Abstract
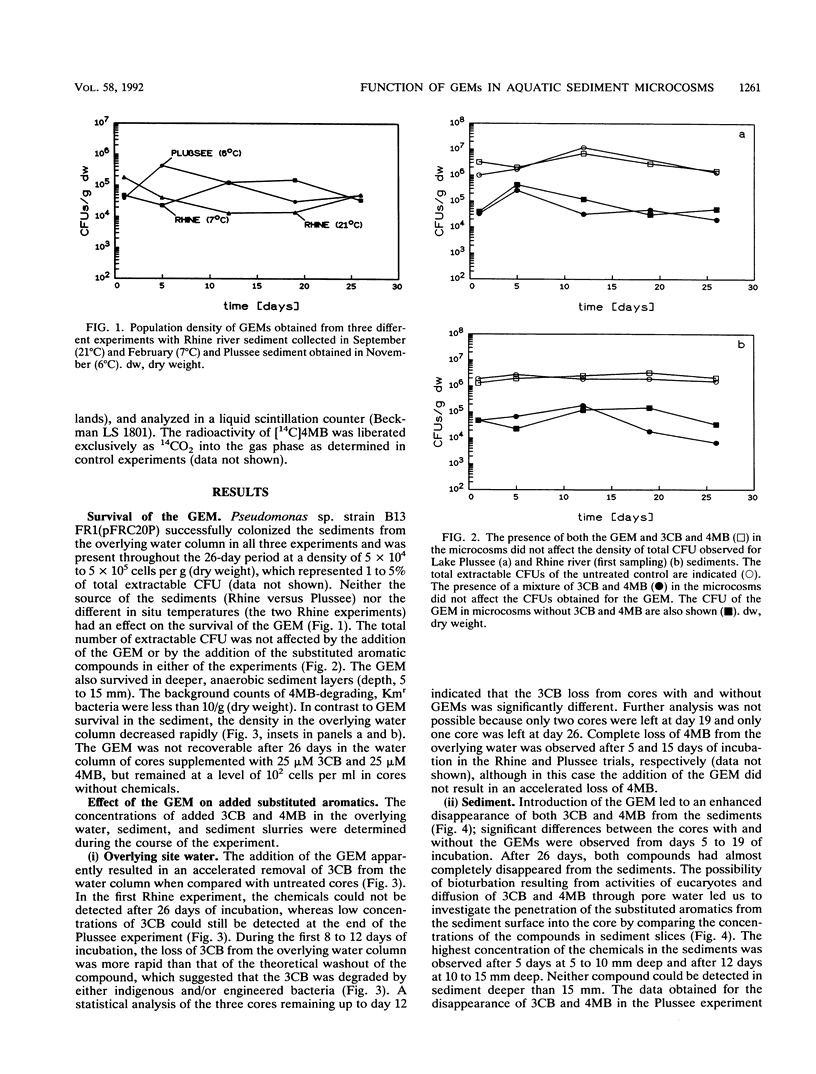
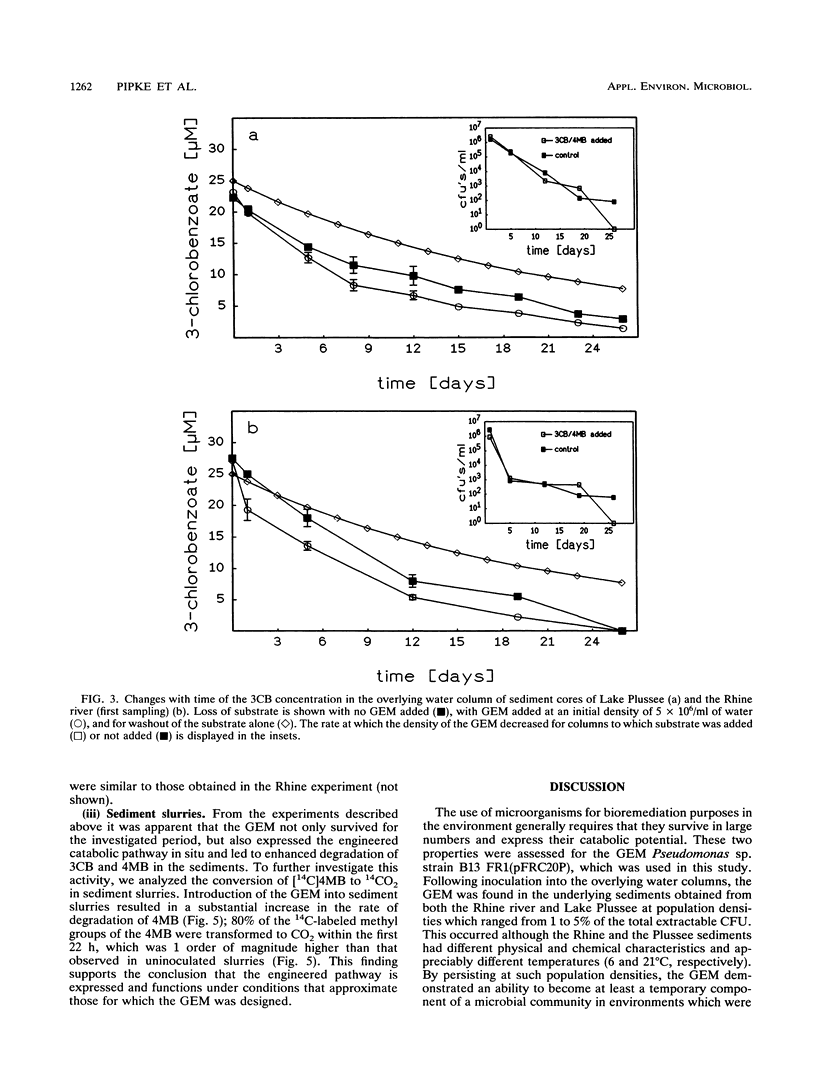
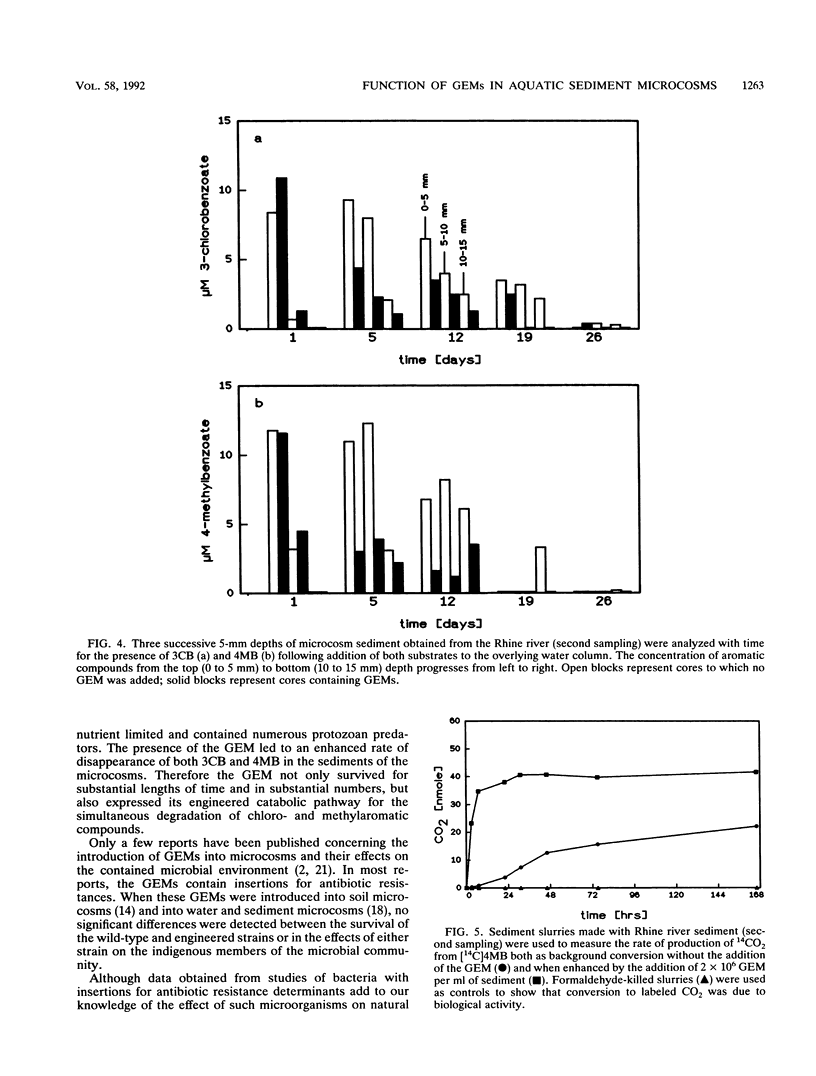
Pseudomonas sp. strain B13 FR1(pFRC20P) is a genetically engineered microorganism (GEM) which is able to degrade chloro- and methylaromatics through a constructed ortho cleavage pathway. The fate of the GEM and its ability to degrade substituted aromatic compounds in two different aquatic sediments was investigated by using a microcosm system which consisted of intact layered sediment cores with an overlying water column. The GEM survived in Lake Plussee and in Rhine river sediments at densities of approximately 10(5) bacteria per g (dry weight) (1 to 5% of the total CFU) throughout a 4-week period of investigation. According to several criteria, the microcosm system was stable and healthy throughout the experiment and the addition of the GEM did not affect the total number of extractable CFU (I. Wagner-Döbler, R. Pipke, K. N. Timmis, and D. F. Dwyer, Appl. Environ. Microbiol. 58:1249-1258, 1992). When compared with uninoculated controls, the presence of the GEM enhanced the rate of degradation of a mixture of 3-chlorobenzoate and 4-methylbenzoate (25 microns each) which had been added to the water column of the sediment cores.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Awong J., Bitton G., Chaudhry G. R. Microcosm for assessing survival of genetically engineered microorganisms in aquatic environments. Appl Environ Microbiol. 1990 Apr;56(4):977–983. doi: 10.1128/aem.56.4.977-983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulthorpe R. R., Wyndham R. C. Survival and activity of a 3-chlorobenzoate-catabolic genotype in a natural system. Appl Environ Microbiol. 1989 Jun;55(6):1584–1590. doi: 10.1128/aem.55.6.1584-1590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Sayler G. S. Problems and potential for in situ treatment of environmental pollutants by engineered microorganisms. Microbiol Sci. 1987 Feb;4(2):59–63. [PubMed] [Google Scholar]
- Kaphammer B., Kukor J. J., Olsen R. H. Regulation of tfdCDEF by tfdR of the 2,4-dichlorophenoxyacetic acid degradation plasmid pJP4. J Bacteriol. 1990 May;172(5):2280–2286. doi: 10.1128/jb.172.5.2280-2286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knackmuss H. J. Xenobiotic degradation in industrial sewage: haloaromatics as target substrates. Biochem Soc Symp. 1983;48:173–190. [PubMed] [Google Scholar]
- McClure N. C., Weightman A. J., Fry J. C. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl Environ Microbiol. 1989 Oct;55(10):2627–2634. doi: 10.1128/aem.55.10.2627-2634.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvos D. R., Lacy G. H., Cairns J. Genetically Engineered Erwinia carotovora: Survival, Intraspecific Competition, and Effects upon Selected Bacterial Genera. Appl Environ Microbiol. 1990 Jun;56(6):1689–1694. doi: 10.1128/aem.56.6.1689-1694.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M. Genetic engineering and biological detoxification of environmental pollutants. Residue Rev. 1981;78:1–11. doi: 10.1007/978-1-4612-5910-7_1. [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Wasserfallen A., Rose K., Timmis K. N. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science. 1987 Jan 30;235(4788):593–596. doi: 10.1126/science.3468623. [DOI] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Scanferlato V. S., Orvos D. R., Cairns J., Lacy G. H. Genetically Engineered Erwinia carotovora in Aquatic Microcosms: Survival and Effects on Functional Groups of Indigenous Bacteria. Appl Environ Microbiol. 1989 Jun;55(6):1477–1482. doi: 10.1128/aem.55.6.1477-1482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerman P. R., Schmidt J. P., Alexander M. Factors affecting the survival and growth of bacteria introduced into lake water. Arch Microbiol. 1988;150(4):320–325. doi: 10.1007/BF00408301. [DOI] [PubMed] [Google Scholar]
- Short K. A., Doyle J. D., King R. J., Seidler R. J., Stotzky G., Olsen R. H. Effects of 2,4-dichlorophenol, a metabolite of a genetically engineered bacterium, and 2,4-dichlorophenoxyacetate on some microorganism-mediated ecological processes in soil. Appl Environ Microbiol. 1991 Feb;57(2):412–418. doi: 10.1128/aem.57.2.412-418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotzky G., Babich H. Fate of genetically-engineered microbes in natural environments. Recomb DNA Tech Bull. 1984 Dec;7(4):163–188. [PubMed] [Google Scholar]
- Strauss H. S., Hattis D., Page G., Harrison K., Vogel S., Caldart C. Genetically-engineered microorganisms: II. Survival multiplication and genetic transfer. Recomb DNA Tech Bull. 1986 Jun;9(2):69–88. [PubMed] [Google Scholar]
- Trevors J. T. Use of microcosms to study genetic interactions between microorganisms. Microbiol Sci. 1988 May;5(5):132–136. [PubMed] [Google Scholar]
- Wagner-Döbler I., Pipke R., Timmis K. N., Dwyer D. F. Evaluation of aquatic sediment microcosms and their use in assessing possible effects of introduced microorganisms on ecosystem parameters. Appl Environ Microbiol. 1992 Apr;58(4):1249–1258. doi: 10.1128/aem.58.4.1249-1258.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


