Abstract
We report the identification of a hypomorphic mouse allele for Foxc1 (Foxc1hith) that survives into adulthood revealing previously unknown roles for Foxc1 in development of the skull and cerebral cortex. This line of mice was recovered in a forward genetic screen using ENU mutagenesis to identify mutants with cortical defects. In the hith allele a missense mutation substitutes a Leu for a conserved Phe at amino acid 107, leading to destabilization of the protein without substantially altering transcriptional activity. Embryonic and postnatal histological analyses indicate that diminished Foxc1 protein expression in all three layers of meningeal cells in Foxc1hith/hith mice contributes to the cortical and skull defects in mutant mice and that the prominent phenotypes appear as the meninges differentiate into pia, arachnoid, and dura. Careful analysis of the cortical phenotypes shows that Foxc1hith/hith mice display detachment of radial glial endfeet, marginal zone heterotopias, and cortical dyslamination. These abnormalities have some features resembling defects in type 2 (cobblestone) lissencephaly or congenital muscular dystrophies but appear later in corticogenesis because of the delay in breakdown of the basement membrane. Our data reveal that the meninges regulate the development of the skull and cerebral cortex by controlling aspects of the formation of these neighboring structures. Furthermore, we provide evidence that defects in meningeal differentiation can lead to severe cortical dysplasia.
Keywords: genetic screen, meninges, migration
The forkhead transcription factor Foxc1 is a crucial regulator of a host of developmental processes including somitic, cardiovascular, calvarial, renal, and ocular development (1–4). Previous studies show that, at least during the formation of the calvarium, Foxc1 exerts its activity through FGF and BMP signaling, both important regulators of cell differentiation and proliferation (5). Targeted deletion of Foxc1 in mice is perinatal or embryonic lethal and produces a wide variety of developmental defects in homozygotes (6). Analysis of compound mutants of Foxc1 and Foxc2, factors with virtually identical DNA-binding domains, demonstrate that in many systems they have redundant function. Interestingly, mutations in FOXC1 were identified in numerous cases as causative for the dominant human disorder Axenfeld–Rieger syndrome (ARS) (7–11) indicating nonredundant functions for Foxc1 in at least some regions. ARS is characterized by dysgenesis of the anterior segment of the eye and is often associated with craniofacial abnormalities and abnormal dentition. ARS has also presented as an autosomal recessive disorder with mental retardation, hydrocephalus, and meningeal calcification (12) or as part of a chromosomal deletion syndrome (including Foxc1 and possibly other genes) associated with mental retardation and hydrocephalus (13–15).
During cephalic development, cranial neural crest cells migrate to cover the head, and these cells ultimately differentiate into the bones of the face and the frontal bone of the skull, cephalic mural cells (i.e., pericytes and blood vessel smooth muscle cells), and the meninges (16, 17). The meninges consist of three distinct mesenchymal cell layers with different functions that form via differentiation of the immature meningeal fibroblasts of neural crest origin. The inner meningeal layer, the pia, produces the basement membrane (BM) covering the cortex and serves as the origin of blood vessels that supply the superficial cerebral cortex (18–20). The middle meningeal layer, the arachnoid, includes specialized structures called arachnoid granulations, which resorb cerebrospinal fluid after circulation through the ventricular system (21). The third and outermost meningeal layer, the dura, is a thick, fibrous, collagenous structure that is tightly associated with the calvarium. Interestingly, the dura has previously been shown to secrete signals that induce calvarial ossification (17, 22). In this study we show that mice with a hypomorphic mutation in the Foxc1 gene have defects in all three meningeal layers with severe consequences for the brain and skull. Our data demonstrate the central importance of Foxc1 in the development of all meningeally based structures and reveal insights into the role of the meninges in controlling the development of adjacent structures: the skull and cerebral cortex. In addition, the mutant mice reveal that defects in meningeal differentiation leading to breakdown of the pial BM at later stages of development can cause a severe cortical dysplasia syndrome associated with marginal zone heterotopias and dyslamination.
Results
In a forward genetic ENU mutagenesis screen (23) we recovered a line of mice (#351) with homozygous mutants characterized by microphthalmia and incomplete skull closure at the most dorsal aspect of the frontal bones (Fig. 1 a and b); we called the mouse line hole-in-the-head (hith). Homozygous animals showed these defects in full penetrance with little variability and were viable and fertile; hith/+ mice were phenotypically indistinguishable from wild-type mice. Examination of the eyes revealed a number of defects similar to anomalies seen in ARS. In whole mount the mutant eye was smaller and the iris was irregularly shaped (Fig. 1a), and in sections the mutant eyes had smaller vitreal cavities, misshapen lenses, anterior chamber defects, and corneal thickening. Bone stains revealed the characteristic frontal bone defects (Fig. 1b). Adult brain sections in hith/hith mice showed dramatic hydrocephalus, dysgenesis of the neocortex and hippocampus, agenesis of the corpus callosum, and marginal zone heterotopias (Fig. 1 c and d). Examination of brain histology over development showed that the most prominent dysplastic features appeared late in corticogenesis between embryonic day 18.5 (E18.5) and postnatal day 7 (P7) and that the hydrocephalus was a late development after P7 (Fig. 1c), consistent with a postnatal defect in cerebrospinal fluid resorption.
Fig. 1.
Phenotype of Foxc1hith/hith mice. (a) Whole mount and cross sections of Foxc1hith/hith eyes highlight the micropthalmalia, dysgenic iris (white arrow), and thickened cornea. The anterior chamber is also lost (the space between the lens and the cornea, black arrow) in the mutants. (Scale bars: 500 μm.) (b) Bony defects are limited to the frontal bone and sagittal suture in the Foxc1hith/hith. (c) Hemisections of cortex at three ages in control and mutant animals shows that major cortical dysplasia develops between E18.5 and P7. The dashed red line in the P7 mutant cortex shows the area of most severe dysplasia in dorsomedial cortical areas. The red arrows show the abnormally formed hippocampal layers, and the asterisk is placed in the dilated lateral ventricle in the adult mutant cortex. Note that there is not prominent ventricular dilatation at earlier ages. (d) Higher-magnification images showing three examples from different brains of heterotopic cells in the marginal zone of mutant neocortex (red arrows).
To determine whether the dysgenesis of dorsomedial neocortical structures was associated with neuronal layering defects, we examined the expression of several layer-specific transcription factors in the adult mutant cortex (as recently reviewed in ref. 24). In P10 control mice Tbr1 prominently stained layer 2/3 and layer 6, with scattered cells in layer 5 (Fig. 2a), and in mutant brain (Fig. 2b) these same layers were identifiable but were clearly disorganized, with areas of stained cells displaced into the marginal zone and significant waviness of the layering organization. Using two different markers specific for deep cortical neurons [layers 5 and 6 for Ctip2 (Fig. 2 c and d) and layer 5 for ER81 (Fig. 2 e and f)] showed that, although these two markers also revealed prominent waviness of the deep cortical layers, they failed to show any collections of stained cells included in the marginal zone heterotopias (Fig. 2 d and f). Thus, it is likely that the superficial heterotopic collections of neurons in the mutant brains predominantly contained neurons of superficial layer fate. Because the neocortex is formed in a characteristic inside-out pattern of neurogenesis (24), with early-born neurons in deeper layers and later-born neurons superficially, it is likely that the cortical dysplasia seen in the hith mutants develops and increases in severity later in corticogenesis. Also, comparing the two hemispheres of the same mutant brain demonstrated variable regional severity of the cortical dysplasia even within the same animal (Fig. 2 b, d, and f).
Fig. 2.
Dyslamination in Foxc1hith/hith mice. (a and b) Tbr1 staining of P10 brains shows labeling of layers 2/3, 5, and 6 in control and mutant brains. The mutant brains are montages generated by merging two images of both brain hemispheres. In mutant brains Tbr1+ cells form heterotopic clusters in the marginal zone (blue arrows in Insets) and reveal significant disorganization of cortical layers (black arrows). (c–f) Ctip2 staining labels cells in layers 5 and 6, and ER81 labels cells in layer 5, and both show significant laminar disorganization in mutant brains (black arrows); however, note that there are no Ctip2- or ER81-stained cells in the marginal zone.
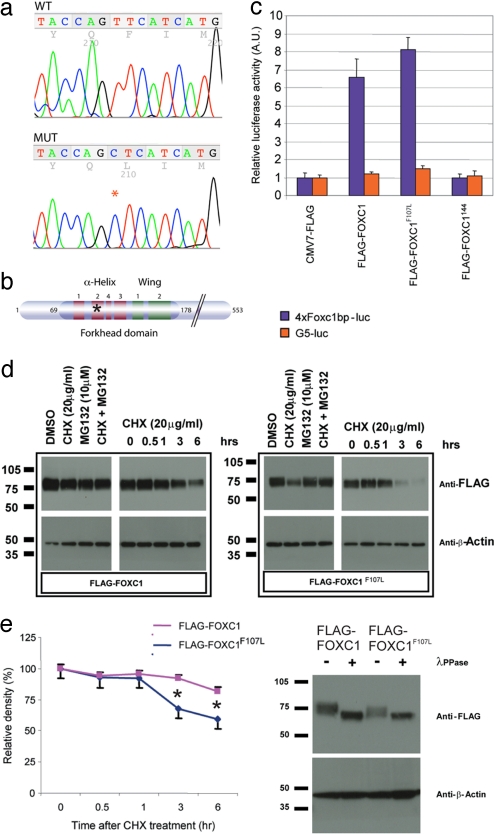
We mapped hith to an interval bounded by markers D13Mit135 and D13Mit275. In Ensembl's latest assembly, NCBIM36, this corresponds to the physical interval between 21.93 Mb and 37.33 Mb on mouse chromosome 13. Based on the resemblance of the eye phenotype to ARS, we sequenced the Foxc1 gene and found a T-to-C transition resulting in a Phe-to-Leu substitution at amino acid 107 within the second helix of the DNA binding forkhead domain (Fig. 3 a and b). Interestingly, in the mouse mutant dysgenetic lens (dyl), a T-to-C transition in the Foxe3 gene results in an amino acid substitution at amino acid 93, the position in the protein homologous to amino acid 107 in Foxc1 (25).
Fig. 3.
Point mutation in the Foxc1 hith allele does not affect transcription but results in destabilization of the protein in vitro. (a) Sequence histograms depicting amino acids 105–109 in the wild-type (WT) and hith mutant (MUT) Foxc1 gene. The asterisk denotes the T-to-C point mutation in amino acid 107 that results in phenylalanine being replaced by leucine. (b) The forkhead domain of Foxc1 is composed of three α-helices, two β-sheets, and two loops that form a “wing-like” formation. The mutation in the hith allele is located in the second α-helix (asterisk). (c) Transcriptional activity of the mutant protein was tested via cotransfection of a control construct (CMV7-FLAG), wild-type Foxc1-containing vector (FLAG-FOXC1), mutant Foxc1 (FLAG-Foxc1F107L), or a transcriptionally inactive truncation mutant of Foxc1 (FLAG-FOXC1144) and a luciferase reporter construct with (4xFoxc1bp-luc) or without (G5-luc) FOXC1 binding sites. Wild-type Foxc1 protein and mutated Foxc1 had similar transcriptional activity when cotransfected with the Foxc1 luciferase reporter, whereas the truncation mutant of Foxc1 showed no specific transcriptional activity. (d) Three hours after application of the protein translation inhibitor CHX (20 mg/ml) to 293 cells transfected with FLAG-tagged wild-type or mutant Foxc1, the mutant Foxc1 protein levels were clearly reduced. This effect was reversed with application of a proteasome inhibitor, MG132 (10 mM), treated 1 h before CHX. A time course of CHX treatment shows a marked decrease in Foxc1 mutant protein by 3 h and further reduction by 6 h after addition of CHX. Results from six independent experiments were plotted (error bars indicate SD). (e) Separation of FLAG-Foxc1 and FLAG-Foxc1F107L protein on an acrylamide gel revealed that both were present in a range of MW species, but mutant protein never attained the highest MW seen in the wild-type protein. Treatment of lysates with the phosphatase λ-PPase indicated that this isoform complexity was due to differences in phosphorylation status of the protein.
Transcriptional assays using a vector with four repeats of the Foxc1 binding motif upstream of a minimal promoter and a luciferase reporter gene showed that F107L-Foxc1 had essentially normal transcriptional activity compared with wild-type Foxc1 whereas truncated Foxc1 without the transcriptional activation domain showed no transcriptional activity (Fig. 3c). Because of suggestions that amino acid 107 is important in regulating the tertiary structure of the forkhead domain (26), we examined the possibility that the mutant protein was unstable. Indeed, protein stability experiments in transfected cells showed that F107L-Foxc1 protein was less stable than Foxc1 (n = 6; P = 0.004) (Fig. 3d). It is likely that the dramatic overexpression of Foxc1 in our transfection assays is the reason for the apparently normal transactivation by mutant protein in our transcriptional assays. Stability of Foxc1 has been linked to the C-terminal phosphorylation of the protein (27), and evaluation of the phosphorylation state of F107L-Foxc1 protein revealed reduced phosphorylation compared with wild-type controls (Fig. 3e).
Because the analysis in transfected cells relies on heterologous expression of nonphysiologic levels of Foxc1, we examined Foxc1 expression in situ as well. Foxc1 mRNA expression was normal in the mutants and, as in the controls and consistent with previous reports (6, 7, 28), was detected in the meninges surrounding the brain and in the head mesenchyme (Fig. 4 a and b). Immunohistochemistry for Foxc1 showed nuclear expression in the meninges and head mensenchyme in unaffected embryos (n = 8) (Fig. 4 c and e). However, in similar areas in mutants, the Foxc1 protein level was dramatically reduced (n = 7) (Fig. 4 d and f). This is consistent with the idea that the F107L mutation in Foxc1 leads to destabilization of the protein and that mice homozygous for this mutation have intermediate levels of Foxc1 function between those of the previously described Foxc1−/− and Foxc1+/− mice (6). Consistent with this, crosses of Foxc1hith/+ mice with Foxc1+/− mice yielded mutant embryos, further establishing that the Foxc1 mutation that we have identified is the pathogenic mutation (data not shown).
Fig. 4.
Foxc1 expression in vivo at E14.5. (a and b) In both Foxc1+/ and Foxc1hith/hith heads, ventral head structures and the meninges (MN) surrounding both the cortex and the midbrain (MB) express Foxc1 mRNA. The Foxc1 signal intensity is similar in control and mutant mice. (c and d) Foxc1 protein is expressed intensely by mesenchymal cells and meningeal cells ventral to the forebrain in Foxc1+/ tissue sections. Expression of Foxc1 is not seen in neural tissue (except in pericytes associated with blood vessels; data not shown). Foxc1 protein expression in Foxc1hith/hith tissue is similarly distributed, but the staining intensity is markedly reduced. (e and f) Foxc1+ cells are distributed throughout the primitive meninges in control mice, but in Foxc1hith/hith meninges expression of Foxc1 in these cells is less. (Scale bars: 1 mm in a and b, 100 μm in c and d, and 50 μm in e and f.)
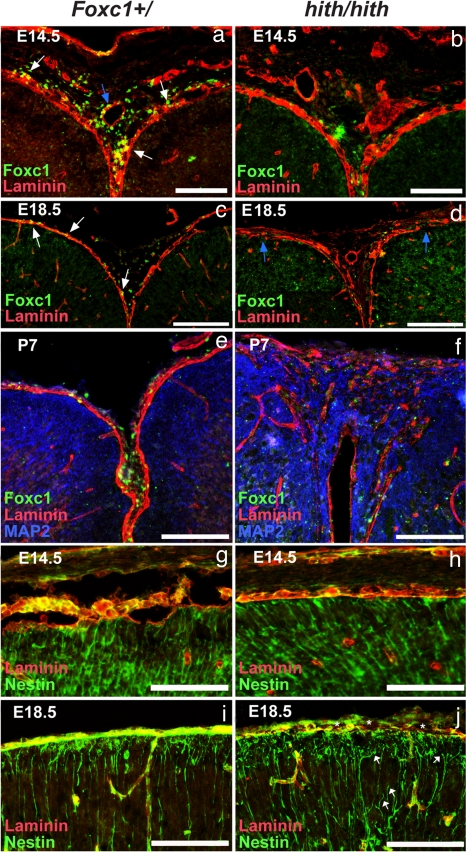
The marginal zone heterotopias and dyslamination seen in the hith mice indicate potential defects in the meningeal-derived pial BM and the interaction of the radial glial cells that serve as a guide for cortical migration and the pial surface where they normally attach. To test this, we examined the continuity of the BM at E14.5, E18.5, and P7 by examining the distribution of laminin and Foxc1. At E14.5 BM integrity was intact (n = 8) (Fig. 5 a and b), but by E18.5 the BM of Foxc1hith/hith brains was less well organized and significant breaches in the BM were evident at the pial surface (n = 3) (Fig. 5 c and d). Soon after birth, areas of BM disruption were associated with heterotopic neurons (Fig. 5 e and f), indicating that disruption in the BM leads to the disorganization of cortical neurons in the marginal zone. When we examined the distribution of Foxc1+ cells in relationship to the BM in the medial cortex there were dramatic differences between mutant and wild-type animals. In the Foxc1+/ brain strongly labeled Foxc1+ cells were distributed widely and in intimate association with the laminin-expressing BM and with the mesenchymal tissue at the dorsal midline of the head, whereas in brains of Foxc1hith/hith mice there were few Foxc1+ cells in this area at any age (Fig. 5 a–f).
Fig. 5.
Progressive breakdown of midline BM and development of cortical dysplasia in Foxc1hith/hith animals. (a and b) Laminin (red) and Foxc1 (green) expression at the cortical midline in Foxc1+/ tissue at E14.5 shows Foxc1+ cells closely associated with the laminin BM (white arrows) and around laminin+ blood vessels (blue arrow). At this early age, the laminin BM is intact in Foxc1hith/hith brains and weakly Foxc1+ cells are associated with both the meninges and blood vessels at the midline. (c and d) At E18.5, a continuous BM surrounds the cortical tissue and Foxc1+ cells are embedded within the pial BM (white arrows). In the Foxc1hith/hith brain, breeches in the laminin BM are apparent at E18.5 (blue arrows), and, although some weakly Foxc1+ cells are observed within in the interhemispheric region, these cells are largely absent in the areas in and around the laminin breeches. (e and f) At P7, severe breaks in the laminin BM are accompanied by infiltrating MAP2+ neural tissue and disorganized laminin deposits within the heterotopias. (g–j) Organized radial glial endfeet (green) overlap with the laminin+ BM at the pial surface in Foxc1+/ brain as an essentially unbroken layer of staining where the endfeet attach to the BM in control and mutants at E14.5 but only in controls at E18.5. In contrast, in the Foxc1hith/hith mice disorganized laminin is associated with detached radial glial endfeet (white arrows) and gaps where no nestin-labeled endfeet are attached to the BM (asterisks) at E18.5. (Scale bars: 500 μm in a and b, 100 μm in c, d, and g–j, and 200 μm in e and f.)
Beyond providing a physical barrier to migrating neurons during brain development, the pial BM serves as a critical attachment point for radial glial endfeet whose fibers span the width of the growing cerebral wall, providing a migratory scaffold for neuronal migration (29). At E14.5, before the loss of BM integrity, radial glial endfeet could be seen properly attaching to the pial laminin+ BM in both control and hith mice (Fig. 5 g and h). However, by E18.5, whereas in the Foxc1+/ brains radial glial endfeet still projected all of the way to the laminin+ BM (Fig. 5i), the mutants had disorganized radial glia with detached endfeet in areas of BM disruption (Fig. 5h). It is likely that these radial glial defects are associated with the dyslamination and neuronal heterotopias seen in the mutant brains.
Foxc1 is strongly expressed in the meninges from very early in development, and Foxc1-null mice demonstrate correspondingly early defects in meningeal development (6). In contrast, as we have shown, hith mice have intact early stages of pial BM development with later breakdown. Importantly, the pial meninges are one of the three meningeal layers that become distinguishable in the last few days of gestation (17). At E18.5 in Foxc1+/ mice, the three distinct meningeal layers were established in the space between ossified calvarium and cortical tissue and Foxc1+ cells occupied all three layers [supporting information (SI) Fig. 6 a and b]. In contrast, the Foxc1hith/hith meninges had defects most apparent at the edge of the holes in the calvarium. In areas of bone formation of hith/hith mutants, weakly Foxc1+ cells populated all layers of the meninges whereas only scattered pial Foxc1+ cells were found where bone had failed to form (SI Fig. 6c). To further clarify the defects in these areas, we examined the expression of Foxc2, a close homolog of Foxc1 also reportedly expressed in the meninges (28, 30). Interestingly, in our higher-resolution analysis we found that only dural cells strongly expressed Foxc2 (SI Fig. 6e), indicating that, whereas Foxc1 is expressed within all layers of meninges, Foxc2 is a dura-specific marker. In mutants, the Foxc2+ dura was intact beneath areas of bone growth but no dura formed within the bone holes (SI Fig. 6f). Because the dura contains signals required to induce ossification, the lack of dura is likely to be critical in the failure to form a continuous skull. In addition, it is likely that the failure to produce differentiated three-layered meninges, including the arachnoid, is responsible for the postnatal development of hydrocephalus due to defects in cerebrospinal fluid resorption.
Discussion
Here we describe the spectrum of phenotypes in mice homozygous for a Foxc1 mutation (hith) resulting in reduced Foxc1 protein stability. Mutants have reduced Foxc1 activity sufficient for early developmental events to proceed normally, uncovering previously unrecognized roles in the maintenance or late development of the meninges. Thus, our mutant mice have features in common with the engineered Foxc1-null mice and the spontaneously occurring congenital hydrocephalus mouse line [also predicted to be a null allele (6)], but with some important differences. Unlike hith mice, the Foxc1−/− mice completely lack skull ossification and produce only a primitive meningeal mesenchyme (6). Consequently, these mice have earlier and more severe defects incompatible with postnatal life or with detailed analysis of cortical development. In the viable Foxc1hith/hith mice we were able to characterize brain malformations first appearing late in gestation and progressing postnatally. Importantly, ARS patients with Foxc1 mutations are generally heterozygous for predicted null mutations and thus are genetically equivalent to Foxc1+/− mice, and in fact a mild version of anterior chamber dysgenesis is seen in Foxc1+/− mice (31). Importantly, cortical dysplasia has not been reported in typical ARS patients and is not reported in Foxc1+/− mice; however, ARS patients with autosomal recessive inheritance have mental retardation and meningeal calcifications (12), and this is reminiscent of the phenotype of the Foxc1hith/hith mice. Thus, in mice the complete loss of Foxc1 leads to a severe early developmental, lethal phenotype, half dosage leads predominantly to anterior segment dysgenesis, and an intermediate between these (the Foxc1hith/hith mice) leads to a viable but severe malformation syndrome in all of the differentiated derivatives of the meningeal mesenchyme, and it is likely that similar human alleles will be found and will expand the spectrum of ARS to include patients with cortical dysplasia.
The cortical dysgenesis and BM defects seen in Foxc1hith/hith animals are quite distinct but have some similarities to defects found in the congenital muscular dystrophies. In the brains of patients with these syndromes (or in engineered mouse models of these diseases), deletion of structural elements of the BM (including laminin subtypes, perlecan, or their cellular receptors dystroglycan, β1, or α6 integrin) or downstream signaling molecules leads to cortical ectopias, detachment of radial glial fibers, and overall laminar disorganization (32–35), reminiscent of, but more severe than, what we have observed here. Our data are consistent with the idea that reduced expression of Foxc1 protein in meningeal cells disrupts the ability of the meninges to produce and maintain the BM as the cortex expands during embryonic and early postnatal development, leading to a later degradation of BM function and causing a distinct later-appearing cortical dysplasia phenotype. This is a mechanism for the production of this cortical phenotype because it appears that meningeal development is sufficiently intact to proceed during early cortical development [unlike the situations where structural BM proteins are mutated and the pial BM is presumably always of low quality from earlier in development (32)] but later fails. It is possible that Foxc1 plays an important role in the extensive remodeling that must occur to maintain a well ordered BM able to cover the growing cortex in late gestation and early postnatal life. In addition, there is currently no literature available addressing whether there are subgroups of patients with cortical dysplasia presenting at the stage of cortical expansion (analogous to early postnatal life in rodents) rather than at earlier stages when radial migration is first ongoing. However, the large numbers of epilepsy patients who present with more limited forms of cortical dysplasia sharing some features with cobblestone lissencephaly (as our mice do) make this quite likely (36).
Materials and Methods
Positional Mapping.
The hith mutation is fully penetrant, and both male and female homozygous mutants can survive until adulthood and are fertile. The polymorphic markers on chromosome 13 used for fine-scale mapping of hith are from proximal to distal as follows: D13Mit218, D13Mit16, D13Mit17, D13Mit135, D13Mit275, D13Mit117, D13Mit177, D13Mit244, and D13Mit91. Meiotic mapping results support this order of markers that corresponds to the one displayed in Ensembl. The hith mutation recombined away from D13Mit135 at least 34 times and recombined away from D13Mit275 at least 27 times. In all cases where one of these two markers segregated from hith the other associated with the mutation. We sequenced all exons (including the UTRs) of six candidate genes within the interval and discovered no other polymorphisms besides the causative mutation in Foxc1. The sequenced candidate genes are as follows: Foxc1 (EntrezGene ID code 17300), Mboat1 (EntrezGene ID code 218121), Dusp22 (EntrezGene ID code 105352), Exoc2 (EntrezGene ID code 66482), Gmds (EntrezGene ID code 218138), and Cdkal (EntrezGene ID code 68916). Sequencing of candidate genes was performed at the Ernest Gallo Clinic and Research Center core sequencing facility using Applied Biosystems (Foster City, CA) Big Dye (3.1) reactions. Sequencing reactions were electrophoresed and analyzed in an Applied Biosystems Genetic Analyzer 3730XL.
Animal Husbandry and Genotyping.
Mice are housed in specific pathogen-free facilities approved by the American Association of Laboratory Animal Care. All animals were handled in accordance with protocols approved by the University of California, San Francisco, Committee on Animal Research. The colony of animals carrying the Foxc1hith allele (induced on C57BL/6 background) is maintained by crossing male carriers with FVB females. This mode of outcross is currently in the fourth generation without any changes in penetrance or variability of the mutant phenotype. Heterozygote animals were determined by genotyping with markers D13Mit135 and D13Mit275. Once Foxc1 had been identified as the mutated gene primers for a polymorphic marker closely linked to the mutation (≈12.8 kb) were devised, and genotyping was based on using this marker. Primer sequences are as follows: D10Egc3 left, ACCTTGTCCTAGTGACTCCGATGG; D10Egc3 right, TTACAGCAGTTCTCCTGACAACTTAATATCTAAG. All phenotypically mutant animals were genotyped by using this marker and were positive for the C57BL/6 allele consistent with the genetic background on which the mutation had been caused.
Transcriptional and Protein Stability Assays.
293 cells were seeded onto polyd-lysine-coated 12-well culture plates 1 day before transfection. Cells were transfected with 1 μg of FLAG-FOXC1 or CMV7-FLAG control construct, 1 μg of luciferase reporter, and 0.7 μg of Renilla luciferase reporter vector with 3 ml of Lipofectamine 2000 for dual luciferase assay using a detection kit (Promega, Madison, WI).
For the protein stability assays, cells grown on a 10-cm dish were transfected with FOXC1 constructs. One day after transfection, cells were divided onto polyd-lysine-coated 12-well culture plates. MG132 (10 mM) was added 1 h before cycloheximide (CHX) treatment. Cells were incubated with CHX (20 mg/ml) for 0 (DMSO vehicle only), 0.5, 1, 3, and 6 h and subjected to Western blotting. Signals from Western blot analysis were analyzed with ImageQuant TL software (GE Healthcare, Fairfield, CT) and plotted using data from six independent experiments.
Immunohistochemistry.
Heads from E14.5 fetuses and P0 eyes were collected and fixed overnight in 4% paraformaldehyde, cryoprotected by using graduated sucrose concentrations (15% and 30%), and then frozen in Tissue-Tek OCT Compound (Sakura Fineteld, Torrance, CA). E18.5, P0, and P7 heads were fresh-frozen in OCT. All tissue was cryosectioned in 10-μm increments. Before antibody application, sections were steamed in 0.01 M citric acid buffer (pH 6.0) for 15 min then cooled in 0.1 M PBS for 15 min at room temperature. Sections were blocked in 1.0% BSA and 0.75% Triton X-100 for 45 min and then incubated in either goat anti-Foxc1 (1:300; Novus Biologicals, Littleton, CO) or goat anti-Foxc2 (1:300; Novus Biologicals) for 1 h. After washing in PBS, horseradish peroxidase-conjugated anti-goat secondary antibody (1:100; Vector Laboratories, Burlingame, CA) was applied for 1 h followed by a short incubation in Alexa Fluor 488-linked tyramide (1:100; Invitrogen, Carlsbad, CA). For double or triple immunolabeling, Foxc1-labeled sections were incubated in rabbit anti-laminin (1:75; Sigma, St. Louis, MO), mouse anti-nestin (1:500; Chemicon, Temecula, CA), and mouse anti-MAP2 (1:300; Chemicon) followed by incubation in Alexa Fluor 594 anti-rabbit secondary and/or Alexa Fluor 633 anti-mouse secondary antibodies (Invitrogen).
Statistical Analysis.
A one-way ANOVA was performed on densitometry data collected from the protein stability experiments using SigmaStat software. To identify statistically significant differences between wild-type and Foxc1 mutant values at specific time points, an all pairwise multiple comparison procedure (Student–Newman–Keuls method) was performed.
Phosphorylation Experiment.
Removal of phosphates of FLAG-FOXC1 with λ-protein phosphatase (NEB, Ipswich, MA) was done with 5 μg of cell extract obtained from 293 cells transfected with FOXC1 expression constructs. Cell extracts were incubated with 400 units of λ protein phosphatase at 30°C for 30 min. The reaction was stopped with SDS gel loading buffer.
In Situ Hybridization.
Foxc1 in situ hybridization was performed as previously described (37) by using an 806-bp35S-UTP-labeled riboprobe. The riboprobe corresponds to base pairs 2192–2997 on the Foxc1 mRNA sequence (GenBank accession no. NM_008592).
Supplementary Material
Acknowledgments
We thank Margaret Robertson and Raymond Chui at the Ernest Gallo Clinic and Research Center core sequencing facility for technical assistance in the sequencing of candidate genes. We also thank Drs. Viktor Kharazia and Holger Rüssig for contributions and assistance in the histological analysis of mutant mice and Drs. Douglas Gould, Jeremy Reiter, and John Rubenstein for helpful discussions. This work was supported by funding from the National Institute on Drug Abuse, Parents Against Childhood Epilepsy, the National Institute of Mental Health, and Autism Speaks.
Abbreviations
- ARS
Axenfeld–Rieger syndrome
- BM
basement membrane
- En
embryonic day n
- Pn
postnatal day n
- CHX
cycloheximide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702618104/DC1.
References
- 1.Kume T, Deng K, Hogan BL. Development (Cambridge, UK) 2000;127:1387–1395. doi: 10.1242/dev.127.7.1387. [DOI] [PubMed] [Google Scholar]
- 2.Kume T, Jiang H, Topczewska JM, Hogan BL. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo S, Fujita H, Nakano A, Kang M, Duarte A, Kume T. Dev Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 4.Wilm B, James RG, Schultheiss TM, Hogan BL. Dev Biol. 2004;271:176–189. doi: 10.1016/j.ydbio.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Rice R, Rice DP, Thesleff I. Dev Dyn. 2005;233:847–852. doi: 10.1002/dvdy.20430. [DOI] [PubMed] [Google Scholar]
- 6.Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. Cell. 1998;93:985–996. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- 7.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, et al. Am J Hum Genet. 1998;63:1316–1328. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. Nat Genet. 1998;19:140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 9.Nishimura DY, Searby CC, Alward WL, Walton D, Craig JE, Mackey DA, Kawase K, Kanis AB, Patil SR, Stone EM, et al. Am J Hum Genet. 2001;68:364–372. doi: 10.1086/318183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panicker SG, Sampath S, Mandal AK, Reddy AB, Ahmed N, Hasnain SE. Invest Ophthalmol Visual Sci. 2002;43:3613–3616. [PubMed] [Google Scholar]
- 11.Honkanen RA, Nishimura DY, Swiderski RE, Bennett SR, Hong S, Kwon YH, Stone EM, Sheffield VC, Alward WL. Am J Ophthalmol. 2003;135:368–375. doi: 10.1016/s0002-9394(02)02061-5. [DOI] [PubMed] [Google Scholar]
- 12.Moog U, Bleeker-Wagemakers EM, Crobach P, Vles JS, Schrander-Stumpel CT. Am J Med Genet. 1998;78:263–266. [PubMed] [Google Scholar]
- 13.Caluseriu O, Mirza G, Ragoussis J, Chow EW, MacCrimmon D, Bassett AS. Am J Med Genet A. 2006;140:1208–1213. doi: 10.1002/ajmg.a.31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Caignec C, De Mas P, Vincent MC, Boceno M, Bourrouillou G, Rival JM, David A. Am J Med Genet A. 2005;132:175–180. doi: 10.1002/ajmg.a.30409. [DOI] [PubMed] [Google Scholar]
- 15.Maclean K, Smith J, St Heaps L, Chia N, Williams R, Peters GB, Onikul E, McCrossin T, Lehmann OJ, Ades LC. Am J Med Genet A. 2005;132:381–385. doi: 10.1002/ajmg.a.30274. [DOI] [PubMed] [Google Scholar]
- 16.Etchevers HC, Vincent C, Le Douarin NM, Couly GF. Development (Cambridge, UK) 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- 18.McLone DG, Bondareff W. Am J Anat. 1975;142:273–293. doi: 10.1002/aja.1001420302. [DOI] [PubMed] [Google Scholar]
- 19.Bauer HC, Bauer H, Lametschwandtner A, Amberger A, Ruiz P, Steiner M. Brain Res. 1993;75:269–278. doi: 10.1016/0165-3806(93)90031-5. [DOI] [PubMed] [Google Scholar]
- 20.Sievers J, Pehlemann FW, Gude S, Berry M. J Neurocytol. 1994;23:135–149. doi: 10.1007/BF01183867. [DOI] [PubMed] [Google Scholar]
- 21.Davson H, Domer FR, Hollingsworth JR. Brain. 1973;96:329–336. doi: 10.1093/brain/96.2.329. [DOI] [PubMed] [Google Scholar]
- 22.Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr, Nakajima A, Shuler CF, Moses HL, Chai Y. Development (Cambridge, UK) 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 23.Zarbalis K, May SR, Shen Y, Ekker M, Rubenstein JL, Peterson AS. PLoS Biol. 2004;2:E219. doi: 10.1371/journal.pbio.0020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- 25.Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. Genes Dev. 2000;14:245–254. [PMC free article] [PubMed] [Google Scholar]
- 26.Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, Walter MA. Am J Hum Genet. 2001;68:627–641. doi: 10.1086/318792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry FB, Mirzayans F, Walter MA. J Biol Chem. 2006;281:10098–10104. doi: 10.1074/jbc.M513629200. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki H, Hogan BL. Development (Cambridge, UK) 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- 29.Marin O, Rubenstein JL. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 30.Rice R, Rice DP, Olsen BR, Thesleff I. Dev Biol. 2003;262:75–87. doi: 10.1016/s0012-1606(03)00355-5. [DOI] [PubMed] [Google Scholar]
- 31.Smith RS, Zabaleta A, Kume T, Savinova OV, Kidson SH, Martin JE, Nishimura DY, Alward WL, Hogan BL, John SW. Hum Mol Genet. 2000;9:1021–1032. doi: 10.1093/hmg/9.7.1021. [DOI] [PubMed] [Google Scholar]
- 32.Hu H, Yang Y, Eade A, Xiong Y, Qi Y. J Comp Neurol. 2007;501:168–183. doi: 10.1002/cne.21238. [DOI] [PubMed] [Google Scholar]
- 33.Halfter W, Dong S, Yip YP, Willem M, Mayer U. J Neurosci. 2002;22:6029–6040. doi: 10.1523/JNEUROSCI.22-14-06029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson EC, Walsh CA. Curr Opin Genet Dev. 2002;12:320–327. doi: 10.1016/s0959-437x(02)00305-2. [DOI] [PubMed] [Google Scholar]
- 35.Beggs HE, Schahin-Reed D, Zang K, Goebbels S, Nave KA, Gorski J, Jones KR, Sretavan D, Reichardt LF. Neuron. 2003;40:501–514. doi: 10.1016/s0896-6273(03)00666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rickert CH. Childs Nerv Syst. 2006;22:821–826. doi: 10.1007/s00381-006-0126-3. [DOI] [PubMed] [Google Scholar]
- 37.Zarbalis K, Wurst W. Mech Dev. 2000;93:165–168. doi: 10.1016/s0925-4773(00)00252-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.