Abstract
Ubiquitination and the degradation of the large subunit of RNA polymerase II, Rpb1, is not only involved in DNA damage-induced arrest but also in other transcription-obstructing events. However, the ubiquitin ligases responsible for DNA damage-independent processes in mammalian cells remain to be identified. Here, we identified Wwp2, a mouse HECT domain ubiquitin E3 ligase, as a novel ubiquitin ligase of Rpb1. We found that Wwp2 specifically interacted with mouse Rpb1 and targeted it for ubiquitination both in vitro and in vivo. Interestingly, the interaction with and ubiquitination of Rpb1 was dependent neither on its phosphorylation state nor on DNA damage. However, the enzymatic activity of Wwp2 was absolutely required for its ubiquitin modification of Rpb1. Furthermore, our study indicates that the interaction between Wwp2 and Rpb1 was mediated through WW domain of Wwp2 and C-terminal domain of Rpb1, respectively. Strikingly, downregulation of Wwp2 expression compromised Rpb1 ubiquitination and elevated its intracellular steady-state protein level significantly. Importantly, we identified six lysine residues in the C-terminal domain of Rpb1 as ubiquitin acceptor sites mediated by Wwp2. These results indicate that Wwp2 plays an important role in regulating expression of Rpb1 in normal physiological conditions.
Posttranslational covalent modification of cellular proteins by ubiquitin has diverse physiological functions. The modification regulates protein-protein interactions, transcriptional activity, and protein subcellular localization, in addition to its major role in targeting proteins for degradation through the 26S proteasome (5, 20). The ubiquitination process is mediated by a sequential action of three enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-protein ligase (E3) (24, 27). Through direct interaction with substrates, E3s play a critical role in determining enzymatic specificity during the ubiquitination reaction. E3s are a large and diverse group of proteins including RING (for really interesting new gene) finger-containing E3 or U-box (a modified RING motif without the full complement of Zn2+ binding ligands) E3, as well as the HECT (for homologous to E6-AP carboxyl-terminus) domain E3. Some HECT domain E3 ligases have a common domain architecture: a N-terminal C2 domain, a WW domain, and a C-terminal catalytic HECT domain (12, 15). The WW domain, having two conserved tryptophan residues, mediates protein-protein interactions and is known to bind specific proteins that contain a proline-rich motif (3). WW domains also recognize phosphoserine- and phosphothreonine-containing substrates (21). The HECT domain contains an active cysteine residue that transfers an activated ubiquitin from E2 onto itself and then onto its substrate, thus providing the ligase enzyme activity (13). Only a few HECT domain E3 ligases have been found in yeast including Rsp5 (11) and Tom1 (36). In contrast, many of these ligases have been identified in mammalian species, including Smurf1/2 (26, 34), Itch (10), Nedd4 (8), WWP1 (19), and WWP2/Wwp2 (28, 42). These E3 ligases regulate many biological processes (15). Human WWP2 was first cloned by Pirozzi et al. (28). Experiments showed that WWP2 binds to and downregulates the epithelial Na(+) channel (22). However, direct evidence for its ubiquitin ligase activity is lacking. Previously, our group identified Wwp2, a murine ortholog of WWP2, as an E3 ligase of the POU domain transcription factor Oct-4. In addition, we demonstrated that Wwp2 has ubiquitin ligase activity both in vitro and in vivo and that an intact HECT domain is essential for its enzymatic activity (42). Like other HECT domain E3 ligases, Wwp2 has an N-terminal C2 domain, a C-terminal HECT domain, and four WW domains in the middle. Since most E3 ligases have more than one substrate, we reasoned that Wwp2 might have substrates in addition to Oct-4. In an effort to identify more substrates of Wwp2, we performed affinity chromatography using a glutathione S-transferase (GST) fusion protein of the WW-HECT domains of mouse Wwp2 and cell lysate from murine F9 embryonal carcinoma cells. Unexpectedly, the large subunit of RNA polymerase II, Rpb1, was found to interact with Wwp2.
Rpb1 is essential for synthesis of mRNA (31, 32). One of its unique features is that it possesses a carboxyl-terminal domain (CTD) that consists of multiple repeats of the consensus heptapeptide sequence YSPTSPS and functions throughout the RNA polymerase II transcription cycle (6). This consensus sequence is highly conserved from yeast to humans. However, the number of repeats and deviations from the consensus sequence in the region near the C terminus seem to have increased through evolution. For example, the heptapeptide sequence is repeated 27 times in budding yeast but 52 times in mammalian CTD. Furthermore, different from the yeast CTD, the mammalian CTD has many variant heptads with substitutions at position 7, particularly in its C-terminal half (heptads 27 to 52) (9, 30). Deletion studies in cultured cells have shown that the CTD plays an essential role in transcription. CTD is important for cell viability and development. Therefore, it is not surprising that CTD is subject to a variety of posttranslational modifications. Multiple modifications and conformations allow CTD to interact with distinct partners and targets (2). During transcription, CTD undergoes characteristic changes in its phosphorylation state. A 220-kDa form of Rpb1 (IIa) with a hypophosphorylated CTD preferentially binds promoters. The IIa form shifts to a 240-kDa form (IIo) with a hyperphosphorylated CTD upon promoter clearance and transcript elongation (7). On the other hand, DNA damage- and transcription inhibition-induced ubiquitination of Rpb1 has been implicated in transcription-coupled repair. In yeast, HECT domain E3 ligase Rsp5 and peptide deformylase Def1 were required for Rpb1 ubiquitination in response to DNA damage (1, 29, 33, 40). In addition to bulky DNA damage-induced Rpb1 ubiquitination, Inukai et al. showed that hydrogen peroxide (H2O2) causes significant ubiquitination and proteasomal degradation of Rpb1 by mechanisms that are distinct from those used after UV irradiation. Most studies on Rpb1 ubiquitination have used DNA damage-induced models. Therefore, experimental data on Rpb1 ubiquitination in the constitutive condition are lacking. In fact, Rsp5 was first found to target Rpb1 ubiquitination in the absence of DNA damage (14, 16). The mammalian counterpart of Rsp5 has not been clearly defined. A human WW domain-containing HECT domain E3 protein, Rpf1/hNedd4, was reported to bind and ubiquitinate both yeast and human Rpb1 in vitro (1). However, a relationship between hNedd4 and hRpb1 in vivo has not been established. In the present study, we investigated the interaction between mouse Rpb1 and Wwp2, a HECT domain E3 ligase closely related to Rsp5. We demonstrated that Wwp2 targets Rpb1 for ubiquitination both in vitro and in vivo. Importantly, we have examined the effect of repressing Wwp2 expression on the steady-state protein levels or ubiquitination of Rpb1 in F9 cells in normal conditions. In addition, the ubiquitin modification sites of Rpb1 mediated by Wwp2 were identified by biochemical study and mass spectrometric analysis. Our results indicate that Wwp2 is indeed an ubiquitin E3 ligase of mammalian Rpb1 and may play an important role in regulating Rpb1 functions.
MATERIALS AND METHODS
Plasmids.
The cDNA sequence of mouse full-length Rpb1 was amplified by reverse transcription-PCR using total RNA from CGR8 mouse embryonic stem (ES) cells (kindly provided by Austin Smith) and cloned into pET-30a(+) (Novagen), pGEX-4T-1 (Amersham Biosciences) or pcDNA3-Flag vectors. Mouse Srg3 full-length cDNA sequence was amplified by PCR and cloned into pET-30a(+) vector. For construction of CTD expression vectors, CTD of Rpb1 was amplified by using cDNA from CGR8 cells, and the PCR product was cloned into pGEX-4T-1 vector, pcDNA3-EGFP-vector or pPyCAGIP vector. To make CTDmk expression vectors, about 400 bp of DNA fragment with lysine residues at 1859, 1866, 1873, 1887, 1908, and 1922 in Rpb1 mutated to arginine residues was synthesized in Shanghai Sangon, Inc., and cloned into pGEX-4T-1-CTD or pPyCAGIP-CTD vector to replace the wild-type sequence. Flag tag and nuclear localization signal of simian virus 40 T antigen sequence were inserted into pPyCAGIP-CTD and pPyCAGIP-CTDmk vectors, respectively. Plasmids used for expression of Wwp2, Wwp2CA mutation, or individual Wwp2 domains have been described previously (42). His-ubiquitin (Ub) expression vector pMT107 was kindly provided by Dirk Bohmann. His-Ub K0 mutation expression vector, kindly provided by Richard Baer, expresses ubiquitin with all lysine residues mutated to arginine residues. The pPyCAGIP-TetR vector was previously described (38). Nineteen-basepair sequences, corresponding to enhanced green fluorescent protein (EGFP; 5′-GGC TAC GTC CAG GAG CGC A-3′), Wwp2-1 (5′-CCG CCA GCC CAG AAT CAA C-3′), and Wwp2-2 (5′-GAT TCC TCT ACC AGT CTT C-3′) were selected to generate pTER+ vectors (kindly provided by Hans Clevers) for RNAi EGFP or RNAi Wwp2, respectively, as described previously (37). The sequences of all constructs were verified by DNA sequencing.
Antibodies.
The antibodies used were as follows: N20 (Santa Cruz), C21 (Santa Cruz), H5 (Covance), H14 (Covance), 8WG16 (Covance), Tubulin (Sigma), and antibodies to Flag epitope (Sigma), His epitope (Santa Cruz), GFP epitope (Roche), and ubiquitin epitope (Cell Signaling). Antibodies to Wwp2 and GST were raised and affinity purified as described previously (42).
Fusion protein expression and affinity purification.
GST and His fusion proteins were expressed and purified according to the manufacturer's instructions from Amersham Biosciences and Novagen, respectively. GST or GST-Wwp2-WW-HECT proteins were bound to glutathione immobilized on Sepharose beads (Amersham Biosciences) in equal molar amounts. Beads were then incubated overnight at 4°C with the whole-cell extract from F9 cells. Bound proteins were washed and boiled in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer. After electrophoresis, the gel was stained with Coomassie blue. Bands, present only in the GST-Wwp2-WW-HECT with F9 cell lysate column, were excised for mass spectrometric analysis (25).
In-gel digestion of proteins and capillary high-performance liquid chromatography mass spectrometric analysis for identification of ubiquitination sites.
Capillary high-performance liquid chromatography mass spectrometric analysis was performed in an integrated system by the method described by Chen et al. (4).
Western blot analysis.
Cells were lysed in coimmunoprecipitation (Co-IP) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 10% [vol/vol] glycerol, 0.5% Nonidet P-40, 1 mM NaF, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The protein concentration was determined by using the BCA protein assay kit (Pierce). For experiments involving transient transfection, cotransfected pSV-β-galactosidase plasmid (kindly provided by Richard Baer) was used to normalize each sample. Western blot analysis was conducted by using enhanced chemiluminescence (Pierce). All experiments were performed at least three times and representative data are shown.
Cell culture and DNA transfection.
HEK 293 cells (kindly provided by Richard Baer) were cultured under standard conditions and transfected by using the calcium phosphate method or Lipofectamine 2000 Reagent (Invitrogen). F9 cells were grown as suggested by ATCC. CGR8, mouse ES cells, were cultured as described previously (42). For tetracycline (Tc)-inducible RNA interference (RNAi) stable cell lines, F9 cells or CGR8 cells were transfected with 40 μg of pPyCAGIP-TetR (Tet repressor) plasmid by electroporation and selected with 1 μg of puromycin/ml. RNAi EGFP or RNAi Wwp2 vectors (40 μg) were subsequently introduced into the cells stably expressing TetR and were selected with 80 μg of zeocin/ml.
Co-IP and pull-down assay.
For Co-IP, cell lysates were prepared in Co-IP buffer and incubated with a specific antibody for 2 h at 4°C, followed by the addition of protein A-Sepharose beads for another 2 h. For GST pull-down experiments, GST fusion proteins bound with glutathione-Sepharose 4B beads in equal molar amounts were incubated with His fusion proteins in TBS-N (20 mM Tris-HCl [pH 7.6], 300 mM NaCl, and 0.1% NP-40) at 4°C for 2 h. Samples from IP or GST pull-down assays were analyzed by Western blotting.
Ubiquitination assay in vivo and in vitro.
For in vivo assays, HEK 293 cells were transfected with pcDNA3-Flag-Rpb1, pPyCAGIP-Flag-CTD, or pPyCAGIP-Flag-CTDmk; pCMV-Wwp2 or pCMV-Wwp2CA; pMT107; and pSV-β-galactosidase. At 48 h after transfection, cells were treated for 8 h with proteasome inhibitor MG132 (20 μM) or dimethyl sulfoxide before harvest. For the Rpb1 full-length ubiquitination assay, cells were lysed in radioimmunoprecipitation assay lysis buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM NaF, and 1 mM PMSF). Cell lysates, normalized by β-galactosidase activity, were adjusted to a final concentration of 1% SDS and boiled for 10 min. After dilution to a final concentration of 0.1% SDS, the sample was immunoprecipitated with M2 beads (Sigma) and analyzed by Western blotting with anti-His antibody. For the Rpb1 CTD ubiquitination assay, cells were lysed in NP-40 lysis buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaH2PO4, 600 mM NaCl, 1% NP-40, 1 mM NaF, and 1 mM PMSF). Cell lysates, normalized by β-galactosidase activity, were adjusted to a final concentration of 8 M urea and 10 mM imidazole. Ubiquitinated proteins were affinity purified on nitrilotriacetic acid His-bind resin (Novagen) and analyzed by Western blotting with anti-Flag antibody. For ubiquitination of endogenous Rpb1, the Tc-inducible Wwp2 RNAi F9 stable cell lines were treated with Tc or ethyl alcohol for 3 days and then treated for 8 h with MG132 (20 μM) before harvest. Cells were lysated in 1% SDS radioimmunoprecipitation assay buffer and boiled for 10 min. After sonication, the lysates were diluted to a final concentration of 0.1% SDS and normalized by protein concentration. The samples were then immunoprecipitated with 8WG16 or H14 antibody and analyzed by Western blotting with anti-ubiquitin antibody.
In vitro ubiquitination assays were carried out as described previously (42) with or without 26S proteasome (Calbiochem). K0 ubiquitin was used in in vitro ubiquitination assays to identify ubiquitination sites by mass spectrometric analysis. For the ubiquitination of CTD mutation in vitro, the GST domain was removed from GST-CTD or GST-CTDmk fusion protein through enzymatic digestion by thrombin (Sigma).
Dephosphorylation of endogenous Rpb1.
To dephosphorylate Rpb1, F9 cells were lysed in Co-IP buffer without NaF and incubated with λ PPase (New England Biolabs) for 30 min at 30°C. Dephosphorylated extracts were used for IPs with H14 or C21 antibodies.
RESULTS
Interaction between HECT domain E3 ligase, Wwp2, and the large subunit of RNA polymerase II, Rpb1.
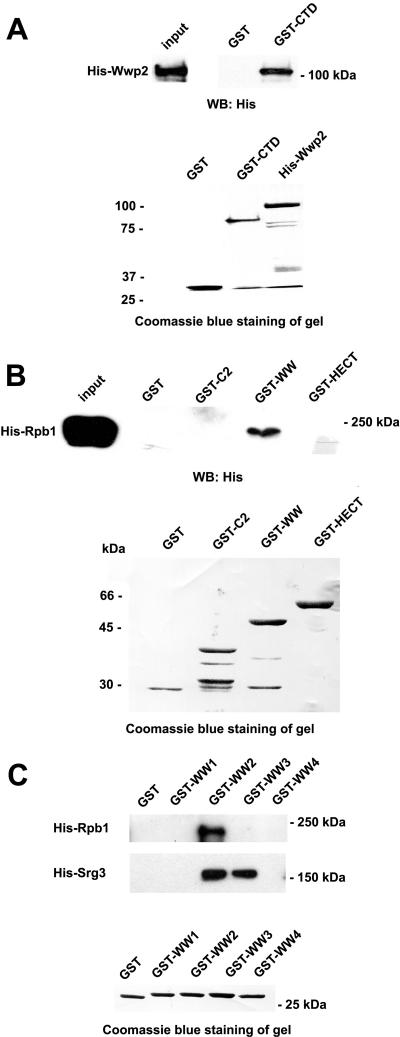
To identify novel substrates of Wwp2, we performed affinity chromatography using bacterially expressed GST-WW-HECT (WW and HECT domains of Wwp2) fusion protein and lysate from murine F9 embryonal carcinoma cells. As shown in Fig. 1A, one band of 220 kDa was present only in the F9 lysate-containing GST-WW-HECT column and was excised for mass spectrometric analysis, which indicated that the band contained the large subunit of RNA polymerase II, Rpb1.
FIG. 1.
Identification and characterization of the interaction between Wwp2 and Rpb1. (A) Coomassie blue staining of an SDS-polyacrylamide gel electrophoresis gel containing the proteins from the indicated columns is shown. (B) GST pull-down assay to show that Wwp2 binds to Rpb1 directly in vitro. (C) Co-IP assay in HEK 293 cells to show that Wwp2 binds to the CTD of Rpb1 specifically in vivo. Whole-cell lysates (WCL) were immunoprecipitated (IP) with anti-Flag antibody and analyzed by Western blotting with antibodies as indicated. (D) Association of endogenous Wwp2 with Rpb1 in F9 cells. The RNAi Wwp2-2 F9 cells were induced with Tc for 3 days and then whole-cell lysates were subjected to immunoprecipitation with anti-Wwp2 antibody or control anti-GST antibody. (E) Association of endogenous Wwp2 with Rpb1 using F9 cell lysate treated with or without λ phosphatase (λ PPase). The whole-cell lysates of F9 cells were pretreated with λ PPase and subjected to IP as D.
To confirm the interaction between Wwp2 and Rpb1, a GST pull-down assay was performed with bacterially expressed GST-Rpb1 fusion protein and His-Wwp2 fusion protein. His-Wwp2CA fusion protein that contains a mutation at residue 838 of cysteine to alanine in the HECT domain of Wwp2 was also included in the assay. We have previously shown that cysteine residue 838 in the HECT domain of Wwp2 is essential for its enzymatic activity (42). Western blot analysis with anti-His antibody indicated that immobilized GST-Rpb1, but not GST alone, was able to pull down His-Wwp2. In addition, the mutation at residue 838 from cysteine to alanine did not affect its association with Rpb1 (Fig. 1B). Theses results indicate that Wwp2 interacts with Rpb1 directly in vitro.
Next, we determined whether Wwp2 and Rpb1 interact in vivo. Coimmunoprecipitation experiments were performed with lysates from HEK 293 cells expressing Flag-Wwp2 or Flag-Wwp2CA and EGFP-CTD or EGFP. The CTD of Rpb1 was used in this experiment because it was easier to express than full-length Rpb1. As shown in Fig. 1C, EGFP-CTD coimmunoprecipitated with Flag-Wwp2 (lane 1, row 1). Mutation of cysteine to alanine at residue 838 of the HECT domain of Wwp2 had no effect on the association of Wwp2 with EGFP-CTD in vivo (lane 5, row 1). As a negative control, EGFP alone did not coimmunoprecipitate with Flag-Wwp2 or Flag-Wwp2CA, suggesting that the interaction between EGFP-CTD and Flag-Wwp2 or Flag-Wwp2CA was mediated specifically by CTD (row 3).
To prove that endogenous Rpb1 specifically coimmunoprecipitates with endogenous Wwp2, we derived stable F9 cell lines that expressed Tc-inducible small interference RNAs (siRNA) specifically targeting Wwp2 coding sequence. The specificity of the siRNA was confirmed by Western blotting, which showed that addition of Tc to the cell culture media dramatically reduced expression of Wwp2 in the cells (Fig. 1D, lane 2 in row 3), whereas the expression level of tubulin was not affected. Next, proteins from F9 cell lysate were immunoprecipitated with anti-Wwp2 antibody or anti-GST antibody, and precipitated proteins were analyzed by using antibody H14, or antibody C21, or anti-Wwp2 antibody and anti-tubulin antibody (Fig. 1D). Interestingly, both hyperphosphorylated and hypophosphorylated Rpb1 were readily detected in Wwp2-antibody-immunoprecipitated complexes without Tc treatment, although it seems that Wwp2 favored to form complex with hypophosphorylated Rpb1 (compare row 1 and row 2 of lane 4 in Fig. 1D). As a negative control, none of the used antibody detected signal in GST-antibody precipitated proteins (lane 3). Moreover, the fact that tubulin antibody did not produce any signal in Wwp2 antibody-immunoprecipitated proteins proves the specificity of our Wwp2 antibody (row 4 of lane 4 and 5). Compellingly, treatment of cells with Tc significantly compromised the association between Wwp2 and Rpb1, especially hypophosphorylated Rpb1. These observations clearly confirm that Wwp2 and Rpb1 specifically form a stable complex in living cells under physiological conditions. Although H14 antibody is claimed to recognize hyperphosphorylated CTD of Rpb1, they may weakly recognize hypophosphorylated CTD as well. To exclude this possibility, F9 cell lysate was treated with λ phosphatase, and the expression levels of hypophosphorylated and hyperphosphorylated Rpb1 were determined by Western blotting. As expected, treatment of cell lysate with the phosphatase completely abolished H14 antibody-recognized signal, although C21-, Wwp2-, and tubulin-antibody-recognized signals were not changed, suggesting that the Western blot band detected by H14 antibody in our experimental conditions is indeed hyperphosphorylated Rpb1 (Fig. 1E, lanes 1 and 2). Similarly, H14 antibody could not detect any signal in Wwp2 antibody-immunoprecipitated proteins after treatment with the phosphatase (row 1 of lane 5), suggesting that the signal recognized by H14 antibody in Wwp2 antibody-immunoprecipitated proteins was indeed from hyperphosphorylated Rpb1 (row 1 of lane 4). In contrast, λ phosphatase treatment did not affect the association of hypophosphorylated Rpb1 with Wwp2. Taken together, our results demonstrate that Wwp2 specifically binds both hyper- and hypophosphorylated Rpb1 in the cells. However, the phosphorylation state of Rpb1 might affect its association with Wwp2, with the hypophosphorylated state of Rpb1 favoring the complex formation.
Mapping the interaction domains in Wwp2 and Rpb1.
GST pull-down assays were performed to identify the domains mediating the interaction between Wwp2 and Rpb1. His-Wwp2 fusion protein and the GST-CTD of Rpb1 fusion protein were expressed in bacteria and purified (Fig. 2A, bottom panel). Western blot analysis showed that GST-CTD, but not GST alone, was capable of binding His-Wwp2 in vitro, suggesting that the CTD domain is sufficient to mediate the interaction between Wwp2 and Rpb1 (Fig. 2A, top panel). On the other hand, WW domain (including four WW domains) of Wwp2 was found to interact with Rpb1 (Fig. 2B). In contrast, the HECT and C2 domains of Wwp2 did not bind Rpb1.
FIG. 2.
Interacting regions between Wwp2 and Rpb1. (A) The ability of the Rpb1 CTD binds to Wwp2 was analyzed by GST pull-down assay (top panel). Bacterially expressed fusion proteins were stained with Coomassie blue (bottom panel). (B) The ability of various truncated Wwp2 fusion proteins to bind His-Rpb1 was tested by GST pull-down assay. (top panel). Various domains of Wwp2 were bacterially expressed and were stained with Coomassie blue (bottom panel). (C) The ability of the individual WW domains of Wwp2 to bind to His-Rpb1 and His-Srg3 was detected by GST pull-down assay (top panel). Bacterially expressed individual WW domains of Wwp2 were stained by Coomassie blue (bottom panel).
In addition, four WW domains of Wwp2 were individually expressed and purified for GST pull-down experiments. The results showed that the second WW domain was the major domain for mediating the association between Wwp2 and Rpb1 in vitro (Fig. 2C). Furthermore, to test whether the negative results obtained from the GST-WW1, -WW3, and -WW4 domains arose from improper folding of these isolated domains, their interaction with Srg3, a known substrate of Wwp2 (unpublished data), was examined. As shown in Fig. 2C, GST-WW3 domain could bind Srg3 with a similar efficiency to that of GST-WW2 domain. The observation indicates that although WW domains have sequence similarity and associate with proline-rich proteins, each domain has a different specificity or affinity for protein interactions.
Wwp2 ubiquitinates Rpb1 both in vitro and in vivo.
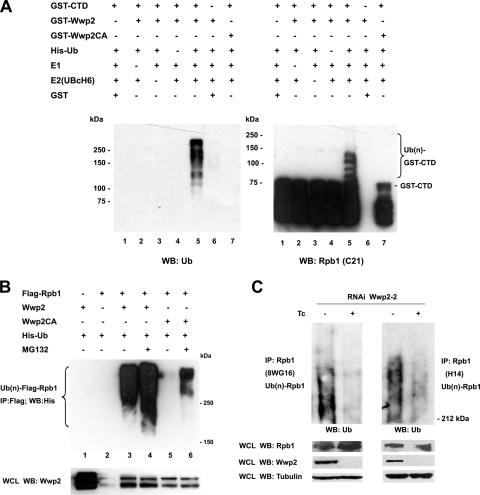
Our group has previously demonstrated that Wwp2 has ubiquitin E3 ligase activity (42). To investigate whether the CTD of Rpb1 can also serve as a substrate for Wwp2, an in vitro ubiquitination assay was performed with purified proteins including GST-Wwp2, GST-Wwp2CA, GST-CTD, His-ubiquitin, E1, E2 (UBcH6), and GST. When ubiquitination reaction products were analyzed by Western blotting with an anti-ubiquitin antibody, higher-molecular-weight species indicative of the addition of ubiquitin moieties to GST-CTD were observed in the presence of E1, E2, ubiquitin, and wild-type Wwp2 (Fig. 3A, left panel, lane 5). These ubiquitinated products were GST-CTD dependent, since the ubiquitination signal was not seen in the absence of CTD (lane 6). In addition, the ubiquitination signal disappeared when wild-type Wwp2 was replaced with Wwp2CA, demonstrating that Wwp2-mediated CTD ubiquitination was dependent upon an intact HECT domain (lane 7), although mutation of the HECT domain did not affect the interaction between Wwp2 and Rpb1, as demonstrated in Fig. 1B and C. To further confirm that the ubiquitinated protein was indeed CTD, the same ubiquitination assay was performed, and the reaction products were examined by Western blotting with anti-Rpb1 antibody (C21). Similarly, the higher-molecular-weight signals were detected in the presence of E1, E2, ubiquitin, wild-type Wwp2, and CTD (Fig. 3A, right panel, lane 5) and were absent when GST-CTD was replaced with GST (lane 6). Again, Wwp2CA did not ubiquitinate CTD (lane 7). These data strongly demonstrate that CTD can serve as a substrate for Wwp2-mediated ubiquitination in vitro and that its ubiquitination requires normal activity of all three enzymes involved in the ubiquitination reaction. Of note, the GST-CTD used in the ubiquitination assay was not modified by phosphorylation. Therefore, Wwp2-mediated ubiquitination of CTD is not dependent upon its phosphorylation.
FIG. 3.
Wwp2 ubiquitinates Rpb1 both in vivo and in vitro. (A) Wwp2 targets CTD of Rpb1 for ubiquitination in vitro. Various purified proteins as indicated were incubated in ubiquitination buffer. Ubiquitinated CTD of Rpb1 was visualized by Western blotting with antibody against ubiquitin (left panel) and CTD of Rpb1 (right panel). (B) Wwp2 enhances ubiquitination of Rpb1 in vivo. HEK 293 cells were transfected with expression vectors as indicated. Cells were treated with or without proteasome inhibitor (MG132) before harvest. Exogenous Rpb1 was immunoprecipitated with anti-Flag antibody from the denatured cell lysate and ubiquitinated Rpb1 was analyzed by Western blotting with anti-His antibody (top panel). Wwp2 expressed in the cells was confirmed by immunoblotting with anti-Wwp2 antibody (bottom panel). (C) Wwp2 protein level regulates the ubiquitination level of endogenous Rpb1 in F9 cells. RNAi Wwp2-2 F9 cells induced with or without Tc for 3 days were treated with MG132 before harvest. Endogenous Rpb1 was immunoprecipitated with its antibodies from the denatured cell lysate, and ubiquitinated Rpb1 was analyzed by Western blotting with anti-ubiquitin antibody. Wwp2 expression levels in the cells were confirmed by immunoblotting with anti-Wwp2 antibody. Tubulin was used as loading control.
The biological relevance of in vitro assays was further investigated by performing in vivo ubiquitination assays. HEK 293 cells were transiently transfected with expression vectors as indicated. Cell lysate was immunoprecipitated with M2 beads and precipitated Rpb1 was analyzed by Western blotting with anti-His antibody. As shown in Fig. 3B, ubiquitination signal was not detected when His-ubiquitin was cotransfected with either Wwp2 or Flag-Rpb1 (lanes 1 and 2, top panel). However, coexpression of His-ubiquitin, Wwp2, and Flag-Rpb1 resulted in Flag-Rpb1 ubiquitination (lane 3). Importantly, the ubiquitin-modified Rpb1 was significantly enhanced when cells were treated with MG132, a 26S proteasome inhibitor, before harvest (lane 4), suggesting that some ubiquitinated Rpb1 is normally degraded through the 26S proteasome. In contrast, we did not detect any ubiquitinated Rpb1 when Wwp2CA was coexpressed (lane 5), although some ubiquitinated Rpb1 was observed when MG132 was used. However, the signal observed with Wwp2CA mutation was much less compared to wild-type Wwp2 (compare lanes 6 and 4) in the presence of MG132. Ubiquitinated Rpb1 observed in lane 6 might result from human WWP2 or other E3 ligases endogenously expressed in HEK 293 cells even in absence of DNA damage. The protein levels for the exogenously expressed Wwp2 were shown in the bottom panel of Fig. 3B. The lower band in Wwp2 Western blot analysis might be degradation product of Wwp2 in HEK 293 cells. These results reveal that Wwp2 can function as an ubiquitin E3 ligase for Rpb1 within cells and provides further evidence for the importance of the cysteine at residue 838 of the HECT domain in the enzymatic reaction.
Having established that Wwp2 can function as an E3 ubiquitin ligase for Rpb1 both in vitro and in vivo, the next question is whether Wwp2 is required for endogenously expressed Rpb1 ubiquitination in physiological conditions. To this end, ubiquitin modification of endogenous Rpb1 in F9 cells expressing Tc-inducible Wwp2 siRNA in the presence of Tc was compared to that in the absence of Tc. The F9 cells were pretreated with 26S proteasome inhibitor to block degradation of ubiquitinated Rpb1, and then Rpb1 in cell lysate was immunoprecipitated with 8WG16 antibody, recognizing hypophosphorylated Rpb1, or H14 antibody, respectively, followed by Western blot analysis with ubiquitin antibody. A high-molecular-weight smear of ubiquitin signal was detected in both 8WG16 and H14 antibody immunoprecipitates when Tc was not present (Fig. 3C). However, treatment of the F9 cells with Tc dramatically knocked down Wwp2 protein level and, importantly, the level of ubiquitinated Rpb1 simultaneously declined. The Rpb1 protein level was not affected by Tc addition due to the stabilizing effect of MG132 pretreatment. The results provide strong evidence for requirement of Wwp2 to ubiquitinate endogenous Rpb1 protein in normal conditions.
Wwp2 promotes degradation of Rpb1 through 26S proteasome.
The best-studied function of protein ubiquitin modification is its role in protein degradation through the 26S proteasome, although other functions for ubiquitin modification have been discovered. To testify that ubiquitinated Rpb1 catalyzed by Wwp2 can be degraded through 26S proteasome, 26S proteasome degradation assay was performed with in vitro Wwp2-ubiquitinated GST-CTD. As shown in Fig. 4A, GST-CTD fusion protein is modified by ubiquitin in the presence of E1, E2, ubiquitin, and GST-Wwp2 (lane 3). The ubiquitin modification of GST-CTD was dependent on the presence of GST-Wwp2 (no signal in lane 1). Interestingly, the ubiquitinated GST-CTD entirely vanished when 26S proteasome was included in the reaction (lane 4). This result clearly shows that ubiquitinated GST-CTD mediated by Wwp2 was degraded through 26S proteasome.
FIG. 4.
Wwp2 promotes degradation of Rpb1 through 26S proteasome. (A) Ubiquitinated GST-CTD catalyzed by Wwp2 can be degraded through 26S proteasome. (B) Knocking down Wwp2 expression elevates the steady-state protein level of Rpb1 in F9 cells. The Tc-inducible RNAi Wwp2 or RNAi EGFP stable cell lines established in F9 cells were treated as indicated. The whole-cell lysate was prepared, and the effect of Wwp2 RNAi on the steady-state level of Rpb1 was analyzed by Western blotting with antibody against Rpb1 (N20) (top row). The protein level of Wwp2 was measured by Western blotting with its antibody (middle row), and tubulin was used as a loading control (bottom row). (C) Wwp2 regulates levels of both hyperphosphorylated and hypophosphorylated Rpb1 in RNAi Wwp2 F9 cells. (D) The steady-state protein level of Rpb1 is upregulated when Wwp2 expression is knocked down in RNAi Wwp2 CGR8 cells.
The physiological significance of the foregoing observations was tested by studying the role of Wwp2 in regulating Rpb1 protein levels in mouse embryonic pluripotent stem cells. For this purpose, we derived both F9 and CGR8 mouse ES cell lines that stably expressed Tc-inducible siRNAs specifically targeting Wwp2 coding sequences. Two regions in the Wwp2 coding sequence were targeted and are referred to as RNAi Wwp2-1 and RNAi Wwp2-2, respectively. Multiple single clones for each targeted region were selected to ensure the specificity of RNA interference. RNAi EGFP stable F9 cell lines were also generated to be used as Tc treatment control. We induced expression of RNAi Wwp2-1, Wwp2-2, and EGFP by the addition of Tc to cell culture media and compared Rpb1 protein levels in the absence or presence of Tc. Western blot analysis showed that addition of Tc significantly reduced Wwp2 protein level for RNAi Wwp2-2 F9 cell line (Fig. 4B, middle row, lanes 3 to 8). Strikingly, total Rpb1 protein levels in the cells, detected by N20 antibody, were inversely enhanced after addition of Tc (lanes 3 to 5, top row), indicating that the downregulation of Wwp2 elevated Rpb1 protein levels. In contrast, addition of the same dosage of Tc to RNAi EGFP F9 cells affected neither Wwp2 protein levels nor total Rpb1 protein levels (Fig. 4B, lanes 1 and 2). These results indicate that elevated Rpb1 protein levels were a result of reduced Wwp2 levels and not the addition of Tc. Of interest, Rpb1 protein levels were much higher when cells were pretreated with MG132, regardless of the absence or presence of Tc (Fig. 4B, lanes 6 to 8, top row). This phenomenon implies that Rpb1 protein in F9 cells is subject to degradation through the 26S proteasome under normal conditions.
The relationship between Wwp2 and Rpb1 was further assessed by using Rpb1 antibodies specifically recognizing the hyperphosphorylated forms, H14 and H5. Intracellular Wwp2 protein levels gradually decreased after induction of RNAi expression in both RNAi Wwp2-1 and RNAi Wwp2-2 F9 cells, and the reduction in Wwp2 levels was more evident with longer induction (Fig. 4C). Meanwhile, protein levels for H14 and H5 antibody-recognized hyperphosphorylated Rpb1 were increased. The similar result was obtained with N20 antibody. These results suggest that Wwp2 may promote degradation of Rpb1 in F9 cells and that the regulatory effect of Wwp2 on Rpb1 levels was not dependent upon its phosphorylation state.
We next sought to determine whether Wwp2 has a similar role in the maintenance of steady-state Rpb1 protein level in another cell line. CGR8 ES cells stably expressing Tc-inducible Wwp2 siRNA were used for this purpose. Seven repeats of Wwp2-1 sequence were inserted into the siRNA expression vector to enhance the efficiency of knocking down Wwp2 expression, while only single sequence was used for Wwp2-2 siRNA expression vector. Similarly to F9 cells, Tc treatment induced significant reduction in Wwp2 protein levels in both Wwp2-1(x7) and Wwp2-2 CGR8 ES cell lines. Inversely, H5 antibody- and H14 antibody-recognized hyperphosphorylated Rpb1 and C21 antibody-recognized hypophosphorylated Rpb1 protein levels were gradually upregulated after Tc addition (Fig. 4D). In contrast, protein level of tubulin remained unchanged with Tc treatment. These data indicate that Wwp2 plays an important role in maintaining Rpb1 protein level at a normal state in embryonic pluripotent stem cells.
Identification of lysine residues in Rpb1 that accept ubiquitin.
It is currently unknown which lysine residues in mammalian Rpb1 are modified by ubiquitination. Therefore, we were keen to identify such modification sites in the GST-CTD fusion protein, which has been demonstrated to be ubiquitinated by Wwp2 in the present study. To this end, we performed mass spectrometric analysis on reaction products from a ubiquitination assay of GST-CTD by Wwp2. Six peptides containing ubiquitin-modified lysine residues were identified (Fig. 5A, the modified lysine residues were italicized and labeled with an asterisk). These were lysine residues at 1859, 1866, 1873, 1887, 1908, and 1922 of murine Rpb1.
FIG. 5.
Identification of ubiquitination sites in CTD. (A) Mass spectrometric analysis identified six peptides containing ubiquitinated lysine residues in the CTD. Numbers represent the positions of these peptides in Rpb1. Modified lysine residues were italicized and labeled with an asterisk. (B) The six identified lysine residues are essential for CTD ubiquitination in vitro. (C) The ability of GST-CTDmk to bind Wwp2 was analyzed by GST pull-down assay (top panel). Bacterially expressed fusion proteins were stained by Coomassie blue (bottom panel). (D) CTD or CTDmk of Rpb1 ubiquitination assay in HEK 293 cells. (E) Wwp2 regulates CTD protein level in HEK 293 cells. The constant amount of Flag-CTD or Flag-CTDmk was cotransfected into HEK 293 cells, together with increasing amount of Wwp2. The protein levels of CTD and CTDmk were determined by Western blot analysis. Cotransfected EGFP was used as a loading control.
To convincingly prove that these lysine residues are ubiquitin acceptor sites for Wwp2-mediated Rpb1 ubiquitination, we constructed bacterial expression vector which carried mutation of the six lysine residues to arginine residues (GST-CTDmk). After GST-CTD and GST-CTDmk fusion proteins were expressed bacterially, the GST domain was removed by enzymatic digestion to avoid lysine residues in the GST domain being modified by ubiquitin. The CTD and CTDmk proteins were purified and examined for their ubiquitin modification by Wwp2. For clarity, a ubiquitin (K0) with its all seven lysine residues mutated to arginine residues was used in this in vitro ubiquitination assay. As shown in Fig. 5B, CTD was efficiently ubiquitinated by Wwp2 when all components for the ubiquitination reaction were present (lane 2). However, little, if any, ubiquitination was detected when CTD was replaced by CTDmk (lane 3). Next, GST pull-down experiment was conducted between His-Wwp2 and GST-CTD or GST-CTDmk to verify that GST-CTDmk was able to associate with His-Wwp2. As shown in Fig. 5C, mutation of the six lysine residues in CTD did not affect its interaction with Wwp2. This observation demonstrates that the lack of ubiquitination for CTDmk was a result of mutation of the lysine residues as the ubiquitin acceptor sites but not due to a lack of interaction between the two proteins. Next, we examined the requirement of the six lysine residues in CTD for its ubiquitin modification in HEK 293 cells. Both Flag-tagged CTD and CTDmk were expressed in HEK 293 cells together with Wwp2 and His-tagged ubiquitin. MG132 at 20 μM was added to culture media 8 h before harvest. The ubiquitinated proteins in cell lysate were isolated by nitrilotriacetic acid affinity beads and analyzed by Western blotting with anti-Flag antibody. Exogenously expressed Wwp2 and Flag-tagged CTD in whole-cell lysate were also measured by Western blot analysis. As shown in Fig. 5D, His-ubiquitin modified Flag-CTD was easily detected as a high-molecular-weight smear. In contrast, there was not any ubiquitin modification signal detectable for Flag-tagged CTDmk. Finally, we wanted to know whether mutations of the six lysine residues in the CTD of Rpb1 indeed lead to increases in the steady level of CTD. To this end, the same amount of Flag-tagged CTD or CTDmk was expressed in HEK 293 cells together with increasing amounts of Wwp2. At 48 h after transfection, the steady-state levels of wild-type CTD and mutated CTD in the cells were analyzed by Western blotting. As shown in Fig. 5E, the steady-state protein levels of Flag-tagged CTD were gradually decreased with increasing protein level of Wwp2. However, no significant reduction in CTDmk protein level was detected when Wwp2 expression level was elevated. This result clearly shows that CTD expression level is closely associated with intracellular Wwp2 level. Overall, our data demonstrate that the six lysine residues in the CTD of Rpb1 are essential for the ubiquitination and degradation of CTD mediated by Wwp2 both in vitro and in vivo. Thus, we established, for the first time, that six lysine residues in the CTD of Rpb1 could serve as modification sites for Wwp2-mediated ubiquitination and degradation.
DISCUSSION
In the present study, we identified Wwp2 as the first mammalian HECT domain ubiquitin E3 ligase that targets the mammalian large subunit of RNA polymerase II, Rpb1, for ubiquitination both in vitro and in vivo. Importantly, our study demonstrates that Wwp2 interacts specifically with Rpb1, independent of DNA damage and phosphorylation state, leading to its ubiquitination and degradation through 26S proteasome. Furthermore, we show here that Wwp2 is essential for the maintenance of Rpb1 steady-state protein levels in embryonic pluripotent stem cells. These results indicate that Wwp2 play an important role in regulating expression of Rpb1 in normal physiological conditions.
The regulation of Rpb1 through posttranslational modification is currently an exciting topic of study. At least three types of posttranslational modifications have been identified for Rpb1, i.e., phosphorylation, glycosylation, and ubiquitination (17, 18, 29, 35). These modifications are likely to play important roles in Rpb1 regulation. Extensive research has focused on modification of Rpb1 by phosphorylation. Two major phosphorylation sites (serine 5 and serine 2) have been identified in the CTD, and phosphorylation state correlates with the transition between transcription initiation and elongation. However, ubiquitination of Rpb1 has been mainly studied in a DNA damage model. The ubiquitination of Rpb1 and its function in normal conditions in mammalian cells have not been documented. It is worthy of note that Rpb1 ubiquitination and degradation are not confined to situations in which there is DNA damage. In budding yeast, Rsp5 binds and ubiquitinates Rpb1 both in the absence and in the presence of DNA damage (1, 14). It has been pointed out that Rpb1 ubiquitination is likely to be a frequent event during transcription and also in the absence of DNA damage (33, 35). Somesh et al. provided evidence for damage-independent transcription arrest leading to Rpb1 ubiquitination in yeast (33). However, the factors responsible for such Rpb1 ubiquitination and degradation in vivo have not been characterized in mammalian cells. In the present study, we demonstrated that Wwp2 specifically binds and ubiquitinates Rpb1 in vitro and in vivo, independently of DNA damage. The easily detected association of Rpb1 with Wwp2 and its ubiquitination in normal F9 cells further supports the notion that at least a subset of Rpb1 is constitutively subjected to ubiquitin modification. Although we show here that Wwp2 specifically formed complex with both hypo- and hyperphosphorylated forms of endogenous Rpb1, the observation that the percentage of hypophosphorylated Rpb1 associated with Wwp2 was higher than that of hyperphosphorylated Rpb1 (Fig. 1D and E) suggests the phosphorylation state of Rpb1 might affect its interaction with Wwp2. Interestingly, Wwp2 shares several features with yeast Rpb1 ubiquitin ligase, Rsp5. The common features include (i) having a C2 domain at the N terminus, a WW domain in the middle, and a HECT domain at the C terminus; (ii) binding and ubiquitinating Rpb1 independent of its phosphorylation state; (iii) WW domain mediating association with Rpb1; (iv) active-site cysteine in HECT domain being essential for ubiquitin ligase activity; (v) 26S proteasome being involved in degradation of Rpb1; (vi) repression of their expression in vivo leading to an elevated steady-state level of Rpb1; and (vii) the CTD of Rpb1 being sufficient for binding to them. These features make Wwp2 a most likely mammalian counterpart of yeast Rsp5 for mediating Rpb1 ubiquitination and degradation constitutively. In present study, we provide convincing evidence to confirm the importance of Wwp2 to Rpb1 protein level and ubiquitination, including the findings that (i) Wwp2-mediated CTD ubiquitination could be degraded through 26S proteasome in vitro (Fig. 4A); (ii) the ubiquitination of endogenous Rpb1 in F9 cells was substantially reduced when Wwp2 expression was repressed (Fig. 3C); and (iii) knocking down expression of endogenous Wwp2 in both F9 and CGR8 ES cells significantly elevated the steady-state Rpb1 protein levels for both hyper- and hypophosphorylated forms (Fig. 4B, C, and D). Nevertheless, our identification of Wwp2 as a novel E3 ligase of mammalian Rpb1 does not preclude that other E3 ligases play a role in the regulation of Rpb1 functions.
The CTD is an essential domain for Rpb1 function and couples transcription with histone modification, mRNA capping, splicing, and polyadenylation. Phosphorylation and glycosylation of Rpb1 have both been mapped to the CTD (2). It was particularly interesting for us to determine whether ubiquitination of Rpb1 mediated by Wwp2 maps to this important domain of Rpb1. The mammalian CTD contains eight lysine residues, which are all located in the C-terminal third of the CTD (comprising heptapeptide repeats 35 to 52) (39). Here, we identified six lysine residues as ubiquitin modification sites in the CTD of mammalian Rpb1 through both mass spectrometric analysis and ubiquitination assay using CTD having six lysine residues mutated to arginine residues (Fig. 5). Identification of the ubiquitinated lysine residues in mammalian Rpb1 will aid in elucidating the molecular mechanisms responsible for Wwp2 regulation of Rpb1 function, as well as the effect of ubiquitination on Rpb1 function. The functional relevance of these six lysine residues is currently under investigation. Consistent with our study, Mitsui et al. showed that a GST-CTD fusion protein of yeast Rpb1, but not GST alone, was phosphorylated and ubiquitinated when incubated with HeLa cell nuclear extract and ATP, suggesting that the CTD was ubiquitinated (23). In contrast, Somesh et al. reported a ubiquitination site at yeast Rpb1 lysine 695, located outside of the CTD, by use of a reconstituted ubiquitination reaction and mass spectrometric analysis (33). Our study provided direct evidence that six lysine residues in the CTD of mammalian Rpb1 can serve as ubiquitin acceptor sites in Wwp2-mediated Rpb1 ubiquitination. However, only CTD was included in our study. Other lysine residues out of the CTD in Rpb1 might also be modified by Wwp2-mediated ubiquitination. Therefore, the possibility that different ubiquitin ligases catalyze ubiquitination at distinct lysine residues under different circumstances deserves further investigation.
Functional consequences of interactions between ubiquitin E3 ligases and Rpb1 may not be restricted to control of Rpb1 levels available for transcription. Interactions of Rpb1 with ubiquitin E3 ligases may also affect its modifications by other factors. For example, it has been known that both Rsp5 and Ess1, an essential yeast prolyl-isomerase, bind Rpb1 directly through their WW domains in such a way that the binding of Rsp5 with Rpb1 antagonizes the binding of Ess1 with Rpb1 (41). More interestingly, E3 ligases associated with Rpb1 may recruit other nuclear proteins, such as kinases, phosphatases, or methylases, to further modify Rpb1, and possibly nearby histone proteins, which in turn would lead to chromatin remodeling. Alternatively, E3 ligases may serve as binding bridges between transcription factors and Rpb1 to regulate specific gene expression. We have demonstrated that Wwp2 interacts with the transcription factors Oct-4 and Rpb1. Therefore, it will be very interesting to know whether Oct-4, Wwp2, and Rpb1 can form a ternary complex at an Oct-4 binding DNA consensus sequence. If such a ternary complex exists, it may play a role during transcriptional activation or repression of Oct-4 downstream genes. Identification of more Wwp2-associated proteins will shed light on the molecular mechanisms by which Wwp2 regulates the functions of Rpb1 and Oct-4.
Acknowledgments
We are deeply grateful to Jun Qin for assistance with the mass spectrometric analysis. We thank Richard Baer, Austin Smith, and Ian Chambers for their kind gifts of vectors and cells and Hans Clevers for generously providing inducible pTER+ RNAi vectors.
This study was supported by grants from the Shanghai Science and Technology Foundations (03DJ14018 and 04DZ14006), the National High Technology Research and Development Program of China (2000CB509900 and 2006CB943900), Shanghai Jiao Tong University School of Medicine, and Shanghai Institutes for Biological Sciences, CAS.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Beaudenon, S. L., M. R. Huacani, G. Wang, D. P. McDonnell, and J. M. Huibregtse. 1999. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10:679-680. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H. I., and M. Sudol. 1995. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA 92:7819-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y., S. W. Kwon, S. C. Kim, and Y. Zhao. 2005. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4:998-1005. [DOI] [PubMed] [Google Scholar]
- 5.Conaway, R. C., C. S. Brower, and J. W. Conaway. 2002. Emerging roles of ubiquitin in transcription regulation. Science 296:1254-1258. [DOI] [PubMed] [Google Scholar]
- 6.Corden, J. L., D. L. Cadena, J. M. Ahearn, Jr., and M. E. Dahmus. 1985. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. Natl. Acad. Sci. USA 82:7934-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahmus, M. E. 1995. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim. Biophys. Acta 1261:171-182. [DOI] [PubMed] [Google Scholar]
- 8.Flores, S. Y., C. Debonneville, and O. Staub. 2003. The role of Nedd4/Nedd4-like dependent ubiquitylation in epithelial transport processes. Pflugers Arch. 446:334-338. [DOI] [PubMed] [Google Scholar]
- 9.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher, E., M. Gao, Y. C. Liu, and M. Karin. 2006. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. USA 103:1717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hein, C., J. Y. Springael, C. Volland, R. Haguenauer-Tsapis, and B. Andre. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18:77-87. [DOI] [PubMed] [Google Scholar]
- 12.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huibregtse, J. M., J. C. Yang, and S. L. Beaudenon. 1997. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 94:3656-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingham, R. J., G. Gish, and T. Pawson. 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23:1972-1984. [DOI] [PubMed] [Google Scholar]
- 16.Kee, Y., N. Lyon, and J. M. Huibregtse. 2005. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 24:2414-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly, W. G., M. E. Dahmus, and G. W. Hart. 1993. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J. Biol. Chem. 268:10416-10424. [PubMed] [Google Scholar]
- 18.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 19.Komuro, A., T. Imamura, M. Saitoh, Y. Yoshida, T. Yamori, K. Miyazono, and K. Miyazawa. 2004. Negative regulation of transforming growth factor-beta (TGF-β) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23:6914-6923. [DOI] [PubMed] [Google Scholar]
- 20.Lipford, J. R., and R. J. Deshaies. 2003. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat. Cell Biol. 5:845-850. [DOI] [PubMed] [Google Scholar]
- 21.Lu, P. J., X. Z. Zhou, M. Shen, and K. P. Lu. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283:1325-1328. [DOI] [PubMed] [Google Scholar]
- 22.McDonald, F. J., A. H. Western, J. D. McNeil, B. C. Thomas, D. R. Olson, and P. M. Snyder. 2002. Ubiquitin-protein ligase WWP2 binds to and downregulates the epithelial Na(+) channel. Am. J. Physiol. Renal Physiol. 283:F431-F436. [DOI] [PubMed] [Google Scholar]
- 23.Mitsui, A., and P. A. Sharp. 1999. Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc. Natl. Acad. Sci. USA 96:6054-6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell. Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 25.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 26.Ogunjimi, A. A., D. J. Briant, N. Pece-Barbara, C. Le Roy, G. M. Di Guglielmo, P. Kavsak, R. K. Rasmussen, B. T. Seet, F. Sicheri, and J. L. Wrana. 2005. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19:297-308. [DOI] [PubMed] [Google Scholar]
- 27.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 28.Pirozzi, G., S. J. McConnell, A. J. Uveges, J. M. Carter, A. B. Sparks, B. K. Kay, and D. M. Fowlkes. 1997. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J. Biol. Chem. 272:14611-14616. [DOI] [PubMed] [Google Scholar]
- 29.Reid, J., and J. Q. Svejstrup. 2004. DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. J. Biol. Chem. 279:29875-29878. [DOI] [PubMed] [Google Scholar]
- 30.Rickert, P., J. L. Corden, and E. Lees. 1999. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18:1093-1102. [DOI] [PubMed] [Google Scholar]
- 31.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 32.Schwartz, L. B., and R. G. Roeder. 1975. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase II from the mouse plasmacytoma, MOPC 315. J. Biol. Chem. 250:3221-3228. [PubMed] [Google Scholar]
- 33.Somesh, B. P., J. Reid, W. F. Liu, T. M. Sogaard, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2005. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121:913-923. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, C., G. Murakami, M. Fukuchi, T. Shimanuki, Y. Shikauchi, T. Imamura, and K. Miyazono. 2002. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 277:39919-39925. [DOI] [PubMed] [Google Scholar]
- 35.Svejstrup, J. Q. 2003. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 116:447-451. [DOI] [PubMed] [Google Scholar]
- 36.Utsugi, T., A. Hirata, Y. Sekiguchi, T. Sasaki, A. Toh-e, and Y. Kikuchi. 1999. Yeast tom1 mutant exhibits pleiotropic defects in nuclear division, maintenance of nuclear structure and nucleocytoplasmic transport at high temperatures. Gene 234:285-295. [DOI] [PubMed] [Google Scholar]
- 37.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, B. B., R. Lu, W. C. Wang, and Y. Jin. 2006. Inducible and reversible suppression of Npm1 gene expression using stably integrated small interfering RNA vector in mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 347:1129-1137. [DOI] [PubMed] [Google Scholar]
- 39.Wintzerith, M., J. Acker, S. Vicaire, M. Vigneron, and C. Kedinger. 1992. Complete sequence of the human RNA polymerase II largest subunit. Nucleic Acids Res. 20:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woudstra, E. C., C. Gilbert, J. Fellows, L. Jansen, J. Brouwer, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2002. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415:929-933. [DOI] [PubMed] [Google Scholar]
- 41.Wu, X., A. Chang, M. Sudol, and S. D. Hanes. 2001. Genetic interactions between the ESS1 prolyl-isomerase and the RSP5 ubiquitin ligase reveal opposing effects on RNA polymerase II function. Curr. Genet. 40:234-242. [DOI] [PubMed] [Google Scholar]
- 42.Xu, H. M., B. Liao, Q. J. Zhang, B. B. Wang, H. Li, X. M. Zhong, H. Z. Sheng, Y. X. Zhao, Y. M. Zhao, and Y. Jin. 2004. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J. Biol. Chem. 279:23495-23503. [DOI] [PubMed] [Google Scholar]