Abstract
Transcription factors are master regulatory switches of differentiation, including the development of specific hematopoietic lineages from stem cells. Here we show that mice with targeted disruption of the CCAAT enhancer binding protein α gene (C/EBPα) demonstrate a selective block in differentiation of neutrophils. Mature neutrophils and eosinophils are not observed in the blood or fetal liver of mutant animals, while other hematopoietic lineages, including monocytes, are not affected. Instead, most of the white cells in the peripheral blood of mutant mice had the appearance of myeloid blasts. We also observed a selective loss of expression of a critical gene target of CCAAT enhancer binding protein α, the granulocyte colony-stimulating factor receptor. As a result, multipotential myeloid progenitors from the mutant fetal liver are unable to respond to granulocyte colony-stimulating factor signaling, although they are capable of forming granulocyte–macrophage and macrophage colonies in methylcellulose in response to other growth factors. Finally, we demonstrate that the lack of granulocyte development results from a defect intrinsic to the hematopoietic system; transplanted fetal liver from mutant mice can reconstitute lymphoid but not neutrophilic cells in irradiated recipients. These studies suggest a model by which transcription factors can direct the differentiation of multipotential precursors through activation of expression of a specific growth factor receptor, allowing proliferation and differentiation in response to a specific extracellular signal. In addition, the c/ebpα−/− mice may be useful in understanding the mechanisms involved in acute myelogenous leukemia, in which a block in differentiation of myeloid precursors is a key feature of the disease.
An important question in hematopoiesis is how the function of master regulatory transcription factors mediate development and differentiation of specific lineages (1–4). Mature myeloid cells, consisting of blood monocytes and tissue macrophages, as well as the neutrophilic and eosinophilic granulocytes, develop from a common myeloid precursor, but the mechanisms controlling the transition from bipotential progenitor myeloblasts to unipotential granulocyte and monocyte precursors have not been identified (5).
The CCAAT enhancer binding protein (C/EBP) family members have a basic leucine zipper structure and bind to similar DNA sequences (6). The founding member of this family, C/EBPα, is highly expressed in liver and adipose tissue (7, 8). C/EBPα transactivates the promoters of hepatocyte and adipocyte specific genes, which are important for energy homeostasis (9–11). C/EBPα is up-regulated in and plays an important role in adipocyte differentiation, and in this cell type it functions in fully differentiated, nonproliferative cells; inhibition of C/EBPα blocks differentiation and expression of C/EBPα induces differentiation (12, 13). c/ebpα knockout mice lack hepatic glycogen stores and die from hypoglycemia within 8 hr of birth (14).
C/EBPα is part of a family of leucine zipper transcription factors, including C/EBPβ and C/EBPδ, which heterodimerize and are differentially regulated during differentiation of adipocytes (12, 15, 16). C/EBPα, C/EBPβ, and C/EBPδ are strongly similar in their C-terminal basic region and leucine zipper domains and diverge in the N-terminal transactivation domain (12, 15, 17–19). The avian homolog of C/EBPβ (NF-M) is a myeloid specific factor that activates the promoter for chicken cMGF, a growth factor related to granulocyte colony-stimulating factor (G-CSF) and interleukin 6 (20–23). In mammalian cells, C/EBPβ (or NF-IL6, the human homolog; ref. 16) has low activity until activated by inflammatory stimuli, which induce phosphorylation and translocation to the nucleus, where it can activate multiple cytokine gene promoters (16, 24, 25). Studies of mice with a targeted disruption of C/EBPβ have indicated the importance of C/EBPβ for macrophage functions related to bacterial killing and tumor cytotoxicity, but myeloid cell development was not adversely affected, consistent with a role of C/EBPβ in cytokine activation rather than lineage development (26, 27).
In the hematopoietic system, C/EBPα is known to be expressed in myeloid cells (28, 29). We have recently reported the transactivation of promoters of myeloid specific receptors for the growth factors G-CSF, macrophage CSF (M-CSF), and granulocyte–macrophage CSF (GM-CSF) by C/EBP family members (30–32). In addition, C/EBPα has been noted to regulate the promoters for several myeloid granule proteins, including myeloperoxidase and neutrophil elastase, which are expressed at the promyelocytic stage (33, 34). These studies suggest that C/EBPα might play a role in myeloid cell development. To understand the role of C/EBPα during hematopoiesis, and specifically in myeloid differentiation, we analyzed the hematopoietic system in mice carrying a targeted disruption of c/ebpα. The c/ebpα−/− mice do not respond to G-CSF growth factor signaling and lack mature granulocytes, but not monocytes and macrophages, in both circulating blood and hematopoietic organs.
MATERIALS AND METHODS
Mice.
C/EBPα +/− mice were generated using targeted AB2.1 embryonic stem cells injected into C57BL/6 blastocysts as reported previously (14). Mice were subsequently bred with C57BL/6 animals. For analysis of response of wild-type and mutant animals to G-CSF, G-CSF (250 μg per kg of body weight) was injected subcutaneously every 12 hr to pregnant mice from the day 17 of pregnancy until birth. Control animals received saline alone.
Hematologic Analysis.
Blood samples were obtained from newborn mice with heparinized microcapillary tubes. The level of hemoglobin was measured with a Hemoglobinometer (Leica, Buffalo, NY). For white blood cell analysis, red blood cells were lysed by hypotonic lysis. White blood cells were cytocentrifuged and then stained with Wright–Giemsa. Analysis of percentage of apoptotic cells in newborn livers and blood was performed by fluorescence microscopy following terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL assay; Boehringer Mannheim). For colony-forming unit (CFU) assays, methylcellulose culture of single-cell suspensions of newborn liver cells were performed in Methocult (Stemcell Technologies, Vancouver), and the mixture was cultured at 37°C in 5% CO2 for 10 days. Colony types were confirmed by dispersion of the cells in the colony and analysis following cytocentrifugation of the cells onto slides and staining with Wright–Giemsa. For G-CFU assays, cells were added to IMDM medium containing 0.8% methylcellulose, 20% fetal bovine serum, and up to 1000 units/ml of human recombinant G-CSF (which is active on murine cells), and colonies were counted daily. G-CFU from wild-type mouse livers were most easily seen 3–5 days later.
Fluorescence-Activated Cell Sorting (FACS).
Flow cytometry analysis was performed on single-cell suspensions of fetal and newborn liver, newborn thymus, and newborn spleen, and blood was collected from bone marrow-transplanted mice. Tissues were broken with syringe plungers, and cells were filtered through 70-μm nylon cell strainers. Cells were stained with fluorescein isothiocyanate- or phycoerythrin-conjugated antibodies and analyzed on a FACScan (Becton Dickinson). Monoclonal antibodies M1/70 (Mac-1 αM chain, CD11b), RB6-8C5 (Gr-1), RA3-6B2 (B220, CD45R), 53-2.1 (Thy-1.2, CD90), H129.19 (CD4), 53-6.7 (CD8a), TER-119, and 104 (CD45.2) were used according to the supplier’s (PharMingen) instructions. Ten thousand cells were analyzed in each FACS assay.
Northern Blot Analysis.
Total RNA was isolated from livers of newborn mice, from brain of adult mice, and from cells collected from 48-hr thioglycollate-stimulated mouse peritoneal cavity. RNA was prepared, and Northern blot analysis performed as described previously (35). Quantitation was performed using imagequant densitometry software on Northern blots exposed in the linear range of the film.
Analysis of Blood Reconstitution in Sublethally Irradiated Mice.
Six- to eight-week-old female C57/BL6J congenic strain C57/B6.SJL PEP3b-BoyJ mice were sublethally irradiated with a single dose of γ irradiation (800 rad) from a 137Cs source (Gammacell 40 Exactor, Nordion Internationa, Kanata, Canada). Liver cells (1 × 106) from newborn C/EBPα +/+, +/−, or −/− C57/BL6J mice in 100 μl of IMDM were injected intravenously into each irradiated recipient animal. Transplanted animals were kept in sterilized cages. The appearance of donor origin blood cells was detected by flow cytometry analyses.
RESULTS
Mutation of the C/EBPα Gene Results in a Selective Loss of Granulocyte Development.
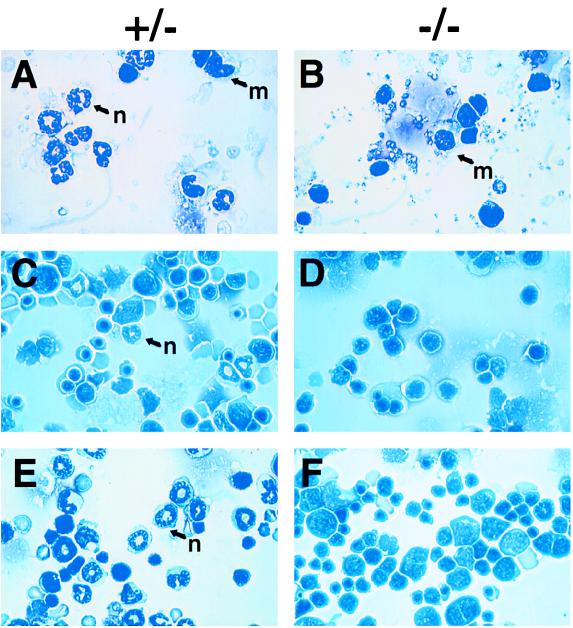
The construction of c/ebpα hemizygous knockout embryonic stem cells and the generation of homozygous mouse strains were described previously (14). c/ebpα is a intronless single-exon gene. The targeted disruption replaced this exon with sequences from the neomycin-resistant gene. Since the homozygous mutant (−/−) mice died within 8 hr of birth, newborn animals were analyzed. No difference between wild-type and heterozygote animals was detected. At birth, −/− mice were also grossly indistinguishable from their littermates. Analysis of peripheral blood from newborn −/− mice showed normal levels of hemoglobin (Table 1) and normal appearance of platelets, indicating normal development of the erythrocytic and megakaryocytic lineages. After lysis of red blood cells, samples were prepared by cytocentrifugation and stained with Wright–Giemsa as shown in Fig. 1. The morphology of white blood cells from normal littermates (+/+ or +/− genotype) and −/− mice displayed a striking difference. The normal littermates showed 87% mature neutrophils, with 0.5% lymphocytes and 12.5% mature monocytes (Fig. 1A). There were no mature neutrophils in the blood of −/− mice, although mature monocytes and lymphocytes can be detected (Fig. 1B). Furthermore, −/− newborns had normal peritoneal macrophages. In addition, no eosinophils could be detected in −/− mice, although they could be easily seen with Fast green staining of peripheral blood from +/− mice. The numbers of monocytes and lymphocytes in −/− mice was indistinguishable from those of normal littermates, but most of the white cells in the blood of −/− mice had the appearance of immature myeloid cells. A majority of these immature cells stained positive for Sudan black, a marker of myeloid progenitors. These cells also had more myeloid CFU activity than that from wild-type or heterozygote blood (see below). These findings indicate that the mature granulocytic population of peripheral blood has been replaced in C/EBPα −/− mice by a progenitor population with myeloid characteristics.
Table 1.
Analysis of blood and liver cells of newborn C/EBPα +/+, +/−, and −/− mice
| Cell components | Percent of cell components
|
|
|---|---|---|
| c/ebpα+/+ and c/ebpα+/− | c/ebpα−/− | |
| Blood | ||
| Hemoglobin | 13.4 ± 1.9* | 12.0 ± 2.2* |
| Total WBC | 4.4 ± 1.5† | 4.0 ± 2.9† |
| Granulocytes | 87.0 ± 7.2 | 0 |
| Monocytes | 12.5 ± 7.4 | 7.8 ± 3.3 |
| Lymphocytes | 0.5 ± 0.9 | 0.5 ± 0.5 |
| Immature cells | 0 | 91.5 ± 2.6 |
| Platelets | 270 ± 96† | 429 ± 63† |
| Liver | ||
| Immature erythroid | 64.6 ± 6.9 | 79.0 ± 3.3 |
| Immature myeloid | 20.5 ± 8.6 | 18.4 ± 2.7 |
| Monocytes | 1.5 ± 1.1 | 2.5 ± 0.2 |
| Granulocytes | 13.6 ± 2.1 | 0 |
Blood (50 μl) was collected from newborn mice into heparinized microcapillary tubes. Total white blood counts (WBC) were determined from six independent +/+ and −/− animals, and platelet counts were determined from two independent +/− and −/− animals using a Coulter counter. For white blood cell differential counts, red blood cells were lysed by hypotonic lysis. White blood cells were cytocentrifuged and then stained with Wright–Giemsa. Percentages were calculated based on cell morphology of a total of 200 white blood cells. Each data point represents the mean ± SD from at least three independent animals. For liver differential counts, hematopoietic cells were analyzed by Wright–Giemsa staining following filtration through nylon cell strainers as described.
Values are in g/dl.
Values are in 103 cells per μl.
Figure 1.
Lack of mature neutrophils in c/ebpα−/− mice. Blood was collected as described in Table 1. After lysing red blood cells, white blood cells from either c/ebpα+/− (A) or c/ebpα−/− (B) newborn mice were cytocentrifuged onto slides and were stained with Wright–Giemsa for analysis of cell morphology. Newborn liver touch preparations from c/ebpα+/− (C) or c/ebpα−/− (D) and G-CSF-administered c/ebpα+/− (E) or c/ebpα−/− (F) mice were stained with Wright–Giemsa. G-CSF (250 μg per kg of body weight) was injected subcutaneously every 12 hr to pregnant mice from day 17 of pregnancy until birth. Mature neutrophils are indicated with an n. Monocytes are indicated with an m.
Analysis of Fetal and Newborn Liver from C/EBPα −/− Mice Reveals a Loss of Granulocytes.
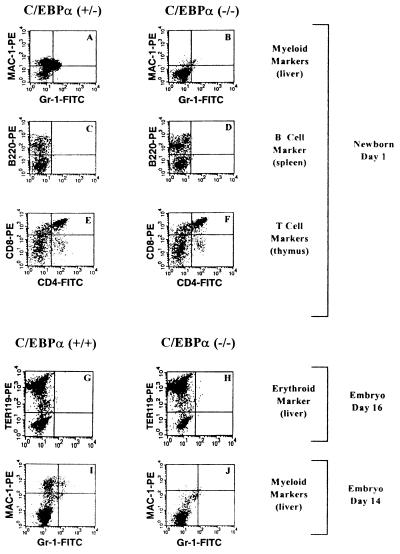
During embryogenesis, definitive blood cells arise mainly from fetal liver, initiating around day 12 of gestation (36). The bone marrow gradually assumes most of the animal’s hematopoietic function between birth and 2 weeks after birth. Hematopoietic cells in different stages of maturation can be found in the normal newborn liver, including myeloblasts, monoblasts, and macrophages, as well as mature granulocytes (Fig. 1C). However, in examining a total of 46 −/− animals, no mature granulocytes were present in newborn liver (Fig. 1D and Table 1). No significant change in percentage of apoptotic cells was observed in C/EBPα −/− livers and blood compared with +/− animals. To quantitatively analyze the difference in granulocyte maturation between control and −/− mice, the monocyte/granulocyte marker Mac-1 and mature granulocyte marker Gr-1 were used in FACS analysis of newborn liver suspension cells. As shown in Fig. 2A, +/− mice showed significant numbers of Mac-1+ cells and Mac-1+/Gr-1+ cells (similar to that of +/+ mice), while only a few single or double positive cells were present in −/− mutant liver (Fig. 2B). B lymphocyte development in the spleen and T lymphocyte development in the thymus were studied with the B cell marker B220 and the T cell markers CD4 and CD8 by FACS analysis. As shown in Fig. 2 C–F, −/− mice have normal B and T lymphocyte development. Mutant mice also have no significant changes in the erythroid marker TER-119 (Fig. 2 G and H). Since C/EBPα deficiency directly affects hepatocyte function, it could alter the environment of blood precursor cells during liver hematopoiesis at a late stage of embryogenesis. Therefore, embryonic day 14 liver suspension cells were also used in FACS analysis, as shown in Fig. 2 I and J. The results were similar to that seen with newborn liver, in that no mature granulocyte markers can be detected from −/− cells. Similar results were obtained with embryonic day 19 fetal livers, which are also not subject to hypoglycemic stress. Therefore, based on both cell morphology and cell surface differentiation marker analyses, C/EBPα deficiency blocks the maturation of granulocytic lineages, with no effect on either erythropoiesis or lymphopoiesis. In addition, the morphology of spleen and thymus from −/− mice appeared normal compared with +/+ or +/− littermates.
Figure 2.
Flow cytometry analysis with cell surface markers. Cells obtained from livers of c/ebpα+/− (A) or c/ebpα−/− (B) newborns and from wild-type (I) or c/ebpα−/− (J) day 14 embryos were stained with antibodies to the mature monocyte/granulocyte surface marker Mac-1 (37) and to the mature granulocyte surface marker Gr-1 (38). Cells obtained from spleens of c/ebpα+/− (C) or c/ebpα−/− (D) newborns were stained with antibodies to the B lymphocyte marker B220. Cells obtained from thymus of c/ebpα+/− (E) or c/ebpα−/− (F) newborns were stained with T lymphocyte surface markers CD4 and CD8. Cells obtained from livers of c/ebpα+/+ (G) or c/ebpα−/− (H) day 16 embryos were stained with antibodies to the erythroid surface marker TER-119.
Mutation of C/EBPα Results in a Selective Loss of G-CSF Receptor mRNA Expression and Lack of Response to G-CSF.
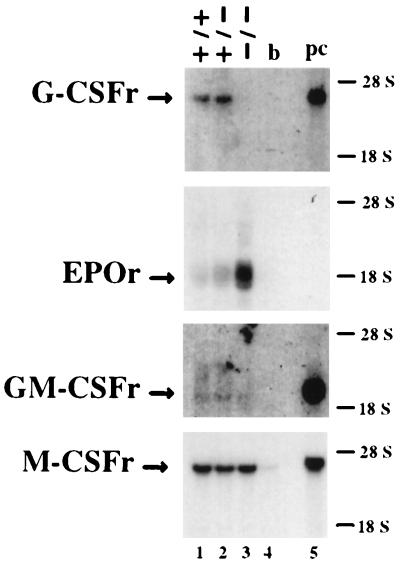
Growth factors play important roles in the complex process of hematopoiesis through signaling via their unique receptors. G-CSF, M-CSF, and GM-CSF are three hematopoietic growth factors specific to myeloid cells due to the specific expression of their receptors on the surface of myeloid precursors and mature cells (39–42). C/EBPα can specifically bind to and transactivate the promoter of each of these receptor genes (30–32). To examine the molecular basis for the abnormal differentiation of granulocytes in c/ebpα−/− mice and the role of C/EBPα in growth factor receptor expression in vivo, Northern blots were used to analyze the expression of these receptors. As shown in Fig. 3, no differences could be detected for M-CSF receptor and GM-CSF receptor mRNA levels between control C/EBPα +/+ and +/− mice and c/ebpα−/− mutant mice. In contrast, the G-CSF receptor mRNA expression in −/− mutant mice was undetectable by Northern blot analysis of newborn liver mRNA. Low levels of G-CSF mRNA could be detected in C/EBPα −/− liver RNA by more sensitive reverse transcriptase-PCR analysis. There was a 4-fold increase in erythropoietin receptor mRNA in livers from −/− mice compared with control animals.
Figure 3.
Northern blot analysis of mRNA of growth factor receptors. Total liver RNA (10 μg) from wild-type (lane 1), c/ebpα+/− (lane 2), or c/ebpα−/− (lane 3) newborn mice, 10 μg of total brain RNA from wild-type mice (lane 4), or 2 μg of total peritoneal cell RNA from thioglycollate-stimulated wild-type mice (lane 5) were electrophoresed in 1% agarose/formaldehyde gels, transferred to nylon membranes, and probed with murine cDNAs of G-CSF receptor (G-CSFr; ref. 43), erythropoietin receptor (EPOr; ref. 44), GM-CSF receptor α (GM-CSFr; ref. 45), or M-CSF receptor (M-CSFr; ref. 46). The position of each signal is indicated on the left side. GM-CSF receptor α mRNA is expressed at relatively low levels in newborn livers, but densitometry demonstrated that the level of GM-CSF receptor α mRNA is not significantly reduced in −/− liver compared with the +/+ or +/− animals.
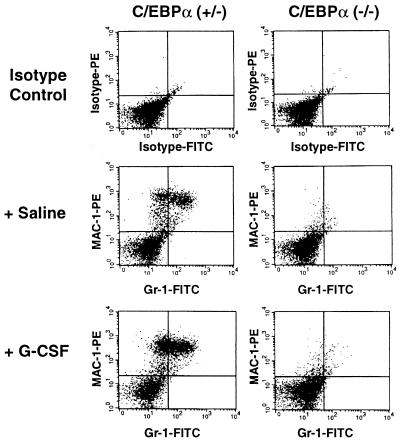
The most striking finding was the absence of detectable mRNA for the G-CSF receptor in c/ebpα−/− mice (Fig. 3). This suggested that lack of G-CSF signaling due to markedly diminished expression of G-CSF receptor could contribute to the block of granulocytic maturation in −/− mice. During hematopoiesis, the G-CSF receptor is specifically expressed on the surface of granulocytic precursors and mature granulocytes (42, 47, 48). It plays an important role in the proliferation, differentiation, and survival of granulocytic cells. To study the effect of G-CSF on in vivo granulocyte differentiation in c/ebpα−/− mice, the pregnant +/− female mice were administered G-CSF subcutaneously (250 μg per kg of body weight) every 12 hr during the last 4 days of pregnancy (49). G-CSF can cross the placental barrier (50). A G-CSF-induced increase in mature granulocytes can be detected in the livers of wild-type newborn mice (Fig. 1 C and E). The population of myeloid marker Mac-1- and granulocyte marker Gr-1-positive cells doubled after G-CSF treatment (Fig. 4). However, there was no response to G-CSF in −/− mutant livers, and no mature neutrophils were induced in the circulating blood and livers of −/− mutant mice (Fig. 1 D and F; Fig. 4). To further confirm the lack of the response to G-CSF stimulation in c/ebpα−/− myeloid precursor cells, liver suspension cells were prepared from both control mice and −/− mice and used for in vitro colony assays in the presence of increasing concentrations of G-CSF. Control cells showed an increase of G-CFU when G-CSF concentrations were increased from 250 to 1000 units/ml in the culture medium; no colonies were observed in the absence of added G-CSF. In assays with −/− cells, no G-CFU colonies could be found, even the highest level of G-CSF (Table 2). When cells were cultured in medium containing factors sufficient for the development of multiple types of colonies, the numbers of erythroid CFU, M-CFU, GM-CFU, and GEMM-CFU from control and −/− newborn liver suspension cells were not significantly different (Table 2). Granulocytes, erythrocytes, monocyte/macrophages, and megaharyocytes (GEMM)-CFU and GM-CFU from C/EBPα −/− mice contained immature granulocytic precursors (blasts, promyelocytes, and metamyelocytes), but not mature granulocytes. In addition, peripheral blood from C/EBPα −/− animals had 5-to 10-fold higher myeloid CFU per ml (GEMM-CFU, GM-CFU, M-CFU) compared with blood from C/EBPα +/− animals (data not shown). Compared with what was observed with C/EBPα +/− mice, Wright–Giemsa analysis of cells from myeloid CFU obtained from the peripheral blood of C/EBPα −/− animals showed a large increase in immature myeloid cells that were similar in morphology to the blasts seen in the peripheral blood of C/EBPα −/− animals (Fig. 1B), with no mature neutrophils observed. No erythroid CFU were detected from the blood of C/EBPα −/− mice. These data indicate that C/EBPα is critical for the differentiation of granulocytic cells in vivo, but is not required for the commitment of pluripotent stem cells to myeloid precursors.
Figure 4.
c/ebpα−/− mutant mice do not respond to G-CSF stimulation. Pregnant mice had been injected subcutaneously with either phosphate-buffered saline or G-CSF (250 μg per kg of body weight) every 12 hr since day 17 of pregnancy. Cells obtained from livers of newborn mice were stained with fluorescein isothiocyanate- and phycoerythrin-conjugated IgG (isotype control) or antibodies to the mature monocyte/granulocyte surface marker Mac-1 and antibodies to the mature granulocyte surface marker Gr-1, as described in Fig. 2 legend.
Table 2.
Analysis of types of CFU colonies in C/EBPα-deficient mice
| C/EBPα Genotype | n | Percent of CFU
|
Total G-CFU | |||
|---|---|---|---|---|---|---|
| E-CFU | M-CFU | GM-CFU | GEMM-CFU | |||
| +/− | 3 | 23.4 ± 6.7 | 29.1 ± 8.1 | 13.0 ± 7.1 | 34.2 ± 13.7 | 19.0 ± 4 |
| −/− | 3 | 12.4 ± 3.6 | 16.5 ± 9.9 | 22.4 ± 12.0 | 48.3 ± 11.9 | 0 |
| P value | 0.06 | 0.16 | 0.31 | 0.25 | ||
For E-CFU, M-CFU, GM-CFU, and GEMM-CFU assays, newborn liver hematopoietic cells (2 × 104 cells per assay) were suspended in 1 ml of methylcellulose as described. Colonies were counted after 10 days of culture. Approximately 50 colonies were observed in each assay using either +/− or −/− liver cells. Colony types were confirmed by morphologic analysis following cytocentrifugation of colonies onto slides and staining with Wright–Giemsa. Results are expressed as mean ± SD. P values were calculated using the Student’s t test. Three animals with each genotype were analyzed, except for results of G-CFU from +/− livers (n = 2). Wild-type (+/+) animals gave results similar to that obtained from heterozygotes (+/−). G-CFU assays were performed as described with 1 × 105 cells per assay and 1000 units/ml of human recombinant G-CSF. No G-CFU were observed in mutant or heterozygote animals in the absence of G-CSF. Results are expressed as total G-CFU per 105 cells. E-CFU, erythroid CFU.
The Defect in Granulocyte Development in C/EBPα Mice Is Intrinsic to the Hematopoietic System.
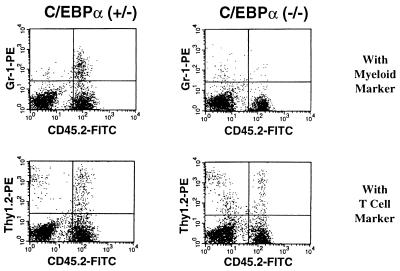
To demonstrate that the defect in granulopoiesis observed in c/ebpα−/− mice is a cell autonomous effect, fetal liver cells from C57/BL6J c/ebpα−/− mice were used in bone marrow transplantation experiments with a congenic strain, which can be distinguished by the cell surface marker CD45 (Ly5; refs. 51 and 52). Liver cells from +/+, +/−, or −/− mutant day 17 embryo C57/BL6J donors (CD45.2+) were transplanted into sublethally irradiated recipient mice (CD45.1+). Eight days after transplantation, mature granulocytes (Gr-1+/CD45.2+) derived from mouse hematopoietic progenitors from +/+ or +/− donors could be detected in the circulating blood of the recipients. However, no such population could be detected in the circulating blood of mice receiving fetal liver cells from −/− mice. Four weeks after transplantation, although a population of donor-derived T lymphocytes (Thy1+/CD45.2+) was present in the recipients, there were still no neutrophils of −/− donor origin in the circulating blood of recipients (Fig. 5). These studies show that the defect in granulopoiesis is intrinsic to the hematopoietic precursors of the c/ebpα−/− mice.
Figure 5.
Hematopoietic early progenitor cells from c/ebpα−/− mice do not undergo granulocytic lineage development in sublethally irradiated recipient mice. Flow cytometry analyses were performed by staining the blood cells collected from mice with antibodies against the mature granulocyte marker Gr-1, the T lymphocyte marker Thy1.2, and the congenic strain marker CD45.2 after 4 weeks of bone marrow transplantation. Donor cells are CD45.2+.
DISCUSSION
Our analysis of the effect of targeted disruption of the C/EBPα gene has demonstrated a selective block in the development of mature granulocytes. Development of other hematopoietic lineages, including monocytes and macrophages, was not affected. In the C/EBPα mutant mice, immature myeloid cells were present in the peripheral blood, and no defects were noted in GEMM-CFU or GM-CFU. These findings demonstrate that normal myeloid precursors are produced in these animals and that disruption of C/EBPα produces a block in differentiation along the granulocytic lineage.
One striking finding was the selective loss of G-CSF receptor mRNA expression in the major hematopoietic organ in the newborn, the liver. G-CSF receptor mRNA could not be detected by Northern blot analysis of C/EBPα −/− newborn liver mRNA (Fig. 3), but low levels were detectable by reverse transcriptase-PCR. This decrease in G-CSF receptor mRNA could be a result of decreased expression of G-CSF receptor mRNA at all stages of myeloid development, consistent with the presence of a functional binding site for C/EBPα in the G-CSF receptor promoter (30). Alternatively, the decrease in G-CSF receptor mRNA could reflect a lack of the cell population (mature neutrophils) expressing highest levels of the receptor. Studies are underway to attempt to distinguish these two explanations. Similarly, the lack of G-CFU could reflect a deficiency of a committed unipotential G-CFU progenitor, although multipotential myeloid progenitors (GEMM-CFU and GM-CFU) are clearly present.
Although previous studies in cell lines demonstrated functional C/EBP binding sites in the promoters for the M-CSF and GM-CSF receptors (31, 32), targeted deletion of C/EBPα did not affect the expression of these two receptors or the development of monocytes and macrophages. These findings could be explained by the expression and/or activation of other C/EBP family members, which are known to be capable of transactivating the M-CSF receptor and GM-CSF receptor promoters (31, 32). We have previously reported that neither C/EBPβ nor C/EBPδ mRNA levels were increased in livers from C/EBPα −/− mice compared with those observed in wild-type animals (14). Alternatively, the expression of these two myeloid growth factors could be activated by other transcription factors thought to be important in monocytic development, including PU.1 (53–56). Therefore, this animal provides an opportunity to study the differential effect of factors affecting development of different myeloid lineages, as well as the role of the G-CSF receptor on granulocyte development.
In addition, absence of C/EBPα increased the relative expression in newborn liver of erythropoietin receptor mRNA, which is critical for erythroid development (57). Morphologic examination of the livers of newborn animals, TER-119 FACS expression, and comparison of the number of erythroid CFU between mutant and wild-type animals did not demonstrate a marked increase in erythroid cells. It will be of interest to investigate a possible negative regulatory role of C/EBPα on erythropoiesis. Granulocyte development may require both up-regulation of C/EBPα as well as down-regulation of genes like GATA-1, which has a negative effect on myelopoiesis in cell lines (53, 58, 59).
In the hematopoietic system, C/EBPα is selectively expressed in myeloid cell lines, and not in erythroid or lymphoid cell lines (28, 60). Recent studies have demonstrated that C/EBPα is expressed at low levels in multipotential human CD34+ cells and is up-regulated as myeloid precursors form G-CFU in vitro (61). Preliminary studies using Northern blot analysis indicate that C/EBPα mRNA is expressed selectively at high levels in human peripheral blood neutrophils but not in peripheral blood monocytes (H. S. Radomska and D.G.T., unpublished data). These studies suggest a pattern of expression in which C/EBPα is expressed in multipotential progenitors and then maintained in granulocytes, but down-regulated in other lineages, including monocytes.
Our results demonstrate the essential role of C/EBPα in the development of mature granulocytes from myeloid precursors through the activation of a critical growth and differentiation receptor, the G-CSF receptor (42, 47, 48). However, targeted disruption of the genes encoding either G-CSF (62) or the G-CSF receptor (63) results in a decrease in peripheral blood neutrophils, but they are still present. In contrast, in the C/EBPα −/− mice, no mature granulocytes are observed, demonstrating that C/EBPα is truly a master regulator of granulocyte development and that critical targets in addition to the G-CSF receptor are likely to exist. Identification of these additional gene targets may provide insights into the mechanism of normal granulocytic development and those involved in acute myelogenous leukemia, in which a block in differentiation of myeloid precursors and persistence of blasts in peripheral blood, similar to the findings in the C/EBPα −/− animals, are key features of the disease.
Acknowledgments
We thank Bernard Mathey-Prevot, Alan D’Andrea, Larry Rohrschneider, and Laura Smith for probes; Donald Yergeau and Steven Ackerman for assistance with Fast green staining for eosinophils; Yuji Yamaguchi and Toshio Suda for assistance with G-CFU assays; Jean Flanagan and Aliki Nichojiannopoulou for technical assistance; Hui-Min Chen, Stuart Orkin, Ramesh Shivdasani, Milton Datta, and Linda Clayton for useful discussions; and Daniel Link and Richard Maki for communication of results regarding G-CSF receptor and PU.1 knockout mice before publication. Supported by National Institutes of Health Grants CA41456, CA59589, and HL56745.
Footnotes
Abbreviations: C/EBP, CCAAT enhancer binding protein; G-, M-, GM-, and GEMM-, granulocyte, macrophage, granulocyte–macrophage, and granulocytes, erythrocytes, monocyte/macrophages, and megaharyocytes, respectively; CSF, colony-stimulating factor; CFU, colony-forming unit; FACS, fluorescence-activated cell sorting.
References
- 1.Shivdasani R A, Orkin S H. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 2.Kehrl J H. Stem Cells. 1995;13:223–241. doi: 10.1002/stem.5530130304. [DOI] [PubMed] [Google Scholar]
- 3.Nichols J, Nimer S D. Blood. 1992;80:2953–2963. [PubMed] [Google Scholar]
- 4.Hromas R, Zon L, Friedman A D. Crit Rev Oncol Hematol. 1992;12:167–190. doi: 10.1016/1040-8428(92)90088-8. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro L H, Look A T. Curr Opin Hematol. 1995;2:3–11. doi: 10.1097/00062752-199502010-00002. [DOI] [PubMed] [Google Scholar]
- 6.Landschulz W H, Johnson P F, McKnight S L. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 7.Johnson P F, Landschulz W H, Graves B J, McKnight S L. Genes Dev. 1987;1:133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- 8.Landschulz W H, Johnson P F, Adashi E Y, Graves B J, McKnight S L. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 9.Costa R H, Grayson D R, Xanthopoulos K G, Darnell J E., Jr Proc Natl Acad Sci USA. 1988;85:3840–3844. doi: 10.1073/pnas.85.11.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman A D, Landschulz W H, McKnight S L. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 11.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu Y, Kelly T J, Lane M D. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 12.Cao Z, Umek R M, McKnight S L. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 13.Lin F T, Lane M D. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N D, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 15.Williams S C, Cantwell C A, Johnson P F. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 16.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams S C, Baer M, Dillner A J, Johnson P F. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman A D, McKnight S L. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 19.Nerlov C, Ziff E B. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas J G, Strobel M, Leutz A, Wendelgass P, Muller C, Sterneck E, Riethmuller G, Ziegler-Heitbrock H W. J Immunol. 1992;149:237–243. [PubMed] [Google Scholar]
- 21.Sterneck E, Muller C, Katz S, Leutz A. EMBO J. 1992;11:115–126. doi: 10.1002/j.1460-2075.1992.tb05034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ness S A, Kowenzleutz E, Casini T, Graf T, Leutz A. Genes Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- 23.Katz S, Kowenzleutz E, Muller C, Meese K, Ness S A, Leutz A. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metz R, Ziff E. Genes Dev. 1991;5:1754–1766. doi: 10.1101/gad.5.10.1754. [DOI] [PubMed] [Google Scholar]
- 25.Tsukada J, Saito K, Waterman W R, Webb A C, Auron P E. Mol Cell Biol. 1994;14:7285–7297. doi: 10.1128/mcb.14.11.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 27.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, Bistoni F, Frati L, Cortese R, Gulino A, Ciliberto G, Costantini F, Poli V. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott L M, Civin C I, Rorth P, Friedman A D. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 29.Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 30.Smith L T, Hohaus S, Gonzalez D A, Dziennis S E, Tenen D G. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 31.Hohaus S, Petrovick M S, Voso M T, Sun Z, Zhang D-E, Tenen D G. Mol Cell Biol. 1995;15:5830–5845. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D-E, Hetherington C J, Meyers S, Rhoades K L, Larson C J, Chen H M, Hiebert S W, Tenen D G. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oelgeschlager M, Nuchprayoon I, Luscher B, Friedman A D. Mol Cell Biol. 1996;16:4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ford A M, Bennett C A, Healy L E, Towatari M, Greaves M F, Enver T. Proc Natl Acad Sci USA. 1996;93:10838–10843. doi: 10.1073/pnas.93.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D-E, Hetherington C J, Gonzalez D A, Chen H M, Tenen D G. J Immunol. 1994;153:3276–3284. [PubMed] [Google Scholar]
- 36.Zon L I. Blood. 1995;86:2876–2891. [PubMed] [Google Scholar]
- 37.Springer T, Galfre G, Secher D S, Milstein C. Eur J Immunol. 1979;9:301–306. doi: 10.1002/eji.1830090410. [DOI] [PubMed] [Google Scholar]
- 38.Hestdal K, Ruscetti F W, Ihle J N, Jacobsen S E W, Dubois C M, Kopp W C, Longo D L, Keller J R. J Immunol. 1991;147:22–28. [PubMed] [Google Scholar]
- 39.Zhang D-E, Hohaus S, Voso M T, Chen H M, Smith L T, Hetherington C J, Tenen D G. Curr Top Microbiol Immunol. 1996;211:137–147. doi: 10.1007/978-3-642-85232-9_14. [DOI] [PubMed] [Google Scholar]
- 40.Metcalf D. Blood. 1993;82:3515–3523. [PubMed] [Google Scholar]
- 41.Sherr C J. Blood. 1990;75:1–12. [PubMed] [Google Scholar]
- 42.Nagata S, Fukunaga R. Growth Factors. 1993;8:99–107. doi: 10.3109/08977199309046930. [DOI] [PubMed] [Google Scholar]
- 43.Fukunaga R, Ishizaka-Ikeda E, Seto Y, Nagata S. Cell. 1990;61:341–350. doi: 10.1016/0092-8674(90)90814-u. [DOI] [PubMed] [Google Scholar]
- 44.D’Andrea A D, Lodish H F, Wong G G. Cell. 1989;57:277–285. doi: 10.1016/0092-8674(89)90965-3. [DOI] [PubMed] [Google Scholar]
- 45.Liboi E, Jubinsky P, Andrews N C, Nathan D G, Mathey-Prevot B. Blood. 1992;80:1183–1189. [PubMed] [Google Scholar]
- 46.Rothwell V M, Rohrschneider L R. Oncog Res. 1987;1:311–324. [PubMed] [Google Scholar]
- 47.Demetri G D, Griffin J D. Blood. 1991;78:2791–2808. [PubMed] [Google Scholar]
- 48.Avalos B R. Blood. 1996;88:761–777. [PubMed] [Google Scholar]
- 49.Molineux G, Pojda Z, Dexter T M. Blood. 1990;75:563–569. [PubMed] [Google Scholar]
- 50.Medlock E S, Kaplan D L, Cecchini M, Ulich T R, del Castillo J, Andresen J. Blood. 1993;81:916–922. [PubMed] [Google Scholar]
- 51.Shen F W, Tung J S, Boyse E A. Immunogenetics. 1986;24:146–149. doi: 10.1007/BF00364741. [DOI] [PubMed] [Google Scholar]
- 52.Komura K, Itakura K, Boyse E A, John M. Immunogenetics. 1975;1:452–456. [Google Scholar]
- 53.Voso M T, Burn T C, Wulf G, Lim B, Leone G, Tenen D G. Proc Natl Acad Sci USA. 1994;91:7932–7936. doi: 10.1073/pnas.91.17.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott E W, Simon M C, Anastai J, Singh H. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 55.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 56.Henkel G W, McKercher S R, Yamamoto H, Anderson K L, Oshima R G, Maki R A. Blood. 1996;88:2917–2926. [PubMed] [Google Scholar]
- 57.Wu H, Liu X, Jaenisch R, Lodish H F. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- 58.Visvader J E, Elefanty A G, Strasser A, Adams J M. EMBO J. 1992;11:4557–4564. doi: 10.1002/j.1460-2075.1992.tb05557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kulessa H, Frampton J, Graf T. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 60.Zhang D-E, Fujioka K I, Hetherington C J, Shapiro L H, Chen H M, Look A T, Tenen D G. Mol Cell Biol. 1994;14:8085–8095. doi: 10.1128/mcb.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng T, Shen H, Giokas D, Gere J, Tenen D G, Scadden D T. Proc Natl Acad Sci USA. 1996;93:13158–13163. doi: 10.1073/pnas.93.23.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lieschke G J, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler K J, Basu S, Zhan Y F, Dunn A R. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 63.Lui F, Wu H Y, Wesselschmidt R L, Kornaga T, Link D C. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]