Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that mediates the toxic effects of its xenobiotic ligands and acts as an environmental checkpoint during the cell cycle. We expressed stably integrated, Tet-Off-regulated AHR variants in fibroblasts from AHR-null mice to further investigate the AHR role in cell cycle regulation. Ahr+/+ fibroblasts proliferated significantly faster than Ahr−/− fibroblasts did, and exposure to a prototypical AHR ligand or deletion of the ligand-binding domain did not change their proliferation rates, indicating that the AHR function in cell cycle was ligand independent. Growth-promoting genes, such as cyclin and cyclin-dependent kinase genes, were significantly down-regulated in Ahr−/− cells, whereas growth-arresting genes, such as the transforming growth factor β1 (TGF-β1) gene, extracellular matrix (ECM)-related genes, and cyclin-dependent kinase inhibitor genes, were up-regulated. Ahr−/− fibroblasts secreted significantly more TGF-β1 into the culture medium than Ahr+/+ fibroblasts did, and Ahr−/− showed increased levels of activated Smad4 and TGF-β1 mRNA. Inhibition of TGF-β1 signaling by overexpression of Smad7 reversed the proliferative and gene expression phenotype of Ahr−/− fibroblasts. Changes in TGF-β1 mRNA accumulation were due to stabilization resulting from decreased activity of TTP, the tristetraprolin RNA-binding protein responsible for mRNA destabilization through AU-rich motifs. These results show that the Ah receptor possesses interconnected intrinsic cellular functions, such as ECM formation, cell cycle control, and TGF-β1 regulation, that are independent of activation by either exogenous or endogenous ligands and that may play a crucial role during tumorigenesis.
The Ah receptor (AHR) is a ligand-activated transcription factor that belongs to the basic region helix-loop-helix/Per-Arnt-Sim (PAS) family of proteins. Prototypical AHR ligands include many polycyclic and halogenated aromatic compounds, such as benzo[a]pyrene (B[a]P) and 2,3,7,8-tetrachloro-dibenzo-p-dioxin (TCDD). The unliganded AHR is considered to be a cytosolic protein that translocates to the nucleus upon ligand binding and dimerizes with a second basic region helix-loop-helix/PAS protein, the AHR nuclear translocator (ARNT), to form a heterodimeric transcription factor (27). The AHR/ARNT complex binds to canonical DNA recognition sequences (termed AhREs, XREs, or DREs) found in the genes coding for many phase I and phase II detoxification enzymes, such as the cytochromes P450 CYP1A1, CYP1A2, and CYP1B1, and initiates gene transcription (68).
Besides its role in the regulation of drug metabolism, the AHR also has an important physiologic role in liver and vascular development, immune system function, and cell growth and differentiation (18, 56). Evidence dating back more than 20 years has shown that TCDD, the prototypical AHR ligand, can inhibit cell proliferation and that this effect is AHR dependent. Confluent mouse epithelial cell cultures exhibited a diminished capacity for DNA replication in the presence of as little as 10 pM TCDD (22). TCDD also inhibited DNA synthesis in rat primary hepatocytes (30) and in rat and mouse liver following partial hepatectomy (3, 46). Inhibition of G1 phase entry by TCDD was first reported for rat 5L hepatoma cells and was found to involve increases in the levels of the CDK2 inhibitor p27KIP1 (37). The activated Ah receptor has also been shown to form protein-protein complexes with the retinoblastoma protein RB (16, 21, 51) to slow down entry into S phase (51) and to silence the expression of S-phase-specific E2F target genes (42).
These observations indicate that the activated AHR slows down cell proliferation, supporting its potential role as a tumor suppressor. On the other hand, evidence from various cell culture model systems is equally strong that the AHR promotes cell cycle progression in the absence of an exogenous ligand. Ma and Whitlock first observed that AHR-defective Hepa 1c1c7 cells proliferated more slowly than wild-type cells due to a prolonged G1 phase and that this effect was reversed by ectopic expression of AHR cDNA (39). AHR-null mouse embryonic fibroblasts (MEFs) were also found to have lower proliferation rates, with G2/M arrest and accumulation of 4N DNA content, than their wild-type counterparts did, possibly due to altered expression of the G2/M kinases Cdc2 and Plk (17). Further supporting the requirement of a functional AHR for normal cell cycle progression, clones derived from human lung epithelial A549 cells overexpressing AHR grew faster than control cells and showed increased DNA synthesis activity (57). Ahr+/+ mouse mammary fibroblasts showed a higher tumorigenic potential than Ahr−/− cells (47), which was suggestive of an AHR oncogenic activity.
A comparison of AHR-positive and AHR-negative cells and tissues derived from wild-type and Ahr−/− mice indicated that transforming growth factor β (TGF-β) may be one of the downstream AHR effectors likely to play a major role in AHR-mediated effects on cell cycle regulation. Ahr−/− mice have increased levels of active TGF-β1 and -β3 proteins associated with elevated liver fibrosis and hepatocyte apoptosis. Primary hepatocytes from these Ahr−/− mice also exhibit increased secretion of active TGF-β, inhibition of cell proliferation and promotion of apoptosis (23, 72), while Ahr−/− MEFs and smooth muscle cells show elevated TGF-β3 mRNA and protein levels (17, 55). AHR activation by TCDD reduced TGF-β1 expression in epithelial and mesenchymal cells during palatogenesis (1) and TGFb2 transcription in human keratinocytes (20). In addition, the repression of TGF-β3 signaling played a role in a TCDD-induced cleft palate (65), further strengthening the concept that AHR activity inhibits TGF-β function. Conversely, TGF-β reportedly down-regulated AHR expression and inhibited CYP1A1 induction by TCDD via a Smad-binding element located in the Ahr promoter region (12, 13, 70). In summary, interplay between TGF-β and AHR signaling pathways may help provide a mechanistic explanation to some of the physiological outcomes of AHR activation.
In the present study, we have used AHR-null fibroblasts expressing stably integrated, Tet-Off-regulated AHR variants to test the hypothesis that the AHR modulates cell cycle progression in the absence of a ligand. We find that the rate of cell proliferation was significantly faster in AHR-positive cells than in AHR-null cells, due to a lengthening of the G1 and S phases of the cell cycle, and that this effect was ligand independent. In AHR-null cells, mRNA levels of growth-promoting genes were significantly decreased and levels of growth-arresting genes, like the TGF-β1 gene and many genes regulated by it, were increased. Unlike the case for Ahr−/− cells, TGF-β1 mRNA levels in Ahr+/+cells were down-regulated by mRNA destabilization resulting from the increased activity of the TTP tristetraprolin protein, an AU-rich element (ARE)-binding protein that destabilizes mRNA (38).
MATERIALS AND METHODS
Reagents, cell culture, and growth conditions.
AHR-null mouse embryo fibroblasts were prepared by standard techniques from 14.5-day-old fetuses and cultured at 37°C in a humidified atmosphere containing 5% CO2 in minimal essential medium alpha (MEM-α) supplemented with 10% fetal bovine serum (FBS; Life Technologies, Inc.), 26 mM NaHCO3, 1% antibiotic/antimycotic solution (Sigma), and 600 μg/ml G418 (GIBCO Invitrogen Corporation) to ensure retention of the AHR-null genotype generated by insertion of the bacterial neo gene. Stable transfectant clones expressing various AHR protein variants (see below) were grown in the same medium containing an additional 3 μg/ml puromycin (A.G. Scientific, Inc.) and 400 μg/ml hygromycin (Calbiochem). Doxycycline (Sigma), a tetracycline analog, was used at a final concentration of 5 μg/ml. When indicated, cultures were treated with TCDD at a final concentration of 5 nM in dimethyl sulfoxide (DMSO) and control cultures were treated with an equivalent volume of DMSO, never to exceed 0.05%.
Generation of vectors for stable AHR expression.
Plasmids pcDNAI/B6AHR and pcDNAI/D2AHR containing 2.7 kb of the full-length cDNA from the high- and low-ligand-binding-affinity variants, the Ahrb1 and Ahrd alleles of C57BL/6 and DB/2 mice, respectively, have been described previously (5). The plasmid pB6AHRΔ323-494 encodes a deletion of 170 amino acids in the PAS B domain and is constitutively active in the absence of ligand (42). Tet-Off-responsive retroviral vectors encoding B6AHR, D2AHR, B6Δ323-494AHR, and ZIP8m were constructed in the pRevTRE vector (Clontech) using standard recombinant DNA techniques. ZIP8m (ZRT- and IRT-like protein 8) carries a loss-of-function mutation from the SLC39A8 zinc/cadmium transporter ZIP8 gene (10) and was used to generate control cells expressing a gene unrelated to the AHR/ARNT signaling pathway.
Construction of cell lines expressing AHR variants.
Stable AHR expression employed the retroviral Tet-Off gene expression system. We first generated an immortalized cell line stably expressing the regulatory protein TetR, by transfection of the pRevTet-Off vector into Ahr−/− mouse embryonic fibroblasts and selection with 3 μg/ml puromycin. A Tet-Off clone with a high dynamic range of doxycycline response, referred to hereafter as Ahr−/−*Tet*Off, was selected for the generation of stable cell lines expressing AHR variants. Ahr−/−*Tet*Off cells were grown to 50% confluence, infected as previously described (59) with retroviral pRevTRE vectors bearing B6AHR, D2AHR, B6Δ323-494AHR, and ZIP8m cDNAs, and selected with 400 μg/ml hygromycin. Clone pools selected for further use showed greater than 95% AHR repression when grown in 5 μg/ml doxycycline for 96 h. Four Tet repressor-regulated cell lines are reported in this work, including Off*Ahr−/−, stably expressing ZIP8m; Off*Ahrb, stably expressing B6AHR; Off*Ahrd, stably expressing D2AHR; and Off*AhrΔ323-494, stably expressing B6Δ323-494AHR.
Cell proliferation and flow cytometry assays.
Cells were synchronized in MEM-α supplemented with 0.1% serum for 96 h in the presence or absence of doxycycline and subsequently harvested and seeded in triplicate at 0.5 × 105 cells/well in a 24-well plate with medium containing 10% fetal bovine serum, doxycycline, or vehicle. Duplicate cultures were treated for 72 h with 5 nM TCDD or DMSO vehicle, and the medium was replenished every 2 days. At 0, 24, 48, and 72 h, cells were washed with phosphate-buffered saline, fixed with cold 75% ethanol, and incubated with 5 μg/ml Hoechst 33258 (Aldrich Chemical Co.) at room temperature in the dark for 30 min. Fluorescence was measured in a Wallac Victor 2 1420 plate reader (PerkinElmer Wallac, Gaithersburg, MD). Cell numbers were determined from a standard curve relating Hoechst fluorescence to cell numbers. For cell cycle studies, cells were maintained in exponential growth in MEM-α containing 10% fetal bovine serum in the presence or absence of doxycycline for 96 h, followed by TCDD or DMSO treatment as described earlier. For some experiments, as indicated, cells were infected with 200 PFU/cell of adeno-Smad7, an adenoviral vector expressing Smad7, the generous gift of Chia-Yang Liu. One million cells were collected and processed for cell cycle analysis using 7-amino-actinomycin D. Data were analyzed with ModFit software (Verity Software House, Topsham, ME). Experiments were performed at least in duplicate, with 10,000 forward-scatter gated events scored for each sample.
Western immunoblotting and immunofluorescence staining.
Whole-cell protein extracts and cytoplasmic and nuclear extracts were prepared as described previously (36). Aliquots were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, blocked, and probed with specific antibodies. Signals were detected with SuperSignal West Pico chemiluminescent substrate or West Femto maximum sensitivity substrate (Pierce, Rockford, IL). For immunofluorescence studies, cells were seeded on 6-mm glass coverslips, fixed in 4% formaldehyde-phosphate-buffered saline, permeabilized, blocked with 5% milk for 0.5 h, and incubated with first antibody at 37°C for 1 h. After washing, coverslips were stained with fluorescence-labeled second antibody and Hoechst solution. Images were obtained using a Nikon Eclipse TE-300 fluorescence microscope. At least five fields were evaluated for each treatment group. Antibodies used were anti-AHR (Biomol Research Laboratories), anti-ARNT (H-172), anti-TTP (N18 and H-120) (Santa Cruz Biotechnology), anti-E2F (E2F-6; Lab Vision), anti-GCLM (a gift from Dan Nebert, University of Cincinnati), anti-MAPKAPK2 (MK2) (Cell Signaling Technology), anti-phospho-MAPKAPK2 (Thr334) (Cell Signaling Technology), anti-Smad4 (Cell Signaling Technology), and anti-β-actin (Sigma).
Nuclear run-on assays.
Nucleus isolation and run-on transcription were carried out as described previously (67) with minor modifications. DNA targets for the run-on assays were PCR amplified from mouse cDNA from Hepa-1 cells. A control yeast (Saccharomyces cerevisiae) intergenic region designated YIG73 was amplified by the same method from yeast genomic DNA. One microgram of each amplified DNA was blotted onto a ZetaProbe membrane, and the blots were hybridized with 1 × 107 cpm of 32P-labeled run-on RNA for 48 h, washed, and exposed to a phosphorimaging plate (Molecular Dynamics, Sunnyvale, CA) for 12 h. The plate was scanned using the Storm 860 PhosphorImager at a 200-μm resolution, and signal intensities for each band were corrected for background and analyzed using ImageQuant 5.2.
High-density microarray hybridization analysis.
Total cellular RNA was isolated with RNeasy (QIAGEN, Valencia, CA). Labeling of DNA targets, preparation of microarray slides, and hybridization reactions were performed by the University of Cincinnati Genomics and Microarray Laboratory Core. The hybridization probes were from the mouse oligonucleotide library (version 3.0; QIAGEN Operon, Alameda, CA), representing 31,769 annotated mouse genes. Hybridization targets were the paired Cy3- and Cy5-labeled control and test cDNAs synthesized from 20 μg of total RNA by an oligo(dT)-primed, reverse transcriptase reaction and were labeled with monofunctional reactive Cy3 and Cy5 (Amersham, Piscataway, NJ). After hybridization under high-stringency conditions, slides were washed and simultaneously scanned at a 10-μm resolution at 635 (Cy5) and 532 (Cy3) nm (GenePix 4000B; Axon Instruments, Inc., Union City, CA). Comparisons were carried out with quadruplicate biological replicates using flipped dye arrays to allow for the removal of gene-specific dye effects. More details of the procedure can be found at http://microarray.uc.edu. Data normalization was performed in three steps for each microarray (35). Complete experimental information and data are available in MIAME compliant format with ID E-MEXP-996 from http://ebi.ac.uk/miamexpress/ (data may be accessed using the following information: user name, Reviewer_E-MEXP-996; password, 1171627806170; special private login page, http://www.ebi.ac.uk/aerep/login).
Reverse transcription and real-time PCR.
cDNAs were synthesized by reverse transcription of 10 μg of total RNA using random hexamers and SuperScript II reverse transcriptase (Life Technologies, Inc.) as described previously (42), followed by amplification with QTaq DNA polymerase master mix (BD). Quantitative real-time PCRs were conducted in a DNA Engine Opticon 2 continuous fluorescence detection system (MJ Research, Bio-Rad) or in a 7500 real time PCR system (Applied Biosystems). The reaction protocol included a 2-min denaturation step at 95°C, followed by 40 cycles of 95°C denaturation for 30 s, annealing for 30 s at 55 to 65°C (depending on each pair of primers [see Table S1 in the supplemental material]) and 72°C extension for 30 s. Data were quantified using threshold cycle (CT) values determined in quadruplicate biological samples, averaged, and normalized to values for β-actin amplification of the same samples. For RNA stability experiments, 18S RNA amplification was used as the internal standard since it is stable compared to β-actin. The severalfold change from the control of each comparison was determined by the ΔΔCT method using the following formula: change = 2ΔΔCT, where ΔΔCT = (CT GeneA − CT standard)Control − (CT GeneA − CT standard)Experiment.
Statistical analyses.
Statistical analyses of microarray data were performed by fitting the mixed-effects linear model for each gene separately, as discussed in detail previously (35). Resulting t statistics from each comparison were then adjusted using a hierarchical empirical Bayes model (58) for calculation of P values and using the expected number of false positives based on the false discovery rate (4). For doubling time calculations, the analysis of covariance model was used to obtain slope estimates of growth curve for each combination of experimental conditions. Doubling time estimates were calculated as the reciprocal of the slope estimate. For an analysis of data from other experiments, group comparisons were made by one-way analysis of variance. A P value of less than 0.05 was considered statistically significant.
RESULTS
Lack of AHR slows down cell growth.
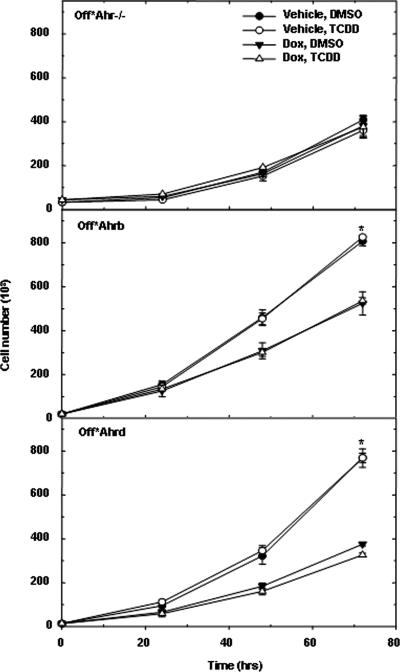
Differences in proliferation rates of Ahr+/+ and Ahr−/− MEFs (17) could be attributed to clonal differences between AHR-positive and AHR-negative cells, which, by virtue of being derived from different mice, are not clonally related. To rule out this possibility, we constructed stable AHR-expressing fibroblast derivatives of Ahr−/− MEFs in which AHR expression was regulated by the Tet-Off Tet receptor. AHR expression and cellular distribution in the Tet-Off cells were similar to the expression and distribution in Ahr+/+ MEFs, as was the induction of CYP1A1 by TCDD. Treatment with doxycycline blocked AHR expression in these cells by as much as 90 to 95% (see Fig. S1 in the supplemental material). The expression of the AHR protein in these cells, which are otherwise identical, depends solely on the presence or absence of doxycycline. Cell proliferation assays in the presence and absence of doxycycline were carried out with Off*Ahrb and Off*Ahrd fibroblasts expressing the high- and the low-affinity AHRs, respectively, and with control Off*Ahr−/− fibroblasts, expressing the unrelated ZIP8m protein. In addition, to test the role of an exogenous AHR ligand, cells were also treated with TCDD or left untreated. Proliferation rates of Off*Ahrb and Off*Ahrd cells were almost identical and were significantly shorter than the growth rate of Off*Ahr−/− cells (Fig. 1 and Table 1). The inhibition of AHR expression by doxycycline significantly increased the proliferation rates of both AHR expressing cells to a rate closer to that of Off*Ahr−/− control cells, providing direct evidence that AHR plays a substantial role in the modulation of cell proliferation. Unexpectedly, TCDD, which functioned normally in Cyp1a1 induction (see Fig. S1C and D in the supplemental material), had no effect on the proliferation rates of any of these cells, suggesting that the AHR role in cell proliferation was independent of its activation by an exogenous xenobiotic ligand (Fig. 1). To confirm this observation, we compared growth rates of Off*Ahr−/−, Off*Ahrb, and Off*AhrΔ323-494 fibroblasts. The latter have a deletion extending into the PAS domain that removes the ligand-binding domain and renders the truncated protein constitutively active and its transactivation activity independent of ligand (42). Just like Off*Ahrb fibroblasts, the doubling time of Off*AhrΔ323-494 fibroblasts was also significantly increased by doxycycline exposure and unaffected by TCDD treatment. Overall, Off*AhrΔ323-494 cells proliferated at the same rates as Off*Ahrb cells and considerably faster than Off*Ahr−/− cells (Table 1). Doxycycline had no significant effect on the growth rate of Off*Ahr−/− cells, ruling out explanations that the observed changes were due to the treatment rather than to the expression of AHR.
FIG. 1.
Growth rate of AHR-expressing fibroblasts. Off*Ahr−/−, Off*Ahrb, and Off*Ahrd fibroblasts were pretreated with 5 μg/ml doxycycline (Dox) or vehicle for 4 days in MEM-α containing 0.1% serum and seeded in 24-well plates at 50,000 cells per well while the fibroblasts continued to be treated with vehicle plus DMSO, vehicle plus 5 nM TCDD, 5 μg/ml Dox plus DMSO, or 5 μg/ml Dox plus 5 nM TCDD in MEM-α containing 10% fetal bovine serum. Cell proliferation was measured by Hoechst assay at 0, 24, 48, and 72 h.
TABLE 1.
Comparison of cell cycle parameters of AHR-negative and AHR-positive fibroblasts
| Cell line and treatment | Ligand | Length of cell cycle phase (hours)c
|
|||
|---|---|---|---|---|---|
| Doubling time | G0/G1 | S | G2/M | ||
| Off*Ahr−/− | |||||
| −Dox | DMSO | 24.6 ± 2.6 | 12.1 ± 0.8 | 9.0 ± 0.5 | 3.5 ± 0.3 |
| TCDD | 26.2 ± 2.7 | 12.4 ± 0.4 | 10.0 ± 0.0 | 3.8 ± 0.4 | |
| +Dox | DMSO | 28.4 ± 3.1 | 14.2 ± 0.4 | 10.3 ± 0.3 | 3.9 ± 0.1 |
| TCDD | 27.2 ± 2.9 | 13.4 ± 0.7 | 10.1 ± 0.4 | 3.7 ± 0.3 | |
| Off*AhrΔ323-494 | |||||
| −Dox | DMSO | 20.3 ± 1.6a | 8.7 ± 0.3a | 8.7 ± 0.1 | 2.9 ± 0.5 |
| TCDD | 20.6 ± 1.7a | 8.2 ± 0.0a | 8.9 ± 0.6 | 3.5 ± 0.6 | |
| +Dox | DMSO | 24.4 ± 2.3b | 9.9 ± 0.4a,b | 11.0 ± 0.2a,b | 3.4 ± 0.6 |
| TCDD | 25.8 ± 2.6b | 9.8 ± 0.1a,b | 12.3 ± 0.7a,b | 3.7 ± 0.6 | |
| Off*Ahrb | |||||
| −Dox | DMSO | 18.2 ± 1.3a | 7.4 ± 0.1a | 8.0 ± 0.5 | 2.8 ± 0.4 |
| TCDD | 18.2 ± 1.3a | 7.1 ± 0.1a | 7.7 ± 0.1a | 3.4 ± 0.1 | |
| +Dox | DMSO | 21.9 ± 1.9a,b | 8.5 ± 0.3a,b | 10.6 ± 0.3b | 2.9 ± 0.1 |
| TCDD | 21.7 ± 1.8a,b | 8.4 ± 0.2a,b | 10.7 ± 0.3b | 2.6 ± 0.1 | |
P was <0.05 for the comparison to Off*Ahr−/− with same treatment and ligand.
P was <0.05 for the comparison between treatments of the same cell line with the same ligand.
The doubling time (T) was calculated as the reciprocal of the slope of the growth curves using the formula slope = 1/T = (log2Nt − log2N0)/t, where Nt represents cell numbers at time t and N0 represents cell numbers at time zero. The analysis of covariance model was used to obtain slope estimates (1/T) for each combination of experimental conditions. The percentages of cells in G0/G1, S, and G2/M phases were determined using the ModFit software program. The length of each phase was calculated as the product of doubling time multiplied by the percentage of cells in each phase.
Flow cytometric analyses showed that G0/G1 and S phases were significantly extended by doxycycline treatment in both AHR-expressing cells relative to its effect on control Off*Ahr−/− cells (Table 1), indicating that the inhibition of AHR expression specifically prolongs the G0/G1 and S phases of the cycle. Furthermore, since all measured parameters were almost identical for Off*Ahrb and Off*AhrΔ323-494 cells, these results strongly indicate that the differences observed between these two Ah receptor variants in response to toxicants do not extend to their ligand-independent roles in cell proliferation.
TGF-β activity determines the growth rate differences between Ahr+/+ and Ahr−/− MEFs.
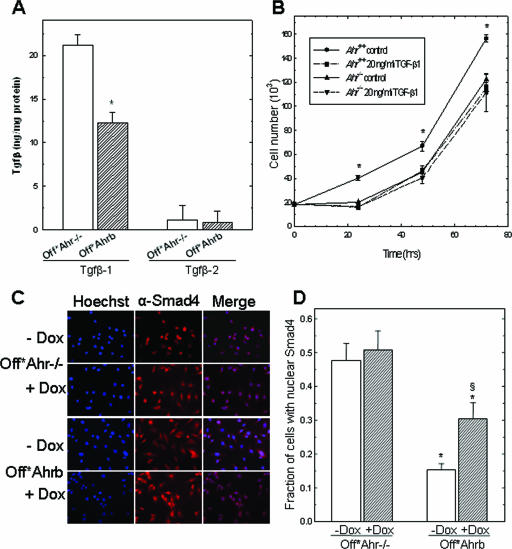
AHR-null cells have also been reported to secrete larger amounts of TGF-β than do their wild-type AHR-positive counterparts (17, 26, 72). To characterize regulatory connections between AHR and TGF-β, we first quantified the secreted levels of TGF-β1 and -β2 in the culture medium of Off*Ahr fibroblasts. Off*Ahr−/− cells, lacking AHR expression, secreted twice as much TGF-β1 as Off*Ahrb cells did, while no significant difference was detected for TGF-β2. In addition, the absolute level of secreted TGF-β1 was 5 to 10 times higher than the level of TGF-β2, suggesting that TGF-β1 contributes most of the functions associated with TGF-β signaling in these cells (Fig. 2A).
FIG. 2.
Effect of TGF-β1 on proliferation of Ahr+/+ and Ahr−/− MEFs. (A) Quantification of TGF-β secreted by Off*Ahrb and Off*Ahr−/− fibroblasts. Medium was collected from cells after 72 h in culture in serum-free Opti-MEM. Latent TGF-β1 and TGF-β2 were activated to their immunoreactive form by acidification and neutralization. Quantification was by enzyme-linked immunosorbent assay using kits specific for TGF-β1 (R&D Systems) and TGF-β2 (Promega). The concentrations of TGF-β1 and TGF-β2 in medium were normalized to total cellular protein. Biological triplicates were analyzed for each cell type. *, P < 0.05. (B) Effect of TGF-β1 on proliferation of Ahr+/+ and Ahr−/− MEFs. Synchronized cells were seeded in quadruplicate at 0.5 × 105 cells/well in a 24-well plate with medium containing 10% FBS. After attachment, cells were treated with 20 ng/ml of recombinant TGF-β1 or with vehicle for the indicated times. hrs, hours. *, P < 0.05. (C) Immunofluorescence detection of Smad4 in Off*Ahr−/− and Off*Ahrb cells. Cells were synchronized in MEM-α containing 0.1% serum in the presence (+) or the absence (−) of 5 μg/ml doxycycline for 4 days. After synchronization, cells were cultured in MEM-α containing 10% serum, while they were continuously treated with or without Dox for another 24 h and immunostained with anti-Smad4, followed by Alexa Fluor 488-labeled goat anti-rabbit immunoglobulin G (H+L) for Smad4 detection and Hoechst for nuclei. Micrographs were taken at ×40 magnification. Shown are representative fields. The green fluorescence of the Smad4 detection system was digitally converted to red to enhance the contrast (blue-red versus blue-green) of the merged panels. (D) Fraction of Off*Ahr−/− and Off*Ahrb cells showing nuclear Smad4 staining. Approximately 500 cells were counted in microscopy fields randomly chosen. *, P value of less than 0.05 for comparison of same treatment between cell lines; §, P value of less than 0.05 for comparison between treatments in the same cell line.
To determine whether differential production of TGF-β in AHR-positive and AHR-negative MEFs could be the reason for the differences in growth rates, we compared growth rates of Ahr+/+ and Ahr−/− MEFs in regular MEM-α with rates in medium supplemented with 20 ng/ml of recombinant TGF-β1. Ahr−/− MEFs grew at the same rate whether or not medium was supplemented with TGF-β1, but Ahr+/+ MEFs grew significantly faster than Ahr−/− MEFs in unsupplemented medium and slowed down to the same growth rate as that of Ahr−/− cells in medium containing TGF-β1 (Fig. 2B). These data suggest that the main determinant of the differences in growth rate between these two cells is their TGF-β levels.
Signaling through activated TGF-β results in the phosphorylation and activation of receptor-regulated R-Smad proteins. R-Smads dimerize with Smad4, translocate to the nucleus, and initiate transcription of TGF-β target genes (45). Hence, Smad4 nuclear translocation is a functional marker of the activation of the TGF-β pathway. To determine whether the TGF-β secreted by Off*Ahr fibroblasts was functionally competent, we assessed the extent of Smad4 nuclear translocation in these cells. Smad4 was found in approximately equal amounts in both cell lines, but it was equally distributed between the nucleus and cytoplasm in Off*Ahr−/− cells treated or not treated with doxycycline, with approximately 50% of the cells from either treatment showing nuclear localization. In contrast, only 15% of untreated Off*Ahrb cells had nucleus-localized Smad4, and this number doubled with AHR repression by doxycycline treatment (Fig. 2C and D).
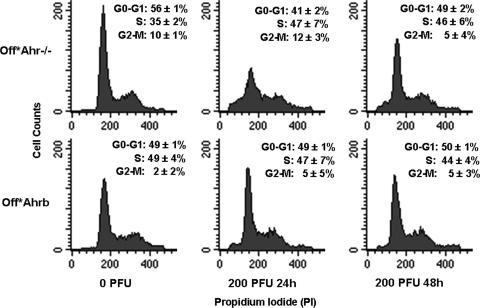
Conversely, the interaction of Smad7 with R-Smads blocks TGF-β signals from reaching the nucleus and activating target gene expression. To test whether the inhibition of TGF-β signaling would make the proliferation pattern of Off*Ahr−/− cells resemble that of Off*Ahrb cells, we used flow cytometry to compare the cell cycle parameters of the two cell lines infected or not with an adenoviral vector expressing Smad7. After 24 and 48 h of adeno-Smad7 infection, the fraction of Off*Ahr−/−cells in G0/G1 decreased from 56% to 41% and 49%, respectively, and the fraction of cells in S phase increased from 35% to 46% to 47%. These values were not significantly different from the corresponding values of Off*Ahrb cells, which were not affected by Smad7 expression (Fig. 3). The combined results of Smad4 translocation and Smad7 inhibition strongly argue in favor of the conclusion that AHR represses the TGF-β signaling pathway, which is activated by AHR repression.
FIG. 3.
Smad7 expression increases the proliferation rate of Off*Ahr−/− cells but not that of Off*Ahrb cells. Cells were infected with 200 PFU per cell of an adenoviral vector expressing Smad7, a multiplicity of infection previously determined to infect greater than 95% of the cells. Flow cytometric determinations were carried out for uninfected cells and in cells infected for 24 and 48 h. The fractions of cells in G0/G1, S, and G2/M are indicated in each panel. The differences between infected and uninfected Off*Ahr−/− cells, but not those of Off*Ahrb cells, are significant (P < 0.05).
AHR controls genes related to cell cycle regulation and TGF-β signaling.
We investigated AHR effects on global gene expression associated with cell cycle regulation by comparing gene expression patterns of untreated Off*Ahrb and Off*Ahr−/− cells. We chose to examine cells in exponential growth and cells synchronized in G0/G1 and S phases, since these were the cell cycle phases most affected by AHR ablation. To generate a cell population of >80% in G0/G1, cells were arrested by serum starvation for 96 h, fed with medium containing 10% FBS, and collected 10 h later; for cells more than 80% in S phase, collection took place 20 h after serum addition. A total of 1,133 transcripts were found to be significantly different using a criterion for differentially expressed genes based on an average fluorescence intensity of at least 100 and an expected number of false positives (66) of 10 or lower in any of the three growth groups, which is approximately equivalent to a false discovery rate of 3%.
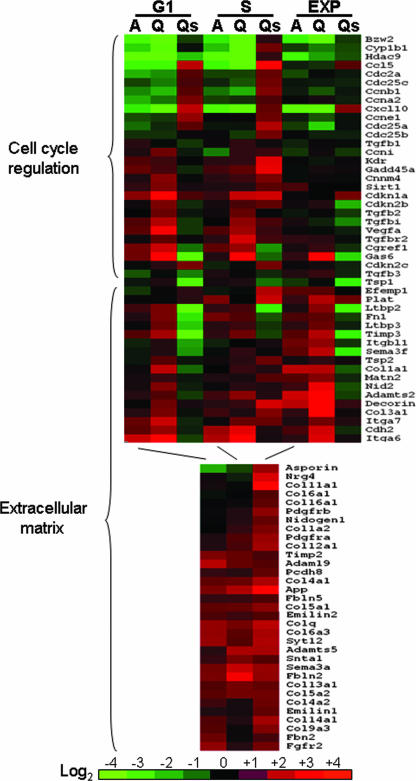
Twenty-seven cell cycle-related genes and genes involved in TGF-β signaling were deregulated in cells lacking AHR and were confirmed by quantitative real-time reverse transcription-PCR (RT-PCR) (Fig. 4; see Table S2 in the supplemental material). Growth-promoting genes, such as those coding for cyclins A, B, and E; Cdc2A and Cdc25 proteins; and the chemokine ligands, Cxcl10 and Ccl5, were significantly down-regulated in Off*Ahr−/− cells relative to those in Off*Ahrb cells, especially in cells synchronized at G1 or S. Bzw2 (basic leucine zipper and W2 domain 2), a gene with a homozygous deletion in Wilms' tumor (60), and Hdac9, encoding a class II histone deacetylase important in the suppression of cardiac hypertrophy (73), were also highly down-regulated in Off*Ahr−/− cells, potentially implicating the AHR in some novel functions. The same was true for several genes responsive to TGF-β signaling, including VEGFa, TGFbi, and Tgfbr2, further pointing to a connection between TGF-β and AHR signaling pathways. On the other hand, growth-arresting genes, such as those coding for the TGF-βs; CDK inhibitors p15, p18, and p21; and several growth-arrest induced genes, including Gas6, Cgref1, and Gadd45a, were significantly up-regulated in Off*Ahr−/− cells compared to those in Off*Ahrb fibroblasts. The expression of Smad7 in Off*Ahr−/− cells reversed the expression patterns of 22 of the 27 cell cycle-related genes that were deregulated in cells lacking AHR, with reversal being most evident in cells synchronized in G0/G1 and S phases (Fig. 4; see Table S2 in the supplemental material). Hence, the inhibition of TGF-β signals in Off*Ahr−/− cells made these cells acquire transcription patterns more similar to those of Off*Ahrb cells.
FIG. 4.
Heat map of gene expression differences between Off*Ahr−/− and Off*Ahrb cells. The color comparison represents gene expression changes in Off*Ahr−/− cells relative to those in Off*Ahrb cells; thus, red represents genes with higher expression levels in Off*Ahr−/− cells and green represents genes with lower expression levels in Off*Ahr−/− cells. The heat map was developed for gene expression values of cells in G0/G1 and S, and exponentially growing cells, as indicated at the top of the graph. “A” denotes the values from microarray hybridization, and “Q” denotes the corresponding values from quantitative real-time RT-PCR. “Qs” denotes the values from quantitative real-time PCR in Off*Ahr−/− cells infected with adeno-Smad7 for 24 h.
Notably, 53 genes involved in extracellular matrix (ECM) formation were significantly up-regulated in Off*Ahr−/− cells (Fig. 4; see Table S3 supplemental material). Eighteen of these genes were selected for quantitative real-time RT-PCR confirmation. With the single exception of S-phase cyclin I, expression changes found by microarray were validated by PCR. For some genes, like those coding for collagen α1(III) chain, decorin, integrin α6, and Adamts2, up-regulation in AHR-null fibroblasts was as high as 20-fold, suggesting a considerable role for the AHR in the suppression of ECM deposition or remodeling. The expression of 13 of the 18 genes validated by quantitative real-time RT-PCR was reversed by adeno-Smad7 infection, strongly indicating that the phenotype of the Off*Ahrb cells is due to the inhibition of TGF-β signaling.
Increased TGF-β1 mRNA stability in cells lacking AHR.
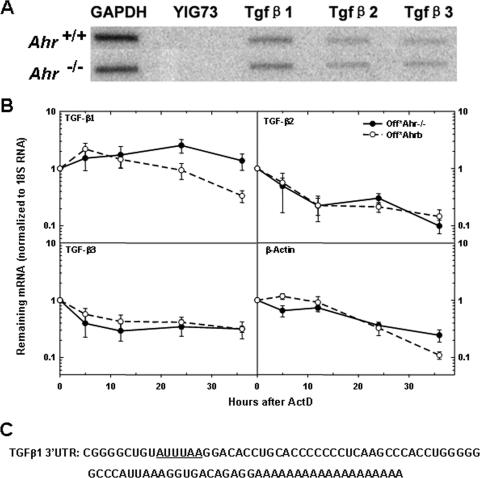
Transcription, pre-mRNA processing, and cytoplasmic mRNA degradation are the three main factors that regulate mRNA accumulation (25). To determine whether the mechanism of down-regulation of TGF-β1 by AHR took place at the transcriptional level, we used nuclear run-on assays to measure the transcription rates of TGF-β genes in Ahr−/− and Ahr+/+ MEFs. We found no significant differences between cell lines in the transcription rates of any of three TGF-β genes (Fig. 5A), indicating that down-regulation was the consequence of some other mechanism. We next measured whether the rate of TGF-β mRNA degradation was affected by AHR status. To obtain mRNA decay curves, we treated Off*Ahr−/− and Off*Ahrb cells with actinomycin D and measured TGF-β1/2/3 and β-actin mRNA levels and 18S rRNA amounts (for normalization) by quantitative real-time RT-PCR. TGF-β1 mRNA in Off*Ahr−/− cells did not appreciably decay over the duration of the experiment, whereas it showed a progressive decline, with a 16-h half-life, in Off*Ahrb cells (Fig. 5B). In contrast, TGF-β2, -β3, and β-actin mRNA levels decayed at approximately the same rate in both Off*Ahrb and Off*Ahr−/− cells (Fig. 5B). These results suggested that the presence of AHR specifically destabilizes TGF-β1 mRNA, but not the other two TGF-βs.
FIG. 5.
AHR destabilizes TGF-β1 mRNA. (A) Nuclear run-on assays of Ahr−/− MEFs and wild-type MEFs. Nuclear run-on assays were conducted as described in Materials and Methods. The hybridization targets were the PCR products of TGF-β1, TGF-β2, and TGF-β3. YIG73 is a yeast intergenic region used as negative control, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a positive control. (B) Analysis of mRNA stability. Quiescent Off*Ahr−/− and Off*Ahrb cells were stimulated to enter the cell cycle with MEM-α containing 10% serum for 40 h and further incubated with actinomycin D (ActD) at a final concentration of 0.05 μg/ml for 36 h. mRNA levels at the various time intervals were determined by quantitative real-time RT-PCR. The results are expressed as the percentage of mRNA remaining relative to the corresponding level before the addition of ActD. Data are the means ± standard deviations (error bars) of three independent measurements, each with technical triplicates of duplicate biological samples for each cell type at each time point. (C) Sequence of 3′UTR of TGF-β1 mRNA, with AUUUAA region underlined.
AHR inhibits the nuclear export of TTP.
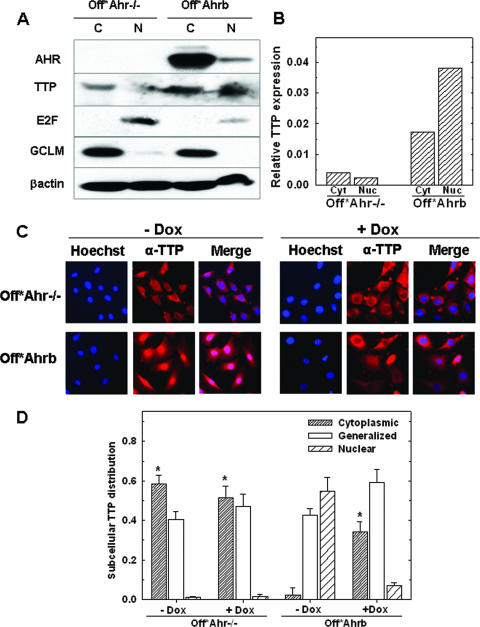
TTP (also known as Zfp36 or HuR) is an ARE-binding protein that destabilizes mRNA via the 3′UTR AREs of many mRNAs (38). TTP activity is regulated through phosphorylation by MK2 (MAPKAPK2), a MAP kinase activated by p38-dependent phosphorylation. Phosphorylation by MK2 promotes TTP nuclear export and a reduction in ARE binding affinity, increasing the mRNA stability of target genes (29). Two lines of evidence suggested that TTP might have a role in the AHR-mediated regulation of TGF-β expression. First, TGF-β1 mRNA, but not TGF-β2 or -β3 mRNA, has one such AUUUA element in its 3′UTR (Fig. 5C); second, microarray data showed that TTP mRNA was down-regulated in Off*Ahr−/− cells relative to that in Off*Ahrb cells. Western immunoblot analyses of TTP expression compared to actin expression showed that considerably more TTP protein was expressed in Off*Ahrb cells than in Off*Ahr−/− cells. Moreover, in Off*Ahrb cells, two-thirds of the TTP protein was nuclear and one-third was cytosolic, while in Off*Ahr−/− cells the reverse was true, with two-thirds of TTP located in the cytoplasmic fraction (Fig. 6A and B). To confirm this observation, we used immunocytochemistry to directly examine TTP subcellular localization. In Off*Ahr−/− cells, there was clear TTP cytoplasmic staining in about 60% of cells, generalized staining in 40% of cells, and nuclear staining in only 1% of cells. Doxycycline treatment had no significant effect on the TTP subcellular localization in these cells. In contrast, in Off*Ahrb cells, the percentage of cells with TTP cytoplasmic staining was 20 times less (<3%) than that in Off*Ahr−/− cells. The inhibition of AHR by doxycycline treatment significantly increased the number of cells with TTP staining in the cytoplasm of Off*Ahrb cells to 34% (Fig. 6B and C), indicating that a significant portion of TTP was exported from the nucleus in the absence of AHR.
FIG. 6.
Subcellular distribution of TTP protein. (A) Western blot analysis of cytoplasmic and nuclear extracts of Off*Ahr−/− and Off*Ahrb cells. One hundred micrograms of cytoplasmic (C) protein fractions or 40 μg of nuclear (N) protein fractions from each cell type was analyzed by SDS-PAGE and immunoblotted with antibodies to AHR; TTP; E2F, a marker of the nuclear compartment; and GCLM, a marker of the cytosolic compartment. (B) Quantitation of the Western blot TTP band intensities relative to the intensity of the actin bands. cyt, cytoplasmic; nuc, nuclear. (C) Representative immunofluorescence micrographs of TTP in Off*Ahr−/− and Off*Ahrb cells. Cells were treated with (+Dox) or without doxycycline (−Dox) for 4 days before being fixed for staining. After fixation, TTP was detected by staining with anti-TTP (N-18) and Alexa Fluro488-labeled rabbit anti-goat immunoglobulin G (H+L) (green). Hoechst was used to stain nuclei. Photographs were taken at ×40 magnification. Shown in the figure are representative fields from approximately 500 cells examined per group. As in Fig. 2, green fluorescence was digitally converted to red to enhance contrast of the merged micrographs. (D) Quantitation of immunofluorescence analysis. Shown is the fraction of cells that have TTP localized to each subcellular compartment. Three categories are classified as cytoplasmic, nuclear, and generalized; in the latter, TTP staining is evenly distributed throughout the cell. Data are presented as means ± SD (error bars) from three groups with 150 to 200 cells examined per group. The asterisks represents a significant difference (P < 0.05) from Off*Ahrb (−Dox) in each category.
Increased MK2 phosphorylation in the absence of AHR.
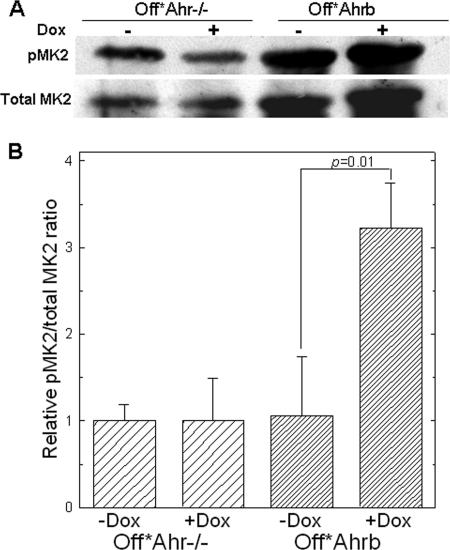
MK2 mRNA levels were also higher in Off*Ahr−/− cells than in Off*Ahrb cells. In the absence of commercial antibodies to phosphorylated TTP, we investigated whether the observed TTP nuclear export in Off*Ahr−/− cells could result from increased MK2-dependent TTP phosphorylation by evaluating the levels of phospho-MK2 (pMK2) and total MK2 using quantitative Western blotting of whole-cell protein extracts from Off*Ahrb and Off*Ahr−/− cells treated with Dox or untreated (Fig. 7A). Ratios of pMK2 to total MK2 were calculated for each cell and treatment condition and expressed relative to the values in Off*Ahr−/− cells (Fig. 7B). pMK2 levels were significantly increased in doxycycline-treated Off*Ahrb cells relative to untreated controls, suggesting that TTP phosophorylation by activated MK2 may be the upstream event of TTP nuclear export that leads to decreased TTP activity and TGF-β1 mRNA stabilization in the absence of AHR.
FIG. 7.
Western immunoblot detection of phosphorylated MK2 in whole-cell extracts of Off*Ahr−/− and Off*Ahrb cells. (A) Cells were cultured in the presence (+) or absence (−) of 5 μg/ml doxycycline for 4 days, and 500 mg of protein extracts were immunoprecipitated with antibody to total MK2. Immunoprecipitates were analyzed by SDS-PAGE and immunoblotted with an antibody to pThr-334-MK2 or with a second anti-total MK2 antibody. This approach was necessary because all of the anti-total MK2 antibodies that we tested had multiple reactivities, but only the reactivity with MK2 was common to all of the antibodies. (B) Quantification of band intensity of pMK2 relative to total MK2 in panel A. pMK2/total MK2 ratios were determined for each protein extract and are expressed relative to the values in Off*Ahr−/− cells.
DISCUSSION
We developed Tet-Off-regulated fibroblast cell lines that stably express AHR variants in an Ahr−/− genetic background and showed that AHR-positive cells proliferate significantly faster than do AHR-negative cells. This result cannot be attributed to a difference in the clonal origins of these cells because AHR-positive and AHR-negative cells arise from the same cell pool, treated or not with doxycycline. Our studies with these cells further strengthen the evidence that the AHR plays a central role in cell cycle regulation. Lack of AHR slows down the G1 and S phases of the cell cycle, which is consistent with previous reports for mouse hepatoma cells (39) but is unlike findings for primary MEFs, where AHR-null cells arrest in G2/M (17). Our cells, although originally derived from MEFs, have been immortalized by continuous passage in culture and may behave more like fibroblast cell lines than primary embryonic fibroblasts.
TCDD has been shown to slow down cell cycle progression in many cell lines (52), yet it had no effect on the growth rate of these AHR-expressing fibroblasts. TCDD-independent AHR effects on cell growth have also been observed in Hepa-1 hepatoma cells (39) and MEFs from wild type-mice (17), perhaps resulting from the recruitment of a fibroblast-specific factor that represses TCDD-dependent activation of endogenous AHR signaling (24). Alternatively, the AHR functions involved in cell growth may not need a xenobiotic ligand, just like its functions in invertebrates. Studies with invertebrate AHR orthologs show that the receptor regulates aspects of embryonic development and does not respond to xenobiotic ligands (53). In this context, it is interesting that high- and low-affinity AHRs and a truncated AHR lacking the ligand-binding domain accelerate cell proliferation equally well, strongly supporting the conclusion that the AHR role in cell cycle regulation is fully ligand independent.
Gene expression profiling with synchronized cells identified a group of proteins governing the G1/S transition, including cyclins A2, B1, and E1, which were down-regulated by the lack of AHR expression. Down-regulation of these cyclins would partly explain the lengthening of G1/S phases observed in Off*Ahr−/− cells. Cdc2 was also significantly down-regulated in Off*Ahr−/− cells, consistent with findings of higher expression levels in AHR-positive MEFs (17). Although Cdc2 is believed to function specifically in promoting the G2/M transition by forming complexes with cyclins A and B, recent evidence shows that it also binds, along with Cdk2, to cyclin E and promotes the G1/S transition (33). Hence, decreased Cdc2 expression may also contribute to G1/S arrest. Lack of AHR expression up-regulated a number of genes associated with cell cycle arrest, including those encoding TGF-βs and several Cdk inhibitors and Gas6, Cgref1, and Gadd45a. Tgfbr2 and Tgfbi, two genes that directly participate in regulation by the TGF-β signaling pathway, were also up-regulated in Off*Ahr−/− cells. These results suggest that one of the AHR roles in the regulation of cell proliferation is the up-regulation of positive and down-regulation of negative effectors of cell cycle progression.
Unexpectedly, the expression of approximately 50 to 55 genes involved in ECM formation was increased, in some cases by as much as 20-fold, in the absence of AHR. The ECM plays a central role in processes affecting cell adhesion, migration, and tissue morphogenesis as well as proliferation and differentiation (15). The up-regulation of procollagen type XIII alpha 1, Timp2, integrin B1, decorin, and thrombospondin 1 has been observed in Ahr−/− vascular smooth muscle cells (26), but none of the other ECM genes found in the current study have been connected with gene regulation through the unliganded Ah receptor, although pathological lesions found in AHR-null mice suggest that the AHR may have a possible role in coordinating tissue/matrix remodeling during tissue morphogenesis (reviewed in reference 28). On the other hand, AHR activation by TCDD has been shown to increase the expression of ECM remodeling genes, such as those for matrix metalloproteinases, plasminogen activator inhibitor 2, and urokinase-type plasminogen activator (19, 63). Other ECM genes identified in our studies, including those for tissue inhibitor of metalloproteinase 2 and 3, integrin α6, integrin α7, matrilin 2, fibronectin, Col1α1, and Col3α1, have also been identified as TCDD targets (2, 28, 64), further supporting a role for the AHR in mediating matrix remodeling and metabolism.
The mechanism by which the AHR regulates the expression of genes related to cell growth and the ECM is unknown. A role for canonical AhRE motifs can be ruled out since these are ligand-independent regulatory events. Recent work has shown that the AHR undergoes active cytoplasm-to-nucleus shuttling in the absence of ligand (49, 50), with its intracellular localization strongly dependent on cell density (31). Shuttling may allow the receptor to fulfill nuclear functions, even though at any one time the majority of it is located in the cytosol. It is possible that AHR undergoes nucleocytoplasmic shuttling in our cell lines, which would directly influence its function as a nuclear protein. More likely, the regulation of ECM and cell cycle genes by AHR results from a pleiotropic effect of TGF-β acting as an AHR downstream mediator. Two of the major functions of TGF-β are the inhibition of cell cycle progression (34) and the control of the degradation and remodeling of matrix proteins (7, 11, 14, 54). The fact that most genes regulated by the AHR in our microarray and RT-PCR data are known TGF-β targets strongly supports the conclusion that AHR promotes cell growth and regulates ECM genes by repressing TGF-β signaling. This conclusion is further strengthened by the observation that the inhibition of TGF-β signaling by Smad7 changes the cell proliferation and gene expression characteristics of AHR-null cells to a phenotype very similar to that of AHR-positive cells. In this context, it is noteworthy that cross talk between the AHR and TGF-β pathways is evolutionarily conserved, reaching as far back in evolution as the invertebrate Caenorhabditis elegans (53).
We do not detect any difference between AHR-null and AHR-positive cells in the transcription rate of any of the three TGF-β genes, unlike findings in human keratinocytes, where TCDD was shown to decrease the transcription rate of the TGF-β2 gene (20). In contrast, TGF-β1 mRNA was stabilized in Off*Ahr−/− cells, indicating that the AHR regulates TGF-β1 levels via posttranscriptional modifications. No differences in mRNA stability were found for TGF-β2 and -β3 between the two cell lines, suggesting that AHR may regulate the three TGF-β isoforms by distinct mechanisms. Although TCDD was shown to stabilize the mRNA of several genes, such as those encoding TGF-α (8, 20), early growth response 1 (44), and plasminogen activator inhibitor 2 (71), effects of AHR on mRNA stability independent of an exogenous ligand have not been reported. Our results imply the existence of a novel mechanism for AHR to regulate mRNA accumulation that does not involve its function as a transcription factor.
Many genes, including several proto-oncogenes and genes for nuclear transcription factors, and cytokines, regulate gene expression through the interaction of specific proteins with AREs located in the 3′UTRs of their mRNAs (6). Of the three TGF-β genes, an ARE is found only in the TGF-β1 mRNA, not in that of TGF-β2 or TGF-β3. TTP has been identified as an ARE-binding protein (40, 41, 69) that destabilizes ARE-containing mRNAs, such as TNF-α (38), vascular endothelial growth factor (9), and interleukin-3 mRNAs (61). TTP activity is negatively regulated by the p38-dependent MK2 (48, 62), which, when activated, phosphorylates TTP and promotes its nuclear export (32). Phosphorylated cytoplasmic TTP has less ARE binding affinity and activity, leading to the increased mRNA stability of its target genes (29). In our work, a marked increase of TTP nuclear export and phospho-MK2 levels was detected in the absence of AHR, suggesting that TTP may regulate the stability of TGF-β1. At this point, how AHR would regulate TTP activity is unclear.
It is widely accepted that most of the toxic and carcinogenic effects of exposure to halogenated and polycyclic aromatic hydrocarbons in animals and humans are mediated by the AHR and that its activation by high-affinity halogenated or polycyclic aromatic hydrocarbon ligands, such as TCDD and B[a]P, may also result in a wide range of cell cycle perturbations, including G0/G1 and G2/M arrest, diminished capacity for DNA replication, and inhibition of cell proliferation (reviewed in reference 43). Massive receptor activation by an unmetabolizable ligand such as TCDD induces its ubiquitination and degradation and effectively creates an AHR-null cellular environment until de novo synthesis builds up receptor levels. Hence, while the endogenous function of the unliganded Ah receptor seems to be to maintain cell cycle progression, the short-term consequences of its ligand-dependent activation are the same as the consequences of its ablation, namely, to promote cell cycle arrest. Our observation that the AHR, through its role in control of TGF-β1 function, can also regulate the expression of ECM genes suggests that it may also play a role in matrix and tissue remodeling during morphogenesis and that there might be loss-of-function mutations in the human AHR gene associated with developmental or invasive diseases.
Supplementary Material
Acknowledgments
We thank Chia-Yang Liu for a gift of adeno-Smad7, D. W. Nebert for a gift of anti-GCLM antibody, and Y. Xia for suggesting the Smad7 experiments and for a critical reading of the manuscript. We also thank the anonymous reviewers whose criticisms have significantly improved the quality of this paper.
This research was supported by NIEHS grants R01 ES06273, R01 ES10807 and the NIEHS Center for Environmental Genetics grant P30 ES06096.
Footnotes
Published ahead of print on 2 July 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abbott, B. D., and L. S. Birnbaum. 1990. TCDD-induced altered expression of growth factors may have a role in producing cleft palate and enhancing the incidence of clefts after coadministration of retinoic acid and TCDD. Toxicol. Appl. Pharmacol. 106:418-432. [DOI] [PubMed] [Google Scholar]
- 2.Andreasen, E. A., L. K. Mathew, and R. L. Tanguay. 2006. Regenerative growth is impacted by TCDD: gene expression analysis reveals extracellular matrix modulation. Toxicol. Sci. 92:254-269. [DOI] [PubMed] [Google Scholar]
- 3.Bauman, J. W., T. L. Goldsworthy, C. S. Dunn, and T. R. Fox. 1995. Inhibitory effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on rat hepatocyte proliferation induced by 2/3 partial hepatectomy. Cell Prolif. 28:437-451. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 5.Chang, C.-Y., and A. Puga. 1998. Constitutive activation of the aromatic hydrocarbon receptor. Mol. Cell. Biol. 18:525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 7.Chipuk, J. E., M. Bhat, A. Y. Hsing, J. Ma, and D. Danielpour. 2001. Bcl-xL blocks transforming growth factor-beta 1-induced apoptosis by inhibiting cytochrome c release and not by directly antagonizing Apaf-1-dependent caspase activation in prostate epithelial cells. J. Biol. Chem. 276:26614-26621. [DOI] [PubMed] [Google Scholar]
- 8.Choi, E. J., D. G. Toscano, J. A. Ryan, N. Riedel, and W. A. Toscano, Jr. 1991. Dioxin induces transforming growth factor-alpha in human keratinocytes. J. Biol. Chem. 266:9591-9597. [PubMed] [Google Scholar]
- 9.Ciais, D., N. Cherradi, S. Bailly, E. Grenier, E. Berra, J. Pouyssegur, J. Lamarre, and J. J. Feige. 2004. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene 23:8673-8680. [DOI] [PubMed] [Google Scholar]
- 10.Dalton, T. P., L. He, B. Wang, M. L. Miller, L. Jin, K. F. Stringer, X. Chang, C. S. Baxter, and D. W. Nebert. 2005. Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. Proc. Natl. Acad. Sci. USA 102:3401-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Iongh, R. U., E. Wederell, F. J. Lovicu, and J. W. McAvoy. 2005. Transforming growth factor-beta-induced epithelial-mesenchymal transition in the lens: a model for cataract formation. Cells Tissues Organs 179:43-55. [DOI] [PubMed] [Google Scholar]
- 12.Dohr, O., and J. Abel. 1997. Transforming growth factor-β1 coregulates mRNA expression of aryl hydrocarbon receptor and cell-cycle-regulating genes in human cancer cell lines. Biochem. Biophys. Res. Commun. 241:86-91. [DOI] [PubMed] [Google Scholar]
- 13.Dohr, O., R. Sinning, C. Vogel, P. Munzel, and J. Abel. 1997. Effect of transforming growth factor-beta1 on expression of aryl hydrocarbon receptor and genes of Ah gene battery: clues for independent down-regulation in A549 cells. Mol. Pharmacol. 51:703-710. [DOI] [PubMed] [Google Scholar]
- 14.Douthwaite, J. A., T. S. Johnson, J. L. Haylor, P. Watson, and A. M. El Nahas. 1999. Effects of transforming growth factor-β1 on renal extracellular matrix components and their regulating proteins. J. Am. Soc. Nephrol. 10:2109-2119. [DOI] [PubMed] [Google Scholar]
- 15.Egeblad, M., and Z. Werb. 2002. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2:161-174. [DOI] [PubMed] [Google Scholar]
- 16.Elferink, C. J., N. L. Ge, and A. Levine. 2001. Maximal aryl hydrocarbon receptor activity depends on an interaction with the retinoblastoma protein. Mol. Pharmacol. 59:664-673. [DOI] [PubMed] [Google Scholar]
- 17.Elizondo, G., P. Fernandez-Salguero, M. S. Sheikh, G. Y. Kim, A. J. Fornace, K. S. Lee, and F. J. Gonzalez. 2000. Altered cell cycle control at the G(2)/M phases in aryl hydrocarbon receptor-null embryo fibroblast. Mol. Pharmacol. 57:1056-1063. [PubMed] [Google Scholar]
- 18.Fernandez-Salguero, P., T. Pineau, D. M. Hilbert, T. McPhail, S. S. Lee, S. Kimura, D. W. Nebert, S. Rudikoff, J. M. Ward, and F. J. Gonzalez. 1995. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science 268:722-726. [DOI] [PubMed] [Google Scholar]
- 19.Gaido, K. W., and S. C. Maness. 1995. Post-transcriptional stabilization of urokinase plasminogen activator mRNA by 2,3,7,8-tetrachlorodibenzo-p-dioxin in a human keratinocyte cell line. Toxicol. Appl. Pharmacol. 133:34-42. [DOI] [PubMed] [Google Scholar]
- 20.Gaido, K. W., S. C. Maness, L. S. Leonard, and W. F. Greenlee. 1992. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-dependent regulation of transforming growth factors-alpha and -beta 2 expression in a human keratinocyte cell line involves both transcriptional and post-transcriptional control. J. Biol. Chem. 267:24591-24595. [PubMed] [Google Scholar]
- 21.Ge, N.-L., and C. J. Elferink. 1998. A direct interaction between the aryl hydrocarbon receptor and retinoblatoma protein. J. Biol. Chem. 273:22708-22713. [DOI] [PubMed] [Google Scholar]
- 22.Gierthy, J. F., and D. Crane. 1984. Reversible inhibition of in vitro epithelial cell proliferation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. 74:91-98. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, F. J., and P. Fernandez-Salguero. 1998. The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug Metab. Dispos. 26:1194-1198. [PubMed] [Google Scholar]
- 24.Gradin, K., R. Toftgard, L. Poellinger, and A. Berghard. 1999. Repression of dioxin signal transduction in fibroblasts. Identification of a putative repressor associated with Arnt. J. Biol. Chem. 274:13511-13518. [DOI] [PubMed] [Google Scholar]
- 25.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 265:11-23. [DOI] [PubMed] [Google Scholar]
- 26.Guo, J., M. Sartor, S. Karyala, M. Medvedovic, S. Kann, A. Puga, P. Ryan, and C. R. Tomlinson. 2004. Expression of genes in the TGF-beta signaling pathway is significantly deregulated in smooth muscle cells from aorta of aryl hydrocarbon receptor knockout mice. Toxicol. Appl. Pharmacol. 194:79-89. [DOI] [PubMed] [Google Scholar]
- 27.Hankinson, O. 1995. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35:307-340. [DOI] [PubMed] [Google Scholar]
- 28.Hillegass, J. M., K. A. Murphy, C. M. Villano, and L. A. White. 2006. The impact of aryl hydrocarbon receptor signaling on matrix metabolism: implications for development and disease. Biol. Chem. 387:1159-1173. [DOI] [PubMed] [Google Scholar]
- 29.Hitti, E., T. Iakovleva, M. Brook, S. Deppenmeier, A. D. Gruber, D. Radzioch, A. R. Clark, P. J. Blackshear, A. Kotlyarov, and M. Gaestel. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 26:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hushka, D. R., and W. F. Greenlee. 1995. 2,3,7,8-Tetrachlorodibenzo-p-dioxin inhibits DNA synthesis in rat primary hepatocytes. Mutat. Res. 333:89-99. [DOI] [PubMed] [Google Scholar]
- 31.Ikuta, T., Y. Kobayashi, and K. Kawajiri. 2004. Cell density regulates intracellular localization of aryl hydrocarbon receptor. J. Biol. Chem. 279:19209-19216. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, B. A., J. R. Stehn, M. B. Yaffe, and T. K. Blackwell. 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 277:18029-18036. [DOI] [PubMed] [Google Scholar]
- 33.Kaldis, P., and E. Aleem. 2005. Cell cycle sibling rivalry: Cdc2 vs. Cdk2. Cell Cycle 4:1491-1494. [DOI] [PubMed] [Google Scholar]
- 34.Kamesaki, H., K. Nishizawa, G. Y. Michaud, J. Cossman, and T. Kiyono. 1998. TGF-beta 1 induces the cyclin-dependent kinase inhibitor p27Kip1 mRNA and protein in murine B cells. J. Immunol. 160:770-777. [PubMed] [Google Scholar]
- 35.Kann, S., C. Estes, J. F. Reichard, M. Y. Huang, M. A. Sartor, S. Schwemberger, Y. Chen, T. P. Dalton, H. G. Shertzer, Y. Xia, and A. Puga. 2005. Butylhydroquinone protects cells genetically deficient in glutathione biosynthesis from arsenite-induced apoptosis without significantly changing their prooxidant status. Toxicol. Sci. 87:365-384. [DOI] [PubMed] [Google Scholar]
- 36.Kann, S., M. Y. Huang, C. Estes, J. F. Reichard, M. A. Sartor, Y. Xia, and A. Puga. 2005. Arsenite-induced aryl hydrocarbon receptor nuclear translocation results in additive induction of phase I and synergistic induction of phase II genes. Mol. Pharmacol. 68:336-346. [DOI] [PubMed] [Google Scholar]
- 37.Kolluri, S. K., C. Weiss, A. Koff, and M. Göttlicher. 1999. p27kip1 induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 13:1742-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma, Q., and J. P. Whitlock, Jr. 1996. The aromatic hydrocarbon receptor modulates the Hepa 1c1c7 cell cycle and differentiated state independently of dioxin. Mol. Cell. Biol. 16:2144-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 41.Ma, W. J., S. Chung, and H. Furneaux. 1997. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 25:3564-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marlowe, J. L., E. S. Knudsen, S. Schwemberger, and A. Puga. 2004. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J. Biol. Chem. 279:29013-29022. [DOI] [PubMed] [Google Scholar]
- 43.Marlowe, J. L., and A. Puga. 2005. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell. Biochem. 96:1174-1184. [DOI] [PubMed] [Google Scholar]
- 44.Martinez, J. M., S. J. Baek, D. M. Mays, P. K. Tithof, T. E. Eling, and N. J. Walker. 2004. EGR1 is a novel target for AhR agonists in human lung epithelial cells. Toxicol. Sci. 82:429-435. [DOI] [PubMed] [Google Scholar]
- 45.Massague, J., S. Cheifetz, M. Laiho, D. A. Ralph, F. M. Weis, and A. Zentella. 1992. Transforming growth factor-beta. Cancer Surv. 12:81-103. [PubMed] [Google Scholar]
- 46.Mitchell, K. A., C. A. Lockhart, G. Huang, and C. J. Elferink. 2006. Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol. Pharmacol. 70:163-170. [DOI] [PubMed] [Google Scholar]
- 47.Mulero-Navarro, S., E. Pozo-Guisado, P. A. Perez-Mancera, A. Alvarez-Barrientos, I. Catalina-Fernandez, E. Hernandez-Nieto, J. Saenz-Santamaria, N. Martinez, J. M. Rojas, I. Sanchez-Garcia, and P. M. Fernandez-Salguero. 2005. Immortalized mouse mammary fibroblasts lacking dioxin receptor have impaired tumorigenicity in a subcutaneous mouse xenograft model. J. Biol. Chem. 280:28731-28741. [DOI] [PubMed] [Google Scholar]
- 48.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H. D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277:3065-3068. [DOI] [PubMed] [Google Scholar]
- 49.Pollenz, R. S., and E. J. Dougherty. 2005. Redefining the role of the endogenous XAP2 and C-terminal hsp70-interacting protein on the endogenous Ah receptors expressed in mouse and rat cell lines. J. Biol. Chem. 280:33346-33356. [DOI] [PubMed] [Google Scholar]
- 50.Pollenz, R. S., S. E. Wilson, and E. J. Dougherty. 2006. Role of endogenous XAP2 protein on the localization and nucleocytoplasmic shuttling of the endogenous mouse Ahb-1 receptor in the presence and absence of ligand. Mol. Pharmacol. 70:1369-1379. [DOI] [PubMed] [Google Scholar]
- 51.Puga, A., S. J. Barnes, T. P. Dalton, C. Chang, E. S. Knudsen, and M. A. Maier. 2000. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J. Biol. Chem. 275:2943-2950. [DOI] [PubMed] [Google Scholar]
- 52.Puga, A., Y. Xia, and C. Elferink. 2002. Role of the aryl hydrocarbon receptor in cell cycle regulation. Chem. Biol. Interact. 141:117-130. [DOI] [PubMed] [Google Scholar]
- 53.Qin, H., and J. A. Powell-Coffman. 2004. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev. Biol. 270:64-75. [DOI] [PubMed] [Google Scholar]
- 54.Rogister, B., P. Delree, P. Leprince, D. Martin, C. Sadzot, B. Malgrange, C. Munaut, J. M. Rigo, P. P. Lefebvre, and J. N. Octave. 1993. Transforming growth factor beta as a neuronoglial signal during peripheral nervous system response to injury. J. Neurosci. Res. 34:32-43. [DOI] [PubMed] [Google Scholar]
- 55.Santiago-Josefat, B., S. Mulero-Navarro, S. L. Dallas, and P. M. Fernandez-Salguero. 2004. Overexpression of latent transforming growth factor-beta binding protein 1 (LTBP-1) in dioxin receptor-null mouse embryo fibroblasts. J. Cell Sci. 117:849-859. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt, J. V., and C. A. Bradfield. 1996. Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 12:55-89. [DOI] [PubMed] [Google Scholar]
- 57.Shimba, S., K. Komiyama, I. Moro, and M. Tezuka. 2002. Overexpression of the aryl hydrocarbon receptor (AhR) accelerates the cell proliferation of A549 cells. J. Biochem. (Tokyo) 132:795-802. [DOI] [PubMed] [Google Scholar]
- 58.Smyth, G. K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article 3. [DOI] [PubMed] [Google Scholar]
- 59.Solis, W. A., N. L. Childs, M. N. Weedon, L. He, D. W. Nebert, and T. P. Dalton. 2002. Retrovirally expressed metal response element-binding transcription factor-1 normalizes metallothionein-1 gene expression and protects cells against zinc, but not cadmium, toxicity. Toxicol. Appl. Pharmacol. 178:93-101. [DOI] [PubMed] [Google Scholar]
- 60.Sossey-Alaoui, K., L. Vieira, D. David, M. G. Boavida, and J. K. Cowell. 2003. Molecular characterization of a 7p15-21 homozygous deletion in a Wilms tumor. Genes Chromosomes Cancer 36:1-6. [DOI] [PubMed] [Google Scholar]
- 61.Stoecklin, G., X. F. Ming, R. Looser, and C. Moroni. 2000. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 20:3753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W. F. Rigby, T. K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutter, T. R., K. Guzman, K. M. Dold, and W. F. Greenlee. 1991. Targets for dioxin: genes for plasminogen activator inhibitor-2 and interleukin-1 beta. Science 254:415-418. [DOI] [PubMed] [Google Scholar]
- 64.Thackaberry, E. A., E. J. Bedrick, M. B. Goens, L. Danielson, A. K. Lund, D. Gabaldon, S. M. Smith, and M. K. Walker. 2003. Insulin regulation in AhR-null mice: embryonic cardiac enlargement, neonatal macrosomia, and altered insulin regulation and response in pregnant and aging AhR-null females. Toxicol. Sci. 76:407-417. [DOI] [PubMed] [Google Scholar]
- 65.Thomae, T. L., E. A. Stevens, and C. A. Bradfield. 2005. Transforming growth factor-beta3 restores fusion in palatal shelves exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Chem. 280:12742-12746. [DOI] [PubMed] [Google Scholar]
- 66.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei, Y. D., K. Tepperman, M. Y. Huang, M. A. Sartor, and A. Puga. 2004. Chromium inhibits transcription from polycyclic aromatic hydrocarbon-inducible promoters by blocking the release of histone deacetylase and preventing the binding of p300 to chromatin J. Biol. Chem. 279:4110-4119. [DOI] [PubMed] [Google Scholar]
- 68.Whitlock, J. P., Jr. 1999. Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 39:103-125. [DOI] [PubMed] [Google Scholar]
- 69.Wilson, G. M., and G. Brewer. 1999. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62:257-291. [DOI] [PubMed] [Google Scholar]
- 70.Wolff, S., P. A. Harper, J. M. Wong, V. Mostert, Y. Wang, and J. Abel. 2001. Cell-specific regulation of human aryl hydrocarbon receptor expression by transforming growth factor-beta(1). Mol. Pharmacol. 59:716-724. [DOI] [PubMed] [Google Scholar]
- 71.Yang, J. H. 1999. Expression of dioxin-responsive genes in human endometrial cells in culture. Biochem. Biophys. Res. Commun. 257:259-263. [DOI] [PubMed] [Google Scholar]
- 72.Zaher, H., P. M. Fernandez-Salguero, J. Letterio, M. S. Sheikh, A. J. Fornace, Jr., A. B. Roberts, and F. J. Gonzalez. 1998. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Mol. Pharmacol. 54:313-321. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.