Abstract
Bleomycin, a radiomimetic drug currently used in human cancer therapy, is a well known carcinogen. Its toxicity is mostly attributed to its potentiality to induce DNA double strand breaks likely arising from the formation of two vicinal DNA strand breaks, initiated by C4-hydrogen abstraction on the 2-deoxyribose moiety. In this work we demonstrate that such a hydrogen abstraction reaction is able to induce the formation of a clustered DNA lesion, involving a 3′ strand break together with a modified sugar residue exhibiting a reactive α,β-unsaturated aldehyde that further reacts with a proximate cytosine base. The lesion thus produced was detected as a mixture of four isomers by HPLC coupled to tandem mass spectrometry subsequent to DNA extraction and enzymatic digestion. The modified nucleosides that constitute new types of cytosine adducts were identified as the likely two pairs of diastereomers of 6-(2-deoxy-β-d-erythro-pentofuranosyl)-2-hydroxy-3(3-hydroxy-2-oxopropyl)-2,6-dihydroimidazo[1,2-c]-pyrimidin-5(3H)-one as inferred from mass spectrometry and NMR analyses of the chemically synthesized nucleosides. We demonstrate that bleomycin, and to a minor extent ionizing radiation, are able to induce significant amounts of the cytosine damage in cellular DNA. In addition, the repair kinetic of the lesion in a human lymphocyte cell line is rather slow, with a half-life of 10 h. The 2′-deoxycytidine adducts thus characterized that represent the first example of complex DNA lesions isolated and identified in cellular DNA upon one radical hit are likely to play an important role in the toxicity of bleomycin.
Keywords: cluster lesions, DNA damage, DNA repair
DNA is continuously exposed to exogenous and endogenous mutagens, including reactive oxygen and nitrogen species, that could alter its integrity (1–3). Regarding the mechanistic aspects of the formation of DNA lesions (3, 4), much attention has focused on base damage. Recent observations suggest that sugar oxidative degradation pathways, which in most cases lead to strand breaks formation (5), may take an important part in the toxicity of ionizing radiation or treatment with so-called radiomimetic drugs. It is well documented that a few radiomimetic molecules are able to oxidize, more or less specifically, different positions of the 2-deoxyribose. These include antitumoral drugs of the bleomycin or enediyne families such as chalicheamicin or neocarzinostatin. Indeed, bleomycin specifically oxidizes the 4-position of 2-deoxyribose, whereas calicheamicin attacks preferentially the 4- and 5-positions, and neocarzinostatin is implicated in 1-, 4-, and 5-oxidation reactions (6). Moreover, cationic metal porphyrins (7) or other artificial nucleases such as bis(1,10-phenanthroline)copper(I) (8) are also able to oxidize the sugar moiety of DNA. Altogether, these data underline the importance of sugar alterations and also the diversity of the oxidation pathways.
Despite this diversity, a feature common to most of the different possible sugar oxidation reactions is the transient generation of reactive aldehydes. Oligonucleotide 3′-phosphoglycaldehydes are produced by C3′-oxidation, whereas base propenals are released from C4′-oxidation, and trans-1,4-dioxo-2-butene is generated subsequently to C5′-oxidation (9) of DNA strands. In addition, it is well established that aldehydic groups, because of their electrophilic character, can react with nucleophilic moieties of DNA bases with subsequent generation of adducts. Thus, malondialdehyde (MDA) has been shown to add to adenine, cytosine, and guanine, the reaction with the latter nucleobase being preponderant, leading to the generation of pyrimido[1,2-α]purin-10-(3H)-one-2-deoxyribose adduct (M1dG) (10). Base propenals, that are structurally analogous to the enol tautomer of MDA (β-hydroxyacrolein), are also able to react with DNA through a mechanism similar to that of MDA (11). It has recently been shown that addition of base propenals to DNA generates M1dG (12, 13). It could be also mentioned that the formation of an adduct between dGuo and phosphoglycolaldehyde has been reported (14). Dedon et al. (15, 16) have highlighted the formation of trans-1,4-dioxo-2-butene DNA adducts that could be generated during γ-irradiation of DNA (17).
The biological relevance of a DNA lesion is directly correlated to its mutagenicity and ability for being repaired. The so-called locally multiply damaged sites (LMDS) or clustered damage are DNA lesions defined as two or more elemental lesions formed within one or two helical turns of DNA (18). It has been shown, by using an indirect approach, that LMDS, including among them double-strand breaks (DSBs) (19), are produced by ionizing radiation in both isolated and cellular DNA, even at low doses of radiation (0.1–1 Gy) (20). It is admitted that LMDS, including also non-DSB lesions represent a signature of the effects of ionizing radiation on DNA (21) as a result of the occurrence of multiple ionization events in double-stranded DNA along the particle track. Repair of LMDS has been shown to be less efficient compared with that of single lesions (22), and it has also been suggested that LMDS could be converted into DSB during the repair process (21, 23). These findings highlight the difficulty for a cell to cope with complex DNA damage, suggesting that LMDS may play an important role in the harmful effects of ionizing radiation.
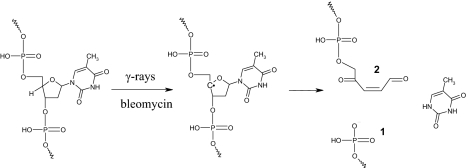
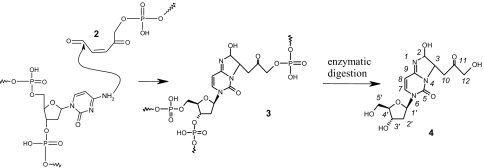
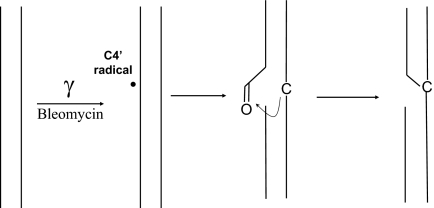
In this work, we report the characterization of a DNA lesion that was initially detected in γ-irradiated isolated DNA as a 2′-deoxycytidine modification exhibiting a 341-Da molecular mass (24). Emphasis was placed on the identification of the DNA lesion and delineation of the mechanism of its formation (Schemes 1 and 2). We demonstrate that C4′-hydrogen abstraction in double-stranded DNA leads to the concomitant formation of a strand break (1) and a reactive aldehyde 2 (Scheme 1) that is able to react with a vicinal cytosine base to generate the adduct 3 (Scheme 2). The identified lesion 4 that is detected by HPLC coupled to tandem mass spectrometry once DNA is extracted and digested could be considered as a clustered DNA lesion, which formation, moreover, requires only one radical hit. Interestingly the lesion that is detected in control cells as a likely contribution of endogenous oxidative processes was found to be significantly generated in the DNA of cells treated with bleomycin, a radiomimetic drug commonly used in human cancer therapy.
Scheme 1.
Mechanism of formation of the aldehydic intermediate 2 mediated by C4′-hydrogen abstraction.
Scheme 2.
Mechanism of formation of the clustered DNA lesion. Reaction of reactive aldehyde 2 with a cytosine base gives rise to the complex DNA lesion 3. Enzymatic hydrolysis of 3 containing DNA produces 4, which formation is detected by HPLC-MS/MS.
Results
Characterization of the Lesion.
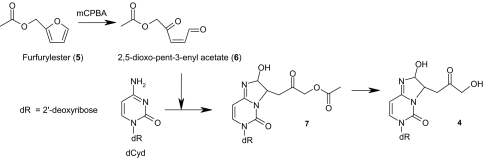
To determine the structure of 4, a chemical synthetic approach was designed to produce larger amounts of the lesion. The strategy we have adopted consisted in the synthesis of the acetylated derivative of the putative ketoaldehyde intermediate (Scheme 3, structure 6) formed by C-4′ hydrogen abstraction that, upon incubation with dCyd, would give rise, after deacetylation, to 4. For that purpose, we have adapted the method described for the synthesis of 4-oxopentenal from 2-methylfuran (25), starting with furfuryl acetate (5) to generate 2,5-dioxo-pent-3-enyl acetate (6) as described in Scheme 3 [see supporting information (SI) Text]. The reaction mixture was then incubated in the presence of dCyd before being analyzed by HPLC-MS/MS after treatment under alkaline conditions to remove the acetyl group. The synthesized products, which appear as four nucleosides, were found to exhibit the same HPLC retention times and mass spectroscopic features as the four isomers of the newly detected lesions 4 generated upon radiation-induced DNA decomposition in aerated aqueous solutions (Fig. 1) (24).
Scheme 3.
Chemical synthesis of 4 (see SI Text). Furfuryl ester (5) is treated with mCPBA to generate the reactive adehyde (6). Reaction of dCyd with 6 produces cytosine adduct 7, which upon deacetylation gives rise to the four isomers of 4 (dR = 2-deoxyribose).
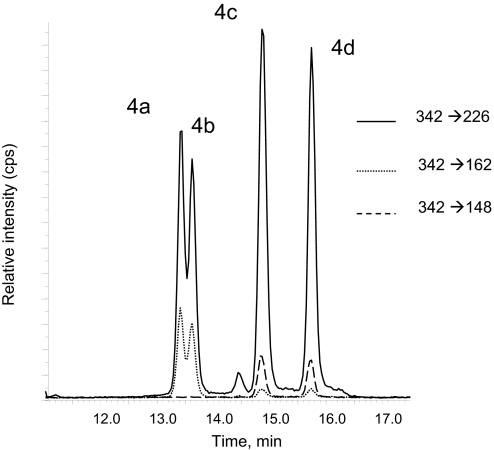
Fig. 1.
Typical chromatogram obtained for the HPLC-MS/MS detection of 4 in digested γ-irradiated isolated ctDNA. The isomers 4a and 4b are detected by using transitions 342 → 226 and 342 → 162, whereas isomers 4c and 4d are detected by using transitions 342 → 226 and 342 → 148.
In the 1H-NMR spectrum of the slowest eluting isomer 4d (SI Text and SI Fig. 5), we note, by comparison with dCyd, the presence of additional resonance signals. The 1H NMR features of isomer 4c are similar to those of 4d. The assignment of all nonexchangeable protons of the 2-deoxyribose moiety of 4d was unambiguously achieved by using 2D COSY experiments (SI Text and SI Fig. 6) starting from H-1′, which is known to resonate at the lowest field among the protons of the 2-deoxyribose moiety. The two aromatic protons of the pyrimidine ring moiety were easily attributed (protons 7 and 8, Scheme 2) because of the occurrence of their strong scalar correlation. Their chemical shifts, together with the strong absorption of 4 at 270 nm confirm that the pyrimidine ring is still aromatic. Starting from proton H-2, which resonates as a doublet because of its scalar coupling with H-3, signals from protons 3 and 10 were attributed by using the 2D COSY experiments. Interestingly, the two protons H-12 and H-12′ were found to be nonequivalent, as observed for H-5′ and H-5″ protons.
The circular dichroism analysis indicates that 4a and 4b on one hand and 4c and 4d on the other hand exhibit an almost opposite Cotton effect at ≈200 nm and, together with UV absorption spectra (SI Text and SI Figs. 7 and 8) data, strongly suggest that 4 exists as likely four diastereomers. Such a relationship may explain the minor differences observed in the mass spectrometric features of the 4 nucleosides, because the two rapidly eluted nucleosides 4a and 4b exhibit similar collision-induced dissociation mass spectra, whose pattern is partly different from that of the two slowest eluted compounds 4c and 4d (Fig. 1). However, we could not completely rule out the possibility that the detected dCyd adducts 4 could be regioisomers. Neither detectable decomposition (in particular, dehydration) nor significant interconversion between the different isomers of 4 was observed upon incubation at pH 4, 7, or 10 for 12 h at 37°C.
Mechanism of Formation of 4.
Insights into the mechanism of formation of 4 were gained from studies involving γ-radiolysis of aqueous aerated solutions of either 2′-deoxycytidine 3′,5′-diphosphate (pdCp), 2′-deoxycytidine 5′-monosphosphate (5′-dCMP), or 2′-deoxycytidine 3′-monosphosphate (3′-dCMP). Levels of 4 were then determined by HPLC-MS/MS (24) subsequent to enzymatic digestion with the combined use of nuclease P1 and alkaline phosphatase. The nucleosides 4 were found to be generated only upon γ-irradiation of aerated aqueous solutions of pdCp or 3′-dCMP (Table 1) indicating that the 3′ hydroxyl group of the 2-deoxyribose must be phosphorylated to generate 4. Then, aerated aqueous solutions of 3′-dTMP were γ-irradiated and subsequently incubated with dCyd. Such a treatment was found to generate 4 even in the absence of enzymatic digestion (Table 1). This indicates that the adducts 4 are formed by the reaction of dCyd with a sugar derivative generated during 3′-dTMP irradiation. This strongly suggests that the sugar derivative, which is stable at least for a few minutes at room temperature, is not a radical but rather the ketoaldehyde 2 produced by the spontaneously β-elimination of the 3′-phosphate group.
Table 1.
Formation (+) or not (−) of 4 upon γ-irradiation of different nucleotide derivatives
| Sample | Digestion | Formation of 4 |
|---|---|---|
| γ-5′-CMP | P1 + Palk | − |
| γ-3′-dCMP | P1 + Palk | + |
| γ-pdCp | P1 + Palk | + |
| γ-3′-TMP + dCyd | P1 + Palk | + |
| γ-3′-TMP + dCyd | None | + |
Formation of 4 was detected by HPLC-MS/MS after either digestion of the samples with both nuclease P1 and phosphatase alkaline (P1 + Palk) or in the absence of hydrolysis (None).
HPLC-MS/MS Measurement of 4.
A quantitative tandem mass spectrometry method involving electrospray ionization in the positive mode combined with the sensitive multiple reaction monitoring detection technique was previously developed for the detection of 4 (see SI Text and SI Fig. 9) based on their characteristic fragmentation (24). Three specific transitions (Fig. 1) are used to detect 4. The sensitivity of the latter method is high because the threshold limit of detection of 4, which is close to 1 fmol injected is ≈15 times lower than that of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo) determined to be ≈15 fmol injected. The transition 342 → 226 was found to be common to the four isomers of 4. Interestingly, the two rapidly eluted nucleosides 4a and 4b could be selectively detected by using the 342 → 162 transition, whereas transition 342 → 148 was used to detect 4c and 4d.
Formation of 4 in Isolated DNA and Inhibition by Methoxyamine.
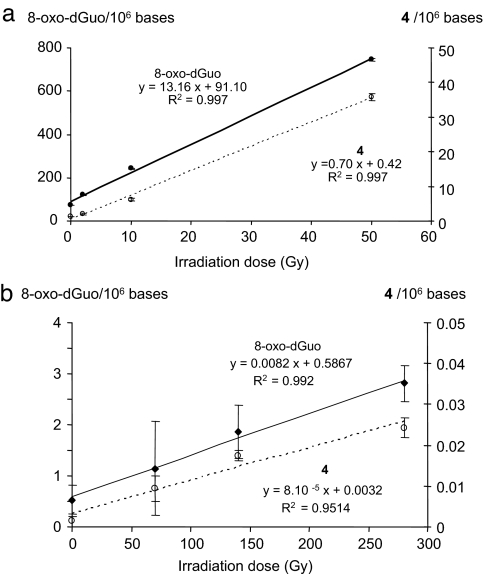
Aerated aqueous solutions of calf thymus DNA were exposed to increasing doses of γ-rays, and the levels of both 8-oxodGuo and 4 were determined by HPLC-MS/MS measurements using an external calibration. As shown in Fig. 2a and as already reported, 8-oxodGuo is generated in a dose-dependent manner (26, 27). Interestingly, a linear increase in the level of 4 was also observed. Comparison of the yields of formation of the two latter lesions revealed that exposure of isolated ctDNA in aerated aqueous solutions to γ-radiation generates ≈20 times more 8-oxodGuo than 4. Experiments were performed by using methoxyamine (MX) to confirm the involvement of a reactive aldehyde in the mechanism of formation of 4. Indeed, MX forms covalent adducts with aldehydes, and such a reaction is commonly used to quantify abasic sites in DNA (28). Levels of 4 generated upon γ-irradiation of aerated aqueous solutions of isolated ctDNA are significantly reduced in the presence of MX, and the decrease efficiency depends on the MX concentration (SI Text and SI Fig. 10). It is important to mention that the observed decrease in the measured levels of 4 is not due to the trapping of hydroxyl radicals by MX because formation of 8-oxodGuo is not decreased in the presence of MX. In addition, such a decrease could not be attributed to a direct reaction of MX with 4 because we have shown that MX is unable to react with purified 4.
Fig. 2.
Determination of the levels of 8-oxodGuo (left scale) and 4 (right scale) in either isolated ctDNA (a) or in the DNA extracted from a human lymphocyte cell line (b) exposed to increasing doses of ionizing radiation.
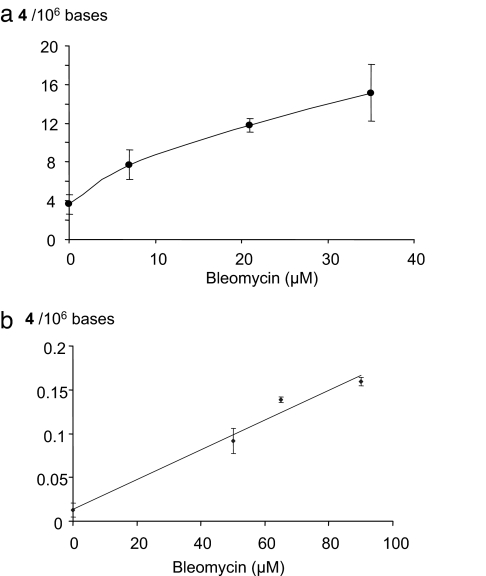
To confirm the origin of the sugar derivative 2 leading to the formation of the cytosine adducts 4, ctDNA was treated with bleomycin, an antitumoral molecule capable of oxidizing the C4-position of 2-deoxyribose in double-stranded DNA (6). Formation of 4 was detected after incubation of isolated ctDNA in the presence of increasing concentrations of bleomycin (Fig. 3a).
Fig. 3.
Formation of 4 in isolated ctDNA (a) and in the DNA extracted from a human lymphocyte cell line (b) upon treatment with increasing concentrations of bleomycin.
Formation of 4 in Cellular DNA.
Human monocytes were either exposed to ionizing radiation or incubated in the presence of bleomycin, and DNA was extracted by using a recently optimized protocol aimed at minimizing artefactual DNA oxidation that could occur during the workup (29). As previously described, 8-oxodGuo was detected in cellular DNA, and its level increases linearly with the applied dose of radiation (Fig. 2b). Interestingly, 4 was detected in untreated cells at a background level determined to be ≈1–3 lesions per 108 nucleosides, indicating a possible endogenous origin. The yield of radiation-induced formation of 4 in cellular DNA represents ≈1% of the level of 8-oxodGuo and, therefore, is almost similar to the yield of formation of double-strand breaks. Moreover, 4 was also detected (SI Text and SI Fig. 9) in the DNA of bleomycin-treated cells (Fig. 3b), whereas the level of 8-oxodGuo remained unchanged.
Rate of Repair of Cytosine Adducts 4 in Cellular DNA.
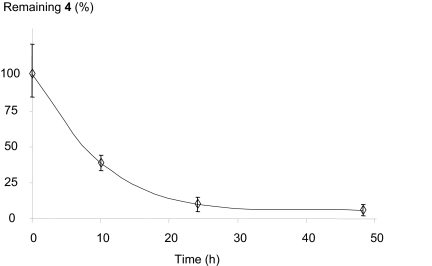
To study the repair rate of cytosine adducts 4 in cellular DNA, cells were first incubated with bleomycin to induce a significant formation of 4. Such a treatment was preferred to ionizing radiation because, under these conditions, cells survival at 24 h, determined by using the MTT test, was >80%. After treatment, cells were washed twice with PBS buffer, and the level of 4 was determined either immediately after incubation with bleomycin or 10, 24, and 48 h after reincubation of the cells in the culture medium. The relative level of 4 reported in Fig. 4, indicates that the half-life of the lesion in the human lymphocyte cell line used is ≈10 h. It may be added that, 24 h after the end of the bleomycin treatment, the measured level of 4 was similar to the background level determined in the control cells. It should be noted that DNA repair studies were performed by using confluent cells to reduce the impact of cell turnover in the disappearance of the DNA lesion.
Fig. 4.
Repair rate of 4 in a human lymphocyte cell line treated with bleomycin. Results are expressed as the percentage of remaining 4 compared with time 0.
Discussion
As a preliminary work, we have previously reported the radiation-induced formation of an as yet unidentified DNA lesion (24). We have shown that the lesion is a cytosine modification exhibiting a molecular mass of 341 Da. In the present work, efforts were made to characterize the lesion and to determine the mechanism of its formation. Consideration of the obtained results allowed us to propose that the cytosine adducts 4, which could be considered as LMDS, consist of the likely four diastereomers of 6-(2-deoxy-β-d-erythro-pentofuranosyl)-2-hydroxy-3(3-hydroxy-2-oxopropyl)-2,6-dihydroimidazo[1,2-c]-pyrimidin-5(3H)-one (Scheme 2).
The proposed mechanism of formation of 4 involves, in the first step, hydrogen abstraction at the 4-position of the 2-deoxyribose moiety of DNA (Scheme 1). This receives further support from the observation that bleomycin is able to induce the formation of 4, upon incubation with either isolated DNA or cells. The carbon-centered radical thus produced could give rise to the unsaturated 4-keto-1-aldehyde precursor 2, as already proposed (30, 31) through β-elimination reaction of the 3′-phosphate group in agreement with the lack of formation of 4 in γ-irradiated 5′-dCMP aqueous solutions. Support for such a spontaneous β-elimination reaction is provided by the observed formation of 4 upon incubation of γ-irradiated 3′-TMP with dCyd. The reaction of the obtained ketoaldehyde 2 with dCyd is similar to that described for 1,4-dioxo-2-butene (32) and 4 presents structural homologies with the oxadiazabicyclo(3.3.0)octaimine adduct formed between dCyd and 1,4-dioxo-2-butene. Trapping of the aldehydic precursor 2 with methoxyamine (MX) provides an explanation for the inhibition of the formation of 4 upon exposure of isolated DNA to ionizing radiation in the presence of increasing MX concentrations. However it should be noted that heretofore, by using photochemical precursors of the C4′ DNA radical, the generation of 2 has not been reported (33, 34), even if its formation has been proposed (30, 31). Therefore, additional work is required to better understand the undergoing reactions initiated by C4′-hydrogen abstraction. In particular, the use of photochemical precursors would be of major help to determine the yield of formation of 2 and its relative conversion to adduct 4. In addition such an approach would certainly allow determining the efficiency of the reaction of 2 with vicinal dCyd according to their relative position in double stranded DNA and to clearly determine the nature of the inter- or/and intra-strand cross-links.
In isolated DNA the relative yield of radiation-induced formation of 4 represents ≈5% of that of 8-oxodGuo. However, this ratio is much lower within cellular DNA, corresponding to ≈1% of that of 8-oxodGuo. The lower efficiency of formation of 4 relatively to 8-oxodGuo in cells compared with isolated DNA may be the result of competitive reactions of 2 with other nucleophiles, including amino acids residues of nuclear proteins. Nucleophilic amino acids are certainly able to react with 2, as shown for lysine residues that are able to form adducts with 1,4-dioxo-butene (35). Such reactions that could give rise to DNA-protein cross-links would decrease the yield of formation of 4 compared with other DNA lesions such as 8-oxodGuo. Interestingly, in cells incubated in the presence of 50 μM bleomycin, a 10-fold increase in the level of 4 is observed (Fig. 3). Under our experimental conditions, incubation of the human monocytes with 50 μM bleomycin generates ≈0.1 lesion per 106 nucleosides, corresponding to ≈600 lesions per cells. The yield of cytosine adducts thus formed appears to be not negligible compared with the number of DNA strand breaks induced by bleomycin as measured by the comet assay (36). We could therefore estimate that the formation of 4 in cells treated with bleomycin is not a minor pathway. The kinetic of removal of 4 in cellular DNA is almost similar to that determined for bulky DNA adduct such as UV-induced DNA bipyrimidine lesions that are known to be repaired by the nucleotide excision repair pathway (37). However, additional experiments are required to clearly define the mechanism(s) and fidelity of repair of 4 that may be different for either intra- or inter-strand cross-links, if the two possible LMDS are produced in cellular DNA.
The presently reported work highlights the putative role of sugar alterations in the indirect biological effects of ionizing radiation and the treatment with radiomimetic drugs. It is now admitted that several diffusible electrophiles are produced by radical oxidation of 2-deoxyribose (38). The sugar derivatives bearing reactive aldehydes, such as butenedialdehyde and 2 are able to form, in the vicinity of strand breaks, stable DNA adducts, the reactions being favored by the secondary structure of DNA. Formation of 4 is even more complex, involving the formation of a DNA cross-link most probably between the two opposite DNA strands, as represented in Scheme 4. Such single-hit generated complex DNA lesions could be considered as LMDS and their formation is likely to be involved in the genotoxicity of bleomycin that is used in human cancer therapy (39).
Scheme 4.
Schematic representation of the formation of 4 in double-stranded DNA. Reaction of the reactive aldehyde with a cytosine base located on the complementary strand gives rise to the formation of an interstrand DNA cross-link in addition to a strand break. Such a lesion represents a LMDS initiated by one radical hit.
Methods
Products.
Calf thymus DNA (ctDNA), 2′-deoxycytidine (dCyd), 2′-deoxycytidine 3′-monophosphate (3′-dCMP), 2′-deoxycytidine 5′-monophosphate (5′-dCMP), thymidine 3′-monophosphate (3′-TMP), bleomycin, and methoxyamine were purchased from Sigma–Aldrich (St. Quentin Fallavier, France). 2′-Deoxycytidine 3′,5′-diphosphate (pdCp) was purchased from Boehringer (Mannheim, Germany).
Irradiations and Digestions.
Either aqueous aerated DNA solutions or cells suspensions in PBS buffer were exposed to the γ-rays of a 60Co source immersed in a water pool. The dose rates were either 2 or 10 Gy/min for experiments involving cells and isolated DNA, respectively. For experiments performed with dCyd derivatives, 2.5 mM aqueous aerated solution of dCyd, 3′-dCMp, 5′-dCMP, or pdCp was γ-irradiated with a dose of 50 Gy. Thereafter levels of 4 were determined by HPLC-MS/MS (see Table 1) following enzymatic digestion as described in detail elsewhere (40). Formation of 4 was also detected upon γ-irradiation of 2.5 mM 3′-TMP (50 Gy) subsequently incubated in the presence of dCyd, without any enzymatic digestion. DNA was extracted from THP1 cells, a human monocyte cell line, by using a recently optimized protocol (29).
Exact Mass Determination.
High-resolution mass spectra were acquired on an electrospray Micromass QToF-2 instrument (Micromass, Manchester, U.K.) as described in detail elsewhere (41). Exact mass determination: 4d ESI+, [M+H]+ = 342.1296 (calculated [M+H]+ = 342.1301).
Calibrated Solutions of 4.
Collected fractions containing 4 were dissolved in water, and a UV absorption spectrum was recorded by using a Hewlett–Packard (Les Ullis, France) model 8453 apparatus. The molecular absorption coefficient, which is identical for at least isomers 4c and 4d, was determined by using 1H NMR as described (42) to be ε280: 13,200 liters·mol−1·cm−1.
NMR Spectroscopy.
The 1H NMR spectra (SI Text) were recorded on a VARIAN Unity 500 spectrometer by using a 5-mm indirect detection probe. The spectrometer was operated at 500.13 MHz at a constant temperature of 283 K.
4c, 1H NMR (D2O, 500 MHz): δ (ppm) = 2.47 (m, H2′, H2″); 2.54 (d, H10, H10′); 3.68 (dd, H12, H12′); 3.76 (q, H5″); 3.85 (q, H5′); 4.10 (m, H4′); 4.45 (m, H3′); 5.25 (t, H3); 6.28 (m, H1′, H8, H2); 8.24 (d, H7)
4d, 1H NMR (D2O, 500 MHz): δ (ppm) = 2.47 (m, H2′, H2″); 2.55 (d, H10, H10′); 3.70 (dd, H12, H12′); 3.81 (q, H5″); 3.90 (q, H5′); 4.12 (m, H4′); 4.51 (m, H3′); 5.14 (t, H3); 6.23 (d, H8); 6.26 (d, H2); 6.35 (t, H1′); 8.00 (d, H7).
HPLC-MS/MS Measurements.
On-line HPLC-MS/MS measurements were carried out by using an Agilent 1100 HPLC system (Agilent Technologies, Massy, France), equipped with a thermostatted autosampler, a binary HPLC pumping system, an oven, and a UV detector connected to a API3000 tandem mass spectrometer (Applied Biosystems, Courtaboeuf, France) through a Turbospray electrospray source (Sciex, Thornil, Canada) as described in detail elsewhere (27, 43). The multiple reaction monitoring mode was applied for the detection of 4, by using transitions 342 → 226, 342 → 168, and 342 → 148 (24). The transition 342 → 226 was monitored with a collision energy of 17 eV and the two others with a collision energy of 40 eV. Under the HPLC conditions used, the four isomers of 4 are eluted at retention times of 13.4, 13.6, 14.9, and 15.8 min, respectively (Fig. 1). Formation of 8-oxodGuo was also monitored as described (43). Unless otherwise indicated, results represent the average and standard deviation of three independent determinations. The relative formation of the different isomers of 4 being constant, reported levels correspond to the sum of the four different isomers as determined by external calibration (SI Text).
Bleomycin Treatment.
Aqueous solutions of calf thymus DNA (ctDNA) were incubated with increasing concentrations of bleomycin (0–35 μM) in the presence of 50 μM Fe(SO4)2(NH4)2 for 1 h at 37°C. Thereafter, DNA was precipitated and washed twice with 70% EtOH before enzymatic digestion. THP1 cells were incubated for 1 h at 37°C with bleomycin (0–120 μM) in PBS buffer before DNA extraction and digestion. For repair studies, cells were treated with 150 μM bleomycin for 1 h and subsequently reincubated in fresh culture medium for 10, 24, and 48 h before the HPLC-MS/MS measurement of 4.
Supplementary Material
Acknowledgments
We thank Dr. Jörg Hau (Nestlé Research Centre, Lausanne, Switzerland) for his contribution to the accurate mass measurement and Dr. Thierry Douki for helpful discussions and critical reading of the manuscript. The work was supported by European Union Marie Curie Training and Mobility Project Number MRNT-CT2003 “CLUSTOXDNA.”
Abbreviation
- LMDS
locally multiple damage sites.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706044104/DC1.
References
- 1.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Ames BN, Shigenaga MK, Hagen TM. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadet J, Douki T, Gasparutto D, Ravanat J-L. Mutat Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Dizdaroglu M. Mutat Res. 1992;275:331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- 5.Rokita SE, Romero-Fredes L. Nucleic Acids Res. 1992;20:3069–3072. doi: 10.1093/nar/20.12.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedon PC, Goldberg IH. Chem Res Toxicol. 1992;5:311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 7.Pitié M, Pratviel G, Bernadou J, Meunier B. Proc Natl Acad Sci USA. 1992;89:3967–3971. doi: 10.1073/pnas.89.9.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Que BG, Downey KM, So AG. Biochemistry. 1980;19:5987–5991. doi: 10.1021/bi00567a007. [DOI] [PubMed] [Google Scholar]
- 9.Chen B, Bohnert T, Zhou X, Dedon PC. Chem Res Toxicol. 2004;17:1406–1413. doi: 10.1021/tx049818e. [DOI] [PubMed] [Google Scholar]
- 10.Del Rio D, Stewart AJ, Pellegrini N. Nutr Metab Cardiovasc Dis. 2005;15:316–328. doi: 10.1016/j.numecd.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Plastaras JP, Dedon PC, Marnett LJ. Biochemistry. 2002;41:5033–5042. doi: 10.1021/bi0113059. [DOI] [PubMed] [Google Scholar]
- 12.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Proc Natl Acad Sci USA. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Taghizadeh K, Dedon PC. J Biol Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 14.Awada M, Dedon PC. Chem Res Toxicol. 2001;14:1247–1253. doi: 10.1021/tx0155092. [DOI] [PubMed] [Google Scholar]
- 15.Gingipalli L, Dedon PC. J Am Chem Soc. 2001;123:2664–2665. doi: 10.1021/ja0056421. [DOI] [PubMed] [Google Scholar]
- 16.Bohnert T, Gingipalli L, Dedon PC. Biochem Biophys Res Commun. 2004;323:838–844. doi: 10.1016/j.bbrc.2004.08.164. [DOI] [PubMed] [Google Scholar]
- 17.Chen B, Vu CC, Byrns MC, Dedon PC, Peterson LA. Chem Res Toxicol. 2006;19:982–985. doi: 10.1021/tx0601197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomax ME, Gulston MK, O'Neill P. Radiat Prot Dosimetry. 2002;99:63–68. doi: 10.1093/oxfordjournals.rpd.a006840. [DOI] [PubMed] [Google Scholar]
- 19.Sutherland BM, Bennett PV, Sutherland JC, Laval J. Radiat Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Proc Natl Acad Sci USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulston M, de Lara C, Jenner T, Davis E, O'Neill P. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison L, Hatahet Z, Wallace SS. J Mol Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 23.Blaisdell JO, Harrison L, Wallace SS. Radiat Prot Dosimetry. 2001;97:25–31. doi: 10.1093/oxfordjournals.rpd.a006634. [DOI] [PubMed] [Google Scholar]
- 24.Regulus P, Spessotto S, Gateau M, Cadet J, Favier A, Ravanat J-L. Rapid Commun Mass Spectrom. 2004;18:2223–2228. doi: 10.1002/rcm.1612. [DOI] [PubMed] [Google Scholar]
- 25.Rentel C, Wang X, Batt M, Kurata C, Oliver J, Gaus H, Krotz AH, McArdle JV, Capaldi DC. J Org Chem. 2005;70:7841–7845. doi: 10.1021/jo050767f. [DOI] [PubMed] [Google Scholar]
- 26.Pouget JP, Frelon S, Ravanat J-L, Testard I, Odin F, Cadet J. Radiat Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 27.Frelon S, Douki T, Ravanat J-L, Pouget JP, Tornabene C, Cadet J. Chem Res Toxicol. 2000;13:1002–1010. doi: 10.1021/tx000085h. [DOI] [PubMed] [Google Scholar]
- 28.Zhou X, Liberman RG, Skipper PL, Margolin Y, Tannenbaum SR, Dedon PC. Anal Biochem. 2005;343:84–92. doi: 10.1016/j.ab.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Ravanat J-L, Douki T, Duez P, Gremaud E, Herbert K, Hofer T, Lasserre L, Saint-Pierre C, Favier A, Cadet J. Carcinogenesis. 2002;23:1911–1918. doi: 10.1093/carcin/23.11.1911. [DOI] [PubMed] [Google Scholar]
- 30.Pogozelski WK, Tullius TD. Chem Rev. 1998;98:1089–1108. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 31.Worth L, Jr, Frank BL, Christner DF, Absalon MJ, Stubbe J, Kozarich JW. Biochemistry. 1993;32:2601–2609. doi: 10.1021/bi00061a018. [DOI] [PubMed] [Google Scholar]
- 32.Byrns MC, Predecki DP, Peterson LA. Chem Res Toxicol. 2002;15:373–379. doi: 10.1021/tx0101402. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Kreller CR, Greenberg MM. J Org Chem. 2005;70:8122–8129. doi: 10.1021/jo0512249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giese B, Beyrich-Graf X, Erdmann P, Petretta M, Schwitter U. Chem Biol. 1995;2:367–375. doi: 10.1016/1074-5521(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 35.Chen LJ, Hecht SS, Peterson LA. Chem Res Toxicol. 1997;10:866–874. doi: 10.1021/tx9700174. [DOI] [PubMed] [Google Scholar]
- 36.Buschini A, Alessandrini C, Martino A, Pasini L, Rizzoli V, Carlo-Stella C, Poli P, Rossi C. Biochem Pharmacol. 2002;63:967–975. doi: 10.1016/s0006-2952(01)00926-1. [DOI] [PubMed] [Google Scholar]
- 37.Courdavault S, Baudouin C, Charveron M, Canguilhem B, Favier A, Cadet J, Douki T. DNA Repair (Amsterdam) 2005;4:836–844. doi: 10.1016/j.dnarep.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Collins C, Zhou X, Wang R, Barth MC, Jiang T, Coderre JA, Dedon PC. Radiat Res. 2005;163:654–662. doi: 10.1667/rr3344. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Stubbe J. Nat Rev Cancer. 2005;5:102–112. doi: 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- 40.Ravanat J-L, Gremaud E, Markovic J, Turesky RJ. Anal Biochem. 1998;260:30–37. doi: 10.1006/abio.1998.2685. [DOI] [PubMed] [Google Scholar]
- 41.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat J-L. J Am Chem Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 42.Ravanat J-L, Remaud G, Cadet J. Arch Biochem Biophys. 2000;374:118–127. doi: 10.1006/abbi.1999.1610. [DOI] [PubMed] [Google Scholar]
- 43.Ravanat J-L, Duretz B, Guiller A, Douki T, Cadet J. J Chromatogr B. 1998;715:349–356. doi: 10.1016/s0378-4347(98)00259-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.