Abstract
In plants, cell-to-cell trafficking of non-cell-autonomous proteins (NCAPs) involves protein–protein interactions, and a role for posttranslational modification has been implicated. In this study, proteins contained in pumpkin (Cucurbita maxima cv Big Max) phloem sap were used as a source of NCAPs to further explore the molecular basis for selective NCAP trafficking. Protein overlay assays and coimmunoprecipitation experiments established that phosphorylation and glycosylation, on both Nicotiana tabacum NON-CELL-AUTONOMOUS PATHWAY PROTEIN1 (Nt-NCAPP1) and the phloem NCAPs, are essential for their interaction. Detailed molecular analysis of a representative phloem NCAP, Cm-PP16-1, identified the specific residues on which glycosylation and phosphorylation must occur for effective binding to NCAPP1. Microinjection studies confirmed that posttranslational modification on these residues is essential for cell-to-cell movement of Cm-PP16-1. Lastly, a glutathione S-transferase (GST)–Cm-PP16-1 fusion protein system was employed to test whether the peptide region spanning these residues was required for cell-to-cell movement. These studies established that a 36–amino acid peptide was sufficient to impart cell-to-cell movement capacity to GST, a normally cell-autonomous protein. These findings are consistent with the hypothesis that a phosphorylation-glycosylation recognition motif functions to control the binding of a specific subset of phloem NCAPs to NCAPP1 and their subsequent transport through plasmodesmata.
INTRODUCTION
The role of endogenous non-cell-autonomously acting proteins in plant development is now well established (Lucas, 1995; Ghoshroy et al., 1997; Jackson and Hake, 1997; Zambryski and Crawford, 2000; Haywood et al., 2002; Wu et al., 2002; Ding et al., 2003; Heinlein and Epel, 2004; Lucas and Lee, 2004; Oparka, 2004; Ruiz-Medrano et al., 2004; Gallagher and Benfey, 2005; Kim et al., 2005; Kurata et al., 2005). Insights are also emerging as to the functions of such non-cell-autonomous proteins (NCAPs) in long-distance signaling, through the phloem, for development, gene silencing, and pathogen defense (Golecki et al., 1998; Jorgensen et al., 1998; Ruiz-Medrano et al., 1999; Kim et al., 2001; Searle et al., 2003; Yoo et al., 2004; Aoki et al., 2005; Haywood et al., 2005; Banerjee et al., 2006; Lough and Lucas, 2006).
The cell-to-cell movement of endogenous and viral NCAPs occurs through plasmodesmata, where two mechanisms appear to operate, namely, a gated pathway for molecular diffusion and selective trafficking in which protein–protein interaction confers specificity (Crawford and Zambryski, 2000; Haywood et al., 2002; Lucas and Lee, 2004; Zambryski, 2004). Interesting structural and functional parallels exist between plasmodesmata and the nuclear pore complex, in terms of selective and nonselective macromolecular trafficking (Lee et al., 2000). A well-characterized mechanism for translocation into the nucleus involves a nuclear localization signal (NLS) on the protein cargo (Pemberton et al., 1998; Cokol et al., 2000; Macara, 2001; Madrid and Weis, 2006), and this transport process can be further regulated by phosphorylation (Jans et al., 2000; Poon and Jans, 2005).
Interestingly, to date, no conserved NCAP feature, equivalent to an NLS, has yet been identified (Lucas et al., 1995; Aoki et al., 2002; Kurata et al., 2005; Trutnyeva et al., 2005; Sasaki et al., 2006). A short, 20–amino acid motif on Cm-Hsc70-1 and Cm-Hsc70-2 allowed these proteins, but not the closely related Cm-Hsc70-3, to function as NCAPs (Aoki et al., 2002). Transfer of this Cm-Hsc70-1 structural motif to the most closely related human Hsp70 chaperone resulted in gain-of-function cell-to-cell movement capacity. However, fusion of this motif to the C terminus of green fluorescent protein (GFP) did not confer movement capacity, suggesting that it does not function as a simple targeting signal. Rather, this 20–amino acid motif appears to operate in the context of the Hsp70 chaperone machinery.
The maize (Zea mays) homeodomain protein KNOTTED1 (KN1) was the first characterized plant NCAP (Lucas et al., 1995). Mutational analysis of the KN1 homeobox domain (S263-S326) coupled with microinjection experiments indicated that this specific region, which interestingly also contained the NLS, was critical for cell-to-cell trafficking. Subsequently, Kim et al. (2005) used an ingenious trafficking assay, based on trichome rescue in Arabidopsis thaliana, to screen for the presence of cis-acting signals involved in KN1 trafficking in vivo. A GFP-YFP-KN1256-359 fusion protein was able to traffic from the L3 to the L2 and L1 layers of the meristem, indicating that the KN1 homeodomain was necessary and sufficient for cell-to-cell movement. Although the manner in which this 100–amino acid region of the KN1 interacts with the NCAP pathway has yet to be established, this finding suggests that some form of secondary structure, rather than an NLS equivalent, may be the basis for recognition and selective transport through plasmodesmata.
A number of studies have reported the involvement of NCAP posttranslational modifications in the regulation of intercellular trafficking, especially with respect to the functioning of viral movement proteins (Waigmann et al., 2000, 2004; Lee and Lucas, 2001). For example, mutations in the phosphorylation sites located in the C-terminal region of the tobacco mosaic virus movement protein (TMV MP) affect both targeting to plasmodesmata and cell-to-cell movement in a plant host dependent manner (Trutnyeva et al., 2005). Here, it is interesting to note that PAPK1, a plasmodesmal-associated protein kinase and a member of the casein kinase I family, was recently shown to recognize and phosphorylate a subset of NCAPs, including the C-terminal residues on the TMV MP (Lee et al., 2005). The manner in which such phosphorylation events influence the ability of an NCAP to move cell to cell has yet to be established.
Although there is only sparse evidence implicating glycosylation in selective NCAP movement in plants, it is interesting to note that an Arabidopsis O-linked N-acetylglucosamine (O-GlcNAc) transferase, SECRET AGENT (SEC), has been implicated in potyvirus infection (Chen et al., 2005). In potyviruses, the capsid protein functions as an MP (Rojas et al., 1997; Carrington et al., 1998; Roberts et al., 1998), and a block to O-GlcNAc modification at the N-terminal region of the Plum pox potyvirus capsid protein results in a reduction in the rate and extent of viral movement (Chen et al., 2005; Scott et al., 2006).
Other circumstantial evidence for the role of glycosylation in NCAP function is derived from the report that GP40, a close homolog of Nicotiana tabacum NON-CELL-AUTONOMOUS PATHWAY PROTEIN1 (Nt-NCAPP1), is modified with O-GlcNAc in vivo (Heese-Peck and Raikhel, 1998). As Nt-NCAPP1 is involved in the entry of a subset of endogenous and viral NCAPs into the plasmodesmal pathway (Lee et al., 2003), its glycosylation state may well serve to regulate access to a plasmodesmal binding site.
Isolation and enrichment of Nt-NCAPP1 was achieved by affinity chromatography using a phloem NCAP, Cm-PP16-1 (Lee et al., 2003). Importantly, Nt-NCAPP1 was also shown to bind to a broad spectrum of additional proteins contained within the pumpkin (Cucurbita maxima) phloem sap; many of these proteins have the capacity for cell-to-cell movement (Balachandran et al., 1997). This raised the possibility that the pumpkin NCAPP1 homolog is involved in the selective trafficking of these phloem NCAPs through the companion cell–sieve element plasmodesmata.
In this study, pumpkin phloem proteins were used as a source of NCAPs to further explore the molecular basis for selective NCAP trafficking in plants. Protein overlay assays and coimmunoprecipitation experiments established that phosphorylation and glycosylation, on both Nt-NCAPP1 and the phloem NCAPs, is essential for protein–protein interaction. Molecular analysis of a representative phloem NCAP, Cm-PP16-1, identified the specific residues on which glycosylation and phosphorylation must occur for effective binding to Nt-NCAPP1. Microinjection studies confirmed that posttranslational modification on these residues is essential for cell-to-cell movement of Cm-PP16-1. Lastly, a glutathione S-transferase (GST)–Cm-PP16-1 fusion protein system was employed to test whether the region spanning these critical residues was sufficient for cell-to-cell movement. These studies established that a 36–amino acid peptide was sufficient to impart cell-to-cell movement capacity to GST, a normally cell-autonomous protein. Our findings are consistent with the hypothesis that a phosphorylation-glycosylation recognition motif functions to control the binding of a specific subset of phloem NCAPs to NCAPP1 and their subsequent transport through plasmodesmata.
RESULTS
Numerous Pumpkin Phloem Sap Proteins Are Phosphorylated/Glycosylated
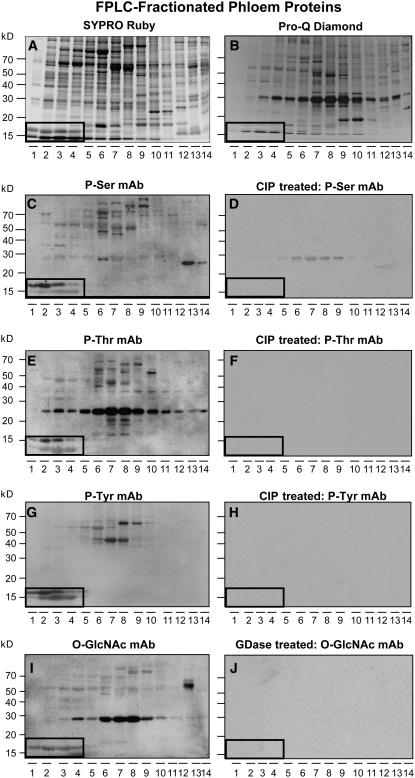
Phloem sap was collected from the stems of 20 6-week-old pumpkin plants and the proteins fractionated using anion-exchange fast protein liquid chromatography (FPLC) (Yoo et al., 2002, 2004). Sufficient aliquots of these fractionated phloem proteins were then frozen for use in all the experiments reported in this study. As shown in Figure 1, the phosphoprotein gel stain Pro-Q Diamond (Schulenberg et al., 2003) detected many of the phloem proteins that were visualized by SYPRO Ruby staining (cf. Figures 1A and 1B). Consistent with this result, phosphoserine, phosphothreonine, and phosphotyrosine monoclonal antibodies recognized a wide range of proteins present within the anion-fractionated phloem sap (Figures 1C, 1E, and 1G). Antigen binding of these phosphoserine, phosphothreonine, and phosphotyrosine monoclonal antibodies was almost completely abolished by treatment with calf intestinal phosphatase (CIP) (Figures 1D, 1F, and 1H), confirming the specificity.
Figure 1.
Subpopulations of Pumpkin Phloem Proteins Are Phosphorylated and Glycosylated.
(A) Pumpkin phloem sap FPLC-fractionated proteins. Proteins were separated on a 13% SDS-PAGE gel and then stained with SYPRO Ruby reagent.
(B) Phosphorylation status of FPLC-fractionated proteins from (A) assessed using Pro-Q Diamond reagent.
(C) Detection of phosphoproteins in FPLC-fractionated phloem sap by an antiphosphoserine monoclonal antibody.
(D) Aliquots of the phloem fractions used in (C) were pretreated with CIP prior to protein gel blot analysis with the antiphosphoserine monoclonal antibody. Absence of signals detected in (C) establishes the specificity of the antibody reaction.
(E) and (F) Detection of phosphoproteins in untreated and CIP-treated FPLC-fractionated phloem proteins by an antiphosphothreonine monoclonal antibody, respectively.
(G) and (H) Detection of phosphoproteins in untreated and CIP-treated FPLC-fractionated phloem proteins by an antiphosphotyrosine monoclonal antibody, respectively.
(I) and (J) Detection of glycosylated proteins in untreated and GDase-treated FPLC-fractionated phloem proteins by an anti-O-GlcNAc monoclonal antibody, respectively.
Boxed areas indicate the location of native Cm-PP16-1 (top band) and Cm-PP16-2 (bottom band).
We next examined the extent to which pumpkin phloem proteins are subjected to O-GlcNAcylation. Protein gel blot analysis performed with an O-GlcNAc monoclonal antibody preparation revealed that a significant number of phloem proteins appeared to be glycosylated with O-GlcNAc (Figure 1I). Pretreatment of these fractionated phloem proteins with β-N-hexoaminidase (GDase) abolished all antigen binding by the O-GlcNAc monoclonal antibody (Figure 1J). These results support the hypothesis that many of the pumpkin phloem proteins, including the well-characterized Cm-PP16-1 and Cm-PP16-2 (highlighted in Figure 1), are posttranslationally modified by phosphorylation and/or glycosylation.
Interaction of Phloem Proteins with NCAPP1 Is Dependent on Their Posttranslational Modification
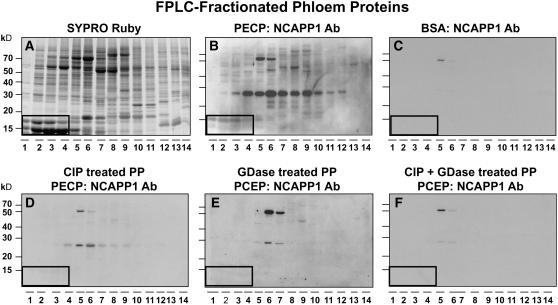
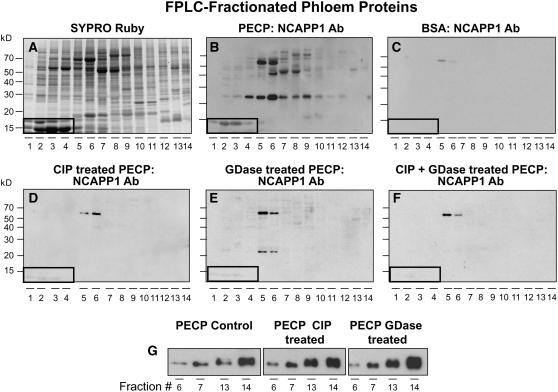
Our previous studies demonstrated that Nt-NCAPP1 is involved in the cell-to-cell movement of specific NCAPs and that it interacts with many of the pumpkin phloem proteins, including Cm-PP16 (Lee et al., 2003). To further explore the involvement of posttranslational modification of phloem proteins in binding to Nt-NCAPP1, we next performed protein overlay assays. As shown in Figures 2A and 2B, native Nt-NCAPP1 contained in a BY-2 plasmodesmal-enriched cell wall protein (PECP) preparation interacted with a subset of phloem proteins. Consistent with our earlier studies, no interaction was detected when the phloem proteins were overlaid with BSA, followed by incubation with the polyclonal antibody preparation (Figure 2C; Lee et al., 2003). Pretreatment of the fractionated phloem proteins with CIP and/or GDase was found to abolish the ability of most proteins to interact with Nt-NCAPP1 (Figures 2D to 2F). These data indicate that a combination of phosphorylation and glycosylation may be necessary for effective interaction between Nt-NCAPP1 and the phloem NCAPs, including Cm-PP16-1 and Cm-PP16-2.
Figure 2.
Posttranslational Modification of Pumpkin Phloem Proteins Is Essential for Nt-NCAPP1 Binding.
(A) Pumpkin phloem sap FPLC-fractionated proteins stained with SYPRO Ruby.
(B) FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with a PECP preparation, and Nt-NCAPP1 interaction partners detected by anti-NCAPP1 polyclonal antibody (Lee et al., 2003).
(C) BSA control. FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with BSA, and then probed with anti-NCAPP1 polyclonal antibodies.
(D) to (F) Nt-NCAPP1 overlay assays performed on FPLC-fractionated phloem proteins (PP) from (A) that were pretreated with CIP, GDase, or both CIP and GDase, respectively; interaction proteins were detected by anti-Nt-NCAPP1 polyclonal antibodies. Note that phosphorylation and glycosylation are necessary for Nt-NCAPP1 binding.
Boxed areas indicate the location of native Cm-PP16-1 (top band) and Cm-PP16-2 (bottom band).
Cm-PP16-1 Binding to Nt-NCAPP1 Requires Phosphorylation and O-GlcNAcylation
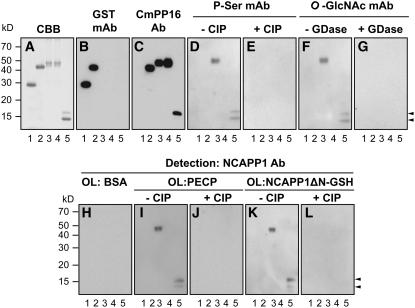
To further evaluate the role for posttranslational modification in NCAP binding to Nt-NCAPP1, we chose Cm-PP16-1 as a representative phloem protein for in depth analysis. To obtain unmodified recombinant Cm-PP16-1, a GST fusion protein was expressed in Escherichia coli. To produce in planta–expressed recombinant Cm-PP16-1, we used an Agrobacterium tumefaciens infiltration system (Voinnet et al., 2003). Here, a GFP construct that contained a StrepII tag and an eight-repeat histidine tag (GSH) was fused to the C-terminal region of the Cm-PP16-1 open reading frame (ORF). Agrobacterium carrying this Cm-PP16-1-GSH construct was infiltrated into young Nicotiana benthamiana leaves. As shown in Figure 3A, both E. coli and in planta–expressed Cm-PP16-1 could be purified to homogeneity using glutathione and StrepII/nickel affinity chromatography, respectively. The identities of these fusion proteins were confirmed using GST and Cm-PP16-1–specific antibody preparations (Figures 3B and 3C). Note that the Cm-PP16-1 antibody recognized Cm-PP16-1, but not Cm-PP16-2, from the native phloem-purified Cm-PP16 preparation (Aoki et al., 2005) (Figure 3C).
Figure 3.
Posttranslational Modification on Cm-PP16-1 Controls Its Binding Capacity with Nt-NCAPP1.
(A) Recombinant Cm-PP16-1 was expressed in and purified from E. coli as a GST-Cm-PP16-1 fusion protein (Lee et al., 2003) or by transient expression in N. benthamiana as a fusion protein with a GFP-Strep-His (GSH) tag (Cm-PP16-1-GSH). Lane 1, GST (27 kD) expressed in E. coli; lane 2, GST-Cm-PP16-1 (43 kD); lane 3, Cm-PP16-1-GSH (46 kD); lane 4, mCmPP16-1-GSH S-all-A (46 kD); lane 5, native phloem-purified Cm-PP16-1/-2. Protein aliquots (1 μg) were separated on 13% SDS-PAGE and purification verified by Coomassie Brilliant Blue (CBB) staining.
(B) and (C) Protein gel blot analyses performed on proteins from (A) using an anti-GST and anti-Cm-PP16-1 polyclonal antibodies, respectively.
(D) and (E) Phosphorylation status of E. coli and in planta–expressed Cm-PP16-1 determined using an antiphosphoserine monoclonal antibody. CIP pretreatment confirms the specificity of the antibody reaction.
(F) and (G) Glycosylation status of E. coli and in planta–expressed Cm-PP16-1 determined using an anti-O-GlcNAc monoclonal antibody. GDase pretreatment confirms the specificity of the antibody reaction.
(H) to (L) Aliquots of proteins from (A) were blotted to membranes and then overlaid with BSA, PECP preparation (100 μg/mL), or NCAPP1ΔN-GSH (5 μg/mL); binding between the various forms of Cm-PP16-1 and NCAPP1 was detected with anti-Nt-NCAPP1 polyclonal antibodies. CIP-treated Cm-PP16-1 controls confirmed the phosphorylation requirement for Cm-PP16-1–Nt-NCAPP1 binding.
Arrowheads indicate position of Cm-PP16-1 (top) and Cm-PP16-2 (bottom).
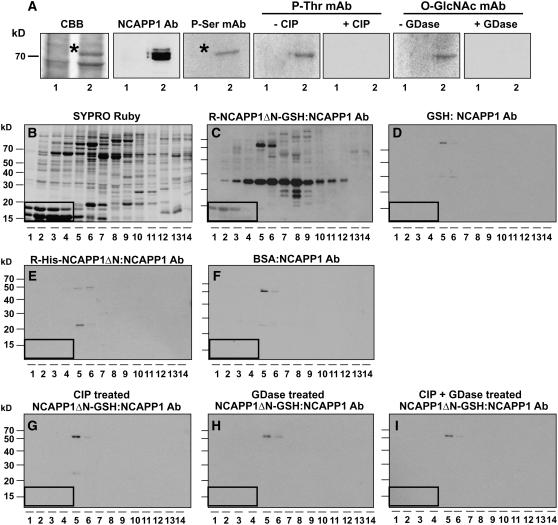
Protein gel blot analysis performed with the phosphoserine and O-GlcNAc monoclonal antibodies confirmed that phloem-purified Cm-PP16-1/-2 and in planta–expressed and purified Cm-PP16-1-GSH were phosphorylated and glycosylated (Figures 3D and 3F, lanes 3 and 5, respectively). As expected, neither antibody recognized the E. coli–expressed and purified Cm-PP16-1-GST (Figures 3D and 3F, lane 2). Treatment of phloem-purified Cm-PP16-1/-2 and Cm-PP16-1-GSH with CIP or GDase abolished the antigen–antibody interactions (Figures 3E and 3G). These data demonstrate the equivalence between native phloem Cm-PP16-1 and our recombinant in planta–expressed and purified Cm-PP16-1-GSH.
Next, Web-based computer programs (http://www.cbs.dtu.dk/services/NetPhos/ and http://www.cbs.dtu.dk/services/YinOYang) were used to identify the putative phosphorylation and gylcosylation sites on Cm-PP16-1. As O-GlcNAcylation could compete with phosphorylation on Ser or Thr residues (Guinez et al., 2005), we focused our attention on putative phosphoserine or phosphothreonine sites. Based on this analysis, Cm-PP16-1 is predicted to have five Ser residues, Ser-12, Ser-41, Ser-66, Ser-108, and Ser-133, that could undergo posttranslational modification. To test for the involvement of these residues in Nt-NCAPP1 interaction, we generated a Cm-PP16-1-GSH mutant, mCm-PP16-1-GSH S-all-A, in which all five Ser residues were substituted with Ala. This mutant was then expressed in and purified from N. benthamiana (Figure 3A, lane 4) and tested for posttranslational modifications. Neither the phosphoserine (Figure 3D) nor the O-GlcNAc monoclonal antibody (Figure 3F) detected the mCmPP16-1-GSH S-all-A, indicating that we had likely identified the appropriate target residue(s).
Purified phloem and recombinant Cm-PP16-1 (in planta and E. coli expressed) were next used in Nt-NCAPP1 overlay experiments. For these studies, we prepared two forms of Nt-NCAPP1, native protein enriched in the PECP prepared from BY-2 suspension cultured cells, and an N-terminal deletion mutant transiently expressed in N. benthamiana as a C-terminal–GSH fusion protein (NCAPP1ΔN-GSH) (Lee et al., 2003). As a negative control for these experiments, we used BSA as the overlay protein, and as shown in Figure 3H, incubation of these blots with Nt-NCAPP1 antibodies failed to yield any evidence of protein–protein interaction. Consistent with our earlier findings, negative results were also obtained when E. coli–expressed Cm-PP16-1-GST was overlaid with either native Nt-NCAPP1 (contained in the PECP preparation) or recombinant Nt-NCAPP1ΔN-GSH (Figures 3I and 3K, lane 2). However, in planta–expressed recombinant Cm-PP16-1-GSH and native phloem–purified Cm-PP16-1/-2 interacted with both native Nt-NCAPP1 (Figure 3I, lanes 3 and 5) and in planta recombinant Nt-NCAPP1ΔN-GSH (Figure 3K, lanes 3 and 5). These Cm-PP16–Nt-NCAPP1 interactions were abolished by CIP treatment (Figures 3J and 3L). Importantly, mCmPP16-1-GSH S-all-A failed to interact with either form of the Nt-NCAPP1 (Figures 3I and 3K, lane 4). Collectively, these results add support for the hypothesis that both phosphorylation and O-GlcNAcylation on the identified Ser residues of Cm-PP16-1 are necessary for its interaction with Nt-NCAPP1.
Nt-NCAPP1 Is Posttranslationally Modified
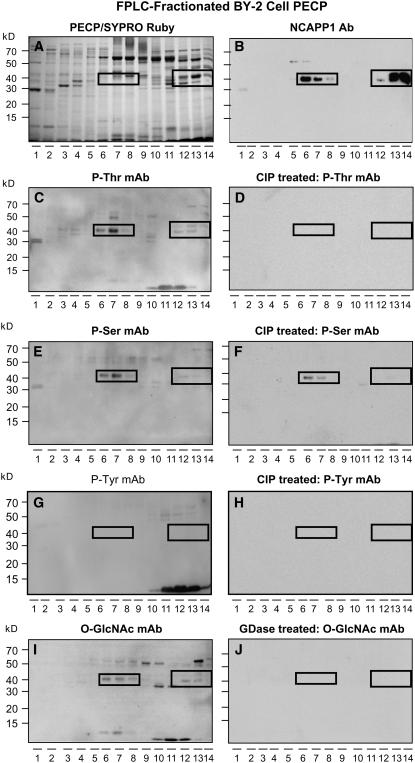
To test whether Nt-NCAPP1 is phosphorylated and/or glycosylated, we first used cation-exchange chromatography to fractionate a BY-2 cell PECP preparation. As shown in Figures 4A and 4B, a comparison of SYPRO Ruby-stained PECPs and protein gel blots performed with Nt-NCAPP1 polyclonal antibodies allowed us to locate the 40-kD Nt-NCAPP1 in fractions 6 to 8 and 12 to 14. The BY-2 cell PECPs were then tested using phosphothreonine (Figure 4C), phosphoserine (Figure 4E), phosphotyrosine (Figure 4G), and O-GlcNAc (Figure 4I) monoclonal antibodies. All antibodies, except for the phosphotyrosine antibody, detected a 40-kD band in fractions 6 to 8 and 12 to 14 of the BY-2 cell PECP preparation. Pretreatment of the BY-2 cell PECP fractions with CIP or GDase abolished, or greatly reduced, the signal obtained with these monoclonal antibodies (Figures 4D, 4F, 4H, and 4J). These results indicate that Nt-NCAPP1 is indeed phosphorylated and O-GlcNAcylated on Ser and Thr residues.
Figure 4.
Native Nt-NCAPP1 Contained in PECP Preparation Is Both Phosphorylated and Glycosylated.
(A) Cation-exchange FPLC-fractionated PECPs separated on a 13% SDS-PAGE gel and stained with SYPRO Ruby.
(B) Protein gel blot analysis of FPLC-fractionated proteins from (A) performed with anti-Nt-NCAPP1 polyclonal antibody preparation. Note the presence of strong Nt-NCAPP1 signal in lanes 6 to 8 and 12 to 14.
(C) to (H) Phosphorylation status of native Nt-NCAPP1 probed using antiphosphothreonine, antiphosphoserine, or antiphosphotyrosine monoclonal antibodies. CIP pretreatment of the FPLC-fractionated proteins from (A) confirmed the specificity of the immunoreaction.
(I) and (J) Glycosylation status of native Nt-NCAPP1 probed using an anti-O-GlcNAc monoclonal antibody. GDase pretreatment of the FPLC-fractionated proteins from (A) confirmed the specificity of the immunoreaction.
Boxed areas indicate the location of Nt-NCAPP1 isoforms on the PECP FPLC fractions.
Nt-NCAPP1 Posttranslational Modification Is Necessary for Interaction with Phloem Proteins
To further investigate whether the posttranslational modification on Nt-NCAPP1 affects the interaction with phloem proteins, we performed protein overlay assay with CIP- and/or GDase-treated BY-2 cell PECP preparations. Pumpkin phloem proteins were overlaid with CIP- and/or GDase-treated BY-2 cell PECPs, and then any interaction with Nt-NCAPP1 was probed using the Nt-NCAPP1 antibody. As shown in Figure 5, CIP and/or GDase pretreatment of the BY-2 cell PECP preparation almost completely abolished the Nt-NCAPP1 interaction with phloem proteins. Here, it is important to note that the CIP and/or GDase pretreatment of our BY-2 cell PECP preparation did not affect the antigenicity of Nt-NCAPP1 to the Nt-NCAPP1 polyclonal antibodies (Figure 5G). Taken together, these findings support the hypothesis that posttranslational modifications on Nt-NCAPP1 are important for its interaction with phloem proteins.
Figure 5.
Posttranslational Modification on Nt-NCAPP1 Controls Its Binding Capacity with Cm-PP16-1 and Other Phloem Proteins.
(A) Pumpkin phloem sap FPLC-fractionated proteins stained with SYPRO Ruby.
(B) FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with a PECP preparation, and Nt-NCAPP1 interaction partners detected by anti-NCAPP1 polyclonal antibodies.
(C) BSA control. FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with BSA, and then probed with anti-NCAPP1 polyclonal antibodies.
(D) to (F) Nt-NCAPP1 overlay assays performed on FPLC-fractionated phloem proteins from (A). PECP preparation was pretreated with CIP, GDase, or both CIP and GDase; interaction proteins were detected by anti-Nt-NCAPP1 polyclonal antibodies. Note that phosphorylation and glycosylation on Nt-NCAPP1 are necessary for binding to Cm-PP16-1 and other phloem proteins.
Boxed areas indicate the location of native Cm-PP16-1 (top band) and Cm-PP16-2 (bottom band).
(G) Antigenicity of Nt-NCAPP1 contained in the PECP preparation was not affected by CIP or GDase treatment. Cation-exchange FPLC fractions 6, 7, 13, and 14 (see Figure 4B) were CIP or GDase treated and then protein gel blot analyses performed with anti-Nt-NCAPP1 polyclonal antibodies.
Recombinant Nt-NCAPP1 Exhibits Equivalent Binding Properties to Native Nt-NCAPP1
Protein gel blot analysis of the BY-2 cell PECP preparation suggested the presence of more than one Nt-NCAPP1 isoform (Figure 4B). Thus, the possibility existed that such isoforms could interact with different subsets of phloem proteins. To explore this possibility, we first purified in planta–expressed recombinant Nt-NCAPP1ΔN-GSH and E. coli–expressed recombinant His-Nt-NCAPP1ΔN (Lee et al., 2003). As shown in Figure 6A, phosphoserine, phosphothreonine, and O-GlcNAc monoclonal antibodies recognized the in planta–expressed Nt-NCAPP1ΔN-GSH, and this cross-reactivity was abolished by CIP or GDase pretreatment. As expected, none of these antibodies recognized the His-ΔN purified from E. coli (data not shown).
Figure 6.
Involvement of Posttranslational Modification of NCAPP1 for Binding to Phloem Proteins.
(A) Phosphorylation and glycosylation status of in planta–expressed and purified recombinant Nt-NCAPP1. Protein gel blot analyses were performed with anti-Nt-NCAPP1 polyclonal antibodies and antiphosphoserine, antiphosphotheronine, and anti-O-GlcNAc monoclonal antibodies. CIP and GDase pretreatment of purified recombinant Nt-NCAPP1 confirmed the specificity of these immunoreactions. Lane 1, partially purified GSH; lane 2, partially purified Nt-NCAPP1ΔN-GSH.
(B) FPLC-fractionated pumpkin phloem proteins stained with SYPRO Ruby.
(C) FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with recombinant Nt-NCAPP1ΔN-GSH, and Nt-NCAPP1 interaction partners detected by anti-NCAPP1 polyclonal antibodies.
(D) GSH control. FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with GSH, and then probed with anti-NCAPP1 polyclonal antibodies.
(E) FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with E. coli–expressed recombinant His-tagged Nt-NCAPP1ΔN (R-His-Nt-NCAPP1ΔN), and Nt-NCAPP1 interaction partners detected by anti-NCAPP1 polyclonal antibodies.
(F) BSA control. FPLC-fractionated phloem proteins were blotted onto membrane, overlaid with BSA, and then probed with anti-NCAPP1 polyclonal antibodies.
(G) to (I) Nt-NCAPP1ΔN-GSH overlay assays performed on FPLC-fractionated phloem proteins. Purified recombinant Nt-NCAPP1ΔN-GSH was pretreated with CIP, GDase, or both CIP and GDase, respectively; interaction proteins were detected by anti-Nt-NCAPP1 polyclonal antibodies. Note that phosphorylation and glycosylation on Nt-NCAPP1 is necessary for binding to Cm-PP16-1 and other phloem proteins.
Boxed areas indicate the location of native Cm-PP16-1 (top band) and Cm-PP16-2 (bottom band).
Next, fractionated pumpkin phloem proteins were overlaid with in planta–expressed Nt-NCAPP1ΔN-GSH, or GSH as a control, and the interacting proteins detected with Nt-NCAPP1 polyclonal antibodies. A very similar binding pattern was observed with both Nt-NCAPP1ΔN-GSH (Figure 6C) and the Nt-NCAPP1 present in the BY-2 cell PECP preparation (Figure 5B). However, in the GSH control, no protein–protein interactions were detected (Figure 6D). Importantly, the recombinant His-Nt-NCAPP1ΔN purified from E. coli displayed only a weak and very limited interaction with the fractionated phloem proteins (Figure 6E). For these experiments, a BSA buffer control was also employed, and this yielded a response equivalent to that of the His-Nt-NCAPP1ΔN treatment (cf. Figures 6E and 6F). Finally, CIP and/or GDase pretreatment of Nt-NCAPP1 abolished its ability to interact with the fractionated phloem proteins (Figures 6G to 6I). Taken together, these results demonstrate the equivalence in properties and patterns of binding, to the fractionated phloem proteins, exhibited by the recombinant Nt-NCAPP1ΔN-GSH and native Nt-NCAPP1. Thus, Nt-NCAPP1 is likely responsible for the majority of the protein–protein interactions detected in our phloem protein overlay assays.
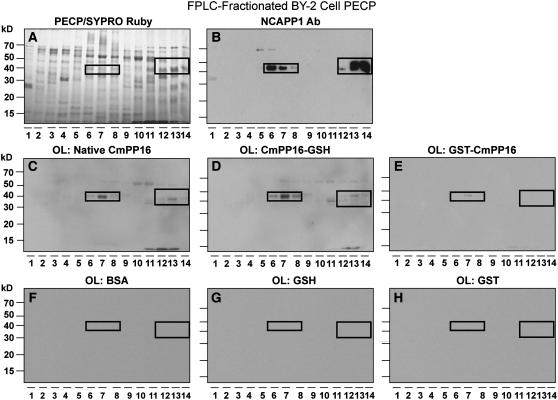
In our initial studies on Nt-NCAPP1, recombinant GST-Cm-PP16-1 expressed in and purified from E. coli was used to develop an affinity column to screen and enrich for proteins involved in the NCAP trafficking pathway (Lee et al., 2003). In view of our findings, we next tested the relative strength of interaction between phloem-purified Cm-PP16, Cm-PP16-1-GSH, and GST-Cm-PP16-1 and the BY-2 cell PECPs. As illustrated in Figure 7, native Cm-PP16-1/-2 interacted with the 40-kD Nt-NCAPP1 present in fractions 6 to 8 and 12 to 14 as well as with a number of other bands. A similar pattern was obtained with overlays performed with in planta–expressed and purified Cm-PP16-1-GSH (Figure 7D). However, the E. coli–expressed and purified GST-Cm-PP16 interacted with only the Nt-NCAPP1 in fraction 7 of the BY-2 cell PECPs (Figure 7E). Controls for these experiments included overlays with BSA (Figure 7F), GSH (Figure 7G), and GST (Figure 7H), and none yield interactions when probed with the Cm-PP16-1 antibody. These results further confirmed the importance of Cm-PP16 posttranslational modification in terms of its strength of interaction with BY-2 cell PECPs, including Nt-NCAPP1.
Figure 7.
Native and in Planta–Expressed, but Not E. coli–Produced, Cm-PP16-1 Interacts Strongly with Nt-NCAPP1.
(A) Cation-exchange FPLC-fractionated PECPs separated on a 13% SDS-PAGE gel and stained with SYPRO Ruby.
(B) Protein gel blot analysis of FPLC-fractionated proteins from (A) performed with anti-Nt-NCAPP1 polyclonal antibody preparation.
(C) to (E) FPLC-fractionated PECP preparations were blotted and then overlaid with native phloem-purified Cm-PP16-1, in planta–expressed recombinant Cm-PP16-1-GSH, and E. coli–expressed recombinant GST-Cm-PP16-1; interaction partners were detected using anti-Cm-PP16-1–specific polyclonal antibodies. Note that the GST-Cm-PP16-1 interaction was limited to Nt-NCAPP1 present in lane 7.
(F) to (H) FPLC-fractionated PECP preparations were blotted and then overlaid with BSA, in planta–expressed GSH, or E. coli–expressed GST. Signals were detected by an anti-Cm-PP16-1–specific polyclonal antibodies.
Boxed areas show the location of Nt-NCAPP1 isoforms.
Native Cm-PP16-1/-2 Coimmunoprecipitates with Nt-NCAPP1
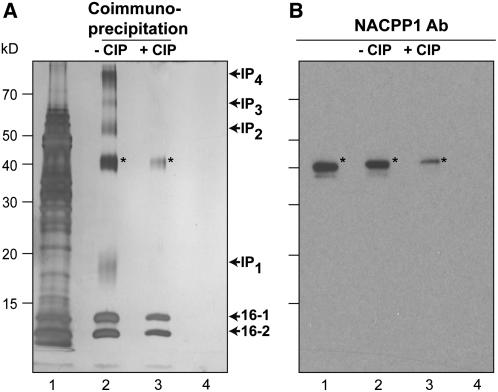
Interaction between Nt-NCAPP1 and Cm-PP16 was also tested by performing coimmunoprecipitation (co-IP) experiments. BY-2 cell PECPs were mixed with phloem-purified native Cm-PP16-1/-2 and then subjected to co-IP using the Cm-PP16-1–specific polyclonal antibodies. As shown in Figure 8A, seven protein bands were coprecipitated; the 40-kD band was identified as Nt-NCAPP1 by protein gel blot analysis (Figure 8B), the two lower molecular weight bands were identified as Cm-PP16-1 and Cm-PP16-2, while proteins in the other four bands remain to be identified. Although we used Cm-PP16-1–specific antibodies, both Cm-PP16-1 and Cm-PP16-2 were pulled down, indicating that Cm-PP16-2 forms a heterodimer with Cm-PP16-1, a finding consistent with earlier results (Aoki et al., 2005).
Figure 8.
Nt-NCAPP1 Coimmunoprecipitates with Cm-PP16-1.
(A) Phloem-purified native Cm-PP16-1 and Cm-PP16-2 were mixed with a PECP preparation (lane 1, input proteins), and co-IP was performed using either anti-Cm-PP16-1 antibody (lanes 2 and 3) or preimmnune serum (lane 4). A comparison of lanes 2 and 3 illustrates the importance of Cm-PP16-1 phosphorylation on protein complex formation. Proteins detected by silver stain. Note that the co-IP complex is comprised of Cm-PP16-1, Cm-PP16-2, interaction protein 1 (IP1), Nt-NCAPP1, IP2, IP3, and IP4.
(B) Presence of Nt-NCAPP1 (asterisks) detected with Nt-NCAPP1 polyclonal antibodies. Lanes are as in (A). These co-IP experiments were performed in duplicate with identical results.
Pretreatment of the phloem-purified Cm-PP16-1/-2 with CIP, prior to incubation with BY-2 cell PECPs, greatly reduced the complexity of the co-IP protein profile (Figure 8A, lane 3). Here, although Nt-NCAPP1 was still coimmunoprecipitated with Cm-PP16-1/-2, the amount of protein was much reduced compared with the native Cm-PP16 co-IP experiment (Figure 8B). Interestingly, the Cm-PP16-2 interaction with Cm-PP16-1 appeared to be unaffected by CIP treatment, suggesting that heterodimer formation is not dependent on phosphorylation. However, as the four additional bands were absent in this CIP-treated Cm-PP16 co-IP experiment, interactions between these prey proteins and the Cm-PP16-1/-2 bait appear to require phosphorylation.
Posttranslational Modification of Cm-PP16-1 Residues Tyr-63 and Ser-66 Is Central for Binding to Nt-NCAPP1
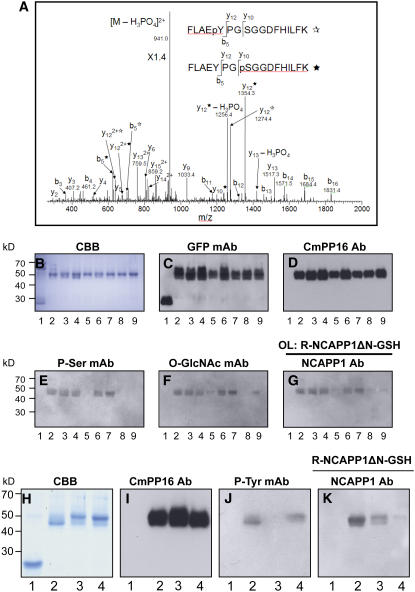
To identify the phosphorylation sites on Cm-PP16-1, phloem-purified protein was digested with trypsin and then subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. An MS/MS spectrum of the identified Cm-PP16-1 phosphopeptides is shown in Figure 9A. Our results indicated phosphorylation of Tyr-63 and Ser-66 on Cm-PP16-1 in vivo. The significance of the identified Cm-PP16-1 phosphorylation sites on Nt-NCAPP1 binding was next assessed by site-directed mutagenesis. First, a series of Cm-PP16-1-GSH mutant constructs was engineered in which codons for Ser-12, Ser-41, Ser-66, Ser-108, and Ser-133 were individually changed to Ala. The resultant recombinant purified mutant proteins (Figure 9B) were confirmed by protein gel blot analysis using anti-GFP (Figure 9C) and anti-Cm-PP16-1 antibodies (Figure 9D). Protein gel blots performed on these various Cm-PP16-1-GSH mutants, using the phosphoserine monoclonal antibody, indicated that only mCmPP16-1-GSH S66A was not detected in this assay (Figure 9E, lane 5).
Figure 9.
Identification and Mutational Analysis of Posttranslational Modification Sites on Cm-PP16-1.
(A) LC-MS/MS analysis of the phosphorylation sites on phloem-purified Cm-PP16-1. The MS/MS spectrum is a mixture of Ser or Tyr phosphorylation of the same peptide, FLAEpYPGSGGDFHILK and FLAEYPGpSGGDFHILK. These two phosphopeptides have the same parent mass-to-charge ratio of 990.1, and most fragments are the same, but fragments y10, y12, and b5 distinguish the two, as denoted by and asterisks for phosphotyrosine and phosphoserine, respectively.
(B) to (G) Analysis of phosphorylation and O-GlcNacylation on Cm-PP16-1 Ser residues.
(B) CBB staining of recombinant purified proteins. Lane 1, GSH; lane 2, Cm-PP16-1-GSH; lanes 3 to 7, Cm-PP16-1-GSH mutants in which Ser-12, Ser-41, Ser-66, Ser-108, and Ser-133, respectively, were replaced with Ala; lane 8, Cm-PP16-1-GSH mutant in which all the above five Ser residues were replaced with Ala (mCmPP16-1-GSH S-all-A); lane 9, Cm-PP16-1-GSH mutant in which Ser-66 was replaced with Asp to produce a phosphorylation mimic (mCmPP16-1-GSH S66D).
(C) Protein gel blot analysis performed with anti-GFP antibody.
(D) Protein gel blot analysis performed with anti-CmPP16-1 polyclonal antibodies.
(E) Protein gel blot analysis performed with an anti-phosphoserine monoclonal antibody.
(F) Protein gel blot analysis performed with an anti-O-GlcNAc monoclonal antibody.
(G) Protein overlay assay (OL) performed with recombinant purified Nt-NCAPP1ΔN-GSH; interacting proteins were detected with anti-Nt-NCAPP1 polyclonal antibodies.
(H) to (K) Analysis of the involvement of Cm-PP16-1 Tyr and Ser residues on binding to Nt-NCAPP1.
(H) CBB staining of the recombinant purified proteins. Lane 1, GSH; lane 2, Cm-PP16-1-GSH; lane 3, Cm-PP16-1-GSH mutant in which Tyr-63 was replaced with Ala (mCmPP16-1-GSH Y63A); lane 4, mCmPP16-1-GSH S-all-A.
(I) Protein gel blot analysis performed with anti-Cm-PP16-1 polyclonal antibodies.
(J) Protein gel blot analysis performed with an antiphosphotyrosine monoclonal antibody.
(K) Protein overlay assay performed with recombinant purified Nt-NCAPP1ΔN-GSH; interacting proteins were detected with anti-Nt-NCAPP1 polyclonal antibodies.
As previously established, mCmPP16-1-GSH S-all-A was also not detected in this protein gel blot assay (Figure 9E, lane 8). In addition, a Cm-PP16-1-GSH mutant in which Ser-66 was replaced with Asp to produce a phosphorylation mimic (mCmPP16-1-GSH S66D) (Trutnyeva et al., 2005) was also not recognized by the phosphoserine antibody (Figure 9E, lane 9). As the other four Ser-to-Ala mutants were detected by the phosphoserine monoclonal antibody (Figure 9E, lanes 3, 4, 6, and 7), these results are consistent with phosphorylation occurring on Cm-PP16-1 Ser-66.
These Cm-PP16-1 mutants were also examined for O-GlcNAcylation using the O-GlcNAc monoclonal antibody. Interestingly, no cross-reactivity was detected with mCmPP16-1-GSH S-all-A (Figure 9F, lane 8), and mCmPP16-1-GSH S66A displayed reduced binding (Figure 9F, lane 5) relative to the other four Ala mutants and wild-type Cm-PP16-1-GSH (Figure 9F, lanes 2 to 4, 6, and 7). These results are consistent with glycosylation of Cm-PP16-1-GSH Ser-66 with O-GlcNAc. Cross-reactivity between the O-GlcNAc monoclonal antibody and mCmPP16-1-GSH S66D (Figure 9F, lane 9) demonstrates that O-GlcNAcylation can also occur on other Cm-PP16-1-GSH Ser residues.
Next, this series of mutants was employed to test the role of Ser-66 modification on Cm-PP16-1–GSH interaction with Nt-NCAPP1. Our overlay assays performed with purified recombinant Nt-NCAPP1ΔN-GSH revealed that mCmPP16-1-GSH S66A, mCmPP16-1-GSH S-all-A, and mCmPP16-1-GSH S66D all showed a greatly reduced level of interaction (Figure 9G, lanes 5, 8, and 9). These findings indicate that phosphorylation or glycosylation of Ser-66 is important to confer binding between CmPP16-1-GSH and Nt-NCAPP1ΔN-GSH. These experiments also indicated that the S66D mutation served as a poor phosphorylation mimic in these binding assays.
The residual weak interaction detected between Nt-NCAPP1ΔN-GSH and mCmPP16-1-GSH S66D may well reflect the influence of phosphorylation on another Cm-PP16 residue, possibly the Tyr-63 that was identified by mass spectrometry (Figure 9A). To test this possibility, purified recombinant mCmPP16-1-GSH Y63A (Figures 9H and 9I) was first analyzed using a phosphotyrosine monoclonal antibody. In contrast with Cm-PP16-1-GSH and mCmPP16-1-GSH S-all-A, mCmPP16-1-GSH Y63A was not recognized by this antibody (Figure 9J, cf. lane 3 with lanes 2 and 4). Next, mCmPP16-1-GSH Y63A was tested in an overlay assay with Nt-NCAPP1ΔN-GSH, and as shown in Figure 9K, the strength of its interaction was intermediate between that for wild-type Cm-PP16-1 and the mCmPP16-1-GSH S-all-A. Thus, our results support the notion that Cm-PP16-1 binding to Nt-NCAPP1 is dependent upon phosphorylation at both Tyr-63 and Ser-66 and that this effect is likely to be cooperative.
Cm-PP16-1 Tyr-63 and Ser-66 Mutants Are Compromised in Plasmodesmal Trafficking
Using microinjection methods, we previously demonstrated that phloem-purified Cm-PP16-1/-2 and E. coli–expressed recombinant Cm-PP16-1 have the capacity to interact with plasmodesmata to induce an increase in size-exclusion limit and move cell to cell (Xoconostle-Cázares et al., 1999). These results suggested that, upon injection into the target cell, the protein can enter the NCAP translocation pathway where it can be posttranslationally modified to allow recognition by NCAPP1. As a test for this hypothesis, we next performed a series of microinjection experiments in which we compared the capacity of wild-type and mutant forms of Cm-PP16-1 to interact with plasmodesmata.
To test for the efficacy of cell-to-cell trafficking, we first labeled phloem-purified Cm-PP16-1 with Oregon Green (OG). After microinjection of OG-labeled Cm-PP16-1 into a target cell, we used confocal laser scanning microscopy to monitor the distribution and intensity of the fluorescent signal. As previously reported (Xoconostle-Cázares et al., 1999; Lee et al., 2003), Cm-PP16-1 underwent efficient and extensive cell-to-cell trafficking (Table 1). With time, the intensity of the fluorescent signal in the injected cell diminished to a low level as Cm-PP16-1 moved into the surrounding cells. Similar results were obtained with recombinant Cm-PP16-1, expressed in and purified from E. coli using a His-Cm-PP16-1 construct and nickel column chromatography. Here, movement was recorded by coinjection with fluorescein isothiocyanate–labeled 10-kD dextran (F-dextran) (Table 1; see Supplemental Figure 1B online).
Table 1.
Cm-PP16-1 Y63A, S66A, and S66D Mutants Are Dysfunctional in Their Capacity for Cell-to-Cell Trafficking
| Microinjection
|
||||
|---|---|---|---|---|
| Movement (n [%])a
|
||||
| Injected Probe | Total | Extensive (10 to 20 Cells) | Limited (One Cell) | None (Injected Cell) |
| Lucifer yellow CH | 18 | 18 (100) | 0 (0) | 0 (0) |
| F-dextranb | 32 | 0 (0) | 2 (6) | 30 (94) |
| OG-labeled phloem-purified Cm-PP16-1c | 46 | 44 (96) | 2 (4) | 0 (0) |
| Phloem-purified Cm-PP16-1 + F-dextran | 41 | 37 (90) | 2 (5) | 2 (5) |
| His-Cm-PP16-1 + F-dextrand | 33 | 31 (94) | 2 (6) | 0 (0) |
| His-Cm-PP16-1 S-all-A + F-dextran | 40 | 0 (0) | 10 (25) | 30 (75) |
| His-Cm-PP16-1 S66A + F-dextran | 40 | 0 (0) | 8 (20) | 32 (80) |
| His-Cm-PP16-1 S66D + F-dextran | 41 | 0 (0) | 13 (32) | 28 (68) |
| His-Cm-PP16-1 Y63A + F-dextran | 66 | 0 (0) | 28 (42) | 38 (58) |
| His-Cm-PP16-1 S12A + F-dextran | 20 | 19 (95) | 1 (5) | 0 (0) |
Cell-to-cell movement of each fluorescent probe was analyzed 2 min after injection into an N. benthamiana mesophyll cell. A Leica confocal laser scanning microscope was used to record the extent of movement; this was categorized as extensive (probe spread out from the injected cell into 10 to 20 neighboring mesophyll cells), limited (probe moved into one neighboring cell), or none (probe remained in injected cell).
10-kD Dextran labeled with fluorescein isothiocyanate was used as a reporter for protein trafficking through plasmodesmata.
Phloem-purified Cm-PP16-1 was labeled with OG for direct analysis of protein trafficking.
Recombinant His-Cm-PP16-1 was expressed in and purified from E. coli.
This level of efficient and extensive trafficking was in marked contrast with that observed when the recombinant His-CmPP16-1 S-all-A mutant was coinjected into target cells along with 10-kD F-dextran. In this situation, fluorescent signal was almost entirely confined to the injected cell (Table 1; see Supplemental Figure 1C online). In a few cases, limited movement occurred as a very weak signal was detected in a neighboring cell(s). Identical results were obtained using the recombinant His-CmPP16-1 S66A, S66D, and Y63A mutants (Table 1; see Supplemental Figures 1D to 1F online). Microinjection experiments performed with recombinant His-CmPP16-1 S12A indicated that the S12A mutation had no effect on cell-to-cell movement capacity (Table 1); these injections served as an additional methodological control. These experiments established that mutations on Tyr-63 and Ser-66 render Cm-PP16-1 dysfunction for cell-to-cell trafficking through plasmodesmata. Taken together, these results support the hypothesis that posttranslational modification of Cm-PP16-1 is required for efficient interaction with NCAPP1, a step that is necessary for subsequent interaction with and movement through plasmodesmata.
Peptide Containing the Cm-PP16-1 Recognition Motif Is Sufficient to Confer Cell-to-Cell Movement Capacity
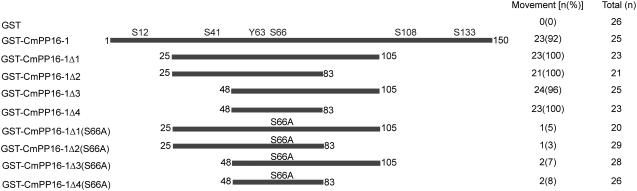
The GST-Cm-PP16-1 fusion protein was previously shown to traffic through plasmodesmata, whereas GST (27-kD protein) alone remained in the injected cells (Lee et al., 2003; Figure 10). This fusion protein system was next employed to test whether the peptide region spanning the Cm-PP16-1 Tyr-63 and Ser-66 residues was sufficient for cell-to-cell movement. As illustrated in Figure 10, GST-Cm-PP16-1Δ1 through GST-Cm-PP16-1Δ4 all exhibited movement capacity equivalent to the GST-Cm-PP16-1 control. By contrast, substitution of Ser-66 to Ala-66 in these GST-Cm-PP16-1 fusion mutants inhibited movement function. These results establish that a 36–amino acid peptide (48 to 83 amino acids of Cm-PP16-1) is sufficient to impart cell-to-cell movement capacity to GST, a normally cell-autonomous protein.
Figure 10.
Cell-to-Cell Movement of GST Is Conferred by a Peptide Containing the Cm-PP16-1 Recognition Motif.
Microinjection experiments were performed with the illustrated series of GST-Cm-PP16-1 fusion proteins. GST-Cm-PP16-1 N- and C-terminal deletion mutant proteins established that a peptide, comprised of 36 amino acids spanning the identified posttranslational modification sites, was sufficient to impart gain-of-function movement capacity to GST. Test proteins were microinjected into N. benthamiana mesophyll cells, and their movement was detected by coinjection of fluorescein isothiocyanate–labeled 10 kD dextran. Total number of injections (n) and percentage of intercellular movement associated with each test probe are shown on the right.
DISCUSSION
In this study, we examined the function of posttranslational modification in terms of the regulation of protein–protein interactions that underlie NCAP recognition and cell-to-cell trafficking through plasmodesmata. First, we established that a significant number of pumpkin phloem sap proteins, many of which likely act as NCAPs, and the Nt-NCAPP1 are phosphorylated and glycosylated (Figures 1 and 4). Next, we established that such posttranslational modifications are critical for efficient interaction between Nt-NCAPP1 and the pumpkin phloem NCAPs (Figures 2, 5, and 7). Detailed analysis of one phloem NCAP, Cm-PP16-1, confirmed the reciprocal requirements of posttranslational modification on Cm-PP16-1 and Nt-NCAPP1 for efficient protein–protein interaction (Figures 3, 6, and 8). Mass spectrometry and site-directed mutagenesis identified Tyr-63 and Ser-66 on Cm-PP16-1 as critical residues for Nt-NCAPP1 binding (Figure 9). In addition, microinjection studies established that mutations on Tyr-63 and Ser-66 render Cm-PP16-1 dysfunction for cell-to-cell movement (Table 1; see Supplemental Figure 1 online). Finally, the GST-Cm-PP16-1 fusion protein system was used to establish that a 36–amino acid peptide spanning the Tyr-63 and Ser-66 of Cm-PP16-1 was sufficient to impart movement capacity to GST, a cell-autonomous protein. Taken together, our findings support the hypothesis that posttranslational modification of phloem NCAPs is required for efficient interaction with NCAPP1, a step that is necessary for subsequent interaction with and movement through plasmodesmata, via the NCAPP1-dependent pathway.
Phloem Phosphoprotein Composition
It is well established that posttranslational modification of proteins is an essential component for many plant regulatory pathways (Shapka et al., 2005; del Pozo et al., 2006; Lindermayr et al., 2006). Our biochemical analysis of the pumpkin phloem proteins indicated that many were phosphorylated on Ser and/or Thr residues. Interestingly, a much smaller population exhibited phosphotyrosine modification (Figure 1). Based on genome analyses, it has been proposed that plants lack bona fide Tyr-specific kinases, and our observed Tyr phosphorylation events may well reflect the action of dual specificity Ser/Thr/Tyr (S/T/Y) kinases (Rudrabhatla et al., 2006). This is interesting in relation to the Cm-PP16-1 Tyr-63, which is not well conserved in other plant Cm-PP16–like proteins, where it is often replaced with Ser or Thr (Xoconostle-Cázares et al., 1999).
The Cm-PP16-1 and other phloem phosphotyrosine substrates may be recognized by members of the casein kinase I (CKI) family; as in animals and yeast, these enzymes have Tyr kinase activity (Knippschild et al., 2005). In this regard, it is interesting to note that we recently identified a CKI isoform, PAPK, that localizes to plasmodesmata and phosphorylates a range of endogenous and viral NCAPs (Lee et al., 2005). Thus, it is possible that PAPK, or a related member, phosphorylates Cm-PP16-1 on Tyr-63. Although a number of protein kinases have been identified in the pumpkin phloem sap (Yoo et al., 2002), much has yet to be learned in terms of their substrates and the biological consequences of such phosphorylation events.
Role of O-GlcNAcylation on Phloem NCAPs
Protein O-GlcNAcylation can occur in the cytosol (Wells et al., 2001; Hart, 2004) and is thought to be involved in many aspects of protein function, including protein–protein interaction and cellular localization (Guinez et al., 2005; Kudlow, 2006; Zachara and Hart, 2006). Importantly, O-GlcNAcylation represents a reversible glycosylation that is catalyzed by OGTase and is thought to counteract phosphorylation on Ser/Thr residues (Lefebvre et al., 2003; Guinez et al., 2005; Yang et al., 2006).
The similarity in the patterns observed for O-GlcNAcylated and Ser/Thr phosphorylated phloem proteins would be consistent with such an O-GlcNAcylation counteraction hypothesis. Our MS/MS and mutation analysis of Cm-PP16-1 established that the Cm-PP16-1 Tyr-63 and Ser-66 residues are phosphorylated in vivo (Figure 9). In addition, the monoclonal antibody against O-GlcNAc on Ser/Thr residues recognized mCmPP16-1 S12A, mCmPP16-1 S41A, mCmPP16-1 S108A, mCmPP16-1 S133A, and native Cm-PP16-1. By contrast, mCmPP16-1 S66A and mCmPP16-1 S66D were recognized only very weakly, and mCmPP16-1 S-all-A was completely nonreactive (Figure 9). These results are consistent with Ser-66 serving as a major O-GlcNAcylation site on Cm-PP16-1. It remains to be determined whether the weak O-GlcNAc signals detected with mCmPP16-1 S66A and mCmPP16-1 S66D reflect minor glycosylation on other Cm-PP16-1 residues. In any event, our findings are consistent with O-GlcNAcylation of Ser-66 on Cm-PP16-1 being critical for effective interaction with Nt-NCAPP1. This requirement for complex formation is consistent with the ineffective nature of the interaction observed between Nt-NCAPP1 and the phosphorylation mimic mutant mCmPP16-1 S66D (Figures 9G and 9K).
In Arabidopsis, two OGTase genes, SPYNDLY (SPY) and SEC, have been identified (Hartweck et al., 2002). Mutant analysis revealed that the sec mutant displays only minor developmental abnormalities, although the double mutant of spy and sec is embryonic lethal. Based on our current findings, it will be interesting to investigate the role of SEC and SPY in O-GlcNAcylation of Arabidopsis phloem NCAPs.
Equivalence in Binding Properties between Phloem-Purified and in Planta–Expressed Recombinant Proteins
The posttranslationally modified forms of both Cm-PP16-1 and Nt-NCAPP1 effectively interacted with each other, and dephosphorylated or deglycosylated forms of these proteins displayed dramatically reduced interactions (Figures 2 and 5). Moreover, the interaction patterns of in planta–expressed recombinant Cm-PP16-1 and Nt-NCAPP1ΔN were similar to those of native Cm-PP16-1 and Nt-NCAPP1 (Figures 3 and 6). However, Cm-PP16-1 and Nt-NCAPP1ΔN expressed in and purified from E. coli did not show significant interaction with each other. These assays suggest that E. coli does not have the appropriate enzymes to posttranslationally modify either Cm-PP16-1 or Nt-NCAPP1ΔN. Importantly, these experiments demonstrate that our in planta protein expression and purification system has direct utility for the biochemical and molecular analysis of phloem proteins. This approach will greatly facilitate the analysis of the less-abundant phloem NCAPs.
We originally isolated Nt-NCAPP1 by affinity chromatography using E. coli–expressed GST-Cm-PP16-1 as bait (Lee et al., 2003). In this study, we found that in BY-2 cell PECP overlay assays, E. coli–expressed GST-Cm-PP16-1 interacted with only Nt-NCAPP1 (Figure 7). Consistent with this finding, only the native Cm-PP16-1 was able to interact with and immunoprecipitate additional proteins contained within the BY-2 cell PECP preparation. Although dephosphorylated native Cm-PP16-1 still immunoprecipitated Nt-NCAPP1, the binding intensity of this dephosphorylated native Cm-PP16 was much weaker than that of native Cm-PP16-1 (Figure 8). Taken together, these results support the hypothesis that posttranslational modification of Cm-PP16-1 is essential for the formation of a stable Cm-PP16-1–Nt-NCAPP1 complex. These posttranslational modifications likely establish specific structures on these proteins that are optimal for their effective interaction (Cao et al., 2006).
Model for Cm-PP16-1 Interaction with NCAPP1 during Cell-to-Cell Movement
In our earlier characterization of the role of Nt-NCAPP1 in cell-to-cell trafficking of NCAPs (Lee et al., 2003), we demonstrated that we could block the trafficking of specific NCAPs, such as Cm-PP16-1 and the TMV MP, by either expression of a dominant-negative mutant form of Nt-NCAPP1 in transgenic plants or coinjection of this mutant along with the fluorescently labeled NCAP. These results were consistent with Cm-PP16-1 and the TMV MP interacting with NCAPP1 to gain access to a putative plasmodesmal docking site. Based on our protein–protein interaction assays, it now appears that Cm-PP16-1 must be phosphorylated on Tyr-63 and glycosylated on Ser-66 to form a stable NCAPP1-Cm-PP16-1 complex. Microinjection of the various Cm-PP16-1 mutants provides further support for this model, as the mCmPP16-1 Y63A, S66A, and S66D proteins were dysfunctional in their capacity to move cell to cell (Table 1). In this regard, it was important that mCmPP16-1 S12A moved through plasmodesmata with the same efficacy as wild-type protein. This finding adds support to the conclusion that the block to trafficking observed with the Y63A, S66A, and S66D mutants reflects a specific dysfunction associated with the change in these critically important residues.
Significance of Reversible Phosphorylation/Glycosylation for Intercellular Movement
In this study, we have demonstrated that a significant number of pumpkin phloem proteins are phosphorylated and/or glycosylated. If we can extrapolate from our results obtained with the phloem NCAP, Cm-PP16-1, it may be that many of these NCAPs similarly require a phosphorylation and glycosylation recognition motif for interaction with NCAPP1 to gain entry into or exit out of the phloem translocation stream. If this were the case, such a recognition motif may serve to regulate the trafficking of NCAPs through the companion cell–sieve element plasmodesmata.
Our finding that phloem-purified Cm-PP16-1 is phosphorylated on Ser-66 (Figure 9A) suggests the presence of competition at this residue between phosphorylation and glycosylation. Given that the Cm-PP16-1 S66A/S66D was incapable of mediating its own cell-to-cell transport, deglycosylation of Ser-66 after cell-to-cell transport, followed by phosphorylation of this same residue- could serve to restrict Cm-PP16-1 to the translocation stream.
In the case of protein localization to the nucleus, the NLS function is often regulated by phosphorylation through masking or revealing the NLS (Jans et al., 2000; Poon and Jans, 2005). It remains to be established whether our proposed NCAP phosphorylation/glycosylation recognition motif functions as a signal for intercellular trafficking, per se, or acts to modulate the activity of yet unidentified intercellular trafficking signals. Our Cm-PP16-1 deletion analysis identified an internal 36–amino acid region that is necessary and sufficient for cell-to-cell movement of GST, a normally cell-autonomous protein. Posttranslational modification of this peptide appears to regulate its capacity to mediate intercellular trafficking because substitution of the Ser-66 residue, a critical residue for Cm-PP16-1 glycosylation, blocked the cell-to-cell movement of these GST-Cm-PP16-1 fusion proteins. This result is important, as it demonstrates that the observed intercellular movement of the GST-Cm-PP16-1Δ1/Δ2/Δ3/Δ4 protein was not caused by passive diffusion of these fusion proteins. Finally, it is interesting to note that the KN1 homeodomain contains five Ser residues. Mass spectrometry of this homeodomain, combined with mutational analysis of these residues, will provide an important test for the involvement of a phosphorylation/glycosylation recognition motif in KN1 targeting to and trafficking through plasmodesmata.
METHODS
Plant Materials
Cucurbita maxima cv Big Max (pumpkin) plants were grown as described previously (Yoo et al., 2004). Nutrients were supplied daily as described (http://greenhouse.ucdavis.edu/materials/nutrients_soil.htm). Pumpkin phloem sap was collected from well-watered pumpkin plants as previously described (Yoo et al., 2004). Maintenance of BY-2 tobacco (Nicotiana tabacum) suspension cultured cells was performed as described previously (Lee et al., 2003). Nicotiana benthamiana plants were grown in a controlled environment chamber (Conviron; model PGR15) under the following conditions: 250 μmol m−2 s−1 PAR, 60% relative humidity, 30°C/20°C day/night temperatures, and a 16-h photoperiod. One-month-old N. benthamiana plants were used for agroinfiltration: after infiltration, plants were kept for 5 d under constant low-light conditions (10 μmol m−2 s−1 PAR) at 23°C.
Protein Expression Vectors
The Cm-PP16-1 ORF was PCR amplified with a primer set of Cm-16-1-FE and Cm-16-1-RSSal (no STOP codon) or Cm-16-1-RSal (with STOP codon) and subcloned into pCR-Blunt II TOPO vector (Invitrogen). All primer sequences used in PCR amplifications are listed in Supplemental Table 1 online. The Nt-NCAPP1 ORF, without the transmembrane region, was also PCR amplified with the primer set of NCAPP1ΔN-FE and NCAPP1-RSSal (no STOP codon) or NCAPP1-RSal (with STOP codon) and subcloned into pCR-Blunt II TOPO vector (Invitrogen).
For purification of recombinant proteins from N. benthamiana, a fusion protein cassette vector, pGSH, was developed in which a C-terminal tag of GFP-StrepII-8xHis (GSH) was engineered and the vector placed under the control of the 35S promoter of Caulifower mosaic virus. To this end, the enhanced GFP (EGFP) coding region of pdGN (Lee et al., 2005) was PCR amplified with a primer set of 35S-Dl and EGFP-rXba-S, digested with BamHI and XbaI, and then subcloned into the BamHI-XbaI site of pdGN to remove the stop codon. Double-stranded oligonucleotides encoding for a short DNA fragment corresponding to the StrepII-8xHis tag was made by annealing two oligonucleotides, Strep-U and Strep-L, and was inserted into the XbaI site of the plasmid to make pGSH. EcoRI-SalI fragments from a pCR-Blunt II TOPO vector containing Cm-PP16-1 or Nt-NCAPP1 were subcloned into the corresponding site of pGSH. NotI fragments of the pGSH derivatives were inserted into the corresponding site of pMLBART (Gleave, 1992), a binary vector in Agrobacterium tumefaciens.
To engineer Cm-PP16-1 mutants, the GeneTailor site-directed mutagenesis system (Invitrogen) was used according to the manufacturer's instructions. The pCR-Blunt II TOPO vector containing Cm-PP16-1 was used as a template and the following primers used for the mutagenesis: mCmPP16-1 S12A (Cm-PP16-S 12A-U and Cm-PP16-S 12A-L); mCmPP16-1 S41A (Cm-PP16-S41A-U and Cm-PP16-S41A-L); mCmPP16-1 Y63A (Cm-PP16-Y63A-U and Cm-PP16-Y63A-L); mCmPP16-1 S66A (Cm-PP16-S66A-U and Cm-PP16-S66A-L); mCmPP16-1 S66D (Cm-PP16-S66D-U and Cm-PP16-S66A-L); mCmPP16-1 S108A (Cm-PP16-S 108A-U and Cm-PP16-S 108A-L); mCmPP16-1 S133A (Cm-PP16-1-S133A-U and Cm-PP16-S133A-L).
To engineer Cm-PP16-1 deletion mutants, PCR cloning was performed with the pCR-Blunt II TOPO vector, containing Cm-PP16-1 as a template, and the following primers: Cm-PP16-1Δ1 (Cm-PP16-P25-F and Cm-PP16-K105-R); Cm-PP16-1Δ2 (Cm-PP16-P25-F and Cm-PP16-G83-R); Cm-PP16-1Δ3 (Cm-PP16-G48-F and Cm-PP16-K105-R); Cm-PP16-1Δ4 (Cm-PP16-G48-F and Cm-PP16-G83-R). The S66A mutant forms were created using the pCR-Blunt II TOPO vector containing Cm-PP16-1 S66A as a template.
pET28a (Novagen) was used for His-tagged protein expression in Escherichia coli. EcoRI-SalI fragments from pCR-Blunt II TOPO vector containing Cm-PP16-1 or Nt-NCAPP1 were recloned into the EcoRI-XhoI sites of pET28a. pGEX6P-1 (GE Healthcare) was used for GST-tagged protein expression in E. coli. EcoRI-SalI fragments from pCRBlunt II TOPO vector containing Cm-PP16-1 or Nt-NCAPP1 were recloned into the EcoRI-XhoI sites of pGEX6P-1.
In all cases, PCR amplification was performed with KOD Hot Start DNA polymerase (Novagen) according to the manufacturer's instructions. All DNA sequences were confirmed by sequencing.
PECP Preparation
BY-2 cell PECP preparations were performed essentially as described previously (Lee et al., 2003). Briefly, BY-2 cells at 7 d after transfer were harvested by filtering a 5-liter culture through Miracloth (EMD Biosciences). Harvested BY-2 cells were first incubated in a gently stirred 75 mM calcium chloride solution, for 1 h at 20°C, and then homogenized with a Bead-Beater using 2.5-mm beads at 4°C in buffer H (40 mM HEPES, pH 6.8, 20 mM KCl, 5 mM KH2PO4, 1 mM EDTA, and 10% glycerol) containing protease inhibitor (10 μg/mL each of leupeptin and aprotinin and 10 mM phenylmethylsulfonyl fluoride). A cell wall pellet was obtained by centrifugation (5 min at 300g) and then washed with buffer H containing 1% CHAPS. PECPs were extracted from this cell wall pellet by overnight incubation at 4°C in the calcium chloride buffer.
Agroinfiltration
To express recombinant proteins in planta, agroinfiltration was performed as previously described (Voinnet et al., 2003). Briefly, Agrobacterium strain C58C1 containing pMLBART was grown overnight, at 30°C, in 2 mL of Luria-Bertani medium containing 200 mg/L of spectinomycin. The suspension was subcultured into 50 mL of Luria-Bertani medium containing 200 mg/L of spectinomycin at 30°C. After an OD600 of 0.5 to 0.8 was attained, the suspension was centrifuged at 5500g for 10 min at 20°C. After removal of supernatant, the bacterial pellet was resuspended in 50 mL of agroinfiltration buffer (50 mM MES, pH 5.6, 10 mM MgC12, and 150 μM acetosyringone) and incubated for 3 h at 20°C. An Agrobacterium culture carrying the p19 silencing suppressor (Voinnet et al., 2003) was also prepared as described above. An aliquot (50 mL) of Agrobacterium suspension containing pMLBART and p19 were mixed together and infiltrated into N. benthamiana leaves. Twenty plants were used per preparative batch, and 5 d after inoculation, the infiltrated leaves were harvested and stored at −80°C until used for protein purification.
Purification of Recombinant Protein
Purification of GSH fusion proteins from agroinfiltrated N. benthamiana leaves was performed as follows. Infiltrated tissue (30 g) was frozen in liquid nitrogen and then extensively ground with a mortar and pestle for 10 min, after which the homogenate was suspended in 30 mL of His extraction buffer (100 mM Tris, pH 8.0, 500 mM NaCl, 0.5% Triton X-100, 10 mM imidazole, 0.5 mM DTT, 10% [v/v] glycerol, 10 mg/L aprotinin, 10 mg/L leupeptin, and 1 mM PMSF). After brief sonication and 1 h incubation on ice, the suspension was centrifuged at 27,700g at 4°C for 30 min. The supernatant was passed through a 0.45-μm syringe filter and then processed for the first round of purification using the HisTrap FF Crude kit (GE Healthcare) according to the manufacturer's instructions. The resultant protein solution was further purified with a StrepTactin Macroprep Cartridge (Novagen) according to the manufacturer's instructions. As a minor modification, 500 μL of the protein solution was added into 4.5 mL of W buffer (100 mM Tris, pH 8.0, 150 mM NaCl, and 1 mM EDTA) containing 100 mg/L of avidine to block the competitive binding of endogeneous biotinylated proteins present in the plant extract. After 30 min incubation on ice, this preparation was used for the second round of purification using a StrepTactin Macroprep cartridge.
For expression of His-tagged or GST-tagged proteins, E. coli strain Rosseta2 (DE3) pLysS (Novagen) was transformed with pET28a or pGEX6P-1 containing the ORF of Nt-NCAPP1, Cm-PP16-1, or the series of Cm-PP16-1 mutants. Protein expression was induced with 1 mM isopropylthio-β-galactoside for 16 h at 22°C. Protein purification was performed with the HisTrap FF Crude (GE Healthcare) or Glutathione Sepharose 4B (GE Healthcare) kit according to the manufacturer's instructions.
Chromatographic Separation of Pumpkin Phloem Proteins and BY-2 Cell PECPs
Anion-exchange chromatography of pumpkin phloem sap proteins was performed as previously described (Yoo et al., 2004). Generally, 30 mL of pumpkin phloem sap (0.1 to 0.2 mg protein/mL) was dialyzed against buffer A (50 mM Tris, pH 7.5, 1 mM EDTA, and 30 mM 2-mercaptoethanol) and then loaded onto a buffer A–equilibrated HiTrap Q column (GE Healthcare) through an FPLC system (GE Healthcare). After washing the column with buffer A, proteins were eluted with a linear gradient of 0 to 500 mM NaCl in buffer A containing 1 M NaCl.
Cation-exchange chromatography of the BY-2 cell PECP preparation was performed as follows. The Centricon Plus-20 system (Millipore) was used to concentrate an initial volume of BY-2 cell PECP preparation to 0.5 to 1 mg protein/mL, followed by dialysis against buffer C (40 mM HEPES, pH 6.8, 2 mM EDTA, and 10% glycerol). Five-milliliter aliquots of the concentrated BY-2 cell PECP preparation were centrifuged at 17,000g for 20 min before being loaded onto a buffer C–equilibrated HiTrap SP column (GE Healthcare) connected to an FPLC system. Proteins were eluted with a linear gradient of 0 to 500 mM NaCl in buffer C containing 1 M NaCl.
Fractionated proteins were separated on SDS-PAGE gels and stained with SYPRO Ruby reagent (Invitrogen). Quantification of proteins was performed by the Bradford method (Bio-Rad).
Protein Gel Blot Analysis
Protein gel blot assays were performed as follows. Briefly, nitrocellulose membrane blots were first blocked for 1 h with 5% nonfat milk made in modified 1× TBS (50 mM Tris-HCl, pH 7.5, and 500 mM NaCl). For protein gel blot analysis with phosphoserine, phosphothreonine, phosphotyrosine (EMD Biosciences), and O-GlcNAc (Pierce) monoclonal antibodies, 1% BSA in 1× TBS was used as blocking agent. Nitrocellulose membrane blots were incubated with the corresponding primary antibodies (antiphosphoserine, antiphosphotheronine, antiphosphotyrosine, anti-O-GlcNAc, anti-Cm-PP16-1, and anti-Nt-NCAPP1 all used at 1:1000 dilution; anti-GFP [Clontech; 1:4000 dilution]), washed three times with 1× TTBS (1× TBS containing 0.5% Tween 20) for 5 min each, followed by 1 h incubation with the secondary antibody (horseradish peroxidase–conjugated anti-rabbit [1:20,000 dilution] or anti-mouse antibodies [1:80,000 dilution]; Sigma-Aldrich). Blots were then washed three times with 1× TTBS for 5 min each and subjected to immunodetection with chemiluminescence reagent (Perkin-Elmer Life Sciences) and film (Kodak Biomax MS; Eastman Kodak).
Protein Overlay Blot Assays
Protein overlay blot assays were performed essentially as described previously (Lee et al., 2003). The FPLC-fractionated pumpkin phloem sap proteins or BY-2 cell PECPs (15 μL) were separated on 13% SDS-PAGE gels and then transferred onto nitrocellulose membranes. Protein blots were overlaid with the probes diluted in BSA buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, and 2 mg/mL BSA) for 40 min at 20°C. Blotted nitrocellulose membranes were washed with 1× TTBS three times for 5 min each and then subjected to protein gel blot analysis procedures, as described above, with appropriate primary antibodies and anti-rabbit horseradish peroxidase–conjugated secondary antibody (Sigma-Aldrich).
LC-MS/MS Analysis of Phloem-Purified Cm-PP16
Gel filtration was used for size separation of pumpkin phloem sap proteins as previously described (Aoki et al., 2005). Cm-PP16-1 and Cm-PP16-2 were further separated on 12% SDS-PAGE gels and then subjected to in-gel digest with trypsin. Extracted peptides were analyzed by LC-MS/MS on a capillary LC system coupled directly to a Thermo Finnigan LTQ ion trap mass spectrometer. Sequence coverage was 84 and 93% for Cm-PP16-1 and Cm-PP16-2, respectively.
Microinjection Experiments
Four-week-old N. benthamiana plants were used for microinjection experiments, and these were performed as described previously (Rojas et al., 1997). Cm-PP16-1 was either labeled directly with OG (Invitrogen) or its movement reported by coinjection with fluorescein isothiocyanate–labeled 10 kD dextran (F-dextran; Sigma-Aldrich). All protein preparations were used at a concentration of 800 μg/mL. Fluorescence analysis of OG-labeled proteins and F-dextran were performed on a confocal laser scanning microscope (model DM RXE 6 TCS-SP2 AOBS; Leica) using an Ar/ArKr laser (488-nm excitation and 525-nm emission). Autofluorescence from chlorophyll was used to establish cellular detail and was obtained using a GHeNe laser (543-nm excitation and 610-nm emission). Images were stacked using Leica LCS 1537 software and then processed with Adobe Photoshop CS2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF307094 (Nt-NCAPP1) and AF079170 (Cm-PP16-1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Posttranslational Modifications on Cm-PP16-1 Are Required for Cell-to-Cell Trafficking through Mesophyll Plasmodesmata.
Supplemental Table 1. Oligonucleotide Primers Used for PCR.
Supplementary Material
Acknowledgments
We thank Young-Jin Lee (UC Davis Genome Center) for expert assistance in phosphoproteomics analysis of Cm-PP16-1. This work was supported by National Science Foundation Grant IOS-0444725. B.X.-C. was supported, in part, by a UCMEXUS-CONACYT Sabbatical Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: William J. Lucas (wjlucas@ucdavis.edu).
Online version contains Web-only data.
References
- Aoki, K., Kragler, F., Xoconostle-Cázares, B., and Lucas, W.J. (2002). A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc. Natl. Acad. Sci. USA 99 16342–16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K., Suzui, N., Fujimaki, S., Dohmae, N., Yonekura-Sakakibara, K., Fujiwara, T., Hayashi, H., Yamaya, T., and Sakakibara, H. (2005). Destination-selective long-distance movement of phloem proteins. Plant Cell 17 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran, S., Xiang, Y., Schobert, C., Thompson, G.A., and Lucas, W.J. (1997). Phloem sap proteins from Cucurbita maxima and Ricinus communis have the capacity to traffic cell to cell through plasmodesmata. Proc. Natl. Acad. Sci. USA 94 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A.K., Chatterjee, M., Yu, Y., Suh, S.G., Miller, W.A., and Hannapel, D.J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18 3443–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Q., Kim, J.H., and Richter, J.D. (2006). CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 13 1128–1134. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Jensen, P.E., and Schaad, M.C. (1998). Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 14 393–400. [DOI] [PubMed] [Google Scholar]
- Chen, D., Juarez, S., Hartweck, L., Alamillo, J.A., Simon-Mateo, C., Perez, J.J., Fernandez-Fernandez, M.R., Olszewski, N.E., and Garcia, J.A. (2005). Identification of secret agent as the O-GlcNAc transferase that participates in Plum Pox virus infection. J. Virol. 79 9381–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, K.M., and Zambryski, P.C. (2000). Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr. Biol. 10 1032–1040. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Diaz-Trivino, S., Cisneros, N., and Gutierrez, C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., Itaya, A., and Qi, Y.J. (2003). Symplasmic protein and RNA traffic: Regulatory points and regulatory factors. Curr. Opin. Plant Biol. 6 596–602. [DOI] [PubMed] [Google Scholar]
- Gallagher, K.L., and Benfey, P.N. (2005). Not just another hole in the wall: Understanding intercellular protein trafficking. Genes Dev. 19 189–195. [DOI] [PubMed] [Google Scholar]
- Ghoshroy, S., Lartey, R., Sheng, J.S., and Citovsky, V. (1997). Transport of proteins and nucleic acids through plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 25–48. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organizational structure conductive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Golecki, B., Schulz, A., Carstens-Behrens, U., and Kollmann, R. (1998). Evidence of graft transmission of structural phloem proteins or their precursors in heterografts of Cucubitaceae. Planta 206 630–640. [Google Scholar]
- Guinez, C., Morelle, W., Michalski, J.C., and Lefebvre, T. (2005). O-GlcNAc glycosylation: A signal for the nuclear transport of cytosolic proteins? Int. J. Biochem. Cell Biol. 37 765–774. [DOI] [PubMed] [Google Scholar]
- Hart, G.W. (2004). Cytoplasmic glycosylation. Biochim. Biophys. Acta 1673: 1. [DOI] [PubMed]
- Hartweck, L.M., Scott, C.L., and Olszewski, N.E. (2002). Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Kragler, F., and Lucas, W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 S303–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42 49–68. [DOI] [PubMed] [Google Scholar]
- Heese-Peck, A., and Raikhel, N.V. (1998). A glycoprotein modified with terminal acetylglucosamine and localized at the nuclear rim shows sequence similarity to aldose-1-epimerases. Plant Cell 10 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein, M., and Epel, B.L. (2004). Macromolecular transport and signaling through plasmodesmata. Int. Rev. Cytol. 235 93–164. [DOI] [PubMed] [Google Scholar]
- Jackson, D., and Hake, S. (1997). Morphogenesis on the move: Cell-to-cell trafficking of plant regulatory proteins. Curr. Opin. Plant Biol. 7 495–500. [DOI] [PubMed] [Google Scholar]
- Jans, D.A., Xiao, C.Y., and Lam, M.H. (2000). Nuclear targeting signal recognition: A key control point in nuclear transport? Bioessays 22 532–544. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A., Atkinson, R.G., Forster, R.L.S., and Lucas, W.J. (1998). An RNA-based information superhighway in plants. Science 279 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., Rim, Y., Wang, L., and Jackson, D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19 788–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289. [DOI] [PubMed] [Google Scholar]
- Knippschild, U., Gocht, A., Wolff, S., Huber, N., Lohler, J., and Stoter, M. (2005). The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell. Signal. 17 675–689. [DOI] [PubMed] [Google Scholar]
- Kudlow, J.E. (2006). Post-translational modification by O-GlcNAc: Another way to change protein function. J. Cell. Biochem. 98 1062–1075. [DOI] [PubMed] [Google Scholar]
- Kurata, T., Okada, K., and Wada, T. (2005). Intercellular movement of transcription factors. Curr. Opin. Plant Biol. 8 600–605. [DOI] [PubMed] [Google Scholar]
- Lee, J.-Y., and Lucas, W.J. (2001). Phosphorylation of viral movement proteins: Regulation of cell-to-cell trafficking. Trends Microbiol. 9 5–8. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Taoka, K., Yoo, B.C., Ben-Nissan, G., Kim, D.J., and Lucas, W.J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Yoo, B.C., and Lucas, W.J. (2000). Parallels between nuclear-pore and plasmodesmal trafficking of information molecules. Planta 210 177–187. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Yoo, B.C., Rojas, M.R., Gomez-Ospina, N., Staehelin, L.A., and Lucas, W.J. (2003). Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299 392–396. [DOI] [PubMed] [Google Scholar]
- Lefebvre, T., Ferreira, S., Dupont-Wallois, L., Bussiere, T., Dupire, M.J., Delacourte, A., Michalski, J.C., and Caillet-Boudin, M.L. (2003). Evidence of a balance between phosphorylation and O-GlcNAc glycosylation of Tau proteins – A role in nuclear localization. Biochim. Biophys. Acta 1619 167–176. [DOI] [PubMed] [Google Scholar]
- Lindermayr, C., Saalbach, G., Bahnweg, G., and Durner, J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281 4285–4291. [DOI] [PubMed] [Google Scholar]
- Lough, T.J., and Lucas, W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57 203–232. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. (1995). Plasmodesmata: Intercellular channels for macromolecular transport in plants. Curr. Opin. Plant Biol. 7 673–680. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 and its mRNA through plant plasmodesmata. Science 270 1980–1983. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., and Lee, J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5 712–726. [DOI] [PubMed] [Google Scholar]
- Macara, I.G. (2001). Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, A.S., and Weis, K. (2006). Nuclear transport is becoming crystal clear. Chromosoma 115 98–109. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J. (2004). Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci. 9 33–41. [DOI] [PubMed] [Google Scholar]
- Pemberton, L.F., Blobel, G., and Rosenblum, J.S. (1998). Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 10 392–399. [DOI] [PubMed] [Google Scholar]
- Poon, I.K.H., and Jans, D.A. (2005). Regulation of nuclear transport: Central role in development and transformation? Traffic 6 173–186. [DOI] [PubMed] [Google Scholar]
- Roberts, I.M., Wang, D., Findlay, K., and Maule, A.J. (1998). Ultrastructural and temporal observations of the potyvirus cylindrical inclusions (CIS) show that the CI protein acts transiently in aiding virus movement. Virology 245 173–181. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R., Zerbini, F.M., Allison, R.F., Gilbertson, R.L., and Lucas, W.J. (1997). Capsid protein and helper component proteinase function as potyvirus cell-to-cell movement proteins. Virology 237 283–295. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cázares, B., and Kragler, F. (2004). The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 7 641–650. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cázares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126 4405–4419. [DOI] [PubMed] [Google Scholar]
- Rudrabhatla, P., Reddy, M.M., and Rajasekharan, R. (2006). Genome-wide analysis and experimentation of plant serine/threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 60 293–319. [DOI] [PubMed] [Google Scholar]
- Sasaki, N., Park, J.W., Maule, A.J., and Nelson, R.S. (2006). The cysteine-histidine-rich region of the movement protein of Cucumber mosaic virus contributes to plasmodesmal targeting, zinc binding and pathogenesis. Virology 349 396–408. [DOI] [PubMed] [Google Scholar]
- Schulenberg, B., Aggeler, R., Beechem, J.M., Capaldi, R.A., and Patton, W.F. (2003). Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J. Biol. Chem. 278 27251–27255. [DOI] [PubMed] [Google Scholar]
- Scott, C.L., Hartweck, L.M., de Jesus Perez, J., Chen, D., Garcia, J.A., and Olszewski, N.E. (2006). SECRET AGENT, an Arabidopsis thaliana O-GlcNAc transferase, modifies the Plum pox virus capsid protein. FEBS Lett. 580 5829–5835. [DOI] [PubMed] [Google Scholar]
- Searle, I.R., Men, A.E., Laniya, T.S., Buzas, D.M., Iturbe-Ormaetxe, I., Carroll, B.J., and Gresshoff, P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299 109–112. [DOI] [PubMed] [Google Scholar]
- Shapka, N., Stork, J., and Nagy, P.D. (2005). Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA binding site affects viral RNA replication. Virology 343 65–78. [DOI] [PubMed] [Google Scholar]
- Trutnyeva, K., Bachmaier, R., and Waigmann, E. (2005). Mimicking carboxy terminal phosphorylation differentially effects subcellular distribution and cell-to-cell movement of Tobacco mosaic virus movement protein. Virology 332 563–577. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Waigmann, E., Chen, M.H., Bachmaier, R., Ghoshroy, S., and Citovsky, V. (2000). Regulation of plasmodesmal transport by phosphorylation of Tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19 4875–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann, E., Ueki, S., Trutnyeva, K., and Citovsky, V. (2004). The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. CRC Crit. Rev. Plant Sci. 23 195–250. [Google Scholar]
- Wells, L., Vosseller, K., and Hart, G.W. (2001). Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science 291 2376–2378. [DOI] [PubMed] [Google Scholar]
- Wu, X.L., Weigel, D., and Wigge, P.A. (2002). Signaling in plants by intercellular RNA and protein movement. Genes Dev. 16 151–158. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, F.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98. [DOI] [PubMed] [Google Scholar]
- Yang, W.H., Kim, J.E., Nam, H.W., Ju, J.W., Kim, H.S., Kim, Y.S., and Cho, J.W. (2006). Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8 1074–1083. [DOI] [PubMed] [Google Scholar]
- Yoo, B.C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, B.C., Lee, J.Y., and Lucas, W.J. (2002). Analysis of the complexity of protein kinases within the phloem sieve tube system. Characterization of Cucurbita maxima calmodulin-like domain protein kinase 1. J. Biol. Chem. 277 15325–15332. [DOI] [PubMed] [Google Scholar]
- Zachara, N.E., and Hart, G.W. (2006). Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761 599–617. [DOI] [PubMed] [Google Scholar]
- Zambryski, P. (2004). Cell-to-cell transport of proteins and fluorescent tracers via plasmodesmata during plant development. J. Cell Biol. 164 165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski, P., and Crawford, K. (2000). Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 16 393–421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.