Torroni et al. [1],[2]. The first four letters of the phylogenetic alphabet for mtDNA haplogroups - A-D - were coined to refer to just four founding haplogroups that exhibit virtually all North and South American mtDNA diversity.
Genetic studies demonstrate that Native Americans inherited their mitochondrial DNA (mtDNA) from a handful of founders who arrived from Asia via Beringia [1],[2]. No more than four major pan American and three minor North American founding mtDNA haplotypes (A2, B2, C1, D1 and X2a, D2, D3, respectively) have been convincingly established in previous studies of control region sequence, RFLP markers and 30 complete mtDNA genomes (Table 1) [1]–[14]. The paucity of established founding mtDNAs suggests that the number of migrants that initially peopled the Americas was relatively low. However, determining the full range of diversity surviving to the present day in the founding population requires high-resolution mtDNA sequence data. Previous estimates of mtDNA diversity are predominantly based on control region sequences representing only a minor fraction of the mtDNA genome. In addition, control region sequences experience a high frequency of recurrent mutations, potentially obscuring the identification of additional founding mtDNAs [14]–[19].
Table 1. Defining mutations for Native American mtDNA haplogroups.
| Hg | HVS I | HVS II | Coding region |
| A | 16223-16290-16319 | 73-235-263 | 663, 1736, 4248, 4824, 8794 |
| A2 | 16111-16223-16290-16319-16362 | 64-73-146-153-235-263 | 8027, 12007 |
| A2a | 16111-16223-16290-16319-16362 | 64-73-146-153-235-263 | 3330 |
| B | 16189 | 73-263 | 8281-8289del |
| B4bd | 16189-16217 | 73-263 | 827, 15535 |
| B4b | 16189-16217 | 73-263 | 499, 4820, 13590 |
| B2 | 16189-16217 | 73-263 | 3547, 4977, 6473, 9950, 11177 |
| C | 16223-16298-16327 | 73-249d-263 | 3552A, 9545, 11914, 13263, 14318 |
| C1 | 16223-16298-16325-16327 | 73-249d-263-290-291d | - |
| C1b | 16223-16298-16325-16327 | 73-249d-263-290-291d | 493 |
| C1c | 16223-16298-16325-16327 | 73-249d-263-290-291d | 1888, 15930 |
| C1d | 16223-16298-16325-16327 | 73-249d-263-290-291d | 7697 |
| C4 | 16223-16298-16327 | 73-249d-263 | 2232iA, 6026, 11969, 15204 |
| C4c | 16223-16245-16298-16327 | 73-263 | 11440, 13368, 14433, 15148 |
| D | 16223-16362 | 73-263 | 4883, 5178A |
| D4 | 16223-16362 | 73-263 | 3010, 8414, 14668 |
| D1 | 16223-16325-16362 | 73-263 | 2092 |
| D2 | 16129-16223-16271-16362 | 73-263 | 3316, 7493, 8703, 9536, 11215 |
| D2a | 16129-16223-16271-16362 | 73-263 | 11959 |
| D2b | 16129-16223-16271-16362 | 73-263 | 9181 |
| D4h3 | 16223-16241-16301-16342-16362 | 73-263 | 3336, 3396, 3644, 5048, 6285, 8949, 9458, 13135 |
| D3 | 16223-16319-16362 | 73-263 | 951, 8020, 10181, 15440, 15951 |
| X | 16189-16223-16278 | 73-153-263 | 6221, 6371, 13966, 14470 |
| X2a | 16189-16213-16223-16278 | 73-153-195-200-263 | 1719, 8913, 12397, 14502 |
The full substitutional motif is shown in control region, the sub-clades defining mutations are indicated in bold.
Even though some additional minor founder types have been later identified in North America, such as X, the hypothesis of just four major founder types in the initial colonization of the New World remains uncontested. However, the timing of their entry remains debated. Previous studies of mtDNA data place estimates for the peopling of the New World in a broad range from 11,000 to over 40,000 years before present (ybp) [reviewed by 20], although more recent estimates range from 20,000–15,000 ybp [21]. Recent archaeological evidence places Homo sapiens in northeastern Siberia at the Yana Rhinoceros Horn Site as early as 30,000 ybp [22] about twice the 15,000 ybp [23] date for humans at the southern end of South America. These archaeological dates suggest two likely scenarios. First, the ancestors of Native Americans peopled Beringia before the Last Glacial Maximum, but remained locally isolated (likely due to ecological barriers) until entering the Americas at 15,000 ybp (Beringian incubation model, BIM) [24]. Second, the ancestors of Native Americans did not reach Beringia until just before 15,000 ybp, and then moved continuously on into the Americas, being recently derived from a larger parent Asian population (direct colonization model, DCM).
The DCM model hypothesizes the presence of founding mtDNA haplotypes that include members from both Northeast Asia and the Americas. It presumes a continuous movement of recently derived migrants across Beringia. In contrast, the BIM model predicts widespread, derived founding haplotypes specific to the Americas that are not found in Asia. This implies that migrants were isolated for an extended period of time before entering the Americas and that the founder haplotypes arose in situ in Beringia. Once in the Americas, these immigrants spread southward. Therefore, the phylogeographic distribution of this diversity can provide insights into the mode of the initial phase of the peopling of the Americas. A nested hierarchy of diversity from north to south in Native American founding haplogroups would reflect a gradual peopling, whereas a uniform distribution of Native American founding haplotypes both in North and South America implies a rapid occupation.
Results
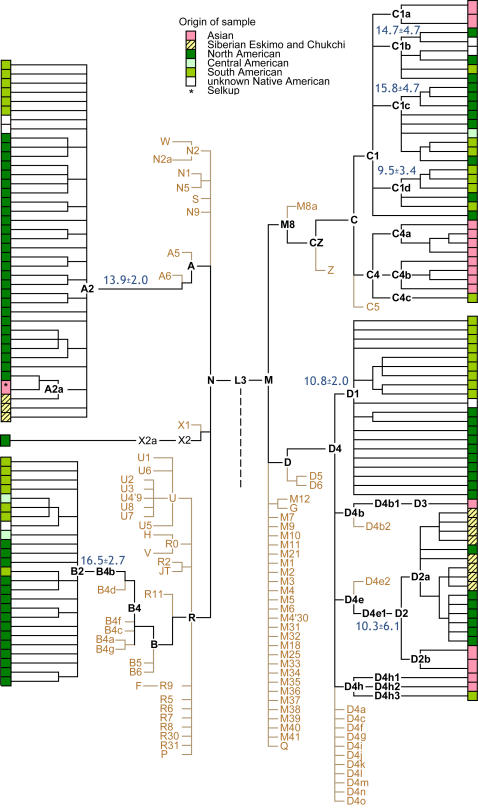
Previous studies of mtDNA variation recognized no major subclade structure within each New World haplogroup [1],[3]–[4],[24]. A few studies of mtDNA variation suggested subclade structure [12],[14],[25], but lacked the power to convincingly demonstrate it. In this study we identified three sub-clades - C1b, C1c and C1d - that incorporate nearly all of Native American haplogroup C mtDNAs. All three are widely distributed in the New World. They are absent in Asia, and show similar coalescence times of approximately 13,900±2,700 years ago (Figure 1). Similar coalescence times were estimated for the other major founder haplogroups - A2, B2 and D1 - suggesting the simultaneous divergence of all founder clades across North and South America. A different C1 sub-clade in Asia - C1a [26]- likely derives from the same ancestral population as the three Native American sub-clades. Thus C1b, C1c and C1d are likely independent New World founders. In addition to C1 sub-clades, we defined two additional founders–D4h3 and C4c. These differ by several mutations from the Asian-derived ancestral clades, D4h and C4, respectively (Figure S1d,c). Haplogroup D4h3 ranges from Alaska to Tierra del Fuego and has recently been identified in Alaskan skeletal remains (10,300 ybp) [27]. We identified haplogroup C4c in two Ijka-speakers from Colombia, but its distribution in the Americas remains poorly characterized.
Figure 1. Schematic representation of phylogeny of human mtDNA outside of Africa.
Branches encompassing Native Americans and their immediate Asian ancestral and sister lineages, represented by complete sequences, are shown in black with coalescence ages indicated and geographic location identified by colours. Lineages in brown correspond to the main haplogroups, found in Eurasia and Oceania, but absent in Native Americans. For complete phylogenetic tree see Supplementary figure 1.
Discussion
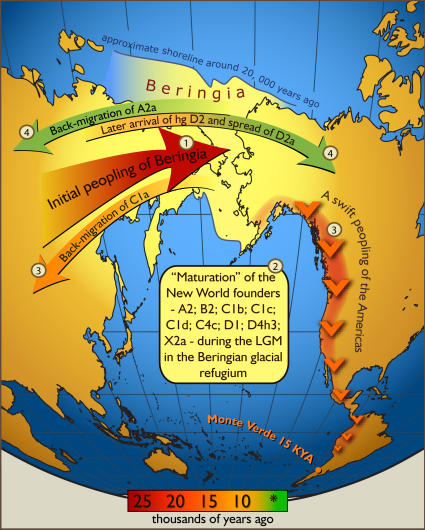
Our phylogeographic analysis of a new mitochondrial genome dataset allows us to draw several conclusions. First, before spreading across the Americas, the ancestral population paused in Beringia long enough for specific mutations to accumulate that separate the New World founder lineages from their Asian sister-clades (Figure 2) [4],[24], [28]–[29]. Second, founding haplotypes are uniformly distributed across North and South America instead of exhibiting a nested structure from north to south (Figure 1). Thus, after the Beringian standstill, the initial North to South migration was likely a swift pioneering process, not a gradual diffusion. This scenario matches the pattern of distribution of the first archaeological sites in Northeast Asia and the Americas [22],[23]. Third, the largely autochthonous pattern of variation seen in Native American mtDNAs suggests that the swift migration was followed by long-term isolation of local populations accompanied with the development of regional haplotypes within continental founder haplogroups [1].
Figure 2. Schematic illustration of maternal geneflow in and out of Beringia.
Colours of the arrows correspond to approximate timing of the events and are decoded in the coloured time-bar. The initial peopling of Berinigia (depicted in light yellow) was followed by a standstill after which the ancestors of the Native Americans spread swiftly all over the New World while some of the Beringian maternal lineages–C1a-spred westwards. More recent (shown in green) genetic exchange is manifested by back-migration of A2a into Siberia and the spread of D2a into north-eastern America that post-dated the initial peopling of the New World.
In addition to illuminating the peopling process during the pioneering phase, the new dataset allows identification of more-recent genetic exchanges around and across Beringia (Figure 2). Specifically, haplogroup D2 consists of two sister clades, one found only in Siberia (D2b) and the other found in northernmost Eskimos, Chukchi, Aleut, and Athapaskans (D2a). While sub-haplogroup D2a is shared between ethno-historically close related Beringian Aleuts and Eskimos, (Figure S1) its sister clade D2b is spread among populations from distantly related linguistic groups (Tungusic, Turkic, Mongolic) (Table S2). A close relationship of matrilineal ancestry between individuals from different linguistic groups may be due to an overlap of geographic range of their ancestors approximately at the time of the Pleistocene-Holocene boundary. Alternatively, some populations may have received the D2b variant through more recent gene flow. It is also worthwhile to note the absence of D2 in all other Native American populations, suggesting that D2 diversified in Beringia after the initial migration into the Americas had occurred. Haplogroup D3 may have also reached America through more recent genetic exchange. It is spread in Nganasans, Mansi, Evenks, Ulchi, Tuvas, Chukchi and Siberian Eskimos [26],[30] and recently reported in Greenland and Canadian Inuit populations [31], but absent in other Native Americans. Additional investigatios of these populations may provide insight into the cause of the phylogenetic connections.
Surprisingly, we also found a Native American sub-type of haplogroup A2 among Evenks and Selkups in southern and western Siberia (Table S2). Previously, this HVS I motif is reported in one Yakut-speaking Evenk in northwestern Siberia [32]. A novel demographic scenario of relatively recent gene flow from Beringia to deep into western Siberia (Samoyedic-speaking Selkups) is the most likely explanation for the phylogeography of haplogroup A2a, which is nested within an otherwise exclusively Native American A2 phylogeny (Figure S1).
The high-resolution sequence data analyzed in this study reveals previously hidden diversity within the Native American mtDNA gene pool. The new data suggest that the initial founders of the Americas emerged from a single source ancestral population that evolved in isolation, likely in Beringia. This scenario is consistent with the unique pattern of diversity from autosomal locus D9S1120 [33] of a private allele in high frequency and ubiquitous in the Americas. The finding that humans were present at the Yana Rhinoceros Horn Site dated to 30,000 ybp [22] suggests that the isolation in Beringia might have lasted up to 15,000 years. Following this isolation, the initial founders of the Americas began rapidly populating the New World from North to South America.
Materials and Methods
The sample-set comprises 601 Native American individuals from 20 populations distributed throughout the Americas (23 Dogribs from Subarctic Canada; 20 Apaches, 20 Northern Paiutes, 11 Zunis from Southwest US; 77 Ngöbes, 34 Kunas from Panama; 39 Emberas, 57 Waunanas from Panama and Colombia; 47 Arsarios, 48 Koguis, 29 Ijkas, 42 Wayuus, 27 Coreguajes, 22 Vaupes from Colombia; 12 Secoyas-Sionas, 32 Cayapas from Ecuador; 9 Tucuman, 18 Salta, 25 Catamarca, 5 Mocovi from Argentina) and 3764 samples from 26 Asian populations (51 Eskimos; 155 Chukchi; 120 Selkups; 66 Kets; 70 Tundra Nenets; 275 Tuvas; 185 Khakas; 339 Altaians; 170 Shors; 71 Koryaks; 85 Nanais; 122 Uyghurs; 406 Kazakhs; 58 Gilyaks; 61 Oroks; 105 Kirghiz; 48 Uzbeks; 38 Tajiks; 201 Buriats; 324 Evenks; 105 Evens; 22 Yukaghirs; 423 Yakuts; 157 Dolgans; 107 Nganasans). A subset of these sequences were reported elsewhere [34]–[42].
First, haplogroup affiliations of the individual samples were determined through RFLP analysis and DNA sequencing of the HVS I region, if not known earlier [34]–[42]. Samples that could not be assigned to haplogroups A–D or X were investigated for evidence of recent admixture, particularly among populations with well established historical accounts of co-existence of Native American and either European or African populations. Samples of European or African origin were excluded from the current study. Further, 20 Native American and 7 Asian samples were selected for complete sequencing of mtDNA genomes. Using these 27 novel and 113 published Native American and relevant Asian complete or coding region sequences [4],[26],[30],[43]–[50], phylogenetic trees were reconstructed based on a maximum parsimony approach (Figure S1, Text S1). From these whole mtDNA genomes coding region markers were selected for screening in the sample set through RFLP analyses or direct sequencing (Table S1). Hierarchical method was used, so, that each Native American sample was first cheked for nucleotide positions, where a polymorphism could be assumed based on the HVS I information and close ethnic, geographic or linquistic affiliation to complete sequenced sample. From Asian populations, samples, which could be relevant to Native American haplogroups, were selected based on HVS I sequence and analyzed for coding region markers (A2–12007, 8027; A2a–3330; C1a–3826, 7598; C4–11969; C4a–12672; C4b–3816; C4c–11440; D2–8703; D2a–4991, 11959; D2b–9181; D4–8414T; D4a–3206; D4e1–3316; D4h–3336, 3644; D4m–9667; D5–5301; D5a–11944, 12026).
DNA was extracted using conventional methods [34]–[42]. Preparation of sequencing templates was carried out following standard protocols, employing FIREPol polymerase (Solis BioDyne). Purified products were sequenced with the DYEnamic™ ET terminator cycle sequencing kit (Amersham Pharmacia Biotech) and analyzed on MegaBace1000 or ABI 3730xl sequencers. Sequences were aligned and analyzed with the Wisconsin Package (GCG) or ChromasPro 1.34.
Coalescence-age calculations and SDs were estimated based on the phylogenies of complete sequences [3],[6]. Given the global propensity of young mtDNA clades showing a significant excess of non-synonymous mutations, application of the raw molecular clock [49] in intra-species data sets is problematic [46]. Therefore, for dating the coalescent times of founder haplogroups we employed only synonymous transitions between the np 577-16023, assuming the rate of 3.5×10-8 (SD 0.1×10-8)/year/position [46]. The complete mtDNA genome data can be found in Genbank.
Supporting Information
Complete Phylogenetic Trees
(0.10 MB XLS)
Genotype data
(0.71 MB XLS)
Haplogroup frequencies.
(0.02 MB XLS)
Acknowledgments
We would like to thank the late John McDonough and the late Dr. Surinder Papiha for their contributions to this research project.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for this work is from NSF BCS#0422144 to RSM and from the University of Tartu #PBGMR06901 to TK.
References
- 1.Torroni A, Schurr TG, Cabell MF, Brown MD, Nell JV, et al. Asian affinities and contiental radiation of the four founding Native American mtDNAs. Am J Hum Genet. 1993;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- 2.Schurr TG, Ballinger SW, Gan YY, Hodge JA, Merriwether DA, et al. Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet. 1990;46:613–623. [PMC free article] [PubMed] [Google Scholar]
- 3.Forster P, Harding R, Torroni A, Bandelt HJ. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 4.Bandelt HJ, Hernnstadt C, Yao YG, Kong QP, Kivisild T, et al. Identification of Native American founder mtDNas through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet. 2003;67:512–524. doi: 10.1046/j.1469-1809.2003.00049.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown MD, Hosseini SH, Torroni A, Bandelt HJ, Allen JC, et al. mtDNA haplgroup X: An ancient link between Europe/Western Asia and North America? Am J Hum Genet. 1998;63:1852–1861. doi: 10.1086/302155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merriwether DA, Rothhammer F, Ferell RE. Distribution of the four founding lineage haplotypes in Native Americans suggests a single wave of migration for the New World. Am J Phys Anthropol. 1995;98:411–430. doi: 10.1002/ajpa.1330980404. [DOI] [PubMed] [Google Scholar]
- 8.Bailliet G, Rothhammer F, Carnese FR, Bravi CM, Bianchi NO. Founder mitochondrial haplotypes in Amerindian populations. Am J Hum Genet. 1994;55:27–33. [PMC free article] [PubMed] [Google Scholar]
- 9.Ward RH, Redd A, Valencia D, Frazier B, Paabo S. Genetic and linguistic differentiation in the Americas. Proc Natl Acad Sci U.S.A. 1993;90:10663–10667. doi: 10.1073/pnas.90.22.10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields GF, Schmiechen AM, Frazier BL, Redd A, Voevoda ML, et al. mtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am J Hum Genet. 1993;53:549–562. [PMC free article] [PubMed] [Google Scholar]
- 11.Lorenz JG, Smith DG. Distribution of four founding mtDNA haplogroups among Native North Americans. Am J Phys Anthropol. 1996;101:307–323. doi: 10.1002/(SICI)1096-8644(199611)101:3<307::AID-AJPA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 12.Santos SE, Ribeiro-Dos-Santos AK, Meyer D, Zago MA. Multiple founder haplotypes of mitochondrial DNA in Amerindians revealed by RFLP and sequencing. Ann Hum Genet. 1996;60:305–319. doi: 10.1111/j.1469-1809.1996.tb01194.x. [DOI] [PubMed] [Google Scholar]
- 13.Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S, et al. Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol Biol Evol. 1993;10:23–47. doi: 10.1093/oxfordjournals.molbev.a039987. [DOI] [PubMed] [Google Scholar]
- 14.Malhi RS, Eshleman JA, Greenberg JA, Weiss DA, Schultz Shook BA, et al. The structure of diversity within New World mitochondrial DNA haplogroups: implications for the prehistory of North America. Am J Hum Genet. 2002;70:905–919. doi: 10.1086/339690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Non AL, Kitchen A, Mulligan CJ. Indentification of the most informative regions of the mitochondrial genome for phylogenetic and coalescent analyses. Mol Phylogenet Evol Epub. 2006 doi: 10.1016/j.ympev.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 16.Wakeley J. Substitution rate variation among sites in hypervariable region 1 of human mitochondrial DNA. J Mol Evol. 1993;37:613–623. doi: 10.1007/BF00182747. [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa M, Di Rienzo A, Kocher TD, Wilson AC. Toward a more accurate time scale for the human mitochondrial DNA tree. J Mol Evol. 1993;37:347–354. doi: 10.1007/BF00178865. [DOI] [PubMed] [Google Scholar]
- 18.Malyarchuk BA, Rogozin IB. Mutagenesis by transient misalignment in the human mitochondrial DNA control region. Ann Hum Genet. 2004;68:324–339. doi: 10.1046/j.1529-8817.2004.00099.x. [DOI] [PubMed] [Google Scholar]
- 19.Sigurğardóttir S, Helgason A, Guicher JR, Stefansson K, Donnelly P. The mutation rate in the human mtDNA control region. Am J Hum Genet. 2000;66:1599–1609. doi: 10.1086/302902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eshleman JA, Malhi RS, Smith DG. Mitochondrial DNA studies of Native Americans: Conceptions and misconceptions of the population prehistory of the Americas. Evo Anth. 2003;12:7–18. [Google Scholar]
- 21.Schurr TG, Sherry ST. Mitochondrial DNA and Y chromosome diversity and the peopling of the Americas: evolutionary and demographic evidence. Am J Hum Biol. 2004;16:420–439. doi: 10.1002/ajhb.20041. [DOI] [PubMed] [Google Scholar]
- 22.Pitulko VV, Nikolsky PA, Girya EY, Basilyan AE, Tumskoy VE, et al. The Yana RHS site: humans in the Arctic before the last glacial maximum. Science. 2003;303:52–56. doi: 10.1126/science.1085219. [DOI] [PubMed] [Google Scholar]
- 23.Dillehay TD. The late Pleistocene cultures of South America. Evol Anth. 1999;7:206–216. [Google Scholar]
- 24.Bonatto SL, Salzano FM. A single and early migration for the peopling of the Americas supported by mitochondrial DNAn sequence data. Proc Natl Acad Sci USA. 1997;94:1866–1871. doi: 10.1073/pnas.94.5.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merriewther DA, Ferrell RE. The four founding lineage hypothesis for the New World: a critical reevaluation. Mol Phylogenet Evol. 1996;5:241–246. doi: 10.1006/mpev.1996.0017. [DOI] [PubMed] [Google Scholar]
- 26.Starikovskaya EB, Sukernik RI, Derbeneva OA, Volodko NV, Ruiz-Pesini E. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann Hum Genet. 2005;69:67–89. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemp BM, Malhi RS, McDonough J, Bolnick DA, Eshleman JA, et al. Genetic analysis of early holocene skeletal remains from Alaska and its implications for the settlement of the Americas. Am J Phy Anth. 2007;132:605–621. doi: 10.1002/ajpa.20543. [DOI] [PubMed] [Google Scholar]
- 28.Szathmary EJ. mtDNA and the peopling of the Americas. Am J Hum Genet. 1993;53:793–799. [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford MH. Cambridge University Press; 1998. The Origins of Native Americans: Evidence from Anthropological Genetics. [Google Scholar]
- 30.Derbeneva OA, Sukernik RI, Volodko NV, Hosseini SH, Lott MT, et al. Analysis of mitochondrial DNA diversity in the Aleuts of the Commander Islands and its implications for the genetic history of Beringia. Am J Hum Genet. 2002;71:415–21. doi: 10.1086/341720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helgason A, Palsson G, Pedersen HS, Angulalik E, Gunnarsdottir ED, et al. MtDNA variation in Inuit populations of Greenland and Canada: migration history and population structure. Am J Phys Anthropol. 2006;130:123–134. doi: 10.1002/ajpa.20313. [DOI] [PubMed] [Google Scholar]
- 32.Pakendorf B, Novgorodov IN, Osakovskija VL, Danilova AP, Protodjakonov AP, et al. Investigating the effects of prehistoric migrations in Siberia: genetic variation and the origin of Yakuts. Hum Genet. 2006;120:334–353. doi: 10.1007/s00439-006-0213-2. [DOI] [PubMed] [Google Scholar]
- 33.Schroeder KB, Schurr TG, Long JC, Rosenberg NA, Crawford MH, et al. A private allele ubiquitous in the Americas. Biol Lett Epub. 2007 doi: 10.1098/rsbl.2006.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolman CJ, Bermingham E. Mitochondrial and nuclear DNA diversity in the Choco and Chibcha Amerinds of Panama. Genetics. 1997;147:1289–302. doi: 10.1093/genetics/147.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolman CJ, Bermingham E, Cooke R, Ward RH, Arias TD, et al. Reduced mtDNA Diversity in the Ngöbe Amerinds of Panama. Genetics. 1995;140:275–283. doi: 10.1093/genetics/140.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rickards O, Martinez-Labarga C, Lum JK, De Stefano GF, Cann RL. mtDNA history of the Cayapa Amerinds of Ecuador: detection of additional founding lineages for the Native American populations. Am J Hum Genet. 1999;65:519–30. doi: 10.1086/302513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaestle FA, Smith DG. Ancient mitochondrial DNA evidence for prehistoric population movement: the Numic expansion. Am J Phys Anthropol. 2001;115:1–12. doi: 10.1002/ajpa.1051. [DOI] [PubMed] [Google Scholar]
- 38.Malhi RS, Mortensen HM, Eshleman JA, Kemp BM, Lorenz JG, et al. Native American mtDNA prehistory in the American Southwest. Am J Phys Anthropol. 2003;120:108–24. doi: 10.1002/ajpa.10138. [DOI] [PubMed] [Google Scholar]
- 39.Fedorova SA, Bermisheva MA, Villems R, Maksimova NR, Khusnutdinova EK. Analysis of mitochondrial DNA lineages in Yakuts. Mol Biol (Mosk) 2003;37:643–53. [PubMed] [Google Scholar]
- 40.Bermisheva MA, Kutuev IA, Spitsyn VA, Villems R, Batyrova AZ, et al. Analysis of mitochondrial DNA variation in the population of Oroks. Russian Journal of Genetics. 2005;41:66–71. [PubMed] [Google Scholar]
- 41.Goltsova TV, Osipova L, Zhadanov S, Villems R. The Effect of Marriage Migration on the Genetic Structure of the Taimyr Nganasan Population. Russian Journal of Genetics. 2005;41:954–965. [PubMed] [Google Scholar]
- 42.Melton PE, Briceno I, Gomez A, Devor EJ, Bernal JE, et al. Biological Relationship Between Central and South American Chibchan Speaking Populations: Evidence From mtDNA. Am J Phys Anthropol Mar 5. 2007 doi: 10.1002/ajpa.20581. [DOI] [PubMed] [Google Scholar]
- 43.Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrnstadt C, Preston G, Howell N. Errors, phantoms and otherwise, in human mtDNA sequences. Am J Hum Genet. 2003;72:1585–6. doi: 10.1086/375406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 46.Kivisild T, Shen P, Wall DP, Do B, Sung R, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–87. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong Q-P, Yao Y-G, Sun C, Bandelt H-J, Zhu C-L, et al. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maca-Meyer N, Gonźalez AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka M, Cabrera VM, Gonzalez AM, Larruga JM, Takeyasu T, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete Phylogenetic Trees
(0.10 MB XLS)
Genotype data
(0.71 MB XLS)
Haplogroup frequencies.
(0.02 MB XLS)