Abstract
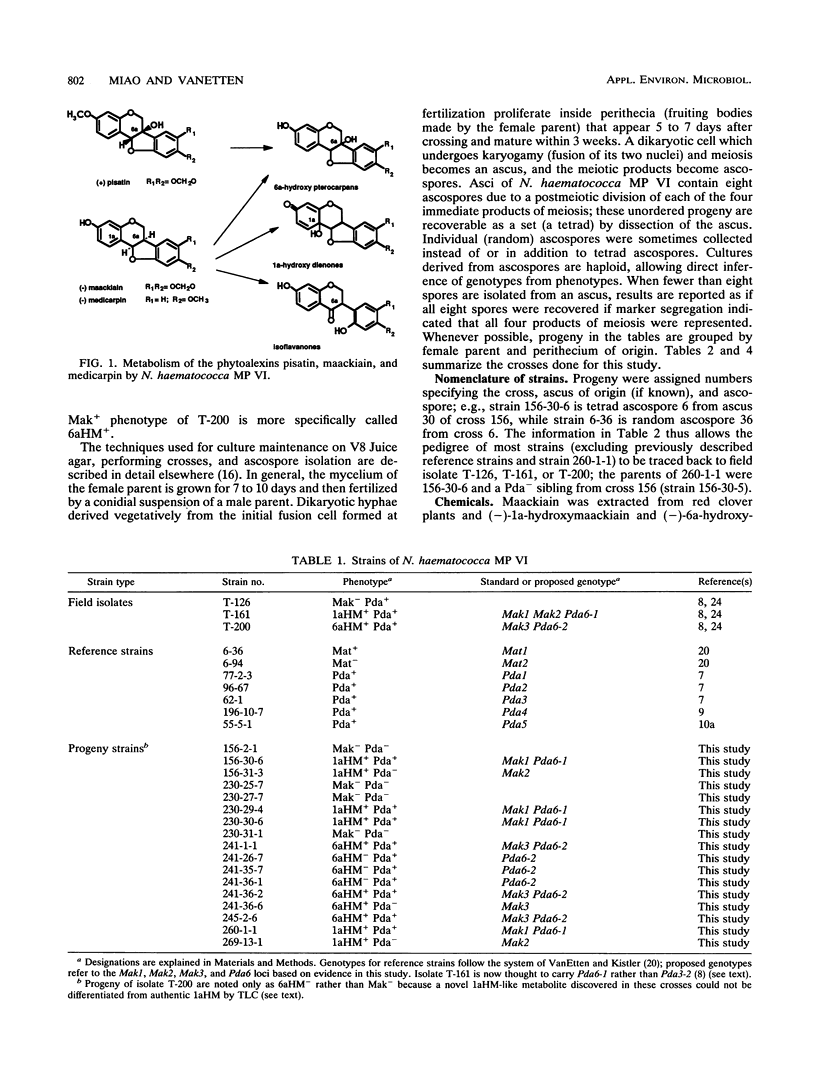
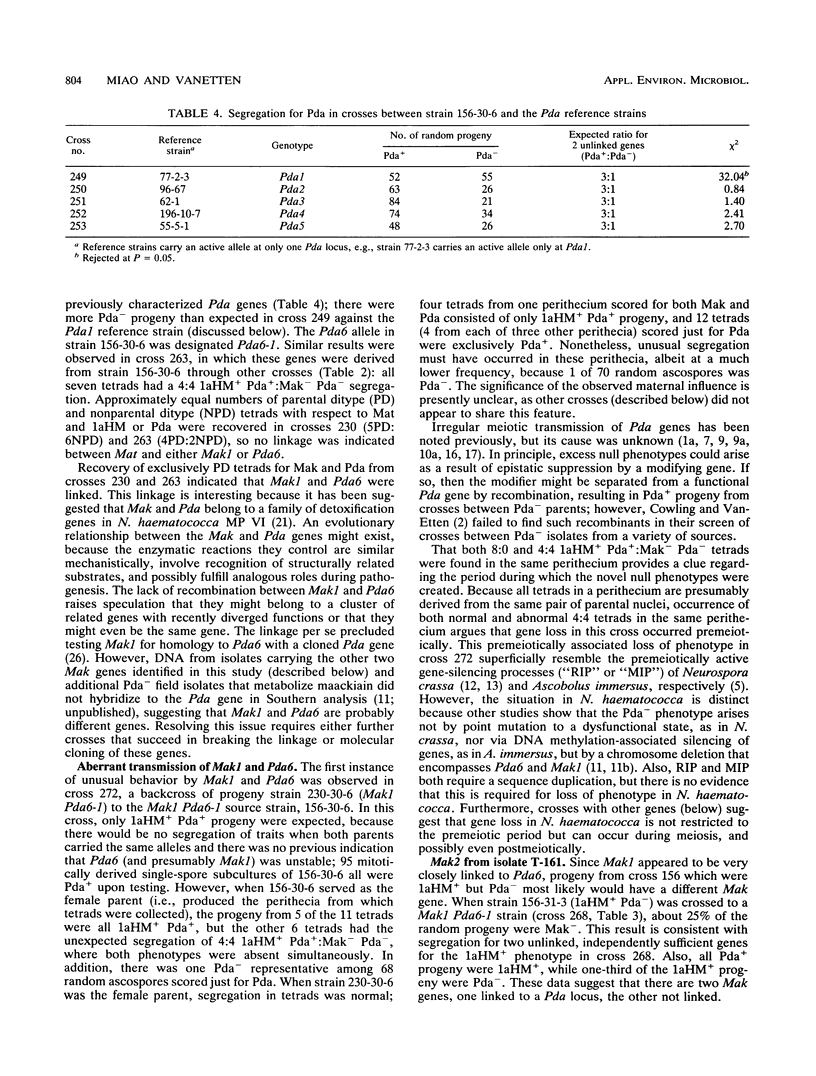
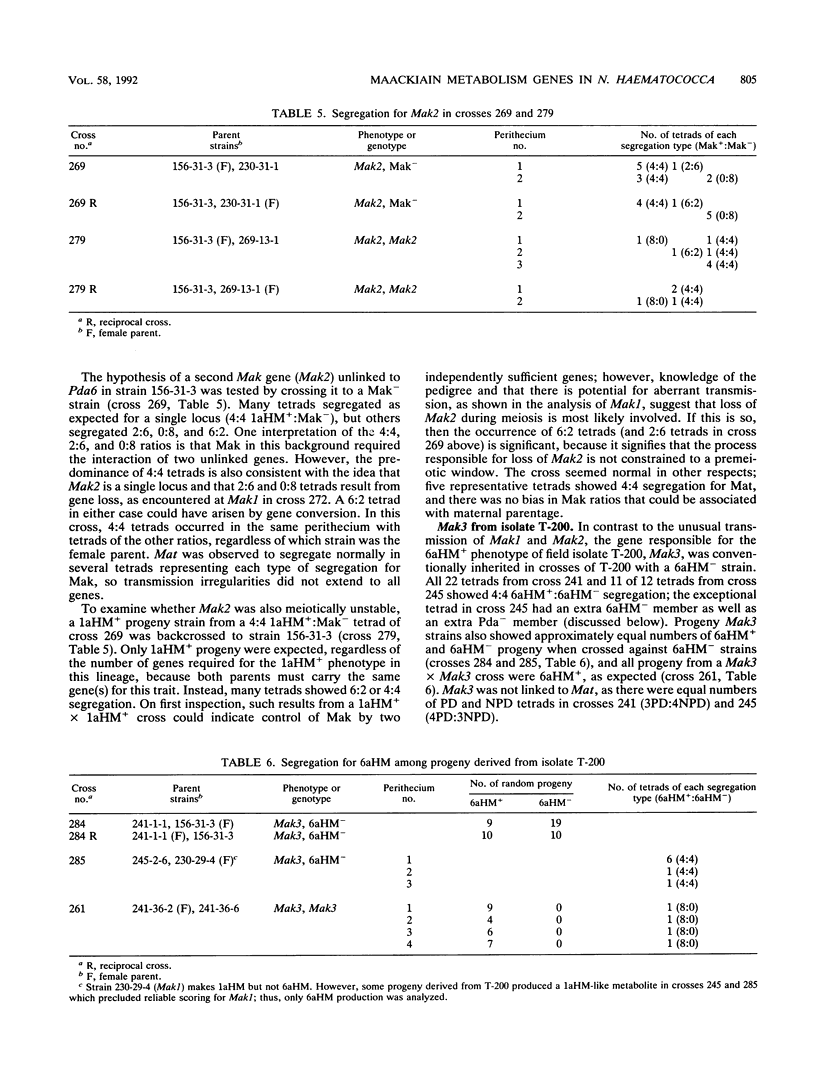
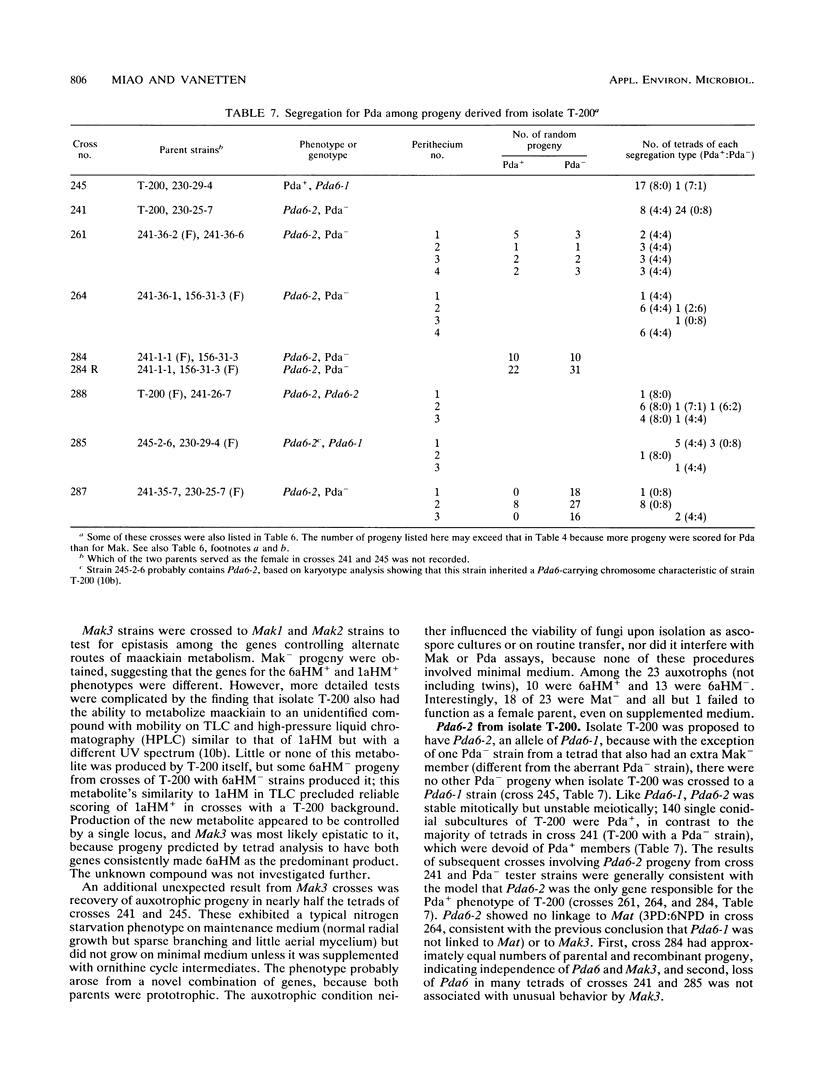
Some isolates of the plant-pathogenic fungus Nectria haematococca mating population (MP) VI metabolize maackiain and medicarpin, two antimicrobial compounds (phytoalexins) synthesized by chickpea (Cicer arietinum L.). The enzymatic modifications by the fungus convert the phytoalexins to less toxic derivatives, and this detoxification has been proposed to be important for pathogenesis on chickpea. In the present study, loci controlling maackiain metabolism (Mak genes) were identified by crosses among isolates of N. haematococca MP VI that differed in their ability to metabolize the phytoalexin. Strains carrying Mak1 or Mak2 converted maackiain to 1a-hydroxymaackiain, while those with Mak3 converted it to 6a-hydroxymaackiain. Mak1 and Mak2 were unusual in that they often failed to be inherited by progeny. Mak1 was closely linked to Pda6, a new member in a family of genes in N. haematococca MP VI that encode enzymes for detoxification of pisatin, the phytoalexin synthesized by garden pea. Like Mak1, Pda6 was also transmitted irregularly to progeny. Although the unusual meiotic behaviors of some Mak genes complicate genetic analysis, identification of these genes should afford a more through evaluation of the role of phytoalexin detoxification in the pathogenesis of N. haematococca MP VI on chickpea.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Faugeron G., Rhounim L., Rossignol J. L. How does the cell count the number of ectopic copies of a gene in the premeiotic inactivation process acting in Ascobolus immersus? Genetics. 1990 Mar;124(3):585–591. doi: 10.1093/genetics/124.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews D. E., Van Etten H. D. Detoxification of the phytoalexin pisatin by a fungal cytochrome P-450. Arch Biochem Biophys. 1983 Jul 15;224(2):494–505. doi: 10.1016/0003-9861(83)90237-0. [DOI] [PubMed] [Google Scholar]
- Miao V. P., Covert S. F., VanEtten H. D. A fungal gene for antibiotic resistance on a dispensable ("B") chromosome. Science. 1991 Dec 20;254(5039):1773–1776. doi: 10.1126/science.1763326. [DOI] [PubMed] [Google Scholar]
- Miao V. P., Matthews D. E., VanEtten H. D. Identification and chromosomal locations of a family of cytochrome P-450 genes for pisatin detoxification in the fungus Nectria haematococca. Mol Gen Genet. 1991 Apr;226(1-2):214–223. doi: 10.1007/BF00273606. [DOI] [PubMed] [Google Scholar]
- Miao V. P., Vanetten H. D. Genetic Analysis of the Role of Phytoalexin Detoxification in Virulence of the Fungus Nectria haematococca on Chickpea (Cicer arietinum). Appl Environ Microbiol. 1992 Mar;58(3):809–814. doi: 10.1128/aem.58.3.809-814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987 Dec 4;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Weltring K. M., Turgeon B. G., Yoder O. C., VanEtten H. D. Isolation of a phytoalexin-detoxification gene from the plant pathogenic fungus Nectria haematococca by detecting its expression in Aspergillus nidulans. Gene. 1988 Sep 7;68(2):335–344. doi: 10.1016/0378-1119(88)90036-4. [DOI] [PubMed] [Google Scholar]


