Abstract
The photosynthetic apparatus is composed of proteins encoded by genes from both the nucleus and the chloroplast. To ensure that the photosynthetic complexes are assembled stoichiometrically and to enable their rapid reorganization in response to a changing environment, the plastids emit signals that regulate nuclear gene expression to match the status of the plastids. One of the plastid signals, the chlorophyll intermediate Mg-ProtoporphyrinIX (Mg-ProtoIX) accumulates under stress conditions and acts as a negative regulator of photosynthetic gene expression. By taking advantage of the photoreactive property of tetrapyrroles, Mg-ProtoIX could be visualized in the cells using confocal laser scanning spectroscopy. Our results demonstrate that Mg-ProtoIX accumulated both in the chloroplast and in the cytosol during stress conditions. Thus, the signaling metabolite is exported from the chloroplast, transmitting the plastid signal to the cytosol. Our results from the Mg-ProtoIX over- and underaccumulating mutants copper response defect and genome uncoupled5, respectively, demonstrate that the expression of both nuclear- and plastid-encoded photosynthesis genes is regulated by the accumulation of Mg-ProtoIX. Thus, stress-induced accumulation of the signaling metabolite Mg-ProtoIX coordinates nuclear and plastidic photosynthetic gene expression.
INTRODUCTION
Proper function of the plant cell depends on the regulated and reciprocal interaction between its different compartments. This includes not only the exchange of metabolic intermediates and energy equivalents but also information. The genetic information of the plant cell is divided between three different compartments, the nucleus, mitochondria, and chloroplasts. However, the majority of the organellar proteins are encoded in the nucleus, and the presence of genes encoding organellar proteins in different cellular compartments presents the complex problem to coordinate the activities of the different genomes of the plant cell (Surpin and Chory, 1997; Surpin et al., 2002; Richly et al., 2003). To achieve this coordination, mechanisms to orchestrate nuclear and organellar gene expression have evolved and these include both anterograde and retrograde controls (Rodermel and Park, 2003). Anterograde mechanisms (nucleus-to-plastid) coordinate gene expression in the plastid with cellular and environmental cues that are perceived and choreographed by genes in the nucleus. This type of traffic includes nuclear-encoded proteins that regulate the transcription and translation of plastid genes. Retrograde (organelle-to-nucleus) signaling, on the other hand, coordinates the expression of nuclear genes encoding plastid proteins with the metabolic and developmental state of the plastid (Susek et al., 1993; Strand et al., 2003).
The photosynthetic apparatus is composed of proteins encoded by genes from both the nucleus and the chloroplast. For example, in the photosynthetic electron transport complexes of the thylakoid membrane, the core subunits are encoded by the chloroplast genome, and the peripheral subunits are encoded by the nuclear genome. In the stroma, the large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) is chloroplastically encoded, whereas the small subunit is nuclear encoded. To ensure that all these photosynthetic complexes are assembled stoichiometrically and to enable their rapid reorganization in response to a changing environment, the plastids emit signals that regulate nuclear gene expression to match the status of the plastids (Mayfield and Taylor, 1984; Burgess and Taylor, 1988; Taylor, 1989; Susek et al., 1993; Beck, 2005). It is now clear that several different plastid processes produce signals influencing nuclear photosynthetic gene expression (Beck, 2005; Nott et al., 2006), and it has been demonstrated that different signals are produced at different developmental stages (Sullivan and Gray, 2002). One of the plastid signals, the chlorophyll intermediate Mg-ProtoporphyrinIX (Mg-ProtoIX) was revealed by characterization of the genome uncoupled (gun) mutants (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003; Strand et al., 2003).
The gun mutants are Arabidopsis thaliana mutants in which the communication between the chloroplast and the nucleus has been disrupted (Susek et al., 1993). Using the genome-uncoupled mutants gun2 and gun5, with restrictions in defined steps in tetrapyrrole biosynthesis, conclusive evidence was provided that Mg-ProtoIX acts as a signaling molecule initiating retrograde communication between the chloroplast and the nucleus (Strand et al., 2003; Nott et al., 2006). Mg-ProtoIX has been shown to accumulate under stress conditions affecting the structure and function of the thylakoid membrane and acts as a negative regulator of nuclear-encoded photosynthesis genes (Strand et al., 2003; Wilson et al., 2003). In the genome-uncoupled mutants gun2 and gun5, Mg-ProtoIX does not accumulate to the same critical amount during stress due to reduced flux through the tetrapyrrole pathway. As a result, the plastid signal is lost in these mutants and expression of a large number of nuclear genes encoding chloroplastic proteins directly associated with the photosynthetic reaction is maintained (Strand et al., 2003).
Although there is clear biochemical (Kropat et al., 1995, 1997, 2000) and genetic (Susek et al., 1993; Larkin et al., 2003; Strand et al., 2003) evidence for Mg-ProtoIX–mediated plastid-to-nucleus communication, it remains unclear how the signal is transmitted from the chloroplast, where it is synthesized, to the nucleus. Is Mg-ProtoIX emitted from the chloroplast itself or does it interact with chloroplast-localized signaling proteins that as a consequence are emitted from the chloroplast and transduce the plastid signal further? Second, does accumulated Mg-ProtoIX also affect the expression of the genes encoded in the plastids and thereby synchronize the photosynthetic gene expression from the two genomes? To test whether Mg-ProtoIX itself is exported from the chloroplast to the cytosol, it was possible to take advantage of the photoreactive property of tetrapyrroles. Thus, Mg-ProtoIX could be visualized in the cells using confocal laser scanning spectroscopy, and our results demonstrate that Mg-ProtoIX accumulates both in the chloroplast and in the cytosol during stress conditions. Furthermore, we tested the expression of plastid-encoded genes in the Mg-ProtoIX over- and underaccumulating mutants copper response defect (crd) and gun5, respectively (Strand et al., 2003; Tottey et al., 2003), and found that expression of the plastid-encoded photosynthesis genes also respond to the accumulation of Mg-ProtoIX, demonstrating that Mg-ProtoIX coordinates nuclear and plastidic photosynthetic gene expression.
RESULTS
Visualization of Tetrapyrroles Using Confocal Laser Scanning Spectroscopy
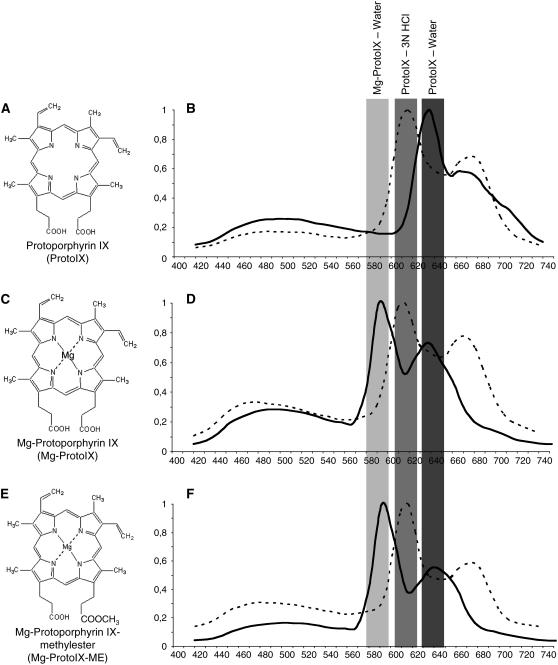
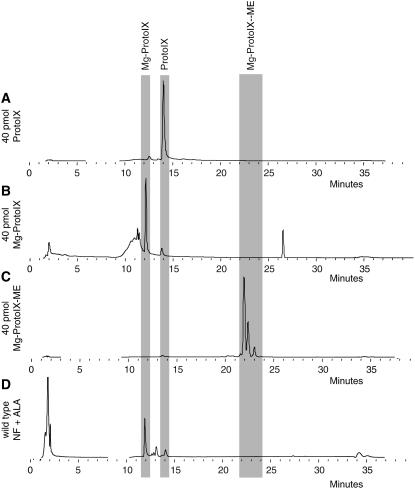
The possibility to use confocal laser scanning spectroscopy to detect emission signals from specific tetrapyrroles was investigated using authentic standards for the tetrapyrroles ProtoIX (Figure 1A), Mg-ProtoIX (Figure 1C), and Mg-protoporphyrinIX-methylester (Mg-ProtoIX-ME) (Figure 1E) (Frontier Scientific). A total of 40 pmol of each tetrapyrrole was applied to 3-mm Whatman paper, and spectral emission was recorded following 405-nm diode laser excitation. ProtoIX showed a clear emission peak between 620 and 640 nm (Figure 1B), whereas Mg-ProtoIX and its methylester Mg-ProtoIX-ME both showed emission peaks between 580 and 600 nm (Figures 1D and 1F). To confirm that the 580- to 600-nm emission peak was associated to the Mg chelated form of ProtoIX, spectral emission of ProtoIX, Mg-ProtoIX, and Mg-ProtoIX-ME in 3 n HCl (Mg removal conditions according to Rebeiz et al., 1970) confirmed that the 580- to 600-nm and 620- to 640-nm emission windows are respectively due to the chelated and unchelated forms of ProtoIX (Figures 1B, 1D, and 1F). In addition to the 580- to 600-nm emission peak, the Mg-ProtoIX and Mg-ProtoIX-ME standards showed emission peaks corresponding to ProtoIX (Figure 1D and 1F). HPLC traces of the Mg-ProtoIX and Mg-ProtoIX-ME standards demonstrated that the standards were not pure and also contained ProtoIX (Figure 2). Using this method we cannot discriminate between Mg-ProtoIX and its methyl ester, Mg-ProtoIX-ME. Mg-ProtoIX-ME has been shown to affect the expression of the nuclear-encoded gene HSP70 in Chlamydomonas to the same extent as Mg-ProtoIX (Kropat et al., 1997). However, in Arabidopsis, Mg-ProtoIX was the only intermediate to significantly accumulate following treatment with norflurazon to initiate plastid-to-nucleus communication (Figure 2D) (Strand et al., 2003). The emission peaks detected from the dissolved tetrapyrrole standards by the confocal microscope were as expected between 620 and 640 nm for ProtoIX and Mg-ProtoIX and between 580 and 600 nm for its methyl ester (Hukmani and Tripathy, 1992). Thus, confocal laser scanning spectroscopy could be used to visualize and discriminate between Mg chelated forms of tetrapyrroles.
Figure 1.
Calibration and Verification of Detection of Tetrapyrroles Using Confocal Laser Scanning Spectroscopy.
Emission spectra detected using confocal laser scanning spectroscopy from authentic standards for tetrapyrroles ProtoIX ([A] and [B]), Mg-ProtoIX ([C] and [D]), and Mg-ProtoIX-ME ([E] and [F]). Emission spectra before (solid line) and after (dotted line) removal of Mg from the protoporphyrin ring by acid from Mg-ProtoIX (D) and Mg-ProtoIX-Me (F).
Figure 2.
HPLC Traces of Tetrapyrrole Standards and Sample Used for Visualization.
HPLC traces for ProtoIX (A), Mg-ProtoIX (B), Mg-ProtoIX-ME (C), and wild-type (D) samples grown on norflurazon (NF) following ALA feeding.
Verification of Emission Signals from Specific Tetrapyrroles Using Mutants Affected in the Synthesis of Tetrapyrroles
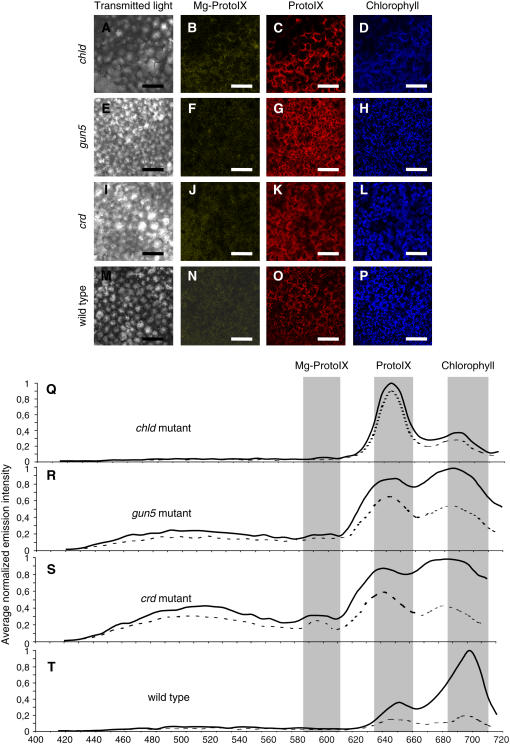
Confocal laser scanning spectroscopy was used to determine tetrapyrrole accumulation in vivo. Aminolevulinic acid (ALA), an early precursor of tetrapyrroles, was fed to Arabidopsis seedlings to amplify tetrapyrrole accumulation (Strand et al., 2003; Tottey et al., 2003). ALA feeding enabled clear and stable visualization of tetrapyrrole intermediates (Figures 3 to 5) that, with our microscope, was not possible without ALA feeding. Mutants with different lesions in the tetrapyrrole pathway (Table 1) were used as controls for the specificity of the emission signals. We used a T-DNA insertion mutant of the D-subunit of Mg-chelatase complex (chld). The homozygote chld mutant is albino and unable to synthesize Mg-ProtoIX and consequently chlorophyll (Strand et al., 2003). In addition, the gun5 mutant has a lesion in the H-subunit of Mg-chelatase, and its accumulation of Mg-ProtoIX following stress conditions is strongly suppressed (Mochizuki et al., 2001; Strand et al., 2003). As a complement, we used the T-DNA insertion mutant crd of the CRD gene, encoding a potential subunit of the cyclase enzyme complex involved in chlorophyll biosynthesis downstream of Mg-ProtoIX (Tottey et al., 2003) (Table 1).
Figure 3.
Verification of Emission Signals from Specific Tetrapyrroles Using Mutants Affected in the Synthesis of Tetrapyrroles.
(A) to (P) Emission using confocal laser scanning spectroscopy from chld ([A] to [D]), gun5 ([E] to [H]), crd ([I] to [L]), and wild-type ([M] to [P]) control seedlings that were ALA fed. Emissions from cotyledons are shown, and representative images were taken at 585 to 615 nm, 627 to 657 nm, and 680 to 710 nm for the specific emission of Mg-ProtoIX ([B], [F], [J], and [N]), ProtoIX ([C], [G], [K], and [O]), and chlorophyll ([D], [H], [L], and [P]), respectively. Bars = 50 μm.
(Q) to (T) Corresponding fluorescence emission spectra are shown from the plastids (solid line) and cytosol (dotted line). Emission spectra were normalized to the maximum value for each measurement, and overall spectrum was calculated by averaging measurements of 10 positions overlapping (solid line) and excluding (dotted line) chloroplasts.
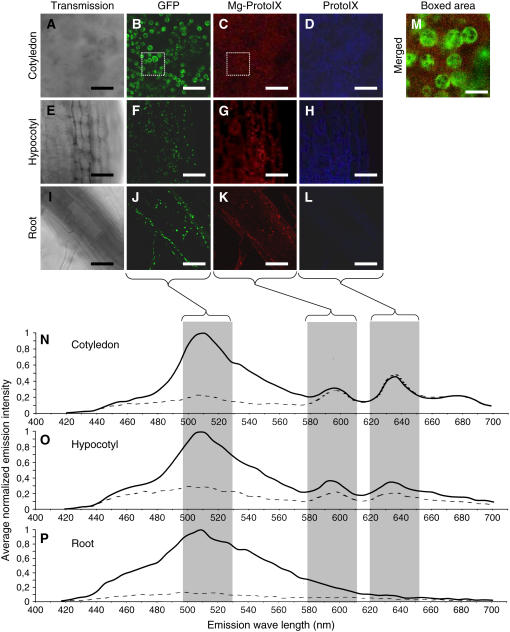
Figure 5.
Visualization of Tetrapyrrole Accumulation.
Accumulation of tetrapyrroles visualized using confocal laser scanning spectroscopy of norflurazon-treated, ALA-fed SCO1:GFP Arabidopsis seedlings. Emission is shown for cotyledon ([A] to [D]), hypocotyl ([E] to [H]), and root ([I] to [L]), and their corresponding fluorescence emission spectra is presented ([N] to [P]). Representative images were retrieved at 507 to 537 nm, 585 to 615 nm, and 627 to 657 nm for specific emission of GFP ([B], [F], and [J]), Mg-ProtoIX ([C], [G], and [K]), and ProtoIX ([D], [H], and [L]), respectively. Emission spectra were normalized to the maximum value for each measurement, and overall spectrum was calculated by averaging measurements of 10 positions overlapping (solid line) and excluding (dotted line) chloroplasts. Merged images of the boxed areas in (B) and (C) with a 1.87-airy pinhole opening are illustrated in (M) (emission windows of 507 to 537 nm for GFP and 585 to 615 nm for Mg-ProtoIX). Bars = 10 μm.
Figure 4.
Verification of Emission Signals from Specific Tetrapyrroles Using Mutants Grown on Norflurazon.
(A) to (L) Emission using confocal laser scanning microscopy from gun5 ([A] to [D]), crd ([E] to [H]), and wild-type ([I] to [L]) norflurazon-grown and ALA-fed seedlings. Emissions from cotyledons are shown and representative images were taken at 585 to 615 nm, 627 to 657 nm, and 680 to 710 nm for the specific emission of Mg-ProtoIX ([B], [F], and [J]), ProtoIX ([C], [G], and [K]), and chlorophyll ([D], [H], and [L]), respectively. Bars = 50μm.
(M) to (O) Corresponding fluorescence emission spectra are shown from the plastids (solid line) and cytosol (dotted line). Emission spectra were normalized to the maximum value for each measurement, and overall spectrum was calculated by averaging measurements of 10 positions overlapping (solid line) and excluding (dotted line) chloroplasts.
Table 1.
Summary of Mutants and Transgenic Lines Used in Our Experiments
| Gene No. | At1g62750 | At5g13630 | At1g08520 | At3g56940 |
| Name | SCO1chloroplast translation elongation factor | GUN5, CHLHMg-chelatase H-subunit | CHLDMg-chelatase D-subunit | CHL27Mg-ProtoIX monomethyl ester (oxidative) cyclase activity |
| Line | SCO:GFP | gun5EMS mutation leaky | chldSALK_150219 insertion in exon | crd1SALK_009052 insertion in exon |
| Chlorophyll content | WT levels | 80% of the wild type | Albino | Variegated20 to 70% of the wild type |
| Reference | Albrecht et al. (2006) | Mochizuki et al. (2001) | Strand et al. (2003) | chl27-as;Tottey et al. (2003) |
Emissions from ALA-fed chld, gun5, crd, and wild-type cotyledons are shown, and representative images were taken at 585 to 615 nm, 627 to 657 nm, and 680 to 710 nm for the specific emission of Mg-ProtoIX (Figures 3B, 3F, 3J, and 3N), ProtoIX (Figures 3C, 3G, 3K, and 3O), and chlorophyll (Figures 3D, 3H, 3L, and 3P), respectively, and corresponding fluorescence emission spectra are presented (Figures 3Q to 3T). In the wild type, clear emission peaks for ProtoIX and chlorophyll were detected (Figures 3O, 3P, and 3T). By contrast, the chld mutant showed almost no accumulation of chlorophyll, but a strong increase in ProtoIX accumulation was detected (Figures 3C, 3D, and 3Q). This emission profile was expected as the chld mutant cannot convert ProtoIX to Mg-ProtoIX. The leaky gun5 mutant showed an emission profile similar to the chld mutant, but with relatively less accumulated ProtoIX and more chlorophyll (Figures 3G, 3H, and 3R). The crd mutant showed a strong accumulation of ProtoIX (Figures 3K and 3S), and a slight accumulation of Mg-ProtoIX could be detected that was not observed in any of the other seedlings (Figure 3S). The spectral traces shift in their emission peak for chlorophyll from 5 to 10 nm in the gun5 and crd mutants. However, this small shift cannot be considered significant as fluorescent emission peaks are influenced by many factors, such as ionic strength and pH; a clear example of this is the spectral analysis of protoporphyrin IX where the emission varies depending on the pH (Figure 1B). Principal component analysis of the emission spectra clearly demonstrated that spectra from the wild type and the mutants are significantly different (see Supplemental Figure 1 online), supporting the specificity of the emission peaks.
To initiate accumulation of Mg-ProtoIX, we used norflurazon, a noncompetitive inhibitor of carotenoid biosynthesis (Figure 2) (Susek et al., 1993; Mochizuki et al., 1996; Strand et al., 2003). When emissions from accumulated tetrapyrrole intermediates were compared between control grown (Figure 3) and norflurazon-treated seedlings, much higher amounts of Mg-ProtoIX could be detected in the norflurazon-treated seedlings (Figures 4B to 4J). This observation confirmed published results demonstrating specific accumulation of Mg-ProtoIX under stress conditions affecting thylakoid structure (Figure 2D) (Strand et al., 2003; Wilson et al., 2003). The crd mutant showed a significant accumulation of Mg-ProtoIX (Figures 4F, 4J, 4N, and 4O), whereas the gun5 mutant demonstrated the opposite effect, underaccumulation of Mg-ProtoIX and overaccumulating ProtoIX, relatively (Figures 4B, 4C, and 4M). We cannot discriminate between the emission from Mg-ProtoIX and Mg-ProtoIX-ME, but HPLC traces showed that no methyl ester accumulated in the wild type (Figure 2) or in the crd and gun5 mutants (data not shown) following norflurazon treatment. Taken together, using the different mutants with specific lesions in the tetrapyrrole pathway grown under different conditions, we could demonstrate that the fluorescence emissions were specific for the tetrapyrroles ProtoIX, Mg-ProtoIX, and chlorophyll.
In Vivo Visualization of Mg-ProtoIX Accumulation in the Cytosol
The snowy cotelydon 1 protein (SCO1) is localized to the stroma of the chloroplast, and a SCO1:green fluorescent protein (GFP) transgenic line (Albrecht et al., 2006) was used for the in vivo localization of Mg-ProtoIX. Seedlings from the SCO1:GFP line were grown on norflurazon and ALA fed. The GFP-tagged SCO1 protein was clearly visualized in the chloroplasts of both hypocotyls and cotyledons and in the plastids of the roots (Figures 5B, 5F, and 5J; see Supplemental Figures 2 to 4 online). Furthermore, the GFP signal was distinctively localized to the chloroplasts both in the green control and the norflurazon-treated seedlings, demonstrating that the envelope membrane of the chloroplast was not disrupted by the norflurazon treatment (see Supplemental Figures 2 to 4 online).
Accumulation of Mg-ProtoIX (Figures 5C, 5G, 5N, and 5O) and ProtoIX (Figures 5D, 5H, 5N, and 5O) was clearly detected in the photosynthetic tissues, cotyledons, and hypocotyls (see Supplemental Figures 2 and 3 online). By contrast, no accumulation of the chlorophyll intermediates could be detected in root tissue (Figures 5K, 5L, and 5P; see Supplemental Figure 4 online). The relative accumulation of the tetrapyrroles in the chloroplast and the cytosol was calculated by averaging the emission from ten positions, overlapping and excluding the chloroplasts (Figures 5N to 5P; see Supplemental Figures 2 to 4 online). The chloroplasts were clearly visualized with the GFP signal in the SCO1:GFP line. In the mutant lines, chloroplastic and cytoplasmic positions were selected by bright-field microscopy (Figure 3 and 4). Bright-field microscopy is not as accurate as using the GFP chloroplast marker, and this may explain why we detect chlorophyll outside the chloroplast in the mutants (Figure 3). The confocal images clearly demonstrated that Mg-ProtoIX and ProtoIX accumulated in the cytosol (Figures 3 to 5; see Supplemental Figures 2 and 3 online). Thus, the signaling metabolite Mg-ProtoIX is exported from the chloroplast to the cytosol. In the cotyledon cells, similar amounts of Mg-ProtoIX accumulated in the chloroplast and the cytosol (Figure 5N; see Supplemental Figure 3K online). The relative cytoplastic accumulation of Mg-ProtoIX was higher in the cotyledons compared with the hypocotyls (Figures 5N and 5O; see Supplemental Figures 2K and 3K online). The cytosolic and chloroplastic accumulation of Mg-ProtoIX was confirmed with three-dimensional UV spectrometry where spectral measurements were acquired in the volume of the cotyledon cells above and through the chloroplasts (see Supplemental Figure 5 online).
The Chloroplast Structures of the gun5 and crd Mutants Are Indistinguishable from the Wild Type after Norflurazon Treatment
To test in planta whether the observed accumulation of Mg-ProtoIX in the chloroplast also has an impact on expression of the genes encoded in the plastid, we used the Mg-ProtoIX under- and overaccumulating mutants gun5 and crd, respectively (Strand et al., 2003; Tottey et al., 2003). First, to eliminate the possibility that the chloroplasts of the mutants are less affected by the norflurazon treatment compared with the wild type, transmission electron micrographs (TEMs) were retrieved from mesophyll cells of the wild type, gun5, and crd from control and norflurazon-treated seedlings. Under control conditions, the chloroplast structure of the gun5 mutant was indistinguishable from the wild type (Figures 6A and 6B). Furthermore, under control conditions, nuclear photosynthetic gene expression is not different in the gun5 mutant compared with the wild type (Susek et al., 1993; Strand et al., 2003), and the gun phenotype is only seen under stress conditions where Mg-ProtoIX accumulates in the wild type (Larkin et al., 2003; Strand et al., 2003). The thylakoid structure was not as condensed in the crd mutant as in the wild type and the gun5 mutant (Figure 6C). This could be explained by the lesion in chlorophyll biosynthesis of the crd mutant (Tottey et al., 2003).
Figure 6.
TEMs of Chloroplasts.
TEMs of Arabidopsis mesophyll cells showing the chloroplast structures in control seedlings grown for 6 d in a 16-h-light/8-h-dark cycle of the wild type (A), gun5 (B), and crd (C) and in seedlings grown for 6 d in a 16-h-light/8-h-dark cycle on 0.5 μM norflurazon for the wild type (D), gun5 (E), and crd (F). Bars = 2 μm.
The norflurazon treatment results in photobleaching, and the seedlings suffer from photooxidation of the thylakoid membranes (Burgess and Taylor, 1988). As a result, the norflurazon-treated seedlings displayed a structure similar to the prolamellar body instead of properly developed thylakoid membranes. By contrast, the envelope membrane was not affected by the norflurazon treatment, demonstrating that the effect of norflurazon is specific to the interior thylakoid membranes (Figures 6D to 6F). This was also demonstrated with the confocal images using the SCO1:GFP line where the GFP signal was distinctively localized to the chloroplasts (Figure 5). Following norflurazon treatment, the chloroplast structures of the gun5 and crd mutants were indistinguishable from the wild type (Figures 6D to 6F).
The crd mutant with a Lesion in the Tetrapyrrole Pathway Downstream of Mg-ProtoIX Does Not Exhibit a gun Phenotype
The accumulation of Mg-ProtoIX results in decreased transcription of nuclear-encoded photosynthetic genes (Burgess and Taylor, 1988; La Rocca et al., 2001; Larkin et al., 2003; Strand et al., 2003). The crd mutant was used to test the effect on photosynthetic gene expression of a lesion in the tetrapyrrole pathway downstream of Mg-ProtoIX. Expression of the nuclear-encoded genes LHCB1 and RBCS was investigated in the wild type and the gun5 and crd mutants in control and norflurazon-treated seedlings (Figures 7A and 7B). Under control conditions, the expression levels of LHCB1 and RBCS were similar in the mutants and wild type (data used in Figure 7). Following norflurazon treatment, the gun5 mutant displayed the previously described derepression of LHCB1 and RBCS compared with the wild type (Mochizuki et al., 2001; Strand et al., 2003), whereas the crd mutant did not display a genome uncoupled, gun phenotype (Figures 7A and 7B). The repression of LHCB1 and RBCS expression in the crd mutant (Figures 7A and 7B) is consistent with what has been reported for barley (Hordeum vulgare) mutants of the cyclase complex (Gadjieva et al., 2005) and confirms the role of Mg-ProtoIX as a negative regulator of nuclear-encoded photosynthesis genes.
Figure 7.
LHCB1 and RBCS Expression Levels.
Expression change following norflurazon treatment compared with the green control in the wild type, gun5, and crd of the nuclear-encoded LHCB1 (A) and RBCS (B) genes. Real-time PCR was used, and 18S rRNA was used as internal standard for the different cDNA samples. Each bar presented as log2 represents the mean (±95% confidence interval [CI]) of at least three independent experiments. Seedlings were grown with and without 0.5 μM norflurazon for 6 d in a 16-h-light/8-h-dark cycle.
Expression of Plastid-Encoded RNA Polymerase–Dependent Genes Responds to Accumulation of Mg-ProtoIX
Plastid-encoded genes are transcribed by at least two types of RNA polymerases: one is the nuclear-encoded plastid RNA polymerase (NEP) and the other is the plastid-encoded RNA polymerase (PEP). NEP mediates the transcription of housekeeping genes (Hanaoka et al., 2005), whereas photosynthesis-related genes are transcribed by PEP (Allison et al., 1996; DeSantis-Maciossek et al., 1999). We investigated the effect on Mg-ProtoIX accumulation on expression of a number of PEP- and NEP-dependent genes (Table 2) following norflurazon treatment.
Table 2.
Plastid-Encoded NEP- and PEP-Dependent Genes Used for Expression Analysis
| NEP | PEP | NEP + PEP | Gene Product | Reference |
|---|---|---|---|---|
| accD | Acetyl-CoA carboxylase β-subunit | Liere et al. (2004) | ||
| rpoB | RNA polymerase β-chain | (Hess et al. (1993) | ||
| psaA | PSI subunit | Meng et al. (1988) | ||
| psaC | PSI subunit | DeSantis-Maciossek et al. (1999) | ||
| psbA | PSII subunit | Allison et al. (1996) | ||
| psbD | PSII subunit | Allison et al. (1996) | ||
| rbcL | Rubisco large subunit | Allison et al. (1996) | ||
| clpP | ATP-dependent protease | Liere et al. (2004) | ||
| rpl33 | Ribosomal protein L33 | Hajdukiewicz et al. (1997) |
Expression levels of the PEP-dependent genes psbA (chlorophyll binding protein D1) and psbD (photosystem II [PSII] protein D2), encoding components of the PSII core, and psaA (photosystem I [PSI] P700 apoprotein A) and psaC (PSI subunit VII) genes, encoding components of PSI, and the rbcL gene, encoding the large subunit of Rubisco, were significantly higher in the gun5 mutant compared with the wild type or the crd mutant when all genotypes were treated with norflurazon (Figures 8A to 8E). Similar to the nuclear-encoded photosynthesis genes (Figure 7), the expression of the plastid-encoded photosynthesis genes psbA, psbD, psaA, psaC, and rbcL was derepressed in gun5 compared with the wild type or the crd mutant following norflurazon treatment (Figures 8A to 8E). RNA gel blots were performed to investigate whether there were any effects on RNA processing following norflurazon treatment in the different genotypes (see Supplemental Figure 6 online). The real-time PCR results could be reproduced using RNA gel blots, and no difference in transcript sizes could be detected between the wild type and the mutants.
Figure 8.
Expression of PEP-Transcribed Plastid-Encoded Genes.
Expression change following norflurazon treatment compared with green control in the wild type, gun, and crd mutants of the plastid-encoded PsbA (A), PsbD (B), PsaA (C), PsaC (D), and RbcL (E) genes. Real-time PCR was used, and 18S rRNA was used as internal standard in the different cDNA samples. Each bar presented as log2 represents the mean (±95% CI) of at least three independent experiments. Seedlings were grown with and without 0.5 μM norflurazon for 6 d in a 16-h-light/8-h-dark cycle.
We also investigated expression levels of the NEP-dependent genes RpoB, which encodes the RNA polymerase β-chain, and AccD, which encodes the acetyl-CoA carboxylase β-subunit (Hess et al., 1993; Liere et al., 2004). The promoters of ClpP encoding an ATP-dependent protease and of Rpl33 encoding ribosomal protein L33 contain both NEP and PEP motifs and are believed to be transcribed by both types of RNA polymerases (Hajdukiewicz et al., 1997; Liere et al., 2004). In the wild type, the expression of these housekeeping genes was not repressed by the norflurazon treatment, and no difference in expression levels could be found between the gun5 mutant and the wild type (Figures 9A to 9D). By contrast, the crd mutant showed repression for the expression of the AccD, Rpl33, and ClpP genes following norflurazon treatment. The repressed expression of the housekeeping genes in the crd mutant could possibly be explained by the observed difference in the thylakoid structure in the crd mutant (Figure 6), suggesting that other plastid signals regulating the housekeeping genes could be activated in the mutant. Thus, unlike the PEP-dependent photosynthesis genes, expression of the plastid-encoded NEP-dependent housekeeping genes could not be linked to the accumulation of Mg-ProtoIX using the over- and underaccumulating mutants crd and gun5, respectively.
Figure 9.
Expression of NEP-Transcribed Plastid-Encoded Genes.
Expression change following norflurazon treatment compared with green control in the wild type, gun5, and crd mutants of the plastid-encoded RpoB (A), AccD (B), ClpP (C), and Rpl33 (D) genes. Real-time PCR was used, and 18S rRNA was used as internal standard in the different cDNA samples. Each bar presented as log2 represents the mean (±95% CI) of at least three independent experiments. Seedlings were grown with and without 0.5 μM norflurazon for 6 d in a 16-h-light/8-h-dark cycle.
Mg-ProtoIX Controls Expression of Plastid-Encoded Photosynthesis Genes by Regulating the Nuclear-Encoded Sigma Factors
PEP consists of core subunits encoded in the plastid genome that assemble with one of the promoter specificity sigma factors (SIG1-6) encoded in the nucleus (Hanaoka et al., 2005). Given the specific derepression of the PEP-dependent genes following norflurazon treatment in the gun5 mutant, we investigated the expression of the nuclear-encoded sigma factor SIG1-6. In the wild type, expression of the SIG1-6 genes was strongly repressed following norflurazon treatment (Figures 10A to 10F). The crd mutant demonstrated the same expression pattern for the SIG1-6 genes as the wild type (Figures 10A to 10F). However, the gun5 mutant did not show any repression of the SIG2-SIG6 genes following norflurazon treatment (Figures 10B to 10F). The SIG1 gene was repressed by the norflurazon treatment in the gun5 mutant but to a lesser extent than the wild type and the crd mutant (Figure 10A). Consequently, the derepression of the plastid-encoded photosynthesis genes in the gun5 mutant could be matched with the maintenance of expression of the nuclear-encoded plastidic sigma factors.
Figure 10.
Expression of SIG1-6 and RPOPT.
Expression change following norflurazon treatment compared with the green control in the wild type, gun5, and crd of the nuclear genes encoding the different sigma factors SIG1 (A), SIG2 (B), SIG3 (C), SIG4 (D), SIG5 (E), and SIG6 (F) genes and the NEP RPOPT (G) using real-time PCR. 18S rRNA was used as internal standard in the different cDNA samples. Each bar presented as log2 represents the mean (±95% CI) of at least three independent experiments. Seedlings were grown with and without 0.5 μM norflurazon for 6 d in a 16-h-light/8-h-dark cycle.
The NEP are encoded by the RPOT genes, and in Arabidopsis, three RPOT genes have been identified: RPOMT and RPOPT are directed to mitochondria and chloroplasts, respectively, whereas the third, RPOT2, has dual targeting properties (Hedtke et al., 1997, 2000; Baba et al., 2004). Expression of RPOPT was not repressed in the wild type or in the gun5 mutant following norflurazon treatment. However, the crd mutant demonstrated a slight repression of the RPOPT expression. The expression pattern for the different genotypes was similar to the expression of the NEP-dependent genes RpoB, AccD, ClpP, and Rpl33 (Figure 9). Expression of the RPOT2 gene could not be detected in seedlings, and it is possible that this gene is expressed later during development. Expression of the NEP gene did not respond to the accumulation of Mg-ProtoIX, whereas the sigma factors necessary for the function of the multisubunit enzyme PEP appears to be controlled by the accumulation of the plastid signal Mg-ProtoIX. Thus, by controlling expression of the sigma factors, the plastid factor Mg-ProtoIX would be able to coordinate nuclear and plastidic photosynthetic gene expression.
DISCUSSION
Taking advantage of the fluorescent properties of tetrapyrroles, we could use confocal laser scanning spectroscopy to visualize in vivo the subcellular localization of Mg-ProtoIX and ProtoIX (Figures 3 to 5; see Supplemental Figures 2 and 3 online). We confirmed the specificity of the emission signals for ProtoIX and Mg-ProtoIX using mutants with specific lesions in the tetrapyrrole pathway (Figures 3 and 4). Furthermore, the emission from Mg-ProtoIX and ProtoIX was specific to photosynthetic tissues, and no emission of the wavelengths corresponding to the tetrapyrroles could be detected in root tissue (Figure 5; see Supplemental Figure 4 online). In addition, emission corresponding to Mg-ProtoIX could only be detected in the norflurazon-grown seedlings (Figures 4 and 5). This observation confirmed published results demonstrating specific accumulation of Mg-ProtoIX under conditions affecting the thylakoid structure (Strand et al., 2003; Wilson et al., 2003).
Mg-ProtoIX accumulation initiates retrograde communication between the chloroplast and the nucleus (Strand et al., 2003; Strand, 2004) and functions as a negative regulator of nuclear photosynthetic gene expression (Beck, 2005). However, until now, it has been unclear how the Mg-ProtoIX signal is transduced from the chloroplast, where it is synthesized, to the nucleus. Our confocal images clearly demonstrated that Mg-ProtoIX accumulated both in the chloroplasts and the cytosol under stress conditions (Figure 5; see Supplemental Figures 2 and 3 online). These results show that the signaling metabolite Mg-ProtoIX is exported from the chloroplast to the cytosol, via a still unknown mechanism, transmitting the plastid signal to the cytosol. The SCO1:GFP line (Albrecht et al., 2006) demonstrated that the GFP signal was restricted to the chloroplasts, confirming that the integrity of the plastid envelope membranes was not affected by norflurazon treatment, ALA feeding, or handling of the material during the microscopy studies (Figure 5; see Supplemental Figures 2 to 4 online). Intact envelope membranes following norflurazon treatment were also demonstrated with the TEMs (Figures 6D to 6F). The intermediate ProtoIX accumulated in the cytosol in all genotypes tested, both under control conditions and following norflurazon treatment (Figures 3 to 5). This further demonstrated that artifacts did not cause the observed export of tetrapyrroles during the experiments. Furthermore, the relative cytoplasmic accumulation of Mg-ProtoIX and ProtoIX was consistently greater in the cotyledons compared with the hypocotyls (Figures 5N and 5O; see Supplemental Figures 2 and 3 online), suggesting that the export mechanism(s) is more active in leaf tissue compared with hypocotyls and that the export of tetrapyrroles is an active and regulated process. Supporting this conclusion, Kropat et al. (1995, 1997, 2000) demonstrated in Chlamydomonas that the light-responsive gene HSP70, encoding a heat shock protein, could be induced in the dark by feeding Mg-ProtoIX to the cells. However, if the cells were fed ProtoIX, the chloroplast-derived Mg-ProtoIX could not access the cytosol in the dark, suggesting that the export mechanism was light regulated (Kropat et al., 2000; Beck, 2005). Efflux of related molecules, such as heme, heme precursors, phytochromobilin, and chlorophyll degradation products, has been observed from chloroplasts (Thomas and Weinstein, 1990; Matile et al., 1992; Jacobs and Jacobs, 1993; Terry et al., 1993), and it is feasible that the export mechanism is promiscuous and that the same transport route(s) could be used by Mg-ProtoIX and ProtoIX. Mutant analysis of Arabidopsis suggests that a plastid-localized ABC transporter–like protein may be involved in the translocation of ProtoIX across the envelope membrane (Moller et al., 2001). Furthermore, a mitochondrial peripheral-type benzodiazepine receptor from Arabidopsis was demonstrated to transport ProtoIX (Lindemann et al., 2004), and the GUN4 protein, which was demonstrated to bind both ProtoIX and Mg-ProtoIX and to be localized to the envelope of the chloroplast (Larkin et al., 2003), may also be involved in the export of tetrapyrroles from the chloroplast. Therefore, while the route(s) for transport of tetrapyrroles or the components involved in this transport is not known, our protocol for visualization of ProtoIX and Mg-ProtoIX accumulation in the plant cell may help identify the components involved in the transport of tetrapyrroles across the envelope membranes.
Identification of signaling metabolites emitted from the organelles used in retrograde communication is novel in plants. However, yeast (Saccharomyces cerevisiae) has been shown to use heme synthesized in the mitochondria to regulate transcription of nuclear genes encoding mitochondrial proteins (Forsburg and Guarente, 1989). Heme synthesis is directly correlated to oxygen levels in the cellular environment and when cells are grown aerobically heme is synthesized in the mitochondria and imported to the nucleus where it activates the transcription factor HAP1 (Kwast et al., 1998). Binding of heme to HAP1 permits HAP1 to bind upstream activation sequences, promoting transcription of many genes required for oxygen use and the ROX1 gene, encoding the aerobic repressor that represses genes encoding proteins required for anaerobic growth (Zhang and Hach, 1999). Our results demonstrated that Mg-ProtoIX accumulated evenly through the cytosol, and our confocal images do not imply that Mg-ProtoIX accumulated specifically in the nucleus. These data suggest that Mg-ProtoIX likely binds a regulatory protein in the cytosol and modifies the activity and/or the translocation of this protein, perhaps through a photoreactive action. As a consequence, expression of nuclear-encoded photosynthetic genes is inhibited. A sequence motif that mediates gene activation by Mg-ProtoIX has recently been identified in Chlamydomonas. Analysis of the HSP70A promoter revealed two regulatory regions that each confers responsiveness to Mg-ProtoIX and light (von Gromoff et al., 2006). Mutational analysis of one of those regulatory regions and an alignment with promoters of other Mg-ProtoIX–inducible genes uncovered the sequence motif (G/C)CGA(C/T)N(A/G)N15(T/C/A)(A/T/G) that may confer Mg-ProtoIX responsiveness (von Gromoff et al., 2006). At this point, the components recognizing the accumulation of Mg-ProtoIX and the trans-acting factors controlling the expression of photosynthetic genes remain to be discovered, and identifying these factors is a challenging task for the future. Nevertheless, our results demonstrate that the subsequent component in the Mg-ProtoIX–mediated plastid-to-nucleus pathway should be a cytosolic protein.
The confocal images demonstrated that a significant amount of Mg-ProtoIX accumulated in the chloroplast in wild-type seedlings following norflurazon treatment (Figures 5N and 5O; see Supplemental Figures 2 and 3 online). The expression of plastid-encoded genes is known to respond to changes in the environment (Pfannschmidt et al., 1999; Tullberg et al., 2000), and when expression of the plastid-encoded genes was investigated following norflurazon treatment, we found that expression of photosynthesis-related genes was maintained in the gun5 mutant compared with the wild type and the crd mutant (Figure 8). Similar to the nuclear-encoded photosynthesis genes, expression of the plastid-encoded psbA, psbD, psaA, psaC, and rbcL genes was severely repressed in the wild type and the crd mutant by the norflurazon treatment, but expression was maintained in the gun5 mutant (Figures 8A to 8E). By contrast, the expression of genes encoding the housekeeping proteins, RpoB, AccD, ClpP, and Rpl33, was not repressed by the norflurazon treatment in the wild type, and the expression was not different in the gun5 mutant compared with the wild type (Figure 9), suggesting that it is not a general effect on chloroplast transcription in the gun5 mutant. Furthermore, the chloroplast structure of the gun5 mutant is indistinguishable from the wild type and the crd mutant following norflurazon treatment (Figure 5), excluding differences in sensitivity to the norflurazon treatment between the wild type and the mutants as the cause for the differences in gene expression. Thus, using the Mg-ProtoIX over- and underaccumulating mutants crd and gun5, respectively, we could link the effect on the photosynthesis-related plastid-encoded genes to the accumulation of Mg-ProtoIX.
Plastid-encoded genes of higher plants are transcribed by at least two types of RNA polymerases: NEP and PEP (Hanaoka et al., 2005). NEP mediates the transcription of housekeeping genes, such as the RpoB, AccD, ClpP, and Rpl33 (Table 2) (Hess et al., 1994; Hajdukiewicz et al., 1997; Xie and Allison, 2002; Liere et al., 2004), whereas photosynthesis-related genes, such as psbA, psbD, and rbcL, are transcribed by PEP (Allison et al., 1996; DeSantis-Maciossek et al., 1999). Our results demonstrated that Mg-ProtoIX specifically affects expression of the PEP-transcribed photosynthesis genes. Expression of RpoB that encodes the PEP β-chain was not repressed by the norflurazon treatment (Figure 9A). However, the difference in expression of the photosynthesis-related genes could be explained by a gun effect on the nuclear-encoded promoter specificity factors of PEP, the sigma factors (SIG1-SIG6) (Figure 10). The expression of the SIG2-SIG6 genes was not repressed in the gun5 mutant following norflurazon treatment; therefore, the expression of the plastid-encoded photosynthesis genes could be maintained. However, at this point we cannot exclude an additional regulatory role for the accumulated Mg-ProtoIX inside the chloroplast.
Thus, in addition to exerting control over nuclear-encoded photosynthesis genes, stress-induced accumulation of Mg-ProtoIX also affects the expression of the plastid-encoded photosynthesis genes by controlling the expression of the sigma factors necessary for the function of the multisubunit enzyme PEP. During the course of evolution, the chloroplasts lost most of their genes to the nucleus and throughout the gradual conversion from endosymbiont to organelle mechanisms have evolved for the plant cell to orchestrate nuclear and organellar gene expression. Our results demonstrate that accumulation of the signaling metabolite Mg-ProtoIX coupled to the export to the cytosol links the functional status of the chloroplast to the nucleus, enabling the plant to synchronize the expression of photosynthetic genes from the nuclear and plastidic genomes.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana wild-type (Columbia) and mutant (gun5, chld, SALK_150219, and crd, SALK_009052) seedlings (Mochizuki et al., 2001; Tottey et al., 2003) were grown on 1× Murashige and Skoog (MS) agar medium (Duchefa) containing 2% sucrose. Seedlings were vernalized for 24 h/4°C in the dark and then grown for 6 d in a 16-h-light/8-h-dark cycle at 23°C. Mg-ProtoIX accumulation was induced by supplementing 1× MS agar medium with 0.5 μM norflurazon.
RNA Isolation and DNase Treatment
Total RNA was isolated using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Total RNA concentration was determined with a Nanodrop ND-1000 spectrophotometer. Before cDNA synthesis, 5 μg of total RNA were DNase treated (Ambion) according to the manufacturer's recommendations.
cDNA Synthesis and Real-Time PCR
Using the iScript cDNA synthesis kit (Bio-Rad), cDNA was synthesized using 1 μg of total RNA according to the manufacturer's instructions. cDNA was diluted 10-fold, and 2 μL of the diluted cDNA was used in a 20-μL iQ SYBR Green Supermix reaction (Bio-Rad). All reactions were performed in triplicates. The following primers were used: chlorophyll a/b binding protein (LHCB1, At1g29920): forward primer, 5′-GGAACGGAGTCAAGTTTGGA-3′; reverse primer, 5′-CAAAATGCTCTGAGCGTGAA-3′; Rubisco small subunit (RBCS, At1g67090): forward primer, 5′-CCTCCGATTGGAAAGAAGAA-3′; reverse primer, 5′-TACACAAATCCGTGCTCCAA-3′; photosystem II protein D2 (psbD): forward primer, 5′-TCATGGTATACTCATGGATTGG-3′; reverse primer, 5′-GACCACCTAATTGACACCAACG-3′; small ribosomal subunit 18 (18SrRNA): forward primer, 5′-TCAACTTTCGATGGTAGGATAGTG-3′; reverse primer, 5′-CCGTGTCAGGATTGGGTAATTT-3′; chlorophyll binding protein D1 (psbA): forward primer, 5′-ATACAACGGCGGTCCTTATG-3′; reverse primer, 5′-AGCAATCCAAGGACGCATAC-3′; photosystem I P700 apoprotein A (psaA): forward primer, 5′-ACTACCACTTGGATCTGGAAC-3′; reverse primer, 5′-AAACGAGCACCGTGGAAATAC-3′; photosystem I subunit VII (psaC): forward primer, 5′-ATAGGATGTACTCAATGTGTCC-3′; reverse primer, 5′-ATCTCTTACAACCAACACAGTC-3′; Rubisco large subunit (rbcL): forward primer, 5′-GTGTTGGGTTCAAAGCTGGT-3′; reverse primer, 5′-GTGGAACTCCAGGTTGAGGA-3′; ATP-dependent Clp protease protein (ClpP): forward primer, 5′-TGACATATAGTGCGACTTGTCAG-3′; reverse primer, 5′-ATACTCCCTGCATTTAATTCCAG-3′; acetyl-CoA carboxylase β-subunit (AccD): forward primer, 5′-AAGCGGAAAGATTCGTGAAA-3′; reverse primer, 5′-TGTCGCATTGAATCCACAAT-3′; RNA polymerase β-chain (RpoB): forward primer, 5′-CGAATAGCCCCTTTTGATGA-3′; reverse primer, 5′-CCTGGATACTCGGGTTCAAA-3′; ribosomal protein L33 (Rpl33): forward primer, 5′-GCCAAGGGTAAAGATGTTCG-3′; reverse primer, 5′-TAGGGGTGTTATGCCGATTC-3′; sigma factor 1 (SIG1) (At1g64860): forward primer, 5′-GGAAGTTGTGCGCTTGTCTA-3′; reverse primer, 5′-GCTGTTCATCAGAAGGCTCA-3′; sigma factor 2 (SIG2) (At1g08540): forward primer, 5′-CCTCAAACCTTCGGAAGTGA-3′; reverse primer, 5′-TTTCTCCCTTGTACCCAACG-3′; sigma factor 3 (SIG3) (At3g53920): forward primer, 5′-TCGGCCACTATTTTAGTGC-3′; reverse primer, 5′-ATGGGGAAAGAAGTTTGGTCA-3′; sigma factor 4 (SIG4) (At5g13730): forward primer, 5′-AGGAGGTTCAGCTTTGCTTG-3′; reverse primer, 5′-TTCTTCTTCCCTCTGCCACTG-3′; sigma factor 5 (SIG5) (At5g24120): forward primer, 5′-TGATATAGTGAGCTTGGACTGG-3′; reverse primer, 5′-CTTGCAGCTCTACCTATTTCG-3′; sigma factor 6 (SIG6) (At2g36990): forward primer, 5′-GCAGCTACTCTTCTCCTTCCTC-3′; reverse primer, 5′-GTGGCTCCTGTGAGAGAACC-3′; RPOPT (At2g24120): forward primer, 5′-TTGGAAGCCGTCTGCTAGAAC-3′; reverse primer, 5′-ATACCTCCTCACCAGCTTAGA-3′; RPOT2 (At5g15700): forward primer, 5′-TACAGTAGGCGATGCCATTG-3′; reverse primer, 5′-CATACTGCTGACAACTTCTG-3′.
Thermal cycling consisted of an initial step at 95°C for 3 min, followed by 40 cycles of 10 s at 95°C, 30 s at 55°C, and 10 s at 72°C. After that a melting curve was performed. RT-PCR was monitored using the MyiQTM single-color real-time PCR detection system (Bio-Rad). The adjustment of baseline and threshold was done according to the manufacturer's recommendations. The relative abundance of all transcripts amplified was normalized to the constitutive expression level of 18S rRNA mRNA. Data were analyzed using LinRegPCR (Pfaffl, 2001; Ramakers et al., 2003). Negative controls for RT-PCR were performed with the RNA samples prior to cDNA synthesis to check for amplification of genomic DNA. No amplification was observed in the samples for any of the primers used.
RNA Gel Blots
RNA gel blots were done according to Strand et al. (2003).
HPLC Analysis
HPLC analysis was done according to the method described by Strand et al. (2003). Leaf material was homogenized in acetone (0.1 M NH4OH, 8:2 [v/v]) and homogenized and centrifuged with ice-cooling. The residue was resuspended again, and the same procedures described above were repeated. The collected supernatants were mixed and centrifuged prior to HPLC analysis. Column eluent was monitored by UV detection, and Mg-ProtoIX was identified and quantified using authentic standards. ProtoIX, Mg-protoIX, and Mg-ProtoIX methylester were purchased from Frontier Scientific.
Confocal Spectroscopy
Reference spectra were obtained by spectral measurement from 410 to 750 nm under 405-nm diode laser excitation of commercially available standard of ProtoIX, MgProtoIX, and MgProtoIX-Me (Frontier Scientific) by dot blot. Three-millimeter Whatman paper (3MMChr) was attached using tape on a microscope slide, and 1 μL containing 40 pmol of the standard was deposited onto the paper, its position marked by pencil, and spectral scan xyz was performed with an SP2 confocal microscope equipped with an inverted DMIRBE II microscope (Leica). Six-day-old seedlings grown with or without norflurazon were fed 3 mL of ALA solution (10 mM 5-aminolevulinate [Sigma-Aldrich], 5 mM MgCl2, and 10 mM KPO4, pH 7.0) under green safelight conditions and incubated for 13 h in the dark to amplify the Mg-ProtoIX accumulation. Confocal images were acquired with an SP2 confocal laser scanning system equipped with an inverted microscope (Leica) and a ×40 water immersion objective (numerical aperture of 0.75). Spectral measurements were performed using a 10-nm window with an airy 1 pinhole opening from 415 to 710 nm with the 405-nm ray line of a UV diode laser. Representative images were taken from 507 to 537 nm, 585 to 615 nm, 627 to 657 nm, and 680 to 710 nm for the specific emission of GFP, Mg-ProtoIX, ProtoIX, and chlorophyll, respectively. Emission spectra were calculated by averaging measurements of 10 positions overlapping and excluding chloroplasts. Spectra normalization was performed by relative readjustment of all the spectral values within a 0 to 1 scale. Principal component analysis and correlated loadings were performed with UV-scaled normalized spectra using SIMCA v10.5 (Umetrics). Three-dimensional UV spectroscopy by XYZλ scans was performed on selected object of an ALA-fed norflurazon-treated hypocotyl SCO1:GFP seedling over a 10-μm depth and every 10 nm between 415 and 705 nm. Spectra normalization was performed as mentioned above.
TEM
Seedlings were grown for 6 d on 1× MS agar medium containing 2% sucrose, with or without 0.5 μM norflurazon. The preparation, embedding, and cutting of the samples were done according to Keskitalo et al. (2005).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Principle Component Analysis (Covariant Matrix) of Cotyledon Spectra from Wild-Type, gun5, crd, and chld Seedlings.
Supplemental Figure 2. Emission from Cotyledons of ALA-Fed Norflurazon and Control SCO:GFP Seedlings.
Supplemental Figure 3. Emission from Hypocotyls of ALA-Fed Norflurazon and Control SCO:GFP Seedlings.
Supplemental Figure 4. Emission from Roots of ALA-Fed Norflurazon and Control SCO:GFP Seedlings.
Supplemental Figure 5. Subcellular Localization of GFP, Mg-ProtoIX, and ProtoIX.
Supplemental Figure 6. RNA Gel Blots of PsbA, PsaC, ClpP, and RpoB.
Supplementary Material
Acknowledgments
We thank Leonore Johansson for her help with the TEM, and Andreas Hansson for providing the crd seeds, and Klaus Apel for providing seeds from the SCO1:GFP line. This work was supported by grants from the Swedish Research Foundation (VR) and by the Foundation for Strategic Research (SSF) (Å.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Åsa Strand (asa.strand@plantphys.umu.se).
Online version contains Web-only data.
References
- Albrecht, V., Ingenfeld, A., and Apel, K. (2006). Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60 507–518. [DOI] [PubMed] [Google Scholar]
- Allison, L.A., Simon, L.D., and Maliga, P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Baba, K., Schmidt, J., Espinosa-Ruiz, A., Villarejo, A., Shiina, T., Gardestrom, P., Sane, A.P., and Bhalerao, R.P. (2004). Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J. 38 38–48. [DOI] [PubMed] [Google Scholar]
- Beck, C.F. (2005). Signaling pathways from the chloroplast to the nucleus. Planta 222 743–756. [DOI] [PubMed] [Google Scholar]
- Burgess, D., and Taylor, W. (1988). The chloroplast affects the transcription of a nuclear gene family. Mol. Gen. Genet. 214 89–96. [Google Scholar]
- DeSantis-Maciossek, G., Kofer, W., Bock, A., Schoch, S., Maier, R.M., Wanner, G., Rudiger, W., Koop, H.U., and Herrmann, R.G. (1999). Targeted disruption of the plastid RNA polymerase gene rpoA, B and C1: Molecular biology, biochemistry and ultrastructure. Plant J. 18 477–489. [DOI] [PubMed] [Google Scholar]
- Forsburg, S.L., and Guarente, L. (1989). Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 5 153–180. [DOI] [PubMed] [Google Scholar]
- Gadjieva, R., Axelsson, E., Olsson, U., and Hansson, M. (2005). Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol. Biochem. 43 901–908. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P.T., Allison, L.A., and Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka, M., Kanamaru, K., Fujiwara, M., Takahashi, H., and Tanaka, K. (2005). Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep. 6 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Borner, T., and Weihe, A. (1997). Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277 809–811. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Borner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, W.R., Muller, A., Nagy, F., and Borner, T. (1994). Ribosome-deficient plastids affect transcription of light-induced nuclear genes: Genetic evidence for a plastid-derived signal. Mol. Gen. Genet. 242 305–312. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., Prombona, A., Fieder, B., Subramanian, A.R., and Borner, T. (1993). Chloroplast rps15 and rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: Evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukmani, P., and Tripathy, B.C. (1992). Spectroscopic estimation of intermediates of chlorophyll biosynthesis: Protopophyrin IX, Mg-protopophyrin and protochlorophyllide. Anal. Biochem. 206 125–130. [DOI] [PubMed] [Google Scholar]
- Jacobs, J., and Jacobs, N. (1993). Porphyrin accumulation and export by isolated barley (Hordeum vulgare) plastids. Plant Physiol. 101 1181–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskitalo, J., Bergquist, G., Gardestrom, P., and Jansson, S. (2005). A cellular timetable of autumn senescence. Plant Physiol. 139 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat, J., Oster, U., Rudiger, W., and Beck, C.F. (1997). Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. USA 94 14168–14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat, J., Oster, U., Rudiger, W., and Beck, C.F. (2000). Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 24 523–531. [DOI] [PubMed] [Google Scholar]
- Kropat, J., von Gromoff, E.D., Muller, F.W., and Beck, C.F. (1995). Heat shock and light activation of a Chlamydomonas HSP70 gene are mediated by independent regulatory pathways. Mol. Gen. Genet. 248 727–734. [DOI] [PubMed] [Google Scholar]
- Kwast, K.E., Burke, P.V., and Poyton, R.O. (1998). Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201 1177–1195. [DOI] [PubMed] [Google Scholar]
- Larkin, R.M., Alonso, J.M., Ecker, J.R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299 902–906. [DOI] [PubMed] [Google Scholar]
- La Rocca, N., Rascio, N., Oster, U., and Rudiger, W. (2001). Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213 101–108. [DOI] [PubMed] [Google Scholar]
- Liere, K., Kaden, D., Maliga, P., and Borner, T. (2004). Overexpression of phage-type RNA polymerase RpoTp in tobacco demonstrates its role in chloroplast transcription by recognizing a distinct promoter type. Nucleic Acids Res. 32 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann, P., Koch, A., Degenhardt, B., Hause, G., Grimm, B., and Papadopoulos, V. (2004). A novel Arabidopsis thaliana protein is a functional peripheral-type benzodiazepine receptor. Plant Cell Physiol. 45 723–733. [DOI] [PubMed] [Google Scholar]
- Matile, P., Schellenberg, M., and Peisker, C. (1992). Production and release of a chlorophyll catabolite in isolated senescent chloroplasts. Planta 187 230–235. [DOI] [PubMed] [Google Scholar]
- Mayfield, S., and Taylor, W. (1984). Carotenoid-deficient maize seedlings fail to accumulate light harvesting chlorophyll a/b binding protein (LHCP) mRNA. Eur. J. Biochem. 144 79–84. [DOI] [PubMed] [Google Scholar]
- Meng, B.Y., Tanaka, M., Wakasugi, T., Ohme, M., Shinozaki, K., and Sugiura, M. (1988). Cotranscription of the genes encoding two P700 chlorophyll a apoproteins with the gene for ribosomal protein CS14: Determination of the transcription initiation site by in vitro capping. Curr. Genet. 14 395–400. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N., Brusslan, J.A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, N., Susek, R., and Chory, J. (1996). An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol. 112 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, S.G., Kunkel, T., and Chua, N.H. (2001). A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 15 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott, A., Jung, H.S., Koussevitzky, S., and Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57 739–759. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397 625–628. [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H., and Moorman, A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339 62–66. [DOI] [PubMed] [Google Scholar]
- Rebeiz, C.A., Abou Haidar, M., Yaghi, M., and Castelfranco, P.A. (1970). Porphyrin biosynthesis in cell-free homogenates from higher plants. Plant Physiol. 46 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F., and Leister, D. (2003). Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep. 4 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel, S., and Park, S. (2003). Pathways of intracellular communication: Tetrapyrroles and plastid-to-nucleus signaling. Bioessays 25 631–636. [DOI] [PubMed] [Google Scholar]
- Strand, A. (2004). Plastid-to-nucleus signalling. Curr. Opin. Plant Biol. 7 621–625. [DOI] [PubMed] [Google Scholar]
- Strand, A., Asami, T., Alonso, J., Ecker, J.R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421 79–83. [DOI] [PubMed] [Google Scholar]
- Sullivan, J.A., and Gray, J.C. (2002). Multiple plastid signals regulate the expression of the pea plastocyanin gene in pea and transgenic tobacco plants. Plant J. 32 763–774. [DOI] [PubMed] [Google Scholar]
- Surpin, M., and Chory, J. (1997). The co-ordination of nuclear and organellar genome expression in eukaryotic cells. Essays Biochem. 32 113–125. [PubMed] [Google Scholar]
- Surpin, M., Larkin, R.M., and Chory, J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14 (suppl.): S327–S338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek, R.E., Ausubel, F.M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799. [DOI] [PubMed] [Google Scholar]
- Taylor, W. (1989). Regulatory interactions between nuclear and plastid genomes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40 211–233. [Google Scholar]
- Terry, M.J., Maines, M.D., and Lagarias, J.C. (1993). Inactivation of phytochrome- and phycobiliprotein-chromophore precursors by rat liver biliverdin reductase. J. Biol. Chem. 268 26099–26106. [PubMed] [Google Scholar]
- Thomas, J., and Weinstein, J. (1990). Measurement of heme efflux and heme content in isolated developing chlroplasts. Plant Physiol. 94 1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey, S., Block, M.A., Allen, M., Westergren, T., Albrieux, C., Scheller, H.V., Merchant, S., and Jensen, P.E. (2003). Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 100 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullberg, A., Alexciev, K., Pfannschmidt, T., and Allen, J.F. (2000). Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the plastoquinone pool. Plant Cell Physiol. 41 1045–1054. [DOI] [PubMed] [Google Scholar]
- von Gromoff, E.D., Schroda, M., Oster, U., and Beck, C.F. (2006). Identification of a plastid response element that acts as an enhancer within the Chlamydomonas HSP70A promoter. Nucleic Acids Res. 34 4767–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K.E., Sieger, S.M., and Huner, N.P.A. (2003). The temperature-dependent accumulation of Mg-protoporphyrin IX and reactive oxygen species in Chlorella vulgaris. Physiol. Plant 119 126–136. [Google Scholar]
- Xie, G., and Allison, L.A. (2002). Sequences upstream of the YRTA core region are essential for transcription of the tobacco atpB NEP promoter in chloroplasts in vivo. Curr. Genet. 41 176–182. [DOI] [PubMed] [Google Scholar]
- Zhang, L., and Hach, A. (1999). Molecular mechanism of heme signaling in yeast: The transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 56 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.