Abstract
Circadian clocks rhythmically coordinate biological processes in resonance with the environmental cycle. The clock function relies on negative feedback loops that generate 24-h rhythms in multiple outputs. In Arabidopsis thaliana, the clock component TIMING OF CAB EXPRESSION1 (TOC1) integrates the environmental information to coordinate circadian responses. Here, we use chromatin immunoprecipitation as well as physiological and luminescence assays to demonstrate that proper photoperiodic phase of TOC1 expression is important for clock synchronization of plant development with the environment. Our studies show that TOC1 circadian induction is accompanied by clock-controlled cycles of histone acetylation that favor transcriptionally permissive chromatin structures at the TOC1 locus. At dawn, TOC1 repression relies on the in vivo circadian binding of the clock component CIRCADIAN CLOCK ASSOCIATED1 (CCA1), while histone deacetylase activities facilitate the switch to repressive chromatin structures and contribute to the declining phase of TOC1 waveform around dusk. The use of cca1 late elongated hypocotyl double mutant and CCA1-overexpressing plants suggests a highly repressing function of CCA1, antagonizing H3 acetylation to regulate TOC1 mRNA abundance. The chromatin remodeling activities relevant at the TOC1 locus are distinctively modulated by photoperiod, suggesting a mechanism by which the clock sets the phase of physiological and developmental outputs.
INTRODUCTION
Many organisms show a cyclic pattern of activity with a period of 24 h (Young and Kay, 2001). These biological rhythms are controlled by an endogenous oscillator or biological clock that temporally coordinates physiology and development in anticipation of the environmental day/night cycles. Such anticipation confers a selective advantage, allowing organisms to increase their fitness and maximize their possibilities of reproductive success and survival (Ouyang et al., 1998; Green et al., 2002; Michael et al., 2003; Dodd et al., 2005). Despite differences in complexity among species, the presence of at least one self-sustained central oscillator has emerged as a common theme in a number of diverse circadian systems (Harmer et al., 2001; Brunner and Schafmeier, 2006). With the exception of the cyanobacteria oscillator that may run independently of transcription (Nakajima et al., 2005; Tomita et al., 2005), the mechanism of clock function is highly conserved in eukaryotes and involves transcriptional autoregulatory loops of positive and negative oscillator components. The negative components feed back to repress their own expression by inhibiting the positively acting oscillator elements (Harmer et al., 2001; Bell-Pedersen et al., 2005). The identification of new components and additional mechanisms of clock regulation have revealed a higher complexity involving various transcriptional/translational interlocked feedback loops that govern the plasticity of the oscillatory activities (Rand et al., 2004; Locke et al., 2006; Zeilinger et al., 2006).
In plants, the first clock components proposed to function at the core of the Arabidopsis thaliana oscillator included the single MYB transcriptional factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) (Wang and Tobin, 1998) and LATE ELONGATED HYPOCOTYL (LHY) (Schaffer et al., 1998). Constitutive overexpression of either gene was found to cause a general arrhythmia, while the loss-of-function mutations retained rhythmicity albeit with a shortened period (Schaffer et al., 1998; Wang and Tobin, 1998; Green and Tobin, 1999). By analyzing mutant phenotypes (Millar et al., 1995), the pseudoresponse regulator TIMING OF CAB EXPRESSION1 (TOC1) (Strayer et al., 2000) was also proposed as a central part of the Arabidopsis oscillator. TOC1 mutant plants exhibited a shortened period phenotype in a number of rhythms (Somers et al., 1998; Strayer et al., 2000), while its overexpression was found to cause arrhythmia (Makino et al., 2002; Más et al., 2003b). Additionally, TOC1 was inferred to have an important function as a molecular link between the environmental signals and the rhythms of circadian and photomorphogenic outputs (Más et al., 2003b). The reciprocal regulation between the three components, CCA1, LHY, and TOC1, was anticipated as one of the loops controlling rhythmicity in Arabidopsis (Alabadí et al., 2001). The partly redundant transcription factors CCA1 and LHY (Alabadí et al., 2002; Mizoguchi et al., 2002) were proposed to repress TOC1 expression (Alabadí et al., 2001) through direct binding to the Evening Element (EE) motif present at the TOC1 promoter (Harmer et al., 2000). Increased TOC1 expression was in turn predicted to directly or indirectly activate the transcription of CCA1 and LHY (Alabadí et al., 2001). Additional clock core components also participate in CCA1/LHY activation. Some of these clock components include GIGANTEA (Fowler et al., 1999; Park et al., 1999), EARLY-FLOWERING4 (Doyle et al., 2002), PHYTOCLOCK1 (Onai and Ishiura, 2005), or LUX ARRHYTHMO (Hazen et al., 2005).
An essential property of clock function is its capacity to maintain rhythms in phase with the environment (Devlin and Kay, 2001). This property highly relies on the clock synchronization by the daily changes in light and temperature that adjust the phase of the rhythms relative to the external day/night cycles. In animals and fungi, the phase adjustments occur through light-dependent changes in the expression and activity of oscillator components that ultimately define the phase of the clock (Crosthwaite et al., 1995; Shigeyoshi et al., 1997; Suri et al., 1998; Akashi and Nishida, 2000; Elvin et al., 2005). In temperate regions, daily rhythms in light and temperature markedly change through the seasons, with variations in daylength or photoperiod providing a predictable indication of the time of year. Daylength perception is particularly important for plants to regulate a number of developmental transitions, including the initiation of flowering (Davis, 2002; Searle and Coupland, 2004). The temporal information provided by the biological clock together with light signals enable plants to regulate the initiation of flowering when light coincides with a sensitive phase of the oscillatory cycle (Carré, 2001; Yanovsky and Kay, 2003). In Arabidopsis, the photoinducible phase is determined by the diurnal transcript oscillation (Suárez-López et al., 2001) and protein stability (Valverde et al., 2004) of the flowering time component CONSTANS (CO) that is able to activate its target, FLOWERING LOCUS T (FT) (Suárez-López et al., 2001). Thus, the coincidence of CO photoperiodic rhythm relative to the light cycle was proposed as the basis for the earlier flowering of Arabidopsis plants under inductive long days (Roden et al., 2002; Yanovsky and Kay, 2002).
Despite the importance of circadian clock function on plant growth and development (Más, 2005; McClung, 2006), the mechanisms responsible for regulation of oscillator expression remain poorly understood. Responses to different environmental signals, including changes in light (Chua et al., 2003; Bertrand et al., 2005; Benhamed et al., 2006) and temperature (Bastow et al., 2004; Sung and Amasino, 2004), have been correlated with the reversible modulation of chromatin folding (Huebert and Bernstein, 2005) that facilitates the transcriptional regulation of inducible genes. The chromatin changes highly rely on posttranslational modifications of histone tails (e.g., methylation, acetylation, phosphorylation, and ubiquitination) (Mellor, 2006). In this sense, histone hyperacetylation has been associated with relaxed chromatin fibers that facilitate transcriptional activation (Grunstein, 1997; Eberharter and Becker, 2002), whereas a compacted chromatin and gene repression have been correlated with hypoacetylated histones (Kuo and Allis, 1998). Here, we demonstrate that the circadian clock regulates the rhythmic changes in histone acetylation-deacetylation at the TOC1 promoter. We also show that the circadian binding of CCA1 facilitates the reversible switch between a transcriptionally permissive chromatin state and a repressive chromatin compaction at the TOC1 locus. Our studies provide evidence that these clock-controlled changes in chromatin structure modulate the expression of the TOC1 gene under different photoperiods. We propose that the photoperiodic phase of TOC1 expression is important for appropriate perception of daylength and regulation of the photoperiod-dependent developmental responses in Arabidopsis.
RESULTS
TOC1 Circadian Expression Correlates with Clock-Controlled Rhythms of Histone Acetylation and Binding of Chromatin Remodeling Factors
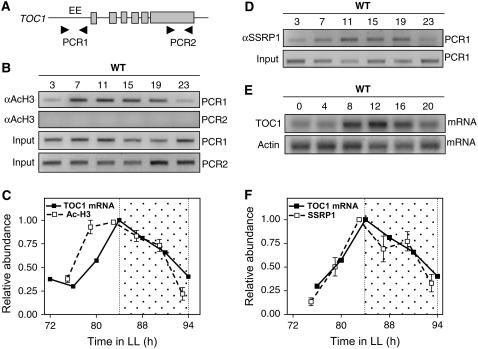
TOC1 exhibits high-amplitude rhythmic mRNA (Strayer et al., 2000) and protein (Más et al., 2003a) oscillations that are central in the control of circadian period by the clock (Más et al., 2003a). Since chromatin remodeling is an important mechanism in the epigenetic regulation of gene expression (Mellor, 2006), we examined whether changes in chromatin structure correlated with the rhythmic oscillation of the TOC1 transcript. To this end, wild-type seedlings were entrained in 12-h-light/12-h-dark cycles (LD) followed by constant light conditions (LL). Chromatin immunoprecipitation (ChIP) assays were performed with an antiacetylated Histone 3 antibody (αAcH3) and subsequent PCR analysis of the TOC1 promoter. The amplified promoter region contains the EE motif (Figure 1A) essential for circadian expression of the TOC1 gene (Alabadí et al., 2001). Our results showed amplified PCR bands throughout the circadian cycle (Figure 1B, PCR1) revealing the in vivo pattern of H3 acetylation at the TOC1 promoter. No amplification was observed with primers flanking a region in the last exon of the TOC1 gene (Figures 1A and 1B, PCR2) or when samples were incubated without antibody during the immunoprecipitation procedure (data not shown). The pattern of acetylated H3 followed a robust circadian oscillation that closely resembled the rhythmic expression of the TOC1 transcript (Figure 1C), while no evident oscillation was observed when ChIP assays were performed with an antibody to unmodified H3 (see Supplemental Figure 1 online). Compared with TOC1 mRNA accumulation, the phase of acetylated H3 was slightly advanced, suggesting that histone acetylation precedes the transcription activity. Together, our results indicate that the circadian transcription of the TOC1 gene is preceded by rhythms of H3 acetylation that are controlled by the biological clock. The oscillations in H3 acetylation might rhythmically change the chromatin structure at the TOC1 promoter conferring differential accessibility to the transcriptional machinery and regulators controlling the expression of the TOC1 gene.
Figure 1.
Circadian Rhythms of H3 Acetylation and SSRP1 Binding to the TOC1 Promoter.
(A) Structure of the TOC1 gene. Gray boxes delimitate exons. Arrowheads indicate the regions for PCR amplification in the ChIP assays.
(B) Representative PCR bands from ChIP assays in wild-type seedlings entrained under LD cycles and subsequently released to LL conditions. Chromatin was immunoprecipitated as specified in Methods using antiacetylated histone H3 (αAcH3) antibody. Input DNA was used as a control.
(C) Relative changes in acetylated H3 and TOC1 mRNA abundance plotted relative to the highest value. Data are represented as mean ± se of three independent experiments.
(D) Representative PCR bands from wild-type seedlings entrained under LD cycles and shifted to LL conditions. Samples were processed by ChIP assays with anti-SSRP1 antibody (Duroux et al., 2004). Input DNA was used as a control.
(E) TOC1 mRNA expression in wild-type seedlings analyzed by RNA gel blots. Actin mRNA was used as a control.
(F) Relative changes in SSRP1 binding and TOC1 mRNA abundance plotted relative to the highest value. Data are representative of at least two independent experiments. Open and dotted boxes indicate the subjective day and subjective night, respectively.
To further investigate the connection between clock-controlled changes in chromatin structure and the regulation of TOC1 expression, we analyzed by ChIP assays the in vivo binding of structure-specific recognition protein1 (SSRP1), one of the components of the FACT (for Facilitates Chromatin Transcription) complex (Duroux et al., 2004). The complex assists transcription by modulating chromatin structure (Grasser, 2005). Consistent with this function, FACT localizes to inducible genes, and its presence was correlated with actively transcribed genes (Duroux et al., 2004). In our experiments, chromatin was immunoprecipitated with the anti-SSRP1 antibody (provided by K.D. Grasser, Aalborg University, Aalborg, Denmark) (Duroux et al., 2004) followed by PCR amplification of the EE-containing region of the TOC1 promoter. The results showed efficiently amplified bands (Figure 1D) consistent with the binding of SSRP1 to the TOC1 locus. The abundance of the amplified bands rhythmically oscillated with a similar circadian phase to that observed for TOC1 mRNA expression (Figures 1D to 1F). ChIP studies with an antibody to the other subunit of the FACT complex (suppressor of Ty 16 [Spt16]) also revealed a rhythmic binding of Spt16 to the TOC1 promoter (see Supplemental Figure 1 online). These results indicate that the circadian transcription of the TOC1 gene correlates with circadian rhythms in the binding of chromatin remodeling factors that function as transcriptional coactivators.
The Biological Clock Regulates the Rhythmic Binding of CCA1 to the TOC1 Promoter
In vitro studies have shown that CCA1 interacts with the EE motif present at the TOC1 promoter (Alabadí et al., 2001). However, the functional relevance of this binding has raised a number of questions due to the lack of in vivo experimental confirmation (Locke et al., 2005). To examine whether CCA1 indeed binds to this region in vivo, we performed ChIP experiments using an anti-CCA1 antibody (kindly provided by E.M. Tobin, University of California, Los Angeles) (Wang and Tobin, 1998) and PCR analysis of the EE-containing region of the TOC1 promoter. Our results showed a rhythmic oscillation in the amplified bands throughout the circadian cycle (see Supplemental Figure 2 online), indicating that the biological clock controls the binding of CCA1 to the TOC1 promoter. No amplification was observed with primers flanking a region in the last exon of the TOC1 gene (data not shown). The CCA1 binding peak occurred between CT23 and CT3, whereas the promoter was minimally occupied during the subjective night, at times when TOC1 expression is maximal. Our results showed that the phase of CCA1 binding strongly correlated with the rhythmic pattern of CCA1 protein abundance (Wang and Tobin, 1998). Together, these results indicate that CCA1 binds in vivo to the TOC1 promoter, and this binding is controlled by the biological clock. Comparisons of CCA1 binding with the pattern of H3 acetylation revealed antiphasic oscillatory waveforms (see Supplemental Figure 2 online), suggesting an antagonistic relationship of these two events at the TOC1 promoter.
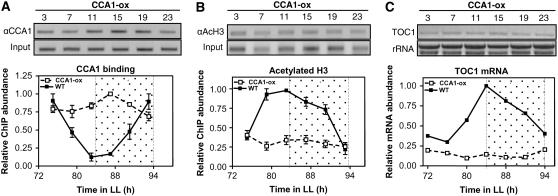
CCA1 Binding Represses Both H3 Acetylation and TOC1 mRNA Expression
Our studies have shown that the pattern of H3 acetylation is out of phase compared with CCA1 binding to the TOC1 promoter. In an attempt to examine the relationship between both mechanisms of regulation, we performed ChIP assays with CCA1-overexpressing plants (CCA1-ox) (Wang and Tobin, 1998) to analyze the effects of constitutive CCA1 expression on both CCA1 binding and H3 acetylation. In ChIP assays using the CCA1 antibody, we observed PCR amplification of the TOC1 promoter at all circadian times examined (Figure 2A) in contrast with the restricted CCA1 circadian binding in wild-type plants. These results indicate that the protein remains bound to the TOC1 promoter throughout the circadian cycle and suggest that CCA1 constant promoter occupancy might be responsible for the clock phenotypes observed in CCA1-ox plants. Conceivably, this could also reflect a similar DNA binding affinity of CCA1 to the TOC1 promoter irrespective of the circadian time. When the ChIP assays were performed using the anti-AcH3 antibody, we observed constant and close to background amplified PCR bands (Figure 2B), suggesting that overexpression of CCA1 decreases H3 acetylation and abolishes its rhythmicity at the TOC1 promoter. Comparisons of CCA1 binding with H3 acetylation and TOC1 mRNA expression in CCA1-ox plants (Figures 2A to 2C) indicate an antagonistic relationship, consistent with the notion that constant CCA1 binding represses H3 acetylation and TOC1 mRNA accumulation. To further confirm the opposed relationship of CCA1 and H3 acetylation, we performed ChIP experiments with the previously described cca1 lhy double mutant plants (Alabadí et al., 2002; Mizoguchi et al., 2002). Our studies showed that H3 acetylation at the TOC1 promoter was significantly increased in cca1 lhy mutants compared with wild-type plants (see Supplemental Figure 3 online). The increment was particularly evident before and after subjective dawn, the proposed times for CCA1/LHY action. Collectively, our results suggest that transcription of the TOC1 gene is regulated by clock-controlled changes in H3 acetylation and SSRP1/Spt16 binding that allow activation of TOC1 transcription (Figure 1; see Supplemental Figure 1 online), while rhythmic binding of CCA1 would function in TOC1 transcriptional repression (Figure 2; see Supplemental Figures 2 and 3 online). Mechanistically, our results suggest a very strong repressing function of CCA1 that impedes H3 acetylation at the TOC1 promoter and results in decreased TOC1 mRNA abundance.
Figure 2.
In Vivo CCA1 Binding, H3 Acetylation, and TOC1 mRNA Accumulation in CCA1-ox Plants.
(A) and (B) Representative PCR bands from ChIP assays with CCA1-ox plants using anti-CCA1 antibody (A) or anti-AcH3 antibody (B). Plants were entrained under 12-h-light/12-h-dark cycles and subsequently released to LL conditions. Input DNA was used as control.
(C) TOC1 mRNA expression in CCA1-ox plants analyzed by RNA gel blots. rRNA was used as a control. In each case, relative changes were plotted relative to the highest value. Data are representative of at least two independent experiments. Open and dotted boxes indicate the subjective day and subjective night, respectively.
Histone Deacetylase Activities Initiate the Declining Phase of TOC1 Rhythmic Expression and Contribute to TOC1 Photoperiodic Regulation
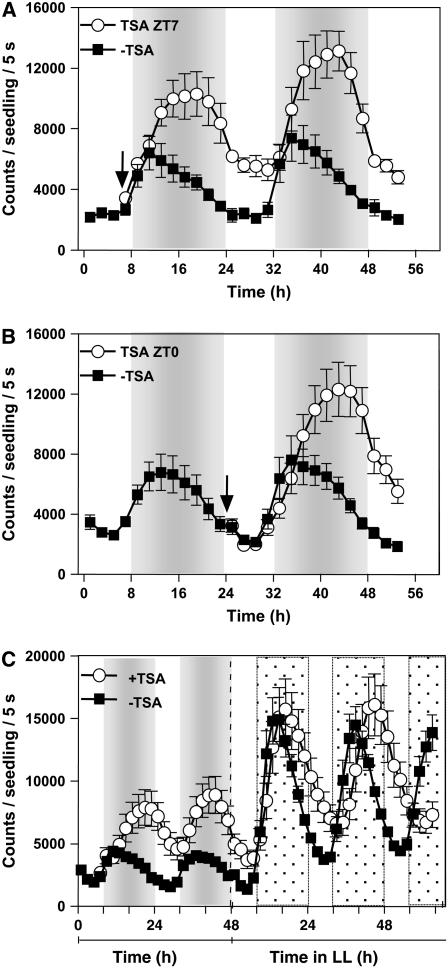
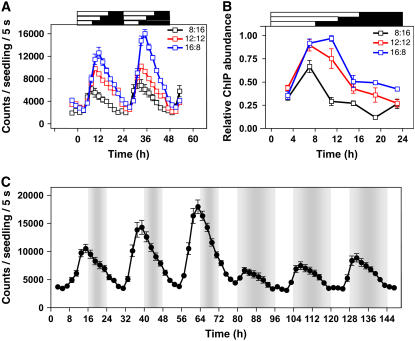
If changes in chromatin structure are important for TOC1 regulation, histone deacetylase (HDAC) activities also might be functionally relevant at the TOC1 promoter. If that is the case, blocking histone deacetylation should affect TOC1 rhythmic expression. To explore this possibility, we analyzed luminescence from plants expressing the TOC1 promoter fused to LUCIFERASE (LUC) as a reporter gene. Luminescence from TOC1:LUC-expressing seedlings was examined under entraining cycles in the absence or in the presence of trichostatin A (TSA), a potent HDAC inhibitor (Chang and Pikaard, 2005). Our results showed that in TSA-treated seedlings, TOC1:LUC expression diurnally oscillated albeit with higher amplitude than the one observed in untreated seedlings (Figure 3). When TSA was applied at Zeitgeber time 7 (ZT7, Figure 3A), a phase shift and a rapid upregulation of TOC1:LUC expression was observed after 3 to 4 h with the inhibitor (i.e., around ZT10 to ZT11). The specific point of TSA action was not due to the time required for the inhibitor to have an effect, as adding TSA at ZT0 (Figure 3B) led to a similar increment in TOC1:LUC luminescence signals also around ZT10. ChIP experiments with TSA-treated plants confirmed that the observed changes in TOC1 expression correlated with an increased pattern of H3 acetylation at the TOC1 promoter (see Supplemental Figure 4 online). Together, these results suggest that HDACs initiate their deacetylation activity mainly at the time when TOC1 expression starts to decline. When seedlings were shifted to LL, a circadian phase delay was observed in TSA-treated seedlings (Figure 3C). However, the amplitude was quite similar to that observed in untreated plants. The comparable amplitudes were due to an evident increment in TOC1:LUC expression after switching the plants to LL conditions. The higher amplitude under LL suggests that the presence of light at subjective dusk modifies the expression of the TOC1 gene. If this is the case, different photoperiods might distinctively affect TOC1 waveform. Indeed, our results showed that longer photoperiods led to higher amplitude (Figure 4A) and delayed phase (Figure 5A) of TOC1:LUC oscillation, indicating a robust photoperiod-dependent regulation. A direct switch from long-day (LgD) to short-day (ShD) conditions resulted in a very rapid decrease in amplitude and advanced phase (Figure 4C), indicating that developmental differences were not responsible for the TOC1 distinctive expression under the various photoperiods. The shape of TOC1:LUC oscillation under LgD was very similar to the one observed in ShD after TSA treatment (Figures 3A and 4A), suggesting that the photic input at the light-to-dark transition modulates HDAC activities relevant for photoperiodic regulation of TOC1 expression. In support of this hypothesis, our ChIP assays revealed that changing daylength had an important effect on the pattern of H3 acetylation at the TOC1 promoter (Figure 4B). This pattern correlated with the different waveforms of TOC1 under each photoperiod (cf. Figures 4A and 4B) and was antiphasic to that observed for CCA1 binding (see Supplemental Figure 5 online). Together, these results indicate that photoperiodic conditions modulate the chromatin structure at the TOC1 locus that in turn controls the transcriptional state of the gene. The complex interplay among histone modifications and CCA1 binding ensures low abundance of TOC1 at dawn and peaks around dusk, which generates a 24-h waveform of TOC1 expression.
Figure 3.
Effects of TSA Treatment on TOC1 Expression.
(A) and (B) Luminescence of transgenic plants carrying the TOC1:LUC transgene examined in the absence or presence of TSA, added at ZT7 (A) or ZT0 (B).
(C) Luminescence signals after switching to LL conditions. Experiments were performed as described in Methods. Dotted line indicates the initiation of free-running conditions in LL.
Data are the means ± se of the luminescence of 6 to 12 individual seedlings. Arrows indicate the Zeitgeber time of TSA treatment. Data are representative of three independent experiments. Open and closed boxes indicate the light and dark periods, respectively.
Figure 4.
Photoperiodic Regulation of TOC1 Expression.
(A) and (B) Luminescence of TOC1:LUC seedlings (A) and in vivo H3 acetylation at the TOC1 promoter (B) after ChIP assays in plants entrained under of 16-h-light/8-h-dark (16:8), 12-h-light/12-h-dark (12:12), or 8-h-light/16-h-dark (8:16) cycles. Samples were processed as described in Methods.
(C) Luminescence of TOC1:LUC plants entrained to LgD cycles (16 h light/8 h dark) and subsequently changed to a ShD regime (8 h light/16 h dark).
Data are the means ± se of the luminescence of 6 to 12 individual seedlings. Data are representative of at least two independent experiments. Open and closed boxes indicate the light and dark periods, respectively.
Figure 5.
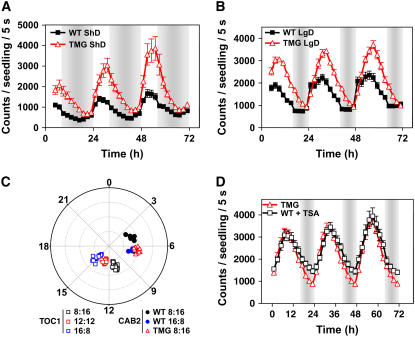
Role of TOC1 in Setting the Phase of CAB2:LUC Expression under Different Photoperiods.
(A) and (B) CAB2:LUC luminescence in wild-type and TMG plants entrained to ShD cycles ([A]; 8 h light/16 h dark) and LgD cycles ([B]; 16 h light:8 h dark). Data are the means ± se of the luminescence of 6 to 12 individual seedlings.
(C) Phase plot of TOC1:LUC and CAB2:LUC expression in wild-type and TMG plants under the indicated photoperiods. Phases (phase/period × 24 h) were plotted against the strength of the rhythm expressed as relative amplitude error. The rhythm strength is graphed from 0 (center of the plot) to 0.8 (periphery of the circle), which indicates robust and very weak rhythms, respectively.
(D) CAB2:LUC expression in TMG and TSA-treated wild-type plants entrained to 16-h-light/8-h-dark cycles. Data are the means ± se of the luminescence of 6 to 12 individual seedlings. Open and closed boxes indicate the light and dark periods, respectively.
Accurate Regulation of TOC1 Expression Is Important for Setting the Photoperiodic Phase of Clock-Controlled CAB2 Gene Expression
Clock synchronization with the environment occurs through adjustments in the expression of clock components that ultimately define the phase of the oscillator (Crosthwaite et al., 1995; Shigeyoshi et al., 1997; Suri et al., 1998; Xu et al., 2000). If the chromatin-dependent phase of TOC1 waveform under different photoperiods is functionally relevant for daylength discrimination, alterations in the regulation of TOC1 expression should correlate with changes in the photoperiodic phase of clock-controlled outputs. To explore this hypothesis, we examined the effects of TSA on the expression of CAB2:LUC, a circadian-regulated output that is modulated by photoperiod (Millar and Kay, 1996). Since CAB2 expression might be also regulated by H3 acetylation (Bertrand et al., 2005; Benhamed et al., 2006), we investigated the role of TOC1 in setting the phase of CAB2 expression using the TOC1 MiniGen (TMG) plants, expressing additional copies of the TOC1 gene (Más et al., 2003b). The basis for this experiment relies on our observations showing that the pattern of TOC1 expression in TSA-treated wild-type plants is highly similar to that previously observed in TMG plants (Más et al., 2003b). Our studies revealed that the alteration of TOC1 expression in TMG plants led to higher amplitude and delayed phase of CAB2:LUC luminescence under both ShD and LgD conditions (Figures 5A and 5B). In TMG plants, CAB2:LUC oscillation under ShD conditions was delayed such that its phase coincided with that of wild-type plants under LgD conditions (Figure 5C). Furthermore, the treatment of wild-type plants with TSA led to a similar CAB2:LUC waveform than the one observed in TMG plants (Figure 5D). The TSA-induced changes on CAB2:LUC expression were clearly reduced in the toc1-2 mutant background and increased in TMG plants (see Supplemental Figure 6 online), suggesting that in addition to the contribution of other possible regulators, the TSA effects are mediated, at least in part, by TOC1. Our results suggest that HDAC activities regulating the phase and amplitude of TOC1 expression at the light-to-dark transitions are important for appropriate photoperiodic regulation of CAB2:LUC expression.
Accurate Regulation of TOC1 Expression Is Important for Setting the Phase of Clock-Controlled Developmental Outputs
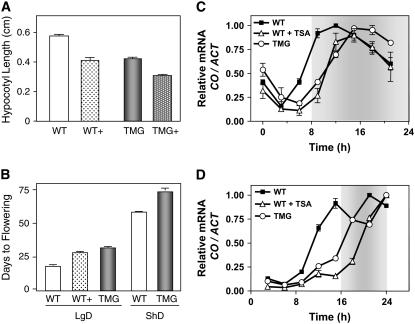
Previous studies have shown that the circadian clock regulates the rhythm of cell expansion controlling hypocotyl elongation (Dowson-Day and Millar, 1999). If changes in the phase of TOC1 are important for setting the phase of the clock, alteration of TOC1 expression by TSA should affect physiological and developmental outputs. To explore this hypothesis, the hypocotyl length of wild-type and TMG seedlings grown in the presence or absence of TSA was measured under ShD conditions. Our results revealed that TSA-treated seedlings displayed significantly shorter hypocotyls than those observed in untreated samples (Figure 6A). The use of the inhibitor in wild-type seedlings led to hypocotyl lengths very similar to the ones observed in untreated TMG plants (Figure 6A), suggesting that in addition to other possible regulators, misexpression of TOC1 contributes, at least in part, to the observed phenotype. Similar to what we had observed for CAB2:LUC expression, the TSA effects on hypocotyl elongation were much more reduced in toc1-2 mutant plants (see Supplemental Figure 7 online), suggesting a role for TOC1 in the TSA-induced changes of hypocotyl elongation.
Figure 6.
Role of TOC1 in Setting the Phase of Clock-Controlled Developmental Outputs.
(A) Hypocotyl lengths of wild-type and TMG plants under ShD (8 h light/16 h dark) conditions. Data are the mean hypocotyl length ± se of 15 to 20 seedlings grown in the presence (+) or absence of TSA.
(B) Transition to flowering in wild-type plants in the presence (+) or absence of TSA and in TMG plants maintained under LgDs (16 h light/8 h dark) or ShDs (8 h light/16 h dark). Flowering time is represented as the number of days to flowering (1-cm-high bolt). The experiments were done twice with similar results.
(C) and (D) Relative changes of CO expression in wild-type seedlings with or without TSA and in TMG seedlings maintained under ShD ([C]; 8 h light/16 h dark) or LgD ([D]; 16 h light/8 h dark) conditions. Relative changes in CO expression were plotted relative to the highest value. Open and closed boxes indicate the light and dark periods, respectively.
Daylength perception in plants is used as an indication of seasonal changes that regulate the developmental transition to flowering (Davis, 2002; Corbesier and Coupland, 2005). We therefore investigated whether the alteration of TOC1 expression affects the photoperiodic flowering pathway. To that end, we examined flowering time in wild-type and TMG plants under both ShD and LgD conditions. We also examined the effects of blocking histone deacetylation by TSA on the initiation of flowering. Our results revealed that both TMG and TSA-treated wild-type plants flowered significantly later than untreated wild-type plants (Figure 6B; see Supplemental Figure 8 online), indicating that in addition to other direct flowering regulators, the proper phase of TOC1 expression is important in the photoperiodic control of flowering time. To conclusively establish the connection between this flowering phenotype and an altered photoperiodic phase of expression, we examined the rhythmic oscillation of CO, a clock-controlled output directly involved in the photoperiodic regulation of flowering time (Hayama and Coupland, 2003). Our results showed that the delayed flowering of TMG and TSA-treated wild-type seedlings correlated with a phase shift in the rhythmic expression of CO under both ShD and LgD (Figures 6C and 6D, respectively). The delayed phase of expression led to a significant proportion of CO mRNA to accumulate in the dark, which rendered a decreased abundance of FT (data not shown). The changes in CO phase of expression relative to the day/night cycles and the low accumulation of FT most likely explain the flowering phenotypes of TMG and TSA-treated wild-type plants. Collectively, our results suggest that in addition to the direct contribution of other regulators, appropriate TOC1 expression is important for setting the phase of clock outputs, including gene expression, hypocotyl elongation, and the timing of the developmental transition to flowering.
DISCUSSION
Eukaryotic genomic DNA is packaged into compacted chromatin that limits the accessibility of regulatory transcription factors (Huebert and Bernstein, 2005). Acetylation of histone N-terminal tails provides a mechanism for reversible modulation of chromatin structure and transcriptional regulation (Jenuwein and Allis, 2001). Our findings show that the circadian transcription of the TOC1 gene is regulated by changes in chromatin structure. The chromatin modifications are controlled by the biological clock and involve a rhythmic pattern of histone acetylation at the TOC1 locus. The activation of a number of mammalian clock genes was also shown to be coupled with changes in histone acetylation (Etchegaray et al., 2003; Curtis et al., 2004; Naruse et al., 2004; Ripperger and Schibler, 2006). These results indicate that despite divergences in oscillator components, a chromatin-dependent mechanism of clock gene regulation is common to both plant and mammal circadian systems. The rhythmic pattern of H3 acetylation at the TOC1 promoter might be indicative of histone acetyl-transferase activities that are circadian regulated by the plant clock. In accordance with this hypothesis, recent studies have shown that the CLOCK protein, an essential component of the mammalian circadian system, is a histone acetyl-transferase (Doi et al., 2006). Compared with TOC1 mRNA accumulation, the circadian phase of histone acetylation is slightly advanced, suggesting that acetylation temporally precedes the transcriptional activity and may favor the accessibility of activators at specific circadian phases. Indeed, proteins of the FACT complex (SSRP1 and Spt16) implicated in transcriptional elongation (Duroux et al., 2004) also bind to the TOC1 promoter in a circadian fashion and with a slightly delayed phase compared with H3 acetylation. The correlation between chromatin changes, FACT binding, and the transcriptional activation of the TOC1 gene is supported by recent findings showing that the chromatin remodeling FACT complex associates with actively transcribed genes in Arabidopsis (Duroux et al., 2004). It would be interesting to extend these studies to other clock components and determine the pervasiveness of chromatin changes on clock gene expression.
In vitro studies with bacterial extracts expressing glutathione S-transferase (GST)-CCA1 or GST-LHY have demonstrated the binding of these single MYB transcription factors to the EE motif present at the TOC1 promoter (Alabadí et al., 2001). The use of ChIP assays has allowed us to conclusively demonstrate that CCA1 binds to the TOC1 promoter in vivo and that this binding is controlled by the circadian clock. Further comparisons of circadian waveforms in CCA1 binding and H3 acetylation suggest that CCA1 might be antagonistic to transcriptionally permissive chromatin folding at the TOC1 promoter. Consistent with a highly repressive function of CCA1, the use of CCA1-overexpressing plants revealed a constant protein binding that correlates with a decreased pattern of both H3 acetylation and TOC1 mRNA accumulation. Studies with cca1 lhy double mutants also revealed an altered pattern of H3 acetylation that correlates with similar changes in TOC1 mRNA expression (Mizoguchi et al., 2002). The mechanism of CCA1 repression might involve recruitment of HDAC activities similar to mammalian CRY1 repressive function involving interaction with the Sin3B, HDAC1, and HDAC2 corepressor complex (Naruse et al., 2004). On the other hand, the fact that CCA1-ox and cca1 lhy double mutant plants affect most, if not all, rhythmic processes, opens up the possibility that the effects we observed are indirect, resulting from the severe clock phenotypes of these plants. However, the fact that CCA1 binds to the TOC1 promoter and that CCA1-ox reduces both H3 acetylation and TOC1 mRNA accumulation while the double cca1 lhy mutation has the contrary effect strongly suggest a direct effect of CCA1 on TOC1 regulation. Furthermore, our results showing that the pattern of acetylation remains antiphasic to that of CCA1 binding under different photoperiods reinforce our conclusions.
In addition to the proposed repressive function of CCA1, our studies with TSA indicate that TOC1 repression is controlled by HDAC activities. An advantage of using chemical inhibitors is that the timing of blockage of HDAC activity can be precisely controlled (Yoshida et al., 1995). Approximately 2% of endogenous mammalian genes are affected by HDAC inhibitors (Richon et al., 2000). In plants, these inhibitors were used to examine the role of HDAC activities in various processes, including, among others, nucleolar dominance (Chen and Pikaard, 1997) and root meristem proliferation (Murphy et al., 2000). Microarray studies in tobacco (Nicotiana tabacum) and Arabidopsis seedlings also showed that TSA induces changes in gene expression and affects the pattern of histone acetylation/deacetylation at specific genes (Chua et al., 2004; Chang and Pikaard, 2005). In our studies with TSA, we were able not only to demonstrate that HDAC activities regulate TOC1 expression but also to determine the timing of HDAC action. The changes from acetylated to deacetylated states of histones are believed to modify chromatin structure to induce transcriptional repression (Wolffe et al., 1997; Kuo and Allis, 1998; Struhl, 1998). Our studies show that HDAC activities contribute to TOC1 declining phase mainly after its peak of expression around the light-to-dark transition. The presence of light at this specific time window seems to importantly modulate the HDAC activities relevant for TOC1 expression. In that sense, the histone acetyltransferase GCN5 and HDAC HD1 have been previously shown to participate in the light regulation of gene expression (Benhamed et al., 2006). Our studies further extend these findings, providing evidence that the biological clock modulates histone-modifying enzymes relevant for regulation of oscillator expression. We also show a previously uncharacterized photoperiodic regulation of TOC1 expression, with increased daylength correlating with higher amplitude and delayed phase of TOC1 rhythmic oscillation. We describe that treatment with TSA changes the ShD shape of TOC1:LUC expression to a LgD waveform, indicating that modulation of HDAC activities contributes to TOC1 photoperiodic regulation. This is also consistent with our results showing a distinct photoperiodic pattern of histone acetylation at the TOC1 promoter. Interestingly, the pattern of acetylation remains antiphasic to that of CCA1 binding under the various photoperiods. The use of daylength cues allows organisms to track time of year and to anticipate predictable annual variations in environmental signals. An accurate photoperiodic perception is essential in maintaining appropriate clock-dependent phase relationships with the external day/night cycles (Saunders, 1997; Oster et al., 2002; Johnson et al., 2003; Merrow et al., 2006). Plants also use the circadian clock to perceive the changing photoperiods and to synchronize gene expression and physiology to the most favorable seasons (Más, 2005; McClung, 2006). Photoperiodic regulation of CAB2 expression (Millar and Kay, 1996), hypocotyl elongation (Dowson-Day and Millar, 1999), and the initiation of flowering (Hayama and Coupland, 2003) are some examples of clock-controlled processes modulated by photoperiodic conditions. In a number of different circadian systems, it has become increasingly clear that the mechanism for daylength measurement relies on the photoperiodic regulation of oscillator components (Messager et al., 1999; Lincoln et al., 2002; Sumova et al., 2003). We propose that plants might use a similar mechanism to appropriately adjust the phase of the clock relative to the different environmental cycles. Based on our experiments, we suggest that in addition to the contribution of other regulators, the HDAC activities relevant in TOC1 expression are important for daylength measurement and subsequent modulation of the photoperiodic phase of CAB2 and CO expression, hypocotyl elongation, and flowering time. Our studies also revealed that the effects of TSA were reduced in the toc1-2 mutant background. Still, we observed higher amplitude of CAB2:LUC expression than the one observed in untreated wild-type plants. Additional factors might contribute to this regulation, including the upregulation by TSA of other clock components and/or histone acetyltransferase and deacetylase activities of GCN5, HD1, and TAF1/HAF2, previously reported to participate in the regulation of a number of light-responsive genes (Bertrand et al., 2005; Benhamed et al., 2006). Among the clock components, CCA1 is a good candidate that might participate in CAB2 regulation. Previous studies have proposed TOC1 as a positive regulator of CCA1 (Alabadí et al., 2001); therefore, the effects on CAB2 expression might be mediated by upregulation of CCA1. However, when CCA1 transcript abundance was examined in TMG plants under LgD (see Supplemental Figure 9 online) or ShD (data not shown), we observed a delay in the phase but not an evident upregulation of CCA1. A phase delay and decreased amplitude of CCA1 was reported in TOC1-overexpressing plants under LL (Makino et al., 2002). These results suggest that the TMG phenotypes are not mediated by upregulation of CCA1 expression.
Although TSA most likely affects other clock components, our studies show that TSA treatment leads to higher amplitude and delayed phase of TOC1 expression. Interestingly, this pattern of TOC1 expression is highly similar to that observed in TMG plants with increased gene dosage (Más et al., 2003b). The reduced effects of the inhibitor in toc1-2 mutant plants and the similar alteration of photoperiodic perception in TSA-treated wild-type and TMG plants suggest that in addition to other possible regulators, misexpression of TOC1 importantly contributes to the observed photoperiodic phenotypes.
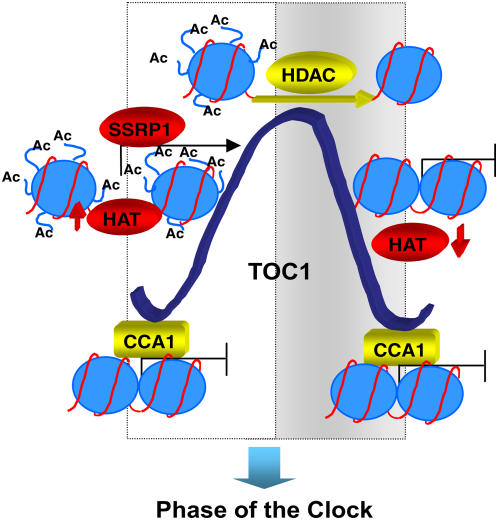
Altogether, our experiments have helped us to dissect TOC1 oscillatory waveform and to identify the mechanisms governing the transcriptional state of the TOC1 gene. The regulatory mechanisms involve activators and repressors that are precisely coordinated to generate 24-h oscillations in TOC1 expression (Figure 7). At dawn, the binding of CCA1 contributes to TOC1 transcriptional repression by impeding histone acetylation at the TOC1 promoter. Decreasing CCA1 protein abundance throughout the day releases the binding and the repression, allowing histone acetyl-transferase activities to acetylate histones at the TOC1 locus and facilitating the accessibility to the transcriptional machinery and activators. After TOC1 peak of expression, a repressive mechanism relying on HDAC activities contributes to the declining phase of TOC1 waveform. The histone deacetylation might facilitate the switch to repressive chromatin structures, promoting the condensation of nucleosomal fibers and/or facilitating the binding of repressive factors (e.g., CCA1). The rhythmic cycle would be again initiated by CCA1 binding around dawn. All these changes in chromatin structure at the TOC1 promoter regulate the transcriptional state of the TOC1 gene under different photoperiods. This regulation facilitates proper phase of entrainment of physiological and developmental outputs in Arabidopsis.
Figure 7.
Schematic Representation Depicting the Rhythmic Regulation of TOC1 Expression.
The circadian expression of TOC1 is controlled by dynamic changes in chromatin structure at the TOC1 locus. TOC1 repression depends on circadian binding of CCA1. Decreased CCA1 binding allows transcriptional activation through rhythmic cycles of histone acetylation and binding of SSRP1 (and Spt16) that favor transcriptionally permissive chromatin structures. HDAC activities after TOC1 peak of expression facilitate the switch to repressive chromatin structures and contribute to the declining phase of TOC1 waveform around dusk. Different photoperiodic conditions distinctively modulate these chromatin remodeling activities, defining a mechanism by which plants might synchronize the phase of the biological clock. Nucleosomes are shown as blue circles with the H3 N-terminal tails as curved lines in pale blue; black arrows indicate transcriptional activation, whereas lines ending in perpendicular dashes indicate repression. Open and shaded boxes indicate the light and dark periods, respectively. HAT, histone acetyl-transferase.
METHODS
Plant Growth Conditions and Bioluminescence Analysis
Arabidopsis thaliana seedlings were grown on Murashige and Skoog (MS) agar with 3% sucrose plates under the indicated LgD cycles with 50 μmol m−2 s−1 of cool white fluorescent light at 22°C. The CCA1-ox plants (Wang and Tobin, 1998) were kindly provided by E.M. Tobin (University of California, Los Angeles). The LUC vector (Millar et al., 1992), TMG (Más et al., 2003b), toc1-2 (Strayer et al., 2000), and cca1 lhy mutant plants (Alabadí et al., 2002) were previously described and provided by S.A. Kay (The Scripps Research Institute, La Jolla, CA). The cca1 lhy double mutant plants (Alabadí et al., 2002) were obtained after crosses of the cca1-1 mutant with the lhy RNA interference lines 48, 50, and 51 as previously described (Alabadí et al., 2002). The TOC1:LUC-expressing plants were generated by Agrobacterium tumefaciens–mediated transformation of plants with a construct containing the TOC1 promoter (from 1558 bp upstream the ATG) fused to LUC, which was used as the reporter gene. In experiments with the inhibitor, 0.1, 1, 3, 5, or 10 μM of TSA (T8552; Sigma-Aldrich) was added to germinated seedlings (∼1 week old) at the indicated time to examine the specific ZT of TSA action. Luminescence was examined and analyzed as previously described (Perales et al., 2006).
ChIP Assays
Two-week-old seedlings were immersed in buffer A (0.4 M sucrose, 10 mM Tris, pH 8, 1 mM EDTA, 1 mM PMSF, 1% formaldehyde, and 0.05% Triton X-100) under vacuum for 10 min followed by an additional 10-min incubation with 0.125 M glycine. Seedlings were ground in liquid nitrogen and resuspended in buffer B (0.4 M sucrose, 10 mM Tris, pH 8, 5 mM β-mercaptoethanol, 1 mM PMSF, 1 μg/mL aprotinin, 1 μg/mL pepstatin A, and 1 μg/mL leupeptin). Nuclei were then collected by centrifugation, resuspended in lysis buffer (50 mM Tris, pH 8, 10 mM EDTA, 1% SDS, 1 mM PMSF, 1 μg/mL aprotinin, 1 μg/mL pepstatin A, and 1 μg/mL leupeptin), and sonicated to ∼400- to 1000-bp fragments. After centrifugation, the supernatants were incubated in dilution buffer (15 mM Tris, pH 8, 150 mM NaCl, 1% Triton-X-100, 1 mM EDTA, 1 mM PMSF, 1 μg/mL aprotinin, 1 μg/mL pepstatin A, and 1 μg/mL leupeptin) overnight at 4°C with sepharose beads conjugated with the indicated antibodies. Anti-H3 and anti-acetyl-histone H3 antibody (Upstate Biotechnology), CCA1 antibody (Wang and Tobin, 1998) (kindly provided by E.M. Tobin, University of California, Los Angeles), and anti-SSRP1 and anti-Spt16 antibodies (Duroux et al., 2004) (kindly provided by K.D. Grasser, Aalborg University, Aalborg, Denmark) were used in this study. The immunocomplexes were washed four times with washing buffer (0.1% SDS, 1% Triton X-100, 1 mM EDTA, 1 mM PMSF, 1 μg/mL aprotinin, 1 μg/mL pepstatin A, and 1 μg/mL leupeptin) and eluted from the beads with 1% SDS and 0.1 M NaHCO3. Cross-links were reversed by incubation at 65°C for 5 to 6 h followed by proteinase K treatment for 1 h at 45°C, phenol/chloroform/isoamyl alcohol extraction, and ethanol precipitation. Pellets were washed with 70% ethanol and resuspended in TE buffer (10 mM Tris, pH 8, and 1 mM EDTA). The primers used for PCR amplification (PCR1) flanked the TOC1 promoter region containing the EE motif (Figure 1A), previously described to be essential for circadian expression of the TOC1 gene (Alabadí et al., 2001). PCR amplification was performed using 33 cycles and 54°C with primers 5′-CTTCTTATCTTGTATCTTACCAC-3′ and 5′-GAATTGGACGGTGGAGATTAAGTC-3′ for PCR1 and primers 5′-TCGGAGAGTCCTCTGCTTTC-3′ and 5′-ATAAGGTACCCAGTTCCCAAAGCATCATCCTGA-3′ for PCR2 (amplification of the last exon at the TOC1 locus). PCR amplification of inputs was performed using 23 cycles and 54°C. Aliquots of the PCR reactions were resolved by electrophoresis on a 2% agarose gel. The ethidium bromide–stained gels were quantitative in the range of DNA concentrations used. Gels were also stained with SYBR green (Molecular Probes) following the manufacturer's recommendations. Similar quantifications to those of ethidium bromide were obtained (see Supplemental Figure 10 online). Images were captured with the Kodak Digital Science System, and quantification was performed with ImageQuant software (Molecular Dynamics) and Scion Image software. The results presented here come from at least two independent experiments meaning independent time-course analysis, with independent chromatin preparations and with duplicate PCR amplification for each time point.
RNA Gel Blots and RT-PCR Analysis
RNA gel blots and RT-PCR analysis were performed with RNA from 12-d-old seedlings grown on MS with 3% sucrose agar plates in the absence or in the presence of TSA (10 μM). RNA was isolated using the RNeasy kit (Qiagen) and separated on 1.2% agarose/formaldehyde gels as previously described (Alabadí et al., 2001; Más et al., 2003b). Analysis was performed on a PhosphorImager using ImageQuant software (Molecular Dynamics) and Scion Image software. For RT-PCR, M-MLV reverse transcriptase (Invitrogen) was used to synthesize first-strand cDNA with oligo(dT32) (Termo Electron) from 2 μg of total RNA at 37°C for 50 min. cDNAs were diluted to 100 μL with TE buffer, and 1 μL of diluted cDNA was used for PCR amplification by TaKaRa Ex Taq. CO was amplified using 25 cycles at 55°C with primers 5′-ACGCCATCAGCGAGTTCC-3′ and 5′-AAATGTATGCGTTATGGTTAATGG-3′. Samples were separated on 1.5% agarose gels, transferred to nitrocellulose membranes (Hybond-N+; Amersham), and subjected to hybridization following standard protocols. The RT-PCR analysis was performed twice and with RNA samples from independent experiments.
Hypocotyl Length and Flowering-Time Assays
For hypocotyl length analysis, seeds were stratified on MS plates with 3% sucrose in the presence or in the absence of TSA. Plates were incubated in the dark at 4°C for 4 d, exposed to white light (50 μmol m−2 s−1) for 6 h, and in the dark for 18 h previous to exposure to ShD conditions. Hypocotyl length was measured after 7 d using the Scion Image software. Flowering time was scored by growing plants under ShD and LgD conditions as previously described (Más et al., 2003b). For TSA treatment, plants were grown under LgD conditions on MS plates in the presence or in the absence of inhibitor. The TSA concentration for gene expression and hypocotyl and flowering-time experiments was carefully chosen considering the type of output examined, the timing of TSA application, and the duration of the treatment. The experimental design was based on using the minimum amount of TSA (in relation to the duration of the treatment) that could elicit a clear effect in each output. In the case of hypocotyl elongation, seeds were grown and germinated on plates supplemented with 0.3 μM TSA, while for the long-term flowering-time experiments, the concentration of TSA was 10 μM.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Pattern of H3 Occupancy and Spt16 Binding to the TOC1 Promoter.
Supplemental Figure 2. Rhythmic Binding of CCA1 to the TOC1 Promoter in Wild-Type Seedlings.
Supplemental Figure 3. In Vivo H3 Acetylation in Wild-Type and cca1 lhy Double Mutant Plants.
Supplemental Figure 4. H3 Acetylation at the TOC1 Promoter in Plants Treated with Different Concentrations of TSA.
Supplemental Figure 5. Rhythmic Binding of CCA1 to the TOC1 Promoter.
Supplemental Figure 6. Comparison of the Effects of TSA Treatment on CAB2:LUC Expression in Wild-Type, TMG, and toc1-2 Mutant Plants.
Supplemental Figure 7. Role of TOC1 in Mediating the TSA-Dependent Effects on Hypocotyl Elongation.
Supplemental Figure 8. Transition to Flowering in Wild-Type Plants in the Presence or Absence of TSA and in TMG Plants Maintained under Long Days or Short Days.
Supplemental Figure 9. Relative Changes of CCA1 Expression in Wild-Type and TMG Seedlings Maintained under Long-Day Conditions.
Supplemental Figure 10. SYBR Green–Stained Gels Displaying PCR Amplifications of the TOC1 Promoter.
Supplementary Material
Acknowledgments
We thank M. Brunner, I. Carré, S. Davis, T. Stratmann, P. Casacuberta, and L. Espinás for discussions and comments on the experiments and the manuscript. We also thank C. Valdivieso for technical support and the greenhouse staff for help with the plant material. We also thank S. Kay for the vector containing the LUC, the TMG, toc1-2, and cca1 lhy mutant plants; E. Tobin for the CCA1 antibody and CCA1-ox seeds, and K. Grasser for the anti-SSRP1 and anti-Spt16 antibodies. This work was supported by grants to P.M. from the Spanish Ministry of Education and Science (BIO2004-02144), by a Marie Curie International Reintegration Grant within the 6th European Community Framework Program (FP6-IRG-012239), and by the European Heads of Research Councils and European Science Foundation through the European Young Investigator Award to P.M. M.P. is supported by an European Union grant.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Paloma Más (pmmgmc@ibmb.csic.es).
Online version contains Web-only data.
References
- Akashi, M., and Nishida, E. (2000). Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14 645–649. [PMC free article] [PubMed] [Google Scholar]
- Alabadí, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Más, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Alabadí, D., Yanovsky, M.J., Más, P., Harmer, S.L., and Kay, S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12 757–761. [DOI] [PubMed] [Google Scholar]
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen, D., Cassone, V.M., Earnest, D.J., Golden, S.S., Hardin, P.E., Thomas, T.L., and Zoran, M.J. (2005). Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed, M., Bertrand, C., Servet, C., and Zhou, D.-X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, C., Benhamed, M., Li, Y.-F., Ayadi, M., Lemonnier, G., Renou, J.-P., Delarue, M., and Zhou, D.-X. (2005). Arabidopsis HAF2 gene encoding TATA-binding Protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 280 1465–1473. [DOI] [PubMed] [Google Scholar]
- Brunner, M., and Schafmeier, T. (2006). Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes Dev. 20 1061–1074. [DOI] [PubMed] [Google Scholar]
- Carré, I.A. (2001). Day-length perception and the photoperiodic regulation of flowering in Arabidopsis. J. Biol. Rhythms 16 415–423. [DOI] [PubMed] [Google Scholar]
- Corbesier, L., and Coupland, G. (2005). Photoperiodic flowering of Arabidopsis: Integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 28 54–66. [Google Scholar]
- Crosthwaite, S.K., Loros, J.J., and Dunlap, J.C. (1995). Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81 1003–1012. [DOI] [PubMed] [Google Scholar]
- Curtis, A.M., Seo, S.-b., Westgate, E.J., Rudic, R.D., Smyth, E.M., Chakravarti, D., FitzGerald, G.A., and McNamara, P. (2004). Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 279 7091–7097. [DOI] [PubMed] [Google Scholar]
- Chang, S., and Pikaard, C.S. (2005). Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J. Biol. Chem. 280 796–804. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J., and Pikaard, C.S. (1997). Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, Y.L., Mott, E., Brown, A.P.C., MacLean, D., and Gray, J.C. (2004). Microarray analysis of chromatin-immunoprecipitated DNA identifies specific regions of tobacco genes associated with acetylated histones. Plant J. 37 789–800. [DOI] [PubMed] [Google Scholar]
- Chua, Y.L., Watson, L.A., and Gray, J.C. (2003). The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S.J. (2002). Photoperiodism: The coincidental perception of the season. Curr. Biol. 12 R841–R843. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F., and Kay, S.A. (2001). Circadian photoperception. Annu. Rev. Physiol. 63 677–694. [DOI] [PubMed] [Google Scholar]
- Dodd, A.N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., Hibberd, J.M., Millar, A.J., and Webb, A.A.R. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. [DOI] [PubMed] [Google Scholar]
- Doi, M., Hirayama, J., and Sassone-Corsi, P. (2006). Circadian regulator CLOCK is a histone acetyltransferase. Cell 125 497–508. [DOI] [PubMed] [Google Scholar]
- Dowson-Day, M.J., and Millar, A.J. (1999). Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17 63–71. [DOI] [PubMed] [Google Scholar]
- Doyle, M.R., Davis, S.J., Bastow, R.M., McWatters, H.G., Kozma-Bognar, L., Nagy, F., Millar, A.J., and Amasino, R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77. [DOI] [PubMed] [Google Scholar]
- Duroux, M., Houben, A., Ruzicka, K., Friml, J., and Grasser, K.D. (2004). The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 40 660–671. [DOI] [PubMed] [Google Scholar]
- Eberharter, A., and Becker, P.B. (2002). Histone acetylation: A switch between repressive and permissive chromatin. EMBO Rep. 3 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin, M., Loros, J.J., Dunlap, J.C., and Heintzen, C. (2005). The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes Dev. 19 2593–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray, J.-P., Lee, C., Wade, P.A., and Reppert, S.M. (2003). Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421 177–182. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser, K.D. (2005). Emerging role for transcript elongation in plant development. Trends Plant Sci. 10 484–490. [DOI] [PubMed] [Google Scholar]
- Green, R.M., Tingay, S., Wang, Z.Y., and Tobin, E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R.M., and Tobin, E.M. (1999). Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96 4176–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein, M. (1997). Histone acetylation in chromatin structure and transcription. Nature 389 349–352. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Panda, S., and Kay, S.A. (2001). Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17 215–253. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2003). Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 6 13–19. [DOI] [PubMed] [Google Scholar]
- Hazen, S.P., Schultz, T.F., Pruneda-Paz, J.L., Borevitz, J.O., Ecker, J.R., and Kay, S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebert, D.J., and Bernstein, B.E. (2005). Genomic views of chromatin. Curr. Opin. Genet. Dev. 15 476–481. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293 1074–1080. [DOI] [PubMed] [Google Scholar]
- Johnson, C.H., Elliott, J.A., and Foster, R. (2003). Entrainment of circadian programs. Chronobiol. Int. 20 741–774. [DOI] [PubMed] [Google Scholar]
- Kuo, M.H., and Allis, C.D. (1998). Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20 615–626. [DOI] [PubMed] [Google Scholar]
- Lincoln, G., Messager, S., Andersson, H., and Hazlerigg, D. (2002). Temporal expression of seven clock genes in the suprachiasmatic nucleus and the pars tuberalis of the sheep: Evidence for an internal coincidence timer. Proc. Natl. Acad. Sci. USA 99 13890–13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C., Kozma-Bognar, L., Gould, P.D., Feher, B., Kevei, E., Nagy, F., Turner, M.S., Hall, A., and Millar, A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C., Southern, M.M., Kozma-Bognar, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino, S., Matsushika, A., Kojima, M., Yamashino, T., and Mizuno, T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: Characterization with APRR1-overexpressing plants. Plant Cell Phisiol. 43 58–69. [DOI] [PubMed] [Google Scholar]
- Más, P. (2005). Circadian clock signaling in Arabidopsis thaliana: From gene expression to physiology and development. Int. J. Dev. Biol. 49 491–500. [DOI] [PubMed] [Google Scholar]
- Más, P., Alabadí, D., Yanovsky, M.J., Oyama, T., and Kay, S.A. (2003. b). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más, P., Kim, W.J., Somers, D.E., and Kay, S.A. (2003. a). Targeted degradation of TOC1 by ZTL modulates circadan function in Arabidopsis. Nature 426 567–570. [DOI] [PubMed] [Google Scholar]
- McClung, C.R. (2006). Plant circadian rhythms. Plant Cell 18 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor, J. (2006). Dynamic nucleosomes and gene transcription. Trends Genet. 6 320–329. [DOI] [PubMed] [Google Scholar]
- Merrow, M., Boesl, C., Ricken, J., Messerschmitt, M., Goedel, M., and Roenneberg, T. (2006). Entrainment of the Neurospora circadian clock. Chronobiol. Int. 23 71–80. [DOI] [PubMed] [Google Scholar]
- Messager, S., Ross, A.W., Barrett, P., and Morgan, P.J. (1999). Decoding photoperiodic time through Per1 and ICER gene amplitude. Proc. Natl. Acad. Sci. USA 96 9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T.P., Salome, P.A., Yu, H.J., Spencer, T.R., Sharp, E.L., McPeek, M.A., Alonso, J.M., Ecker, J.R., and McClung, C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302 1049–1053. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., and Kay, S.A. (1996). Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 93 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Short, S.R., Chua, N.H., and Kay, S.A. (1992). A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2 629–641. [DOI] [PubMed] [Google Scholar]
- Murphy, J.P., McAleer, J.P., Uglialoro, A., Papile, J., Weniger, J., Bethelmie, F., and Tramontano, W.A. (2000). Histone deacetylase inhibitors and cell proliferation in pea root meristems. Phytochemistry 55 11–18. [DOI] [PubMed] [Google Scholar]
- Nakajima, M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T., and Kondo, T. (2005). Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308 414–415. [DOI] [PubMed] [Google Scholar]
- Naruse, Y., Oh-hashi, K., Iijima, N., Naruse, M., Yoshioka, H., and Tanaka, M. (2004). Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 24 6278–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai, K., and Ishiura, M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10 963–972. [DOI] [PubMed] [Google Scholar]
- Oster, H., Maronde, E., and Albrecht, U. (2002). The circadian clock as a molecular calendar. Chronobiol. Int. 19 507–516. [DOI] [PubMed] [Google Scholar]
- Ouyang, Y., Andersson, C.R., Kondo, T., Golden, S.S., and Johnson, C.H. (1998). Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA 95 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. [DOI] [PubMed] [Google Scholar]
- Perales, M., Portolés, S., and Más, P. (2006). The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J. 46 849–860. [DOI] [PubMed] [Google Scholar]
- Rand, D.A., Shulgin, B.V., Salazar, D., and Millar, A.J. (2004). Design principles underlying circadian clocks. J. R. Soc. Interface 1 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon, V.M., Sandhoff, T.W., Rifkind, R.A., and Marks, P.A. (2000). Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA 97 10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger, J.A., and Schibler, U. (2006). Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38 369–374. [DOI] [PubMed] [Google Scholar]
- Roden, L.C., Song, H.R., Jackson, S., Morris, K., and Carré, I.A. (2002). Floral responses to photoperiod are correlated with the timing of rhythmic expression relative to dawn and dusk in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 13313–13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, D.S. (1997). Insect circadian rhythms and photoperiodism. Invert. Neurosci. 3 155–164. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229. [DOI] [PubMed] [Google Scholar]
- Searle, I., and Coupland, G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO J. 23 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeyoshi, Y., Taguchi, K., Yamamoto, S., Takekida, S., Yan, L., Tei, H., Moriya, T., Shibata, S., Loros, J.J., Dunlap, J.C., and Okamura, H. (1997). Light-induced resetting of a mammalian clock is associated with rapid induction of the mPer1 transcript. Cell 91 1043–1053. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A.R., Pearson, M., and Kay, S.A. (1998). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125 485–494. [DOI] [PubMed] [Google Scholar]
- Strayer, C.A., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Más, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Struhl, K. (1998). Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12 599–606. [DOI] [PubMed] [Google Scholar]
- Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Sumova, A., Jac, M., Sladek, M., Sauman, I., and Illnerova, H. (2003). Clock gene daily profiles and their phase relationship in the rat suprachiasmatic nucleus are affected by photoperiod. J. Biol. Rhythms 18 134–144. [DOI] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159–164. [DOI] [PubMed] [Google Scholar]
- Suri, V., Qian, Z., Hall, J.C., and Rosbash, M. (1998). Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron 21 225–234. [DOI] [PubMed] [Google Scholar]
- Tomita, J., Nakajima, M., Kondo, T., and Iwasaki, H. (2005). No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307 251–254. [DOI] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wolffe, A.P., Wong, J., and Pruss, D. (1997). Activators and repressors: Making use of chromatin to regulate transcription. Genes Cells 2 291–302. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Mori, T., and Johnson, C.H. (2000). Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 19 3349–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2003). Living by the calendar: How plants know when to flower. Nat. Rev. Mol. Cell Biol. 4 265–275. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., Horinouchi, S., and Beppu, T. (1995). Trichostatin A and trapoxin: Novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17 423–430. [DOI] [PubMed] [Google Scholar]
- Young, M.W., and Kay, S.A. (2001). Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2 702–715. [DOI] [PubMed] [Google Scholar]
- Zeilinger, M.N., Farre, E.M., Taylor, S.R., Kay, S.A., and Doyle, F.J. (2006). A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst. Biol. 2 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.