Abstract
Hypoxia and nutrient deprivation are environmental stresses governing the survival and adaptation of tumor cells in vivo. We have identified a novel role for the Rb tumor suppressor in protecting against nonapoptotic cell death in the developing mouse fetal liver, in primary mouse embryonic fibroblasts, and in tumor cell lines. Loss of pRb resulted in derepression of BNip3, a hypoxia-inducible member of the Bcl-2 superfamily of cell death regulators. We identified BNIP3 as a direct target of pRB/E2F-mediated transcriptional repression and showed that pRB attenuates the induction of BNIP3 by hypoxia-inducible factor to prevent autophagic cell death. BNIP3 was essential for hypoxia-induced autophagy, and its ability to promote autophagosome formation was enhanced under conditions of nutrient deprivation. Knockdown of BNIP3 reduced cell death, and remaining deaths were necrotic in nature. These studies identify BNIP3 as a key regulator of hypoxia-induced autophagy and suggest a novel role for the RB tumor suppressor in preventing nonapoptotic cell death by limiting the extent of BNIP3 induction in cells.
Programmed cell death plays an important role in normal developmental processes and in eliminating potentially pathological cells from the organism, and increased resistance to apoptotic cell death is a hallmark of cancer (13). The role of nonapoptotic cell death, including autophagic cell death and programmed necrosis, in cancer is less clear (24, 30, 56). Autophagy is a well-conserved mechanism activated in response to nutrient deprivation, and it involves the catabolic degradation of macromolecules and organelles by autophagosomes to regenerate metabolites for energy and growth (24, 30). Excess or prolonged autophagy can lead to autophagic cell death (29, 40), whereas inhibition of autophagy appears to promote necrotic cell death (7, 12). In contrast to processes of apoptosis or autophagic cell death that have tumor suppressor functions, necrosis promotes tumor progression through induction of inflammatory responses that initiate regenerative proliferation and invasion (50). Thus, the factors determining whether a cell undergoes a specific type of cell death (apoptotic, autophagic, or necrotic) in response to a given stress are significant for understanding the progression of cancer.
The RB tumor suppressor functions as a negative regulator of the cell cycle through inhibition of E2F transcription factors (49), but it plays other less well defined roles in promoting cell survival (5). In particular, loss of pRB has been shown to sensitize tumor cells to apoptosis induced by chemotherapeutic agents, and certain viral oncoproteins promote apoptosis through the inactivation of pRB (5). However, pRB previously has not been implicated in the regulation of nonapoptotic cell death.
Mice lacking the function of pRb die in midgestation and exhibit various developmental defects, including increased cell death in the nervous system, lens, and liver (5). Although cell death in the lens and peripheral nervous system likely is due to intrinsic defects in cell cycle exit during terminal differentiation, apoptotic cell death in the central nervous system appears to be the consequence of hypoxia resulting from abnormalities outside the nervous system (11, 33). Hypoxia in the Rb null mouse has been attributed to the combined effects of defective red blood cells (33, 46) and abnormal placental development (52).
The apoptotic mechanisms underlying cell death in the lens and central nervous system of Rb null mice are well characterized, but the extensive cell death that becomes apparent in the Rb null fetal liver (FL) from embryonic day 13.5 (E13.5) onwards is not well understood. In the present study, we examined cell death in the Rb null FL and determined that hypoxia-associated cell death in the FL was nonapoptotic. In addition, we observed that RB-deficient tumor cells and Rb null mouse embryonic fibroblasts (MEFs) also were hypersensitive to nonapoptotic cell death in vitro. Loss of RB resulted in derepression of BNIP3, a Bcl-2-related BH3-only protein that is induced by hypoxia in cultured cardiomyocytes (15, 26) and ischemic regions of tumors (38, 45). Here, we have demonstrated that BNIP3 is essential for hypoxia-induced autophagy and that BNIP3 is a direct RB/E2F target gene. Knockdown of BNIP3 inhibited autophagy and promoted necrotic cell death of RB-deficient tumor cells. These results have significance for understanding the roles of pRB and BNIP3 in responses to hypoxia and in determining the type of cell death induced by nutrient deprivation.
MATERIALS AND METHODS
Mice and TUNEL assays.
All mice were maintained in a specific-pathogen-free barrier facility. Timed matings of Rb heterozygous mice were set up to generate MEFs from E13.5 embryos. Embryos were fixed in 10% neutral buffered formalin and sagittally sectioned for terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays as described previously (31).
Electron microscopy.
Whole FLs or cultured cell pellets were fixed in 2.5% glutaraldehyde, and sections were analyzed by using a Philips CM120 transmission electron microscope.
Cell culture, green fluorescent protein (GFP)-light chain 3 (LC3) staining, and other cell death assays.
Tumor cell lines and MEFs were grown in high-glucose Dulbecco's modified Eagle's medium, 2 mM l-glutamine, and penicillin-streptomycin in either 10% or 0.1% fetal calf serum. Passage 1 to 4 MEFs were used for all experiments. Hypoxia studies were carried out at 0.5% oxygen; desferroxamine (DFO) was used at a final concentration of 260 μM, and dimethyl-oxallyl glycine (DMOG) was used at a final concentration of 1 mM. The caspase inhibitor Boc-D-FMK was used at a final concentration of 20 μM. Tumor necrosis factor alpha (TNF-α) was used at a final concentration of 1,000 U/ml. Flow cytometric analysis of FLs was carried out as described previously (46). Flow cytometric analysis of tumor cell lines was carried out after cells were stained in the culture dish with 10 μg/ml propidium iodide (PI), which was added 15 min prior to harvesting. All cells (adherent and floating) were harvested and washed once in cold phosphate-buffered saline for analysis of PI uptake. Cells were transfected with pGFP-LC3 (a kind gift from T. Yoshimori, Japan) prior to visualization by fluorescence microscopy.
In situ hybridization and RNase protection assay.
In situ hybridization was carried out as described previously (31), using full-length BNip3 cDNA as a template to generate an antisense probe. For RNase protection assays, approximately 100 ng of gel-purified antisense template linearized to BNip3 or to an acidic ribosomal phosphoprotein PO (ARPPO) control was transcribed using a riboprobe in vitro transcription kit (Promega) incorporating [32P]CTP (Amersham). Specific RNA transcripts were detected using the RPAII RNase protection assay kit (Ambion, Austin, TX).
Quantitative real-time PCR.
Relative quantitation of real-time PCR products was performed using QuantiTect SYBR Green PCR and the Applied Biosystems 7900 fast real-time PCR system, followed by analysis with the associated SDS 2.3 software. Samples were amplified in triplicate and normalized by subtracting threshold cycle values for 18S rRNA.
Western blotting and gel shift procedures.
Whole-cell protein extracts were prepared in radioimmunoprecipitation assay buffer (1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 10 mM Tris, pH 8.0, 0.14 M NaCl, and inhibitors). Samples were separated by SDS-13% polyacrylamide gel electrophoresis. Membranes were incubated overnight at 4°C in primary antibody to BNip3 (Abcam) at 1:500 in a mixture of Tris-buffered saline, 0.1% Tween 20, and 5% milk.
Gel shifts to measure E2F DNA binding activity were carried out as described previously (31), except that nuclear extracts were used instead of whole-cell lysates. The sequences of oligonucleotides used for the E2F site probes were the following: E2F consensus forward, ATT TAA GTT TCG CGC CCT TTC TCA; E2F consensus reverse, TGA GAA AGG GCG CGA AAC TTA AAT; BNip3 E2F site forward, GCC GCC CTC CCG CGC ACT CCT; BNip3 E2F site reverse, AGG CAG GAG TGC GCG GGA GGG; mutant BNip3 E2F site forward, GCC GCC CTC CTG CGT ACT CCT; mutant BNip3 E2F site reverse, AGG CAG GAG TAC GCA GGA GGG.
Hypoxia-inducible factor 1α (HIF-1α) gel shifts were carried out using nuclear lysates. Protein samples were in incubated in sample buffer [20 mM HEPES-KOH, pH 7.9, 100 mM KCl, 6 mM MgCl2, 0.5 mM EDTA, 5% glycerol, 1.5 μg poly(dI-dC)] in the absence or presence of a 100-fold excess of cold probe or anti-HIF-1α antibody (NB 100-123; Novus Biologicals) for 30 min at 4°C. Radiolabeled probes were added and incubated for a further 5 min at 4°C. Complexes were separated by nondenaturing polyacrylamide gel electrophoresis (4.4% acrylamide, 0.5× Tris-borate-EDTA) for 2 h 45 min at 4°C. The sequences of oligonucleotides used for the HIF gel shift probes were the following: HIF consensus forward, AAA GGG AAA GAA CTA AAC ACA CAG C; HIF consensus reverse, CAA CTT GAA TAC ATT GAC CAT ATC G; BNip3 HIF site forward, GCG CCG CAC GTG CCA CAC; BNip3 HIF site reverse, GGA GCG TGT GGC ACG TGC; mutant BNip3 HIF site forward, GCG CCG CAG TTG CCA CAC; mutant BNip3 HIF site reverse, GGA GCG TGT GGC AAC TGC.
Chromatin immunoprecipitation assays (ChIPs).
Cells were harvested following cross-linking with 1% (vol/vol) formaldehyde, which was stopped by addition of glycine to a final concentration of 125 mM. Cells were washed and harvested, and isolated nuclei were lysed in nuclear lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% [wt/vol] SDS). DNA was sheared by sonication on ice, and cleared supernatant was incubated with antibody (1 μg of one of the following antibodies: for E2F-1, sc-193; E2F-3, sc-878; E2F-4, sc-1082 [all from Santa Cruz]; pRb, G3-245 [Pharmingen]; and HIF-1α, NB 100-123 [Novus Biologicals]) overnight at 4°C. Immune complexes were collected with 50% protein A-Sepharose for 1 h at 4°C.
Complexes were washed, and antibody-DNA-protein complexes were released with elution buffer (1% SDS, 0.1 M NaHCO3). Cross-links were reversed, and DNA was recovered by phenol-chloroform extraction and precipitated in the presence of glycogen. Immunoprecipitated DNA or input DNA was amplified by 30 cycles of PCR using the following primer sequences: for the BNip3 promoter, BNIP3P-FOR (5′ CCT AGC TAG CCG GTC CAC TTC TGC ATT AGA CC) and BNIP3P-REV (5′ CGG AGG ATC TTG CGC CGC TCA GTT CTG AGG CCA GG3′); for the luciferase reporter plasmid, pGL1-FOR (5′ TGT ATC TTA TGG TAC TGT AAC TG 3′) and pGL2-REV (5′ CTT TAT GTT TTT GGC GTC TTC CA 3′).
Reporter constructs and luciferase assays.
Site-directed mutagenesis of the mouse BNip3 promoter cloned into the pGL2 vector (Promega) was carried out as described previously (32) with primers for the E2F and HIF binding sites carrying the mutations described above for gel shift analysis. Cells (1.3 × 105) were transfected with 1 mg of plasmid DNA using Lipofectamine 2000 according to the manufacturer's protocol. Cells were harvested for luciferase assays 20 h later. Light emission was immediately measured using a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
Immunofluorescence.
BNIP3 immunofluorescence was determined using a 1:1,000 dilution of primary antibody to BNip3 (Abcam). HMG-B1 antibody (Abcam) was used to stain cells as described previously (43).
ATP assay.
For the ATP assay, cells were treated for 24 h, counted, harvested at 4°C, and resuspended at 1,000 cells/μl in sterile double-distilled H2O. Cells were boiled for 10 min, cooled on ice for 30 s, and spun at 10,000 × g for 5 min at 4°C. Cleared supernatant was used to determine the ATP concentration by using an ATP determination kit (Molecular Probes).
Knockdown of BNip3.
For transient knockdown experiments, Saos2 cells were transfected with specific short interfering RNA (siRNA) for BNip3 or with control, nonsilencing siRNA (Dharmacon) using HiPerFect transfection reagent (QIAGEN). The medium was changed 5 h after transfection, and cells were exposed for 48 h to 0.5% oxygen or were treated with DMOG 48 h after transfection.
RESULTS
Nonapoptotic cell death in the Rb null FL.
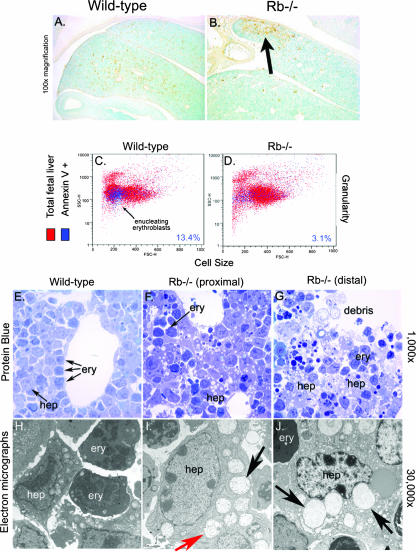
Cell death in the Rb null FL previously was shown to be independent of the apoptosome (18). However, the mechanistic basis of this cell death was not further explored. We set out to determine the nature of cell death in the developing FLs of Rb null mice (Fig. 1). The pattern of TUNEL staining seen for E13.5 FLs was markedly different between wild-type and Rb null embryos. Individual TUNEL-positive cells were dispersed throughout the wild-type FL at E13.5 (Fig. 1A; also see Fig. S1D in the supplemental material), whereas in the Rb null mouse, large clusters of TUNEL-positive cells were localized to the ventral end of the FL (Fig. 1B; also see Fig. S1F in the supplemental material), and little or no cell death was apparent in proximal regions of the Rb null FL (Fig. 1B; also see Fig. S1E in the supplemental material). The localized nature of cell death in peripheral regions of the Rb null FL that are farthest from the hepatic vasculature is consistent with ischemia as the causative agent (46, 52).
FIG. 1.
Nonapoptotic cell death in the midgestational FLs of Rb null mice. (A and B) TUNEL assays to detect cell death in wild-type and Rb null E13.5 FLs. (C and D) Flow cytometric analysis of annexin V-fluorescein isothiocyanate-stained FL cells. (E to G) Light microscopic analysis of protein-blue-stained FL sections showing increased vacuolation in Rb null FL (F and G, arrows) compared to the level of vacuolation in wild-type FL (E). Dark-staining erythroblasts (F, ery) were not overtly affected. Cellular debris is evident in distal regions of Rb null FL (G). (H to J) Electron microscopic analysis of wild-type (H) and Rb null FLs (I and J). Extensive vacuolation in Rb null hepatocytes (hep) is associated with increased mitochondrial swelling (I and J, black arrows). Double-membrane-bound vesicles also are seen in Rb null hepatocytes (I and J, red arrow).
Surprisingly, more annexin V staining was seen with wild-type FL (13.4% annexin V-positive cells) (Fig. 1C) than with Rb−/− FL (3.1% annexin V-positive cells) (Fig. 1D). However, this may be explained by phosphotidylserine externalization during erythroblast enucleation (53) that normally takes place in wild-type FL but that is blocked in Rb null embryos (46) and not by increased apoptosis. Nevertheless, failure of Annexin V to stain Rb−/− FL cells was consistent with nonapoptotic cell death.
We observed higher numbers of highly vacuolated cells with the Rb null FL (Fig. 1F and G) than with the wild-type FL (Fig. 1E). There was no evidence of nuclear condensation, membrane blebbing, or reduced cell size in dying cells of the Rb null FL; rather, affected cells had enlarged nuclei and highly vacuolated cytoplasm (Fig. 1F, G, I, and J), consistent with nonapoptotic cell death. Vacuolation and cell death was detected primarily in large cells with prominent nucleoli and not in smaller electron-dense cells of the Rb null FL (Fig. 1F and G), indicating that hepatocytes, as opposed to erythroblasts (both of which make up the FL at this stage of development), were specifically affected (Fig. 1I and J).
The ultrastructural features of the vacuoles observed in Rb null FL were consistent with swollen mitochondria (Fig. 1I and J; also see Fig. S1K, L, N, and O in the supplemental material). We observed a gradual increase in mitochondrial volume and disruption of mitochondrial cristae in Rb null FL, progressing from the healthier end of the liver (Fig. 1I; also see Fig. S1K and N in the supplemental material) to the most badly affected distal tip (Fig. 1J; also see Fig. S1L and O in the supplemental material). Increased organelle and cell volume in the absence of chromatin condensation is most consistent with necrotic cell death, and we observed increased numbers of inflammatory cells in the regions of the greatest cell death (see Fig. S1I in the supplemental material). However, in addition to swollen mitochondria, we also observed double-membrane-bound vesicles that resembled autophagosomes (Fig. 1I), suggesting that cell death in parts of the FL was autophagic. Thus, cell death in the Rb null FL is nonapoptotic, occurs preferentially in fetal hepatocytes, is localized to regions of the liver most distant from the hepatic vasculature, and exhibits features of both autophagic and necrotic cell death.
BNip3 expression is derepressed in the Rb null FL.
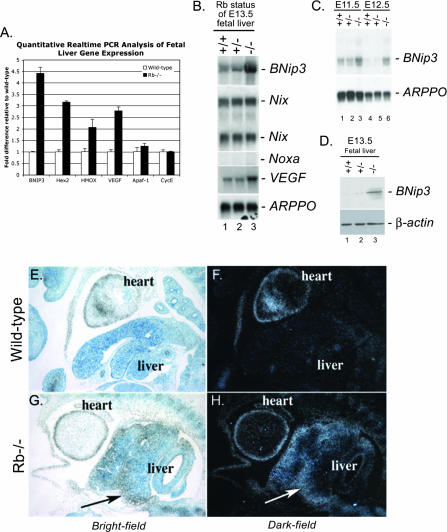
To determine whether increased cell death observed in the Rb null FL was associated with hypoxia, we compared the expression of four characterized HIF response genes (encoding BNIP3, hexokinase 2, heme oxygenase, and vascular endothelial growth factor [VEGF]) between wild-type and Rb null E13.5 FLs by quantitative real-time PCR (Fig. 2A). We observed that all four HIF target genes were expressed at significantly higher levels in the Rb null FL than in the wild-type FL, consistent with increased hypoxia in the Rb null embryo, as previously reported (33, 52). BNip3 was of particular interest, since it is one of the few Bcl-2 family members implicated in nonapoptotic cell death (3, 51). We confirmed by Northern blotting that BNip3 mRNA was expressed at increased levels in Rb null FL (Fig. 2B, lane 3) compared to the level of expression in either wild-type (Fig. 2B, lane 1) or Rb heterozygous (Fig. 2B, lane 2) FL. Expression of VEGF similarly was elevated in Rb null FL (Fig. 2B, lane 3) compared to expression in wild-type FL (Fig. 2B, lane 1). Other BH3-only proteins, such as Noxa and Nix (BNip3L), also are reported to be hypoxia inducible (23, 45), but our analysis revealed that Noxa was not expressed in either wild-type or Rb null FL (Fig. 2B) and that Nix levels were the same in Rb null FL and wild-type FL (Fig. 2B). BNip3 failed to be repressed during FL development in Rb null mouse embryos (Fig. 2B), in contrast to wild-type FL, in which BNip3 mRNA expression was silenced between E11.5 and E12.5 (Fig. 2C). Increased levels of BNip3 mRNA in E13.5 Rb null FL were reflected in increased BNip3 protein levels (Fig. 2D).
FIG. 2.
BNip3 expression is up-regulated in the Rb null FL. (A) Quantitative real-time PCR for known HIF target genes (encoding BNIP3, hexokinase 2, heme oxygenase, and VEGF) and two known E2F target genes (encoding Apaf-1 and cyclin E2 [CycE]) carried out on cDNA derived from E13.5 wild-type and Rb null FLs. (B) Total RNA from FLs from wild-type (lane 1), Rb heterozygous (lane 2), or Rb null (lane 3) E13.5 mice was examined by Northern blotting for expression of BNip3, Nix, Noxa, VEGF, and ARPPO (a control for loading). (C) Northern blot analysis of BNip3 mRNA expression in wild-type (lanes 1 and 4), Rb heterozygous (lanes 2 and 5), and Rb null (lanes 3 and 6) FLs at E11.5 (lanes 1 to 3) or E12.5 (lanes 4 to 6). (D) BNip3 protein expression in wild-type (lane 1), Rb heterozygous (lane 2), and Rb null (lane 3) FLs determined by Western blotting. (E to H) BNip3 mRNA was detected in wild-type (E and F) or Rb null (G and H) E13.5 mouse embryo sections by in situ hybridization. BNip3 mRNA was visualized at 100× magnification by bright-field (E and G) and by dark-field (F and H) microscopy on toluidine blue-counterstained sections.
On examining the expression of BNip3 mRNA by in situ hybridization, we observed that while BNip3 is barely detectable in wild-type FL at E13.5 (Fig. 2E and F), there is high-level expression in the Rb null FL (Fig. 2G and H). Furthermore, we noted that the expression of BNip3 was greatest in peripheral areas of the Rb null FL (Fig. 2G and H), where we detected the highest levels of cell death (Fig. 1B). These observations implicate BNip3 in the nonapoptotic cell death observed in the Rb null FL.
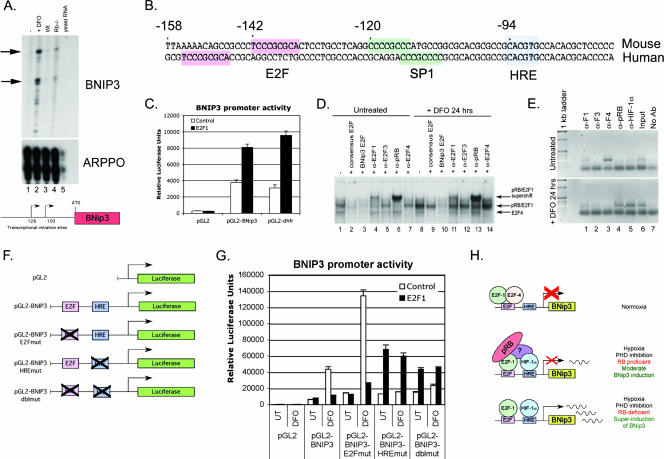
Functional repression of the BNIP3 promoter by pRB/E2F.
Other BH3-only proteins previously have been shown to be up-regulated by overexpression of E2F-1 (17). To determine whether BNip3 was a direct target of RB/E2F-mediated repression, we mapped the transcriptional initiation site of the mouse BNip3 promoter by RNase protection (Fig. 3A) and then aligned the mouse and human promoter sequences to determine what transcriptional regulatory elements were conserved between species (Fig. 3B). We observed a high degree of homology in the proximal 200 bp, with a conserved HIF response element (HRE) mapping to −94 bp (relative to the start site of transcription) in the mouse and human promoters (Fig. 3B). In addition, we observed a conserved E2F binding site mapping 48 bp upstream of the HRE in the mouse promoter and 61 bp upstream of the HRE in the human promoter (Fig. 3B).
FIG. 3.
The BNIP3 promoter contains a functional E2F binding site that interacts functionally with the HRE. (A) Transcriptional initiation sites in the mouse BNIP3 promoter were determined by RNase protection assays using RNA from untreated (lane 1) or DFO-treated (lane 2) MEFs and wild-type (lane 3) FL (Wt), Rb null (lane 4) FL, or control yeast (lane 5) RNA. A diagrammatic representation of the two major initiation sites in the mouse BNIP3 promoter is shown. (B) Sequence alignment of the proximal regions of the mouse and human BNIP3 promoters highlighting conserved regulatory elements that include an HRE (blue) at −94 bp relative to the start site of transcription and an E2F site (pink) at −142 bp (mouse) and −155 bp (human). (C) The proximal 500 bp of the mouse BNIP3 promoter was cloned upstream of the luciferase reporter gene and tested for its activity in U2OS cells in response to overexpression of E2F-1, and its activity then was compared to that of the control reporter construct pGL2 lacking a promoter and that of the characterized E2F-responsive mouse dhfr promoter. (D) The binding activity of the BNIP3 promoter E2F site was tested by EMSA using nuclear lysates from untreated (lanes 1 to 7) or DFO-treated (lanes 8 to 14) U2OS cells in the presence or absence of excess cold oligonucleotide competitor encoding either the consensus E2F recognition sequence (lanes 2 and 9) or the BNip3 E2F site (lanes 3 and 10). The effect of supershifting antibodies to E2F-1 (α-E2F1) (lanes 4 and 11), E2F-3 (α-E2F3) (lanes 5 and 12), pRB (α-pRB) (lanes 6 and 13), or E2F-4 (α-E2F4) (lanes 7 and 14) on DNA-protein complex migration also was examined. (E) The identity of E2Fs bound to the endogenous BNIP3 promoter in untreated (top panel) or DFO-treated (bottom panel) U2OS cells was examined by ChIPs using antibodies to E2F-1 (α-F1), E2F-3 (α-F3), E2F-4 (α-F4), pRB (α-pRB), or HIF-1α (α-HIF-1α) and compared to results with no-antibody negative control samples (No Ab) or to those with input positive control samples (Input). (F) Diagrammatic representation of mutations introduced into the mouse BNIP3 promoter to test the functional interaction between E2F and HIF in regulating BNip3 transcription. (G) Reporter (luciferase) gene assay for BNIP3 promoter activity in U2OS cells following DFO treatment, E2F-1 overexpression, or mutation of either the E2F site, the HRE, or both sites in the context of the proximal 500 bp of the mouse BNIP3 promoter. UT, untreated. (H) Schematic representation of the proposed interaction between pRB, E2F family members, and HIF at the BNIP3 promoter.
We cloned 500 bp of proximal BNIP3 promoter sequence upstream of the firefly luciferase reporter gene and compared the effect of E2F-1 overexpression on BNIP3 promoter activity to its effect on the activity of a characterized E2F target gene promoter, the dhfr promoter (Fig. 3C). Overexpression of E2F-1 increased BNIP3 promoter activity by approximately 2.5-fold over basal BNIP3 promoter activity in U2OS cells, similar to what was seen with the induction of the dhfr promoter, a characterized E2F target gene (Fig. 3C).
By electrophoretic mobility shift assays (EMSA) (Fig. 3D), we observed strong E2F binding to the mouse BNip3 promoter E2F site (lane 1) in nuclear extracts from U2OS cells that could be competed out with excess cold oligonucleotide encoding either the E2F consensus DNA binding site (lane 2) or the BNip3 E2F site (lane 3). Binding to this site was supershifted with antibodies to E2F-1 (lane 4), pRB (lane 6), and E2F-4 (lane 7) but not with antibody to E2F-3 (lane 5). The complex that supershifted with antibodies to E2F-1 and pRB (lanes 4 and 6) was distinct from the complex that supershifted with antibody to E2F-4 (lane 7). When U2OS cells were treated with the prolyl hydroxylase (PHD) inhibitor DFO to stabilize HIF, we observed increased overall binding activity in nuclear lysates (lane 8) of the complex that supershifts with antibodies to E2F-1 (lane 11) and pRB (lane 13) and reduced levels of the E2F-4-containing complex (lanes 8 and 14) compared to the binding activity and levels of E2F-4-containing complex in lysates from untreated U2OS cells (lane 1).
Although EMSA is a particularly sensitive method to detect protein-DNA complexes, it does not always reflect complex formation in vivo in the context of whole chromatin. To determine whether the BNIP3 promoter binds E2Fs and pRB in vivo, we carried out ChIPs on live cells before and after HIF stabilization (Fig. 3E) by using antibody to E2F-1 (lane 1), E2F-3 (lane 2), E2F-4 (lane 3), pRB (lane 4), or HIF-1α (lane 5). We failed to observe any BNIP3 promoter sequence immunoprecipitating with antibody to E2F-3, pRB, or HIF-1α in untreated U2OS cells (lanes 2, 4, and 5 in the top panel), and only a very weak signal was observed with antibody to E2F-1 (lane 1 in the top panel). However, we observed a strong PCR product generated from E2F-4 immunoprecipitates (lane 3 in the top panel), indicating that, in untreated U2OS cells, E2F-4 was bound to the BNIP3 promoter and that E2F-3, pRB, and HIF-1α were not, while E2F-1 was bound in limited amounts. By contrast, on repeating ChIPs with lysates from DFO-treated U2OS cells, we observed a loss of the E2F-4 interaction (Fig. 3E, lane 3 in the bottom panel) but increased association of pRB and HIF-1α with the BNIP3 promoter (lanes 4 and 5 in the bottom panel). These results suggested that in untreated U2OS cells, where BNIP3 is transcriptionally silent, E2F-4 is bound at the promoter and may be involved in its repression but that, following induction of BNIP3 transcription by treatment of U2OS cells with DFO, we observe increased binding of pRB and HIF-1α at the BNIP3 promoter. Thus, the results of ChIPs (Fig. 3E) were largely consistent with data from EMSA (Fig. 3D), showing reduced E2F-4 binding and increased pRB bound at the BNIP3 promoter following DFO treatment.
The proximity of the E2F site and HRE in the mouse and human BNIP3 promoters, combined with the effect of RB loss on BNIP3 expression, suggested that there might be a functional interaction between pRB/E2F-1 and HIF in regulating BNIP3 transcription. To test this, we carried out site-directed mutagenesis of the E2F and HIF binding sites in the mouse BNip3 promoter (Fig. 3F). Overexpression of E2F-1 attenuated BNip3 promoter activity following DFO treatment (Fig. 3G), supporting our hypothesis that pRB/E2F-1 complexes limit HIF-mediated induction of the BNIP3 promoter. Mutation of the E2F site increased the basal activity of the BNIP3 promoter but caused a much more substantial increase in promoter activity following DFO treatment of U2OS cells, confirming that the complex bound to the E2F site is an effective repressor of HIF-mediated induction of BNIP3. As expected, mutation of the E2F site reduced induction of the BNIP3 promoter brought about by E2F-1 overexpression (Fig. 3G). Intriguingly, mutation of the E2F site did not prevent E2F-1 overexpression from inhibiting DFO-induced activity of the BNIP3 promoter, suggesting that E2F-1 influences promoter activity both in cis and in trans.
Further support for a functional interaction between the E2F site and the HRE in the BNIP3 promoter came from our observations that mutation of the HRE resulted in increased E2F-1 responsiveness of the BNIP3 promoter (Fig. 3G). However, this effect was independent of DFO treatment, indicating that it may be mediated by factors other than HIF. We noted that the BNIP3 HRE generates a much more complex banding pattern by EMSA (see Fig. S2A, lanes 1 to 6, in the supplemental material) than does the HIF consensus (see Fig. S2A, lanes 13 to 14, in the supplemental material), which points to binding of the BNIP3 HRE by other factors in addition to HIF-1α. Specifically, binding to the BNIP3 HRE by these factors is not competed out with the HIF consensus oligonucleotide competitor (see Fig. S2A, lanes 2 and 8, in the supplemental material) or supershifted with antibody to HIF-1α (see Fig. S2A, lanes 5 and 11, in the supplemental material) or HIF-2α (see Fig. S2A, lanes 6 and 12, in the supplemental material), suggesting that these factors are unrelated to HIF-1α or HIF-2α. The effect of mutating the BNIP3 HRE for regulation of the BNIP3 promoter by E2F-1 overexpression therefore may be mediated through altered binding of these additional factors.
Given that the E2F site mediates repression of the BNIP3 promoter under basal conditions but that this repressive activity is potentiated by DFO treatment, we explored the possibility of a direct physical interaction between HIF-1α, E2F-1, and/or pRB. Previous reports have suggested that pRB directly interacts with HIF-1α (4), but as shown in Fig. S2B in the supplemental material, we have been unable to demonstrate such an interaction under basal conditions or after DFO treatment. Furthermore, mutagenesis of the E2F site abolished the association of pRB with the BNIP3 promoter, but mutation of the HRE had no significant effect on pRB association with the promoter under either basal or DFO-induced conditions. Thus, we conclude that the association of pRB with the BNIP3 promoter is dependent on the E2F site and that the functional interaction between the E2F site and the HRE is mediated by other unidentified factors.
In summary, our results demonstrate a direct role for pRB/E2F in repressing the induction of BNIP3 promoter activity by HIF (Fig. 3H).
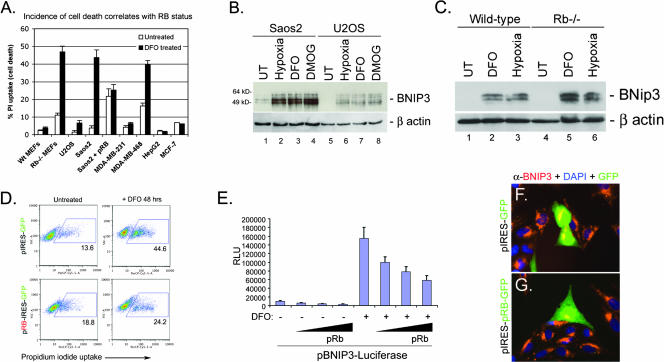
RB-deficient cells show increased sensitivity to cell death induced by PHD inhibitors.
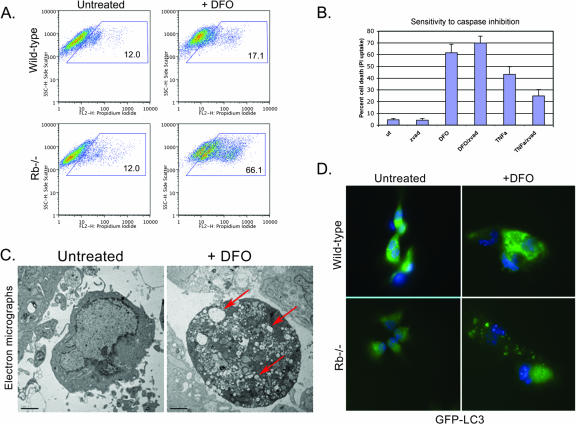
Given our observations that loss of Rb in the developing FL induced high levels of BNip3 and was associated with increased nonapoptotic cell death, we set out to determine whether loss of Rb in other contexts also led to increased BNip3 levels and cell death. We examined the effect of treating primary wild-type and Rb null MEFs, or a range of human tumor cell lines that differed in their RB statuses, with the PHD inhibitor DFO, which mimics aspects of hypoxia by promoting HIF stabilization (16). We observed a complete correlation between RB status and the extent of cell death induced by 48 h of treatment with DFO (Fig. 4A). Wild-type MEFs were resistant to DFO, whereas Rb null MEFs showed a substantial increase in cell death. Similarly, tumor cells with functional pRB (U2OS, MDA-MB-231, HepG2, and MCF-7) were resistant to DFO-induced cell death, while RB-deficient tumor cells (Saos2 and MDA-MB-468) were sensitive to cell death induced by DFO (Fig. 4A). Furthermore, BNIP3 protein was induced by hypoxia, DFO, or DMOG (an alternative PHD inhibitor) to much higher levels in RB-deficient Saos2 cells (Fig. 4B, lanes 2 to 4) than in U2OS cells (Fig. 4B, lanes 6 to 8). The level of BNIP3 induction in Saos2 cells exposed to hypoxia (Fig. 4B, lane 2) was similar to that detected in cells treated with either DFO (Fig. 4B, lane 3) or DMOG (Fig. 4B, lane 4). Similarly, we observed that BNip3 was induced to higher levels in Rb null MEFs treated with DFO (Fig. 4C, lane 5) or exposed to 0.5% oxygen (Fig. 4C, lane 6) for 48 h than in wild-type MEFs (Fig. 4C, lanes 2 to 3). These results are consistent with a role for pRB in repression of BNIP3 expression.
FIG. 4.
RB status and BNIP3 expression correlate with cell death. (A) The incidence of cell death of primary MEFs and different human tumor cell lines was assessed after 48 h of treatment with DFO by measuring uptake of PI, which was taken up only by dying cells. Wt, wild type. (B) Western blot analysis of BNIP3 induction in Saos2 and U2OS cells by hypoxia, DMOG treatment, or DFO treatment. (C) Western blot analysis of BNip3 induction in wild-type and Rb null MEFs by DFO treatment or hypoxia. UT, untreated. (D) Saos2 cells were transiently transfected with pIRES-GFP or pRB-IRES-GFP and treated with DFO for 48 h. The incidence of cell death was determined by flow cytometric analysis of PI uptake. (E) The effect of pRB overexpression for BNIP3 promoter activity was determined for Saos2 cells by a luciferase assay for reporter gene expression. Results presented are the averages of at least three independent experiments. RLU, relative luciferase units. (F and G) The effect of pRB expression on the levels of BNIP3 protein was determined by immunofluorescent staining for BNIP3 (red) in cells that expressed GFP by virtue of transfection with either pIRES-GFP (F) or pRB-IRES-GFP (G). DAPI, 4′,6′-diamidino-2-phenylindole.
Reintroduction of pRB into RB-deficient Saos2 cells reduced the overall levels of cell death in DFO-treated cultures to those observed for untreated control cultures (Fig. 4D). Furthermore, overexpression of pRB in Saos2 cells attenuated induction of the BNIP3 promoter by DFO in a dose-dependent manner, as determined by reporter gene activity (Fig. 4E), and reduced the levels of BNIP3 protein induced by DFO treatment (Fig. 4G). These results show that the sensitivity of Saos2 cells to PHD inhibition could be abrogated by pRB and was associated with increased BNIP3 levels due to derepression of the BNIP3 promoter.
Cell death induced by PHD inhibition is nonapoptotic and is associated with ATP depletion.
We set out to determine whether the cell death taking place in Rb null MEFs (Fig. 5) or RB-deficient Saos2 cells (Fig. 6) treated with PHD inhibitors was nonapoptotic, as was the case with the developing FL. Rb null MEFs were markedly more sensitive to cell death induced by treatment with DFO (Fig. 5A) than were wild-type MEFs, but when we examined whether this death could be blocked by pretreatment with a generic caspase inhibitor (z-vad-fmk) we failed to block death, indicating that death was not caspase dependent (Fig. 5B). Control cultures of Rb null MEFs induced to undergo apoptosis by treatment with TNF-α were protected by pretreatment with z-vad-fmk (Fig. 5B). When we examined the morphology of DFO-treated Rb null MEFs by electron microscopy (Fig. 5C), we observed that dying cells showed increased levels of vesicle formation compared to those of wild-type MEFs, and we also observed features of autophagic cell death. Redistribution of LC3 from diffuse expression into a punctate pattern that colocalizes with autophagosomes is widely used as a marker of autophagy (12, 22), and on examining the pattern of GFP-LC3 staining in untreated and DFO-treated MEFs, we observed that DFO induced punctate LC3 focus formation much more extensively in DFO-treated RB null MEFs than in wild-type untreated MEFs (Fig. 5D). These results suggest that Rb null MEFs undergo autophagic cell death in response to DFO treatment.
FIG. 5.
Nonapoptotic cell death of Rb null MEFs. (A) Wild-type and Rb null MEFs were cultured in the presence of 260 μM DFO for 48 h, and the viability of MEFs was compared to that of untreated controls as determined by flow cytometric analysis of PI uptake. (B) DFO-induced cell death of Rb null MEFs is resistant to the pancaspase inhibitor z-vad-fmk, in contrast to death induced by TNF-α and cycloheximide. (C) Electron microscopy of untreated and DFO-treated Rb null MEFs reveals small double-membrane-bound vesicles in hypoxic cells (red arrow), indicative of autophagy. (D) GFP-LC3 staining of untreated and DFO-treated (48 h) wild-type and Rb null MEFs reveals increased punctate GFP-LC3 staining in Rb null MEFs, consistent with increased autophagy.
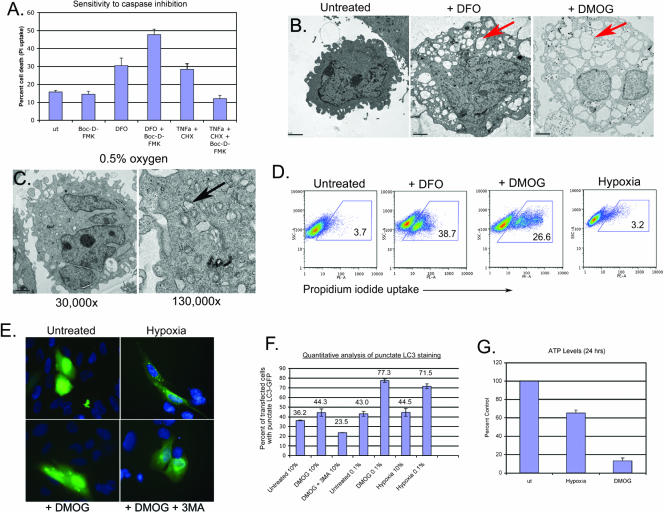
FIG. 6.
Nonapoptotic cell death induced by PHD inhibition. (A) DMOG-induced cell death of Saos2 cells is resistant to the pancaspase inhibitor Boc-D-FMK, in contrast to death induced by TNF-α and cycloheximide. (B) Electron microscopy of Saos2 revealed extensive vacuolation in DFO- and DMOG-treated cells (red arrow) compared to the level of vacuolation seen with untreated Saos2 cells. (C) Electron microscopy of Saos2 cells revealed small double-membrane-bound vesicles in hypoxic cells (black arrow), indicative of autophagy. The vacuolation observed with DMOG (B) is not apparent. (D) Viability of Saos2 cells exposed to 260 μM DFO, 1 mM DMOG, or 0.5% oxygen (hypoxia) for 48 h was determined by flow cytometric analysis of PI uptake. (E) GFP-LC3 staining of untreated Saos2 cells or Saos2 cells exposed to 0.5% oxygen, 1 mM DMOG, or 1 mM DMOG plus 3-methyladenine (3MA). (F) The incidence of autophagy in either hypoxic or DMOG-treated Saso2 cell cultures was quantified microscopically by counting the number of cells that exhibited punctate GFP-LC3 staining and expressing that as a percentage of the total number of cells that expressed GFP-LC3 (punctate and diffuse). (G) ATP levels in Saos2 cells were determined after treatment with DMOG or exposure to 0.5% oxygen for 24 h.
Similarly, we observed that cell death induced by DFO treatment of Saos2 cells was not blocked by the generic caspase inhibitor Boc-D-FMK, unlike death induced by treatment with TNF-α and cycloheximide (Fig. 6A). Similarly, DMOG-induced cell death was resistant to caspase inhibitors (data not shown). Saos2 cells exhibited extensive cytoplasmic vacuolation following 48 h of treatment with either DFO or DMOG (Fig. 6B), an enlarged nucleus, and increased cell volume in the absence of chromatin condensation (Fig. 6B), consistent with nonapoptotic cell death, such as that observed with the Rb null FL (Fig. 1). Interestingly, DFO-induced death of Rb null MEFs also was accompanied by cytoplasmic vacuolation and other features of nonapoptotic cell death (Fig. 5C). In contrast, hypoxic Saos2 cells did not show evidence of abnormal vacuolation (Fig. 6C), and surprisingly, hypoxic Saos2 cells did not undergo cell death (Fig. 6D), again in contrast to the effects of DFO or DMOG. At a higher magnification, double-membrane vesicles were apparent in hypoxic Saos2 cells (Fig. 6C), suggesting that Saos2 cells induced autophagy in response to hypoxia. These observations indicate that Saos2 cells undergo autophagy in response to hypoxia but undergo nonapoptotic cell death in response to DFO or DMOG.
To determine whether the vesicles observed in hypoxic cells (Fig. 6C) were likely to be autophagosomes, we examined the effect of hypoxia on the localization of LC3 (Fig. 6E). Exposure of Saos2 cells to 0.5% oxygen or treatment with DMOG induced punctate GFP-LC3 staining in the cytoplasm (Fig. 6E) that was potentiated by serum starvation (Fig. 6F) and blocked by 3-methyladenine (Fig. 6E and F), consistent with cells undergoing autophagy in response to both hypoxia and PHD inhibition. Quantitative analysis of GFP-LC3 puncta confirmed that hypoxia and DMOG treatments induced autophagy to similar extents, and they did so in a manner that synergized with serum starvation (Fig. 6F).
Nonapoptotic cell death is associated with ATP depletion, resulting in apoptosome inhibition and reactive oxygen species (ROS) accumulation (56). When we measured ATP levels in Saos2 cells exposed to either 0.5% oxygen or 1 mM DMOG, we observed a much more dramatic depletion of ATP in DMOG-treated cells than in cells grown under hypoxic conditions (Fig. 6G). Thus, nonapoptotic cell death induced by PHD inhibition is associated with increased ATP depletion.
These results indicated that hypoxia and PHD inhibition have similar effects in terms of BNIP3 induction and the incidence of autophagy. However, PHD inhibition had the added effect of promoting ATP depletion and nonapoptotic cell death of RB-deficient cells.
BNIP3 is required for hypoxia-induced autophagy and is not sufficient for cell death induced by DFO/DMOG.
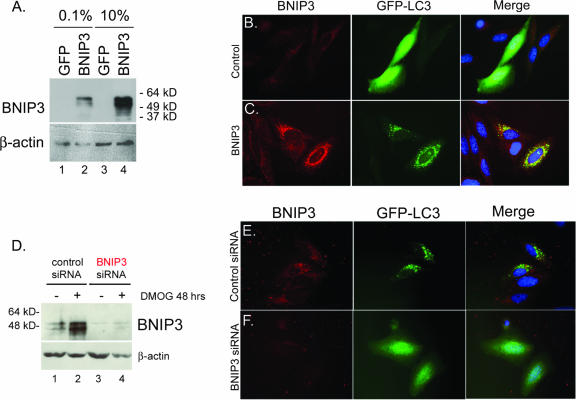
Given the ability of hypoxia, DFO, and DMOG to induce BNIP3 (Fig. 4B) and to promote autophagy (Fig. 5D and 6E), we sought to determine whether BNIP3 was required or was sufficient for the induced autophagy. BNIP3 overexpression (Fig. 7A) induced a marked increase in the amount of LC3 puncta in BNIP3-expressing cells following serum starvation (Fig. 7C; also see Fig. S3B in the supplemental material) compared to that in control transfected cells (Fig. 7B; also see Fig. S3A in the supplemental material). Consistent with the role of serum deprivation in promoting hypoxia-induced autophagy (Fig. 6F), the ability of BNIP3 to promote autophagy required serum deprivation. Furthermore, knockdown of BNIP3 expression (Fig. 7D and F) inhibited punctate LC3 staining following exposure to 1 mM DMOG treatment (Fig. 7F) or to 0.5% oxygen (see Fig. S3D in the supplemental material). These results demonstrate that BNIP3 is essential for autophagy induced by either hypoxia or DMOG treatment.
FIG. 7.
BNIP3 is required for hypoxia-induced autophagy. (A) Western blot analysis of BNIP3 in lysates from Saos2 cells that had been transfected with the control plasmid (pIRES-GFP) or with the BNIP3 expression plasmid (pBNIP3-IRES-GFP) under conditions of low-concentration (0.1%) or high-concentration (10%) serum. (B and C) The effect of BNIP3 overexpression (C, red) for the incidence of punctate GFP-LC3 staining (green) was assessed in Saos2 cells (grown in low-concentration serum) and compared to that observed in control cultures transfected with empty vector (B). (D) Western blot analysis of BNIP3 in lysates from Saos2 cells that had been transfected with either control siRNA or BNIP3 siRNA from untreated cells or cells treated with DMOG for 48 h. (E and F) Knockdown of BNIP3 (F, red) in hypoxic, serum-starved Saos2 cells blocked autophagy, as determined by loss of punctate GFP-LC3 staining (green), whereas no effect was observed with control siRNA.
Previous reports have presented evidence that overexpression of BNIP3 was sufficient to induce cell death (14, 25), while others demonstrated that BNIP3 was not sufficient to kill cells without further posttranslational modifications (15, 26, 39). The fact that hypoxia and PHD inhibitors induced BNIP3 expression and autophagy to equivalent levels (Fig. 4B and C and 6E) but that DFO/DMOG additionally promoted nonapoptotic cell death (Fig. 4A and 6D) suggested to us that BNIP3 primarily was involved in regulating autophagy as a survival mechanism, as opposed to regulating cell death. When we overexpressed BNIP3 in cells in the presence or absence of serum (Fig. 8A and B), we failed to observe any decrease in viability compared to that of control transfected cells. Similar results were obtained with U2OS and MCF-7 cells (see Fig. S4 in the supplemental material), which previously had been reported to be sensitive to cell death induced by BNIP3 overexpression (25). Thus, our results demonstrate that BNIP3 expression is not sufficient to induce cell death.
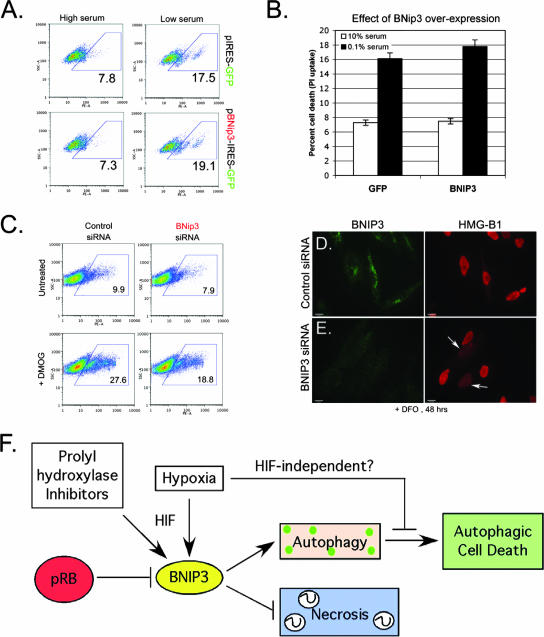
FIG. 8.
BNIP3 is necessary, but not sufficient, for cell death induced by DFO or DMOG. (A and B) The effect of BNIP3 overexpression on the incidence of cell death induced by 48 h of DFO treatment, in the presence or absence of serum, was determined by flow cytometric analysis of PI uptake. (C) Flow cytometric analysis of the effect of BNIP3 knockdown on cell death induced by PHD inhibition, as determined by PI uptake. (D and E) Immunofluorescent staining for HMG-B1 release from DFO-treated cells that had been transfected with either control siRNA (D) or BNIP3 siRNA (E) reveals increased release of the nuclear protein HMG-B1 from cells in which BNIP3 is knocked down (E, arrows). (F) Model summarizing the role of RB and BNIP3 in responses to hypoxia and the chemical inhibitors of PHDs (DFO and DMOG).
To determine whether BNIP3 was required for DMOG-induced cell death, even if it was not sufficient to cause cell death by itself, we examined the effect of knocking down BNIP3 expression in Saos2 cells, as depicted in Fig. 7D. We observed that knockdown of BNIP3 reduced the level of cell death induced by DMOG treatment (18.8%) compared to that of the controls (27.7%). Given that previous research found that inhibiting autophagy promoted necrosis (7, 12), we speculated that the reason knockdown of BNIP3 was not achieving a more dramatic rescue of cell death was that the loss of BNIP3 and autophagy (Fig. 7F), while preventing autophagic cell death, was now inducing necrotic cell death. To determine if blocking autophagy by knocking down BNIP3 (Fig. 7F) had switched the type of cell death taking place to necrotic cell death, we examined the cellular localization of the nuclear protein HMG-B1. Under conditions of apoptosis or autophagy, HMG-B1 remains tightly associated with chromatin and localized to the nucleus (43). However, under conditions of necrosis, HMG-B1 is released from the nucleus and exhibits increased cytoplasmic and extracellular staining (55). When we examined Saos2 cells that had been treated with DFO for 48 h for release of HMG-B1, we observed that cultures in which BNIP3 had been knocked down showed higher numbers of cells that had released HMG-B1 from the nucleus (Fig. 8E, arrows) than DFO-treated cultures that had been transfected with the control siRNA (Fig. 8D). These results suggested that the reason knockdown of BNIP3 did not appear to protect cells completely from death was due to the requirement for BNIP3 and autophagy to prevent necrotic death. Thus, BNIP3 levels appear to modulate the balance between autophagy and autophagic cell death and also between autophagy and necrosis. Too much BNIP3, such as that observed in cells lacking RB, causes autophagic death, whereas too little BNIP3, such as that observed in cells in which BNIP3 is knocked down, leads to necrosis.
In summary, we have shown that BNIP3 is superinduced by hypoxia in RB-deficient cells, that BNIP3 is a direct target for transcriptional repression mediated by pRB/E2F, and that pRB acts to attenuate induction of BNIP3 by HIF. BNIP3 was required for autophagy induced by hypoxia or PHD inhibition, and knockdown of BNIP3 switched the type of cell death induced by treatment with PHD inhibitors from autophagic to necrotic.
DISCUSSION
We previously demonstrated a critical role for the Rb tumor suppressor in protecting against apoptotic cell death in the developing nervous system (31), and here we identify a novel function for pRb in protecting against nonapoptotic cell death in the developing FL. We identify BNIP3 as a direct target of transcriptional repression by pRB/E2F and show that BNIP3 is required for autophagy induced by hypoxia and PHD inhibition and that loss of BNIP3 resulted in necrotic cell death, consistent with previous reports that inhibiting autophagy promoted necrosis (7, 12).
The role of pRb in protecting against apoptosis has been attributed to its role in repressing expression of critical cell death regulatory genes, such as those encoding Apaf-1, caspases, and p73 (19, 35, 37). We have identified a novel role for pRb in regulating nonapoptotic cell death and identify BNip3 as a target gene involved in promoting autophagy and nonapoptotic cell death. Various reports have commented on the difficulty of detecting pRB bound to E2F target gene promoters in vivo (48), and we speculate that pRB may associate only with E2F-regulated promoters under specific stress conditions, such as hypoxia, oxidative stress, and DNA damage. Our data show that E2F-1, and not E2F-3, is associated with the BNIP3 promoter under stress conditions, and previous studies have pointed to a unique role for E2F-1 in promoting cell death in response to certain stresses (9, 10, 27). We have shown that E2F-1 displaces E2F-4 as the major E2F bound to the BNIP3 promoter under such conditions and that recruitment of pRB to the BNIP3 promoter is associated with E2F-1 binding (Fig. 3H). Intriguingly, displacement of E2F-4 from the p73 promoter by E2F-1 in response to DNA damage was reported to be dependent on E2F-1 acetylation (41). Similarly, selective phosphorylation of E2F-1 by ATM (28) and Chk2 (47) may explain aspects of its unique role in cell death induced by DNA damage. Thus, E2F-1 and pRB may play unique roles in cell death due to posttranslational modifications induced by specific stresses that target them to stress response genes, such as BNIP3.
We have demonstrated that pRB/E2F interacts functionally with HIF-1α to regulate the BNIP3 promoter. Specifically, pRB/E2F-1 bound to the E2F site acted to reduce the level of induction of BNIP3 transcription promoted by HIF-1α interaction with the HRE in the BNIP3 promoter. Similarly, HIF-1α bound at the HRE appeared to limit the responsiveness of the promoter to E2F-1 overexpression. We were unable to detect any direct interaction between HIF-1α and either pRB or E2F-1. Furthermore, the ability of pRB to interact with the BNIP3 promoter was dependent on the E2F site and was not blocked by mutation of the HRE. Thus, we speculate that other factors, possibly chromatin remodeling enzymes, act as intermediaries in the functional interaction between pRB/E2F-1 and HIF-1α in the BNIP3 promoter (Fig. 3H).
We also postulate that the functional interaction between pRB/E2F and HIF that occurs at the BNIP3 promoter is a conserved transcriptional motif that may explain the attenuated expression of other hypoxia-regulated genes in normal cells that is not seen in tumor cells. Interestingly, E2Fs recently have been implicated in the hypoxia-induced down-regulation of BRCA1 and Rad51 expression (1, 2). Our preliminary analysis of published array-based data for the purpose of finding E2F-regulated genes (42) that are also known HIF target genes (44) has identified other genes that also may be coordinately regulated by E2Fs and HIF, including the genes encoding VEGF, the transferrin receptor, Stra-13/Dec-1, telomerase reverse transcriptase, heme oxygenase, and others (K. Tracy, B. J. C. Dibling, and K. F. Macleod, unpublished data). The significance of this is not clear, although several of those genes that we predict to be regulated in this manner have characterized functions in tumorigenesis, such as the VEGF gene (16).
The role of BNIP3 in cell death, autophagic or necrotic, is controversial, with several groups proposing a role for BNip3 as an inducer of cell death (25, 51) but others showing that BNIP3 expression was not sufficient to kill cells; our data agree with the findings of the latter studies (26, 39). How might the differences in the reported activity of BNIP3 be reconciled? We noted that the methods used to measure cell viability in earlier studies consisted of counting nuclei that stained positive with acridine orange (25) or involved clonogenic assays (34), but given that autophagy promotes uptake of acridine orange by acidic vesicles and induces growth arrest, these assays may reflect increased autophagy as opposed to cell death. The BH3 domain of BNIP3 is highly divergent, with only 3 out of 11 amino acids conserved, in contrast to BIM, for example, in which 6 out of 11 amino acids in its BH3 domain are conserved relative to those in other Bcl-2 family members (54). This divergence in primary sequence may explain the inability of BNIP3 to kill cells when overexpressed. Thus, additional signals may be required to induce the killing potential of BNIP3. For instance, acidosis was required to activate BNIP3 protein to kill ischemic cardiomyocytes (26), although the mechanistic basis of this still is not understood.
Autophagy has been proposed to inhibit tumor progression by promoting growth arrest, by preventing ROS and DNA damage through elimination of oxidized molecules and organelles (particularly mitochondria), and by preventing necrosis and associated inflammatory responses (6, 12, 24). BNIP3 expression is induced during early stages of human cancer (36, 38, 45) and is down-regulated at late stages, coincident with progression to metastasis (34, 38, 45). In addition to inhibiting tumor progression through the prevention of DNA damage associated with accumulation of oxidized proteins and damaged organelles or through its ability to promote growth arrest and autophagic cell death (24), we suggest that autophagy also may have a role in blocking tumor progression by preventing necrotic cell death (Fig. 8F). Our data show that inhibition of autophagy associated with BNIP3 expression resulted in necrosis. Thus, we predict that autophagy in vivo will delay the onset of necrosis and inflammation that promote metastasis (50). Our work shows that the RB tumor suppressor modulates the levels of BNIP3 in cells, and we propose that this allows the cell to engage in effective autophagy in response to hypoxia and nutrient deprivation. In other words, by attenuating the induction of BNIP3 by HIF, pRB is maintaining BNIP3 levels within a range that promotes autophagy and prevents both autophagic cell death (too much BNIP3) and necrotic cell death (too little BNIP3). This is consistent with its role as a tumor suppressor, since autophagy promotes reduced growth rates as well as the elimination of oxidized molecules and organelles that promote DNA damage (8, 20, 21).
In summary, our work has identified a key role for BNIP3 downstream of the RB tumor suppressor in regulating the levels of autophagy induced by hypoxia and nutrient stress.
Supplementary Material
Acknowledgments
We thank Nic La Thangue for providing the pdhfr-luciferase reporter gene construct and Xin Lu for the E2F-1 overexpression plasmid.
Work on this project was supported by an American Cancer Society institutional pilot project grant.
Footnotes
Published ahead of print on 18 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bindra, R. S., S. L. Gibson, A. Meng, U. Westermark, M. Jasin, A. J. Pierce, R. G. Bristow, M. K. Classon, and P. M. Glazer. 2005. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 65:11597-11604. [DOI] [PubMed] [Google Scholar]
- 2.Bindra, R. S., and P. M. Glazer. 2007. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene 26:2048-2057. [DOI] [PubMed] [Google Scholar]
- 3.Bruick, R. K. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA 97:9082-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budde, A., N. Schneiderhan-Marra, G. Petersen, and B. Brune. 2005. Retinoblastoma susceptibility product pRB activates hypoxia-inducible factor (HIF-1). Oncogene 24:1802-1808. [DOI] [PubMed] [Google Scholar]
- 5.Chau, B. N., and J. Y. J. Wang. 2003. Coordinated regulation of life and death by RB. Nat. Rev. Cancer 3:130-138. [DOI] [PubMed] [Google Scholar]
- 6.Degenhardt, K., G. Chen, T. Lindsten, and E. White. 2002. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cells 2:193-203. [DOI] [PubMed] [Google Scholar]
- 7.Degenhardt, K., R. Mathew, B. Beaudoin, K. Bray, K. L. Anderson, G. Chen, C. Mukherjee, Y. Shi, C. Gelinas, Y. Fan, D. A. Nelson, S. Jin, and E. White. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation and tumorigenesis. Cancer Cells 10:51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGregori, J. 2002. The genetics of the E2F family of transcription factors. Biochim. Biophys. Acta 1602:131-150. [DOI] [PubMed] [Google Scholar]
- 9.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denchi, E. L., and K. Helin. 2005. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 6:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson, K. L., J. L. Vanderluit, J. M. Hebert, W. C. McIntosh, E. Tibbo, J. G. MacLaurin, D. S. Park, V. A. Wallace, M. Vooijs, S. K. McConnell, and R. S. Slack. 2002. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 21:3337-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gozuacik, D., and A. Kimchi. 2004. Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23:2891-2906. [DOI] [PubMed] [Google Scholar]
- 13.Green, D. R., and G. I. Evan. 2002. A matter of life and death. Cancer Cells 1:19-30. [DOI] [PubMed] [Google Scholar]
- 14.Guo, K., G. Searfoss, D. Krolikowski, M. Pagnoni, C. Franks, K. Clark, K. T. Yu, M. Jaye, and Y. Ivashchenko. 2001. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Diff. 8:367-376. [DOI] [PubMed] [Google Scholar]
- 15.Hamacher-Brady, A., N. R. Brady, S. E. Logue, M. R. Sayen, M. Jinno, L. A. Kirschenbaum, R. A. Gottlieb, and A. B. Gustafsson. 2006. Response to myocardial ischemia/reperfusion injury involves BNip3 and autophagy. Cell Death Diff. 13:1-12. [DOI] [PubMed] [Google Scholar]
- 16.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 17.Hershko, T., and D. Ginsberg. 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F-1 mediates apoptosis. J. Biol. Chem. 279:8627-8634. [DOI] [PubMed] [Google Scholar]
- 18.Ho, A. T., Q. H. Li, R. Hakem, T. W. Mak, and E. Zacksenhaus. 2004. Coupling of caspase-9 to Apaf1 in response to loss of pRb or cytotoxic drugs is cell-type specific. EMBO J. 23:460-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelam, D. I. Smith, W. Liu, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin. 2000. Role for the p53 homologue p73 in E2F-1 induced apoptosis. Nature 407:645-648. [DOI] [PubMed] [Google Scholar]
- 20.Jin, S. 2006. Autophagy, mitochondrial quality control and oncogenesis. Autophagy 2:80-84. [DOI] [PubMed] [Google Scholar]
- 21.Jin, S., and E. White. 2007. Role of autophagy in cancer. Autophagy 3:28-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. Y., H. J. Ahn, J. H. Ryu, K. Suk, and J. H. Park. 2004. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia inducible factor 1a. J. Exp. Med. 199:113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, Y., T. Kanzawa, R. Sawaya, and S. Kondo. 2005. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 5:726-734. [DOI] [PubMed] [Google Scholar]
- 25.Kothari, S., J. Cizeau, E. McMillan-Ward, S. J. Israels, M. Bailes, K. Ens, L. A. Kirshenbaum, and S. B. Gibson. 2003. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene 22:4734-4744. [DOI] [PubMed] [Google Scholar]
- 26.Kubasiak, L. A., O. M. Hernandez, N. H. Bishopric, and K. A. Webster. 2002. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc. Natl. Acad. Sci. USA 99:12825-12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C. Park, P. Giangrande, L. Wu, H. I. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. H. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8:105-113. [DOI] [PubMed] [Google Scholar]
- 28.Lin, W., F. Lin, and J. R. Nevins. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15:1833-1844. [PMC free article] [PubMed] [Google Scholar]
- 29.Lum, J. J., D. E. Bauer, M. Kong, M. H. Harris, C. Y. Li, T. Lindsten, and C. B. Thompson. 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120:237-249. [DOI] [PubMed] [Google Scholar]
- 30.Lum, J. J., R. J. DeBerardins, and C. B. Thompson. 2005. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Biol. 6:439-448. [DOI] [PubMed] [Google Scholar]
- 31.Macleod, K., Y. Hu, and T. Jacks. 1996. Loss of Rb activates both p53-dependent and -independent cell death pathways in the developing mouse nervous system. EMBO J. 15:6178-6188. [PMC free article] [PubMed] [Google Scholar]
- 32.Macleod, K., and M. Plumb. 1991. Derepression of mouse β-globin gene transcription during erythroid differentiation. Mol. Cell. Biol. 11:4324-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacPherson, D., J. Sage, D. Crowley, A. Trumpp, R. T. Bronson, and T. Jacks. 2003. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 23:1044-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manka, D., Z. Spicer, and D. E. Millhorn. 2005. Bcl-2/adenovirus E1B 19 kDa interacting protein-3 knockdown enables growth of breast cancer metastases in the lung, liver and bone. Cancer Res. 65:11689-11693. [DOI] [PubMed] [Google Scholar]
- 35.Moroni, M. C., E. S. Hickman, E. L. Denchi, G. Caprara, E. Colli, F. Cecconi, H. Muller, and K. Heliin. 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3:552-558. [DOI] [PubMed] [Google Scholar]
- 36.Murai, M., M. Toyota, H. Suzuki, A. Satoh, Y. Sasaki, K. Akino, M. Ueno, F. Takahashi, M. Kusano, H. Mita, K. Yanagihara, T. Endo, Y. Hinoda, T. Tokino, and K. Imai. 2005. Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin. Cancer Res. 11:1021-1027. [PubMed] [Google Scholar]
- 37.Nahle, Z., J. Polakoff, R. V. Davuluri, M. E. McCurrach, M. D. Jacobson, M. Narita, M. Q. Zhang, Y. Lazebnik, D. Bar-Sagi, and S. W. Lowe. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4:859-864. [DOI] [PubMed] [Google Scholar]
- 38.Okami, J., D. M. Simeone, and C. D. Logsdon. 2004. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 64:5338-5346. [DOI] [PubMed] [Google Scholar]
- 39.Papandreou, I., C. Krishna, F. Kaper, D. Cai, A. J. Giaccia, and N. C. Denko. 2005. Anoxia is necessary for tumor cell toxicity caused by a low-oxygen environment. Cancer Res. 65:3171-3178. [DOI] [PubMed] [Google Scholar]
- 40.Pattingre, S., A. Tassa, X. Qu, R. Garuti, X. H. Linag, N. Mizushima, M. Packer, M. D. Schneider, and B. Levine. 2005. Bcl-2 anti-apoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927-939. [DOI] [PubMed] [Google Scholar]
- 41.Pediconi, N., A. Ianari, A. Costanzo, L. Belloni, R. Gallo, L. Cimino, A. Porcellini, I. Screpanti, C. Balsano, E. Alesse, A. Gulino, and M. Levrero. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5:552-558. [DOI] [PubMed] [Google Scholar]
- 42.Polager, S., Y. Kalma, E. Berkovich, and D. Ginsberg. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21:437-446. [DOI] [PubMed] [Google Scholar]
- 43.Scaffidi, P., T. Misteli, and M. E. Bianchi. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418:191-195. [DOI] [PubMed] [Google Scholar]
- 44.Semenza, G. L. 2003. Targeting HIF for cancer therapy. Nat. Rev. Cancer 3:721-732. [DOI] [PubMed] [Google Scholar]
- 45.Sowter, H. M., M. Ferguson, C. Pym, P. Watson, S. B. Fox, C. Han, and A. L. Harris. 2003. Expression of the cell death genes BNip3 and Nix in ductal carcinoma in situ of the breast; correlation of BNip3 levels with necrosis and grade. J. Pathol. 201:573-580. [DOI] [PubMed] [Google Scholar]
- 46.Spike, B. T., A. Dirlam, B. C. Dibling, J. Marvin, B. O. Williams, T. Jacks, and K. F. Macleod. 2004. The Rb tumor suppressor is required for stress erythropoiesis. EMBO J. 23:4319-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens, C., L. Smith, and N. B. La Thangue. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5:401-409. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 49.Trimarchi, J., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 50.Vakkila, J., and M. T. Lotze. 2004. Inflammation and necrosis promote tumour growth. Nat. Rev. Immunol. 4:641-648. [DOI] [PubMed] [Google Scholar]
- 51.Vande Velde, C., J. Cizeau, D. Dubik, J. Alimonti, T. Brown, S. Israels, R. Hakem, and A. H. Greenberg. 2000. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 20:5454-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, L., A. de Bruin, H. I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J. C. Thompson, M. C. Ostrowski, T. J. Rosol, L. A. Woollett, M. Weinstein, J. C. Cross, M. L. Robinson, and G. Leone. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421:942-947. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida, H., K. Kawane, M. Koike, Y. Mori, Y. Uchiyama, and S. Nagata. 2005. Phosphotidlyserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature 437:754-758. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, H. M., P. Cheung, B. Yanagawa, B. M. McManus, and D. C. Yang. 2003. BNips: a group of pro-apoptotic proteins in the Bcl-2 family. Apoptosis 8:229-236. [DOI] [PubMed] [Google Scholar]
- 55.Zong, W., D. Ditsworth, D. E. Bauer, Z. Q. Wang, and C. B. Thompson. 2004. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 18:1272-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zong, W., and C. B. Thompson. 2006. Necrotic death as a cell fate. Genes Dev. 20:1-15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.