Abstract
RGS proteins constitute a newly appreciated and large group of negative regulators of G protein signaling. Four members of the RGS family act as GTPase-activating proteins (GAPs) with apparent specificity for members of the Giα subfamily of G protein subunits. We demonstrate here that two RGS proteins, RGS4 and GAIP, also act as GAPs for Gqα, the Gα protein responsible for activation of phospholipase Cβ. Furthermore, these RGS proteins block activation of phospholipase Cβ by guanosine 5′-(3-O-thio)triphosphate-Gqα. GAP activity does not explain this effect, which apparently results from occlusion of the binding site on Gα for effector. Inhibitory effects of RGS proteins on G protein-mediated signaling pathways can be demonstrated by simple mixture of RGS4 or GAIP with plasma membranes.
The α subunits of signal-transducing, heterotrimeric G proteins are molecular switches and clocks, and these functions are imparted to the proteins by conformational changes that result from the binding and hydrolysis of GTP. Gα proteins are inactive as GDP-bound species because of reduced affinity (compared with the GTP-bound protein) for downstream effectors and, in addition, enhanced affinity for the G protein βγ subunit complex. Binding of Gα-GDP to βγ occludes sites for interaction with downstream effectors on both α and βγ. G proteins are activated by appropriate plasma membrane-bound, heptahelical receptors, which catalyze exchange of GDP for GTP, and they are deactivated as a result of their intrinsic GTPase activity.
The last few years have brought heightened appreciation of regulation of Gα-catalyzed GTP hydrolysis by proteins known as GTPase-activating proteins (GAPs). Such GAP activities were first demonstrated with certain effectors for G protein action—notably phospholipase Cβ and the γ subunit of retinal cyclic GMP phosphodiesterase (1–7). Genetic studies (particularly in yeast and worms) have now resulted in discovery of a large family (at least 20 members in mammals) of negative regulators of G protein signaling (RGS proteins), and biochemical characterization of a few of the family members has demonstrated that they, too, act as GAPs, especially toward members of the Gi subfamily of Gα proteins (8, 9).
Mammalian RGS proteins are presumed to play an important role in the down-regulation or desensitization of G protein-mediated signaling pathways, as one such protein (Sst2p) does in yeast (10–16). However, much remains to be learned, including such obvious and crucial questions as mechanisms of acceleration of GTPase activity, cellular distribution of the family members, the specificity of their interactions with Gα subunits, mechanisms of regulation of RGS protein activity, and the importance of RGS protein-mediated inhibition of G protein signaling compared with other mechanisms (particularly receptor-directed kinases) that have been documented over the past two decades.
In the relatively few reports that have appeared to date, RGS proteins have been shown to interact preferentially with members of the Gi subfamily of α subunits and to have no apparent effect on the GTPase activity of Gsα or G12α, representatives of two of the remaining three subfamilies of Gα proteins (17–20). Technical complexities have delayed examination of interactions of RGS proteins with members of the important Gq class of α subunits. We report here that two RGS proteins, RGS4 and GAIP, accelerate the GTPase activity of Gqα and, furthermore, that RGS proteins can interfere directly with the interactions of activated G protein α subunits with at least one effector, apparently inhibiting G protein function by acting as effector antagonists as well as GAPs.
MATERIALS AND METHODS
Preparation of Proteins.
Recombinant RGS4 and GAIP were synthesized in Escherichia coli and purified as described (17, 18). Recombinant Gqα and Gzα were purified after expression in Sf9 cells (21, 22), while myristoylated Goα was purified after expression in E. coli (23). Phospholipase Cβ1 was purified from bovine brain (24).
Measurement of GAP Activity.
Values of Km and Vmax for RGS4 and GAIP were determined with GTP-Goα as substrate (18). Briefly, myristoylated Goα was incubated with 10 μM [γ-32P]GTP and 10 mM EDTA at 20°C for 20 min to prepare substrate. Varying concentrations of GTP-Goα were then incubated at 4°C for 5 min prior to addition of 15 mM MgSO4, 150 μM GTP, and either 2.2 nM RGS4 or 49 nM GAIP. Aliquots of this reaction mixture were withdrawn at 10-sec intervals during the first 40 sec, and the reaction was stopped by addition of 5% (wt/vol) Norit A charcoal in 50 mM NaH2PO4. The supernatant containing 32Pi was counted, and the initial rate of GTP hydrolysis was analyzed as a function of substrate (GTP-Goα) concentration. Similar assays were performed at a fixed concentration of GTP-Gzα (18).
Reconstitution of M1 Muscarinic Receptors and Gqα or Gqα and Phospholipase Cβ1.
M1 muscarinic receptors and phospholipase Cβ1 were purified from baculovirus-infected Sf9 cells as described (2) and generously provided by Gloria Biddlecome and Elliott Ross (this department). Gqα and the G protein β1γ2 complex were purified from baculovirus-infected Sf9 cells as before (21). M1 receptors, Gqα, and βγ were reconstituted into phospholipid vesicles as described, and receptor-dependent, steady-state GTP hydrolysis by Gqα was measured (Fig. 1) (1, 2, 22). Assays (50 μl total volume) were performed with 5 μM [γ32P]GTP (6000 cpm/pmol) for 8 min at 30°C, and phospholipid vesicles contained 7 fmol of the M1 receptor and 24 fmol of Gqα per assay. The capacity of guanosine 5′-(3-O-thio)triphosphate (GTPγS)-Gqα to activate phospholipase Cβ1 was assessed as described previously (see Fig. 3) (22).
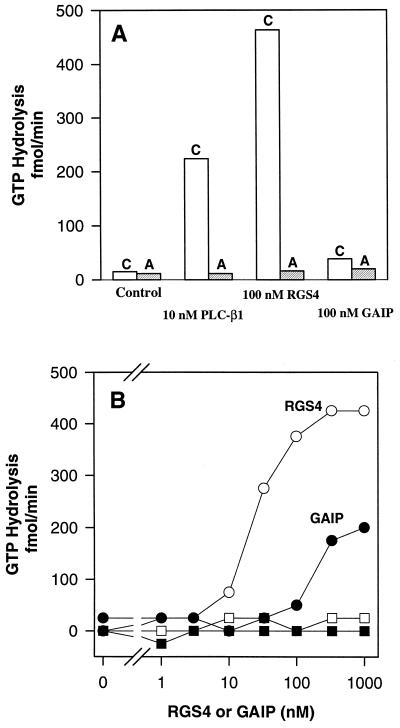
Figure 1.
Activation of the GTPase activity of Gqα by RGS4 and GAIP. M1 muscarinic cholinergic receptors and αqβ1γ2 were reconstituted into phospholipid vesicles as described in Materials and Methods. The indicated concentrations of RGS4, GAIP, or phospholipase Cβ1 were then added to the vesicles, and steady-state GTPase activity was assayed for 8 min at 30°C. (A) Assays were performed in the presence of 1 mM carbachol (C, open bar) or 10 μM atropine (A, shaded bar). (B) Assays were performed in the presence of the indicated concentrations of RGS4 (open symbols) or GAIP (filled symbols) and 1 mM carbachol (circles) or 10 μM atropine (squares).
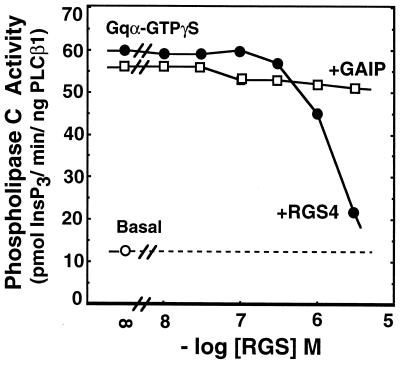
Figure 3.
The effect of RGS4 and GAIP on activation of phospholipase Cβ1 by GTPγS-Gqα. Purified Gqα was activated with 1 mM GTPγS for 1 h at 30°C. Activated Gqα (10 nM) was then mixed with purified phospholipase Cβ1 (1 ng) and [3H]phosphatidylinositol-4,5-bisphosphate-containing phospholipid vesicles in the presence of the indicated concentrations of RGS4 or GAIP. Synthesis of [3H]inositol 1,4,5-trisphosphate was measured.
Preparation of Cell Membranes.
Confluent NG-108 cells were grown at 37°C in DMEM supplemented with 10% fetal calf serum, 0.1 mM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine and harvested in buffer containing 50 mM Na·Hepes (pH 8), 1 mM EDTA, 150 mM NaCl, and 0.1 mM phenylmethylsulfonyl fluoride. Homogenates were prepared by nitrogen cavitation and centrifuged at 500 × g to remove nuclei and unbroken cells. Supernatants were centrifuged at 100,000 × g, and membranes were suspended in buffer A (50 mM Na·Hepes, pH 7.2/1 mM EDTA/3 mM EGTA/5 mM MgCl2/150 mM NaCl/2 mM dithiothreitol/0.1 mM phenylmethylsulfonyl fluoride) prior to storage at −80°C.
Measurement of Phospholipase C Activity in NG-108 Cell Membranes.
Reactions were carried out as described (22, 25, 26) in a final volume of 60 μl. NG-108 membranes (5 μg/assay) in 10 μl of buffer A were mixed with an equal volume of RGS4 or GAIP in 10 μl of buffer B (50 mM Na·Hepes, pH 7.2/3 mM EGTA/100 mM NaCl/2 mM dithiothreitol/80 mM KCl) at 4°C for 30 min. Membranes and RGS proteins were then mixed with 20 μl of sonicated vesicles containing [3H]phosphatidylinositol 4,5-bisphosphate and phosphatidylethanolamine (25, 27) and the indicated hormone and/or guanine nucleotide in buffer B. Reactions were initiated by addition of 10 μl of 9 mM CaCl2 (25) in buffer B. Assays were carried out for 30 min at 30°C. Reactions were stopped and processed as described (26).
Measurement of Adenylyl Cyclase Activity in NG-108 Cell Membranes.
NG-108 membranes (22 μg per assay), prepared as described above, were mixed with the indicated concentrations of RGS4 or GAIP and incubated at 4°C for 15 min. Adenylyl cyclase activity was assayed (28) in the presence of 50 mM Na·Hepes (pH 8.0), 4 mM MgCl2, 1 mM EDTA, 100 μM GTP, 500 μM [α-32P]ATP (80 cpm/pmol), 3 mM dipotassium phosphoenolpyruvate, and 10 μg/ml pyruvate kinase for 20 min at 30°C.
Binding of Gqα to RGS4.
Gqα (50 nM) and hexa-histidine-tagged RGS4 (500 nM) were mixed in 100 μl of buffer C (50 mM Na·Hepes, pH 8.0/5 mM MgCl2/10 mM 2-mercaptoethanol/0.1% C12E10) containing 100 mM NaCl and incubated on ice for 10 min in the presence or absence of 200 nM phospholipase Cβ1. NaF (5 mM) and AlCl3 (30 μM) were included in the buffer to prepare Gqα-GDP-AlF4−. Ni-NTA resin (Qiagen, Chatsworth, CA) was equilibrated with buffer C containing 100 mM NaCl and 25 μl was added to the mixture of proteins, followed by further incubation on ice for 5 min. The resin was collected by brief centrifugation, and the supernatant was saved as the flowthrough fraction. The resin was washed three times with 150 μl of buffer C containing 400 mM NaCl, and 10 mM imidazole. NaF and AlCl3 were included in the wash buffer when they were present initially. The supernatant from the third wash was saved as the wash fraction. The resin was finally eluted with 50 μl of buffer C containing 150 mM imidazole. Fractions (10 μl) were analyzed by immunoblotting after PAGE in the presence of SDS using anti-Gqα serum Z811 (26).
RESULTS
RGS4 and GAIP Are GAPs for Gqα.
RGS4 and GAIP markedly accelerate GTP hydrolysis by Giα subfamily members, but not by Gsα or G12α (17–19). These Gα proteins can be prepared in their GTP-bound forms by incubation with the nucleoside triphosphate in the absence of Mg2+, and the GTP-Gα substrates can then be used in single-turnover assays. Conventional assays that assess steady-state hydrolysis of GTP are irrelevant because the rate of catalysis is limited by product (GDP) dissociation. It has not been possible to prepare sufficient quantities of GTP-Gqα for single-turnover assays. Accordingly, Gq must be reconstituted with an appropriate guanine nucleotide exchanger (receptor) to regenerate substrate for steady-state GTPase assays (by catalyzing dissociation of product, GDP-Gqα). Thus, Gqα was reconstituted with purified G protein βγ subunits and M1 muscarinic cholinergic receptors in phospholipid vesicles, and steady-state rates of carbachol-stimulated GTPase activity were measured following addition of potential GAPs (Fig. 1). RGS4 and GAIP both stimulated this rate dramatically. Appropriately, the effects of these GAPs were dependent on the presence of an agonist (carbachol) for the receptor and were invisible when the antagonist atropine was present. The effect of RGS4 was evident at 10-fold lower concentrations than was that of GAIP. RGS4 also appeared to be a more efficacious GAP than was either GAIP or a saturating concentration of phospholipase Cβ1, although it was not possible to achieve sufficiently high concentrations of GAIP to assess its maximal effect unequivocally. The maximal rate of GTPase activity observed in these assays may have been limited by the rate of receptor-catalyzed nucleotide exchange. Other members of the Gqα subfamily (G11α, G14α, G16α) were not available for testing.
RGS4 and GAIP Inhibit Receptor- and G Protein-Directed Activation of Phospholipase Cβ.
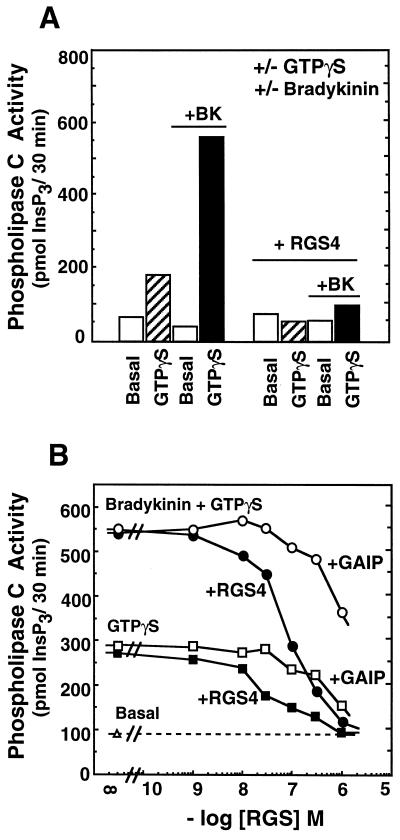
The Gq subfamily of G proteins couples cell-surface receptors to activation of phospholipase Cβ. NG-108 cells express both Gqα and G11α (27), and bradykinin activates phospholipase Cβ in these cells and membranes derived therefrom to stimulate synthesis of Ins(1,4,5)P3 (29). However, for unknown reasons this effect of bradykinin is not observed when GTP is present (not shown) and requires a nonhydrolyzable guanine nucleotide analog such as GTPγS (Fig. 2A); this anomaly is consistent with previous observations using membranes derived from mammalian cells (30, 31). Despite the fact that Gα proteins do not hydrolyze GTPγS, even in the presence of RGS4 or GAIP, both RGS4 and GAIP inhibited bradykinin plus GTPγS-mediated activation of phospholipase C (Fig. 2). Consistent with the data shown in Fig. 1, RGS4 was roughly 10-fold more potent than GAIP. Since the data of Fig. 1 demonstrate that RGS4 and GAIP do not inhibit receptor-mediated nucleotide exchange on Gqα, this activity of the RGS proteins implies their capacity to inhibit directly the stimulation of phospholipase Cβ by activated Gqα. Such inhibition is demonstrated in Fig. 3 using purified GTPγS-Gqα and phospholipase Cβ1, although higher concentrations of RGS4 were required to mediate the effect in this reconstituted system compared with NG-108 membranes (and the effect of GAIP in this assay was not apparent at the concentrations achieved).
Figure 2.
The effect of RGS4 and GAIP on GTPγS ± bradykinin-activated synthesis of inositol 1,4,5-trisphosphate by NG-108 cell membranes. (A) NG-108 membranes were incubated as described under Materials and Methods with 10 μM GDPβS (basal) or 3 μM GTPγS in the presence or absence of 1 μM bradykinin and/or 1 μM RGS4, as indicated, and production of [3H]inositol 1,4,5-trisphosphate (InsP3) was measured. (B) NG-108 membranes were incubated as described for A in the presence of bradykinin + GTPγS (circles) or GTPγS (squares) and the concentration of RGS4 (filled symbol) or GAIP (open symbols) was varied as indicated.
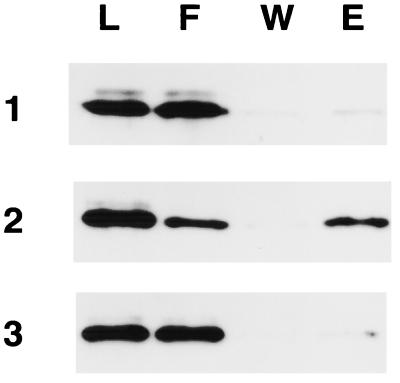
The most obvious mechanism for RGS4-mediated blockade of activation of phospholipase Cβ1 by Gqα would involve overlapping binding sites on Gqα for the effector and the RGS protein. The results shown in Fig. 4 indicate that this may be true. Gqα activated with GDP-AlF4− associated specifically with hexa-histidine-tagged RGS4 bound to Ni-NTA resin (compared with the GDP-bound form of Gqα). This interaction was prevented by inclusion of phospholipase Cβ1 (in excess of Gqα) in the incubation mixture. The AlF4−-activated form of Gsα did not bind to RGS4 under the same conditions (data not shown). We suspect that RGS4 and phospholipase Cβ1 cannot interact with Gqα simultaneously (see Discussion).
Figure 4.
Inhibition of Gqα binding to RGS4 by phospholipase Cβ1. Gqα-GDP (50 nM; row 1) or Gqα-GDP-AlF4− (50 nM; rows 2 and 3) was incubated with hexa-histidine-tagged RGS4 (500 nM) in the absence (rows 1 and 2) or presence (row 3) of phospholipase Cβ1 (200 nM). Ni-NTA resin was added and processed as described under Materials and Methods, and fractions were analyzed by immunoblotting (L, load; F, flowthrough; W, wash; E, eluate).
RGS4 and GAIP Inhibit Receptor- and G Protein-Directed Inhibition of Adenylyl Cyclase.
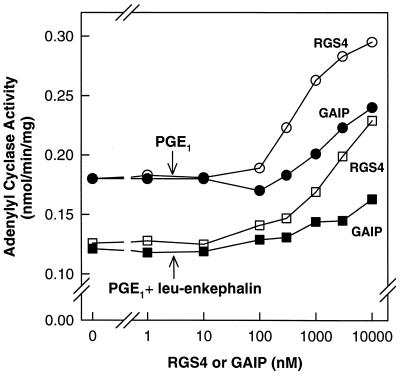
The data shown above represent the first demonstration of attenuation of G protein-mediated signaling by simple addition of an RGS protein to an endogenous, membrane-bound signaling system. We have tested the generality of this observation by examination of the effects of exogenously added RGS4 and GAIP on hormone-sensitive adenylyl cyclase activity in NG-108 membranes. Members of the Gi subfamily of G proteins couple cell-surface receptors to inhibition of adenylyl cyclase activity. NG-108 cells contain several Giα subfamily members, including Giα2, Giα3, GoαA, GoαB, and Gzα (32–34), and opiate receptors inhibit cyclic AMP synthesis in these cells or membranes derived therefrom. In NG-108 membranes, the μ-opioid receptor agonist [Leu]enkephalin inhibited (by 33%) prostaglandin E1 (PGE1)-stimulated adenylyl cyclase activity (Fig. 5). Inclusion of either RGS4 or GAIP reversed enkephalin-mediated inhibition of cyclic AMP synthesis and enhanced PGE1-stimulated activity significantly. The RGS proteins also elevated basal adenylyl cyclase activity modestly (not shown). RGS4 was more potent than GAIP in these assays.
Figure 5.
The effect of RGS4 and GAIP on adenylyl cyclase activity in NG-108 membranes. NG-108 membranes and the indicated concentrations of RGS4 (open symbols) or GAIP (filled symbols) were incubated for 15 min at 4°C. Adenylyl cyclase activity in these membranes was then measured as described under Materials and Methods in the presence of 10 μM PGE1 (circles) or 10 μM PGE1 + 2 μM [Leu]enkephalin (squares). The data shown are averages of duplicates determinations from a single experiment, which is representative of three experiments.
DISCUSSION
The observations described above extend knowledge of the specificity of RGS protein action from the Giα subfamily to at least one member of the Gqα subfamily of G protein subunits. This was not anticipated, since the receptors and effectors associated with Gi- and Gq-mediated signaling pathways are in general quite distinct. However, there is some cross-talk, evidenced, for example, by Gi-mediated activation of phospholipase Cβ; this effect appears to result from release of G protein βγ subunits from Giα-containing oligomers (35). Despite this, it seems premature to speculate on the physiological significance of this pattern of specificity in the absence of a great deal of other missing information. However, these data do highlight the need to assess, quantitatively, the specificity of the many members of the RGS protein family, and we thus wonder how to approach this question most appropriately.
As noted above, the effect of RGS4 on the GTPase activity of Gqα was evident at 10-fold lower concentrations than was that of GAIP. Examination of RGS protein specificity by variation of RGS concentration may have some relevance, since transcriptional or translational regulation of at least some of these proteins seems likely (15, 36). We thus compared increasing concentrations of RGS4 and GAIP for their capacity to stimulate the GTPase activity of a fixed concentration of GTP-Gzα and noted that the effect of GAIP was evident at a 100-fold lower concentration than was that of RGS4 (not shown), suggesting differential effects of RGS4 and GAIP on Gqα and Gzα. More conventionally, we took advantage of the ready availability of Goα to present increasing concentrations of GTP-Goα to RGS4 or GAIP. We thus estimated that GAIP has a relatively high apparent affinity for GTP-Goα (Km = 0.2 μM) but turned over substrate slowly (Vmax = 4/min at 4°C) (not shown). By contrast, RGS4 has a high Km for GTP-Goα (2 μM) but an estimated Vmax in excess of 500/min (18). We have chosen only to mention these analyses because of recent findings that GAIP is palmitoylated in vivo (37); we also have noted that our preparation of GAIP appears aggregated on gel filtration. We thus believe that attempts to perform quantitative assessment of the specificity of RGS protein action using purified proteins in vitro is premature because of limited knowledge of patterns of cellular expression, subcellular localization, and potential covalent modifications. Questions of specificity should probably be approached with intact cells, although this may be difficult.
Another novel finding is that RGS4 and GAIP can block GTPγS-mediated activation of phospholipase Cβ by Gqα. Since RGS proteins do not act as guanine nucleotide dissociation inhibitors (17) and they cannot stimulate hydrolysis of GTPγS by Gα proteins (unpublished observations), we presume they must interfere directly with the interaction between Gqα and phospholipase Cβ1 (Fig. 4). Preliminary examination of the crystal structure of a complex of RGS4 with Giα1 (bound to AlF4− and GDP) indicates that the so-called RGS box of RGS4 forms a four-helix bundle that contacts the flexible switch I, II, and III domains of the Gα subunit (J. G. Tesmer, D.M.B., A.G.G., and S. R. Sprang, unpublished observations). Since these regions of Gα have been implicated in interactions with effectors, binding of an RGS protein at these sites should interfere with effector binding. We thus envision that RGS proteins can act as true effector antagonists, a novel mechanism of inhibition of these signaling pathways.
We have shown previously that RGS4 interacts preferentially with Gα-GDP-AlF4− complexes, which are transition state analogs (18). However, Hunt et al. (20) demonstrated that RGS10 has a relatively high affinity for GTPγS-Gα complexes. It is possible that some RGS proteins will bind preferentially to the transition state for GTP hydrolysis while others may also have high affinity for the ground state (GTP-Gα). Such considerations could dictate whether an RGS protein would predominantly act catalytically as a GAP or stoichiometrically as an effector antagonist.
Finally, we have demonstrated that RGS proteins can be mixed with appropriate membranes to demonstrate their inhibitory effects on G protein-mediated signaling—at least with phospholipase Cβ and adenylyl cyclase. Such manipulations may provide useful tools for evaluation of the role of G proteins in poorly characterized signaling systems. Of interest, RGS4 and GAIP both stimulated basal adenylyl cyclase activity modestly and enhanced PGE-stimulated (unpublished observations) enzymatic activity by >50%, in addition to their anticipated effect to overcome [Leu]enkephalin-mediated inhibition of adenylyl cyclase. The simplest explanation is that some level of tonic inhibition of adenylyl cyclase is evident in these preparations and is relieved by the interaction between the RGS protein and a Giα subfamily member or that the receptor for PGE1 can activate both Gs and Gi (38). Similar results have been obtained after treatment of cells and membranes with pertussis toxin, which ADP-ribosylates Giα proteins and blocks their interactions with receptors (39).
Acknowledgments
We thank Pamela Sternweis, Linda Hannigan, and Jeffery Laidlaw for excellent technical assistance; Drs. Gloria Biddlecome and Elliott M. Ross for assistance with reconstitution of M1 receptors and Gq; and Drs. John Tesmer and Stephen Sprang for helpful discussions. D.M.B. is a member of the Cell Regulation Graduate Program of the University of Texas Southwestern Graduate School of Biomedical Sciences and was supported by the Medical Scientist Training Program and Pharmacological Sciences Training Grant GM07062. This work was supported by National Institutes of Health Grant GM 34497 and the Raymond and Ellen Willie Distinguished Chair of Molecular Neuropharmacology.
Footnotes
Abbreviations: GAP, GTPase-activating protein; GTPγS, guanosine 5′-(3-O-thio)triphosphate; PGE1, prostaglandin E1.
References
- 1.Berstein G, Blank J L, Jhon D Y, Exton J H, Rhee S G, Ross E M. Cell. 1992;70:411–418. doi: 10.1016/0092-8674(92)90165-9. [DOI] [PubMed] [Google Scholar]
- 2.Biddlecome G H, Berstein G, Ross E M. J Biol Chem. 1996;271:7999–8007. doi: 10.1074/jbc.271.14.7999. [DOI] [PubMed] [Google Scholar]
- 3.Arshavsky V Y, Bownds M D. Nature (London) 1992;357:416–417. doi: 10.1038/357416a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arshavsky V Y, Dumke C L, Zhu Y, Artemyev N O, Skiba N P, Hamm H E, Bownds M D. J Biol Chem. 1994;269:19882–19887. [PubMed] [Google Scholar]
- 5.Angleson J K, Wensel T G. Neuron. 1993;11:939–949. doi: 10.1016/0896-6273(93)90123-9. [DOI] [PubMed] [Google Scholar]
- 6.Angleson J K, Wensel T G. J Biol Chem. 1994;269:16290–16296. [PubMed] [Google Scholar]
- 7.Pages F, Deterre P, Pfister C. J Biol Chem. 1993;268:26358–26364. [PubMed] [Google Scholar]
- 8.Dohlman, H. G. & Thorner, J. (1996) J. Biol. Chem., in press. [DOI] [PubMed]
- 9.Neer, E. J. (1996) Curr. Biol., in press.
- 10.Chan R K, Otte C A. Mol Cell Biol. 1982;2:11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan R K, Otte C A. Mol Cell Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiner J L, Guttierez-Steil C, Blumer K J. J Biol Chem. 1993;268:8070–8077. [PubMed] [Google Scholar]
- 13.Dohlman H G, Apaniesk D, Chen Y, Song J P, Nusskern D. Mol Cell Biol. 1995;15:3635–3643. doi: 10.1128/mcb.15.7.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koelle M R, Horvitz H R. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 15.Druey K M, Blumer K J, Kang V H, Kehrl J H. Nature (London) 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 16.Siderovski D P, Hessel A, Chung S, Mak T W, Tyers M. Curr Biol. 1996;6:211–212. doi: 10.1016/s0960-9822(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 17.Berman D M, Wilkie T M, Gilman A G. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 18.Berman D M, Kozasa T, Gilman A G. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- 19.Watson N, Linder M E, Druey K M, Kehrl J H, Blumer K J. Nature (London) 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 20.Hunt T W, Fields T A, Casey P J, Peralta E G. Nature (London) 1996;383:175–177. doi: 10.1038/383175a0. [DOI] [PubMed] [Google Scholar]
- 21.Kozasa T, Gilman A G. J Biol Chem. 1995;270:1734–1741. doi: 10.1074/jbc.270.4.1734. [DOI] [PubMed] [Google Scholar]
- 22.Hepler J R, Biddlecome G H, Kleuss C, Camp L A, Hofmann S L, Ross E M, Gilman A G. J Biol Chem. 1996;271:496–504. doi: 10.1074/jbc.271.1.496. [DOI] [PubMed] [Google Scholar]
- 23.Linder M E, Pang I-H, Duronio R J, Gordon J I, Sternweis P C, Gilman A G. J Biol Chem. 1991;266:4654–4659. [PubMed] [Google Scholar]
- 24.Kozasa T, Hepler J R, Smrcka A V, Simon M I, Rhee S G, Sternweis P C, Gilman A G. Proc Natl Acad Sci USA. 1993;90:9176–9180. doi: 10.1073/pnas.90.19.9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smrcka A V, Hepler J R, Brown K O, Sternweis P C. Science. 1991;251:804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 26.Hepler J R, Kozasa T, Smrcka A V, Simon M I, Rhee S G, Sternweis P C, Gilman A G. J Biol Chem. 1993;268:14367–14375. [PubMed] [Google Scholar]
- 27.Gutowski S, Smrcka A V, Nowak L, Wu D, Simon M I, Sternweis P C. J Biol Chem. 1991;266:20519–20524. [PubMed] [Google Scholar]
- 28.Smigel M D. J Biol Chem. 1986;261:1976–1982. [PubMed] [Google Scholar]
- 29.Yano K, Higashida H, Inoue R, Nozawa Y. J Biol Chem. 1984;259:10201–10207. [PubMed] [Google Scholar]
- 30.Uhing R J, Prpic V, Jiang H, Exton J H. J Biol Chem. 1986;261:2140–2146. [PubMed] [Google Scholar]
- 31.Hepler J R, Harden T K. Biochem J. 1986;239:141–146. doi: 10.1042/bj2390141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilk-Blaszczak M A, Gutowski S, Sternweis P C, Belardetti F. Cell. 1994;12:109–116. doi: 10.1016/0896-6273(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 33.Garibay J L R, Kozasa T, Itoh H, Tsukamoto T, Matsuoka M, Kaziro Y. Biochim Biophys Acta. 1991;1094:193–199. doi: 10.1016/0167-4889(91)90008-l. [DOI] [PubMed] [Google Scholar]
- 34.Mullaney J, Magee A I, Unson C G, Milligan G. Biochem J. 1988;256:649–656. doi: 10.1042/bj2560649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blank J L, Brattain K A, Exton J H. J Biol Chem. 1992;267:23069–23075. [PubMed] [Google Scholar]
- 36.Dietzel C, Kurjan J. Mol Cell Biol. 1987;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Vries L, Elenko E, Hubler L, Jones T L Z, Farquhar M G. Proc Natl Acad Sci USA. 1996;93:15203–15208. doi: 10.1073/pnas.93.26.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asano T, Katada T, Gilman A G, Ross E M. J Biol Chem. 1984;259:9351–9354. [PubMed] [Google Scholar]
- 39.Katada T, Amano T, Ui M. J Biol Chem. 1982;257:3739–3746. [PubMed] [Google Scholar]